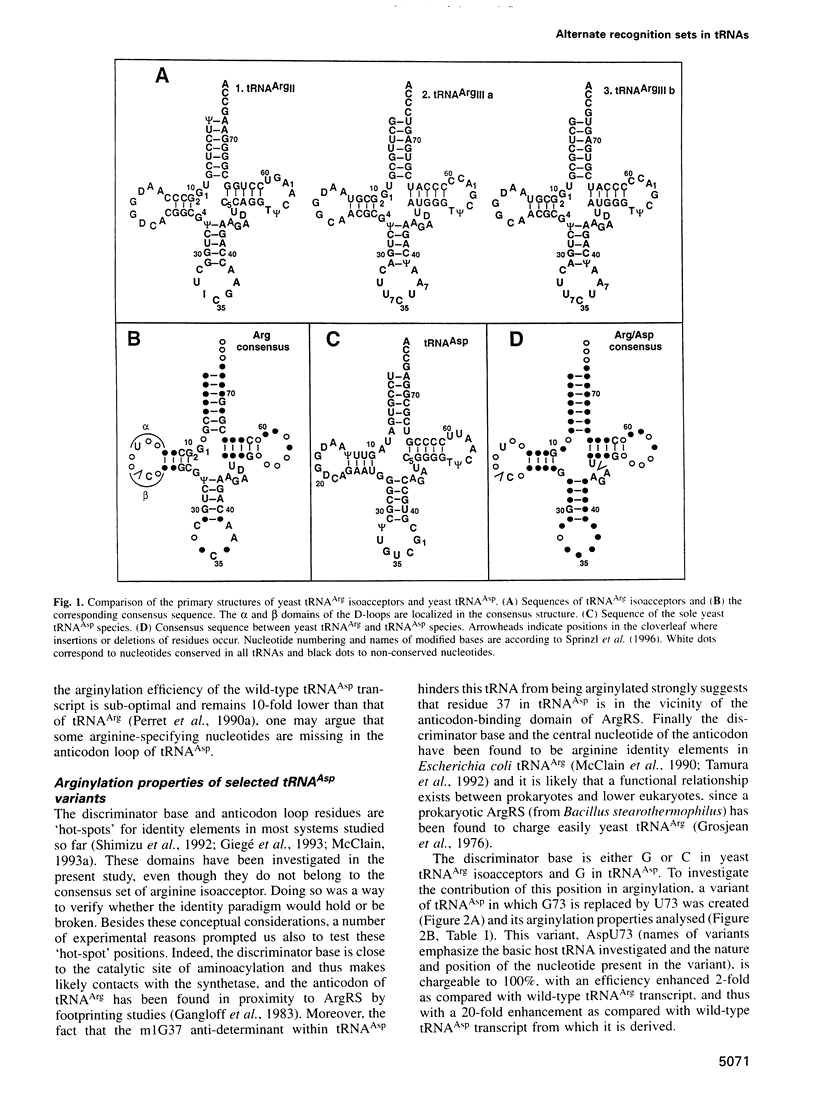
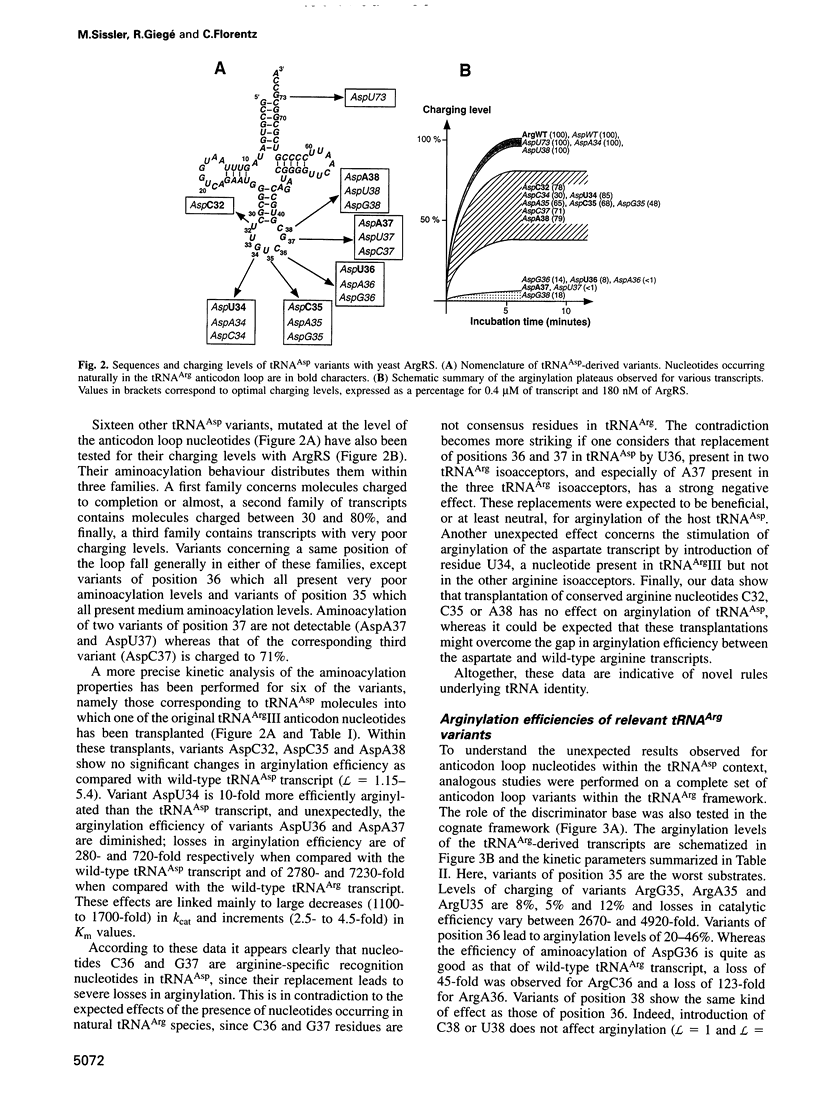
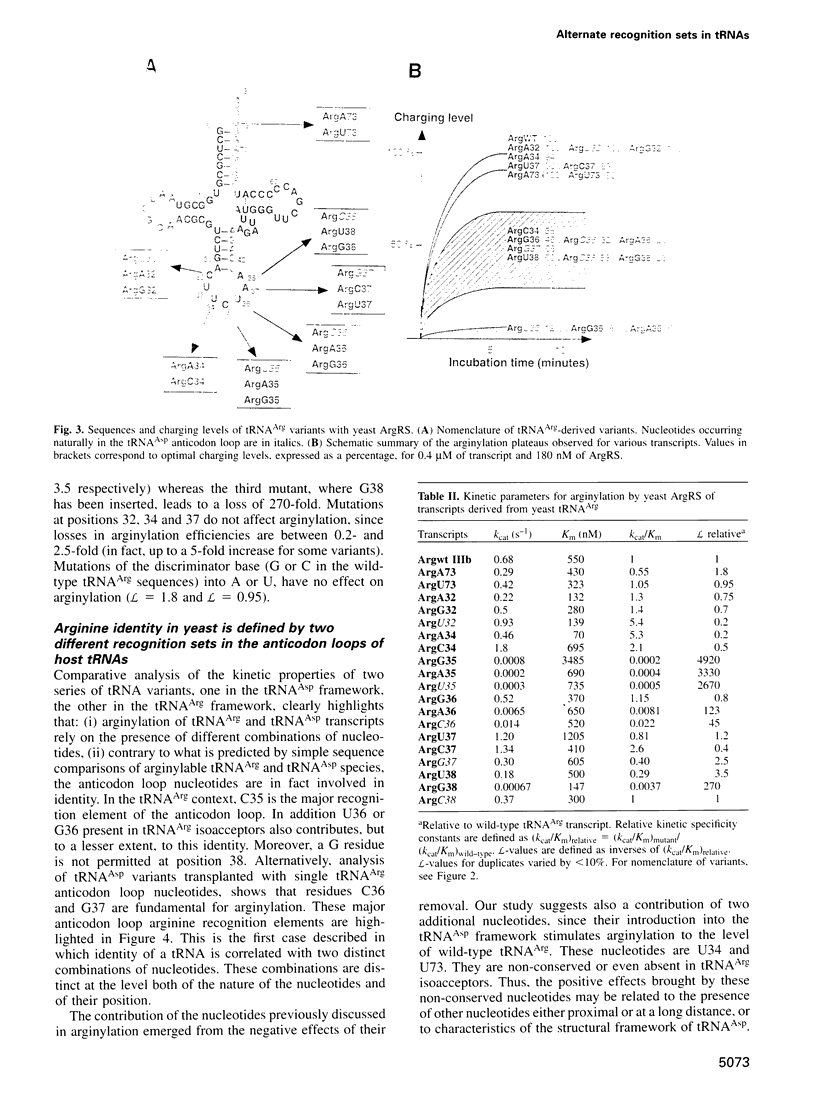
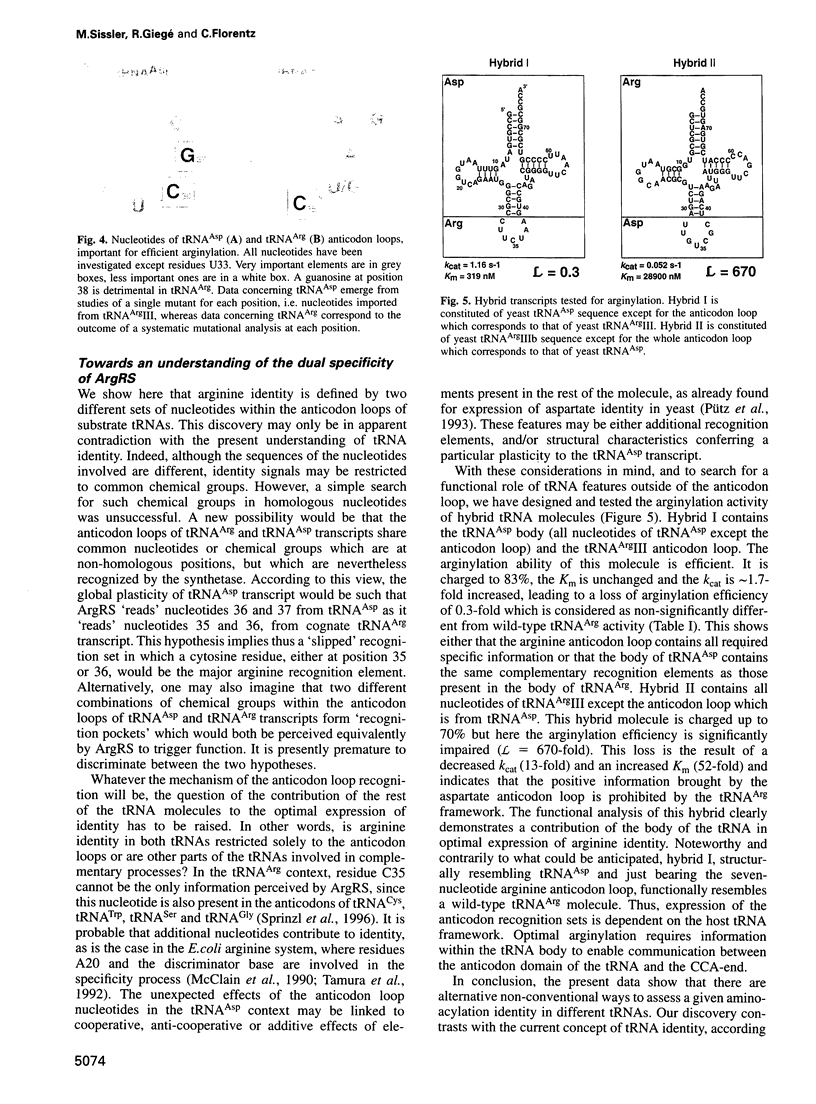
Abstract
Yeast arginyl-tRNA synthetase recognizes the non-modified wild-type transcripts derived from both yeast tRNA(Arg) and tRNA(Asp) with equal efficiency. It discriminates its cognate natural substrate, tRNA(Arg), from non-cognate tRNA(Asp) by a negative discrimination mechanism whereby a single methyl group acts as an anti-determinant. Considering these facts, recognition elements responsible for specific arginylation in yeast have been searched by studying the in vitro arginylation properties of a series of transcripts derived from yeast tRNA(Asp), considered as an arginine isoacceptor tRNA. In parallel, experiments on similar tRNA(Arg) transcripts were performed. Unexpectedly, in the tRNA(Arg) context, arginylation is basically linked to the presence of residue C35, whereas in the tRNA(Asp) context, it is deeply related to that of C36 and G37 but is insensitive to the nucleotide at position 35. Each of these nucleotides present in one host, is absent in the other host tRNA. Thus, arginine identity is dependent on two different specific recognition sets according to the tRNA framework investigated.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cavarelli J., Rees B., Ruff M., Thierry J. C., Moras D. Yeast tRNA(Asp) recognition by its cognate class II aminoacyl-tRNA synthetase. Nature. 1993 Mar 11;362(6416):181–184. doi: 10.1038/362181a0. [DOI] [PubMed] [Google Scholar]
- Crick F. H. The origin of the genetic code. J Mol Biol. 1968 Dec;38(3):367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Derrick W. B., Horowitz J. Probing structural differences between native and in vitro transcribed Escherichia coli valine transfer RNA: evidence for stable base modification-dependent conformers. Nucleic Acids Res. 1993 Oct 25;21(21):4948–4953. doi: 10.1093/nar/21.21.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekland E. H., Szostak J. W., Bartel D. P. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science. 1995 Jul 21;269(5222):364–370. doi: 10.1126/science.7618102. [DOI] [PubMed] [Google Scholar]
- Frugier M., Florentz C., Schimmel P., Giegé R. Triple aminoacylation specificity of a chimerized transfer RNA. Biochemistry. 1993 Dec 21;32(50):14053–14061. doi: 10.1021/bi00213a039. [DOI] [PubMed] [Google Scholar]
- Gangloff J., Jaozara R., Dirheimer G. Study of the interaction of yeast arginyl-tRNA synthetase with yeast tRNAArg2 and tRNAArg3 by partial digestions with cobra venom ribonuclease. Eur J Biochem. 1983 May 16;132(3):629–637. doi: 10.1111/j.1432-1033.1983.tb07410.x. [DOI] [PubMed] [Google Scholar]
- Gangloff J., Keith G., Ebel J. P., Dirheimer G. Structure of aspartate-tRNA from brewer's yeast. Nat New Biol. 1971 Mar 24;230(12):125–126. doi: 10.1038/newbio230125a0. [DOI] [PubMed] [Google Scholar]
- Gangloff J., Schutz A., Dirheimer G. Arginyl-tRNA synthetase from baker's yeast. Purification and some properties. Eur J Biochem. 1976 May 17;65(1):177–182. doi: 10.1111/j.1432-1033.1976.tb10403.x. [DOI] [PubMed] [Google Scholar]
- Giegé R., Puglisi J. D., Florentz C. tRNA structure and aminoacylation efficiency. Prog Nucleic Acid Res Mol Biol. 1993;45:129–206. doi: 10.1016/s0079-6603(08)60869-7. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Charlier J., Darte C., Dirheimer G., Giege R., de Henau S., Keith G., Parfait R., Takada V. Purification, characterization and mechanism of action of several aminoacyl-tRNA synthetases from Bacillus stearothermophilus. Experientia Suppl. 1976;26:347–362. doi: 10.1007/978-3-0348-7675-9_28. [DOI] [PubMed] [Google Scholar]
- Hall K. B., Sampson J. R., Uhlenbeck O. C., Redfield A. G. Structure of an unmodified tRNA molecule. Biochemistry. 1989 Jul 11;28(14):5794–5801. doi: 10.1021/bi00440a014. [DOI] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. Functional compensation of a recognition-defective transfer RNA by a distal base pair substitution. Biochemistry. 1992 Oct 27;31(42):10310–10314. doi: 10.1021/bi00157a019. [DOI] [PubMed] [Google Scholar]
- Keith G., Dirheimer G. Reinvestigation of the primary structure of brewer's yeast tRNA 3 Arg. Biochem Biophys Res Commun. 1980 Jan 15;92(1):116–119. doi: 10.1016/0006-291x(80)91527-2. [DOI] [PubMed] [Google Scholar]
- Martinis S. A., Schimmel P. Microhelix aminoacylation by a class I tRNA synthetase. Non-conserved base pairs required for specificity. J Biol Chem. 1993 Mar 25;268(9):6069–6072. [PubMed] [Google Scholar]
- McClain W. H., Foss K., Jenkins R. A., Schneider J. Nucleotides that determine Escherichia coli tRNA(Arg) and tRNA(Lys) acceptor identities revealed by analyses of mutant opal and amber suppressor tRNAs. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9260–9264. doi: 10.1073/pnas.87.23.9260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Nicholas H. B., Jr Differences between transfer RNA molecules. J Mol Biol. 1987 Apr 20;194(4):635–642. doi: 10.1016/0022-2836(87)90240-3. [DOI] [PubMed] [Google Scholar]
- McClain W. H. Rules that govern tRNA identity in protein synthesis. J Mol Biol. 1993 Nov 20;234(2):257–280. doi: 10.1006/jmbi.1993.1582. [DOI] [PubMed] [Google Scholar]
- McClain W. H. Transfer RNA identity. FASEB J. 1993 Jan;7(1):72–78. doi: 10.1096/fasebj.7.1.8422977. [DOI] [PubMed] [Google Scholar]
- Muramatsu T., Nishikawa K., Nemoto F., Kuchino Y., Nishimura S., Miyazawa T., Yokoyama S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988 Nov 10;336(6195):179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- Nicholas H. B., Jr, McClain W. H. An algorithm for discriminating sequences and its application to yeast transfer RNA. Comput Appl Biosci. 1987 Sep;3(3):177–181. doi: 10.1093/bioinformatics/3.3.177. [DOI] [PubMed] [Google Scholar]
- Perret V., Florentz C., Puglisi J. D., Giegé R. Effect of conformational features on the aminoacylation of tRNAs and consequences on the permutation of tRNA specificities. J Mol Biol. 1992 Jul 20;226(2):323–333. doi: 10.1016/0022-2836(92)90950-o. [DOI] [PubMed] [Google Scholar]
- Perret V., Garcia A., Grosjean H., Ebel J. P., Florentz C., Giegé R. Relaxation of a transfer RNA specificity by removal of modified nucleotides. Nature. 1990 Apr 19;344(6268):787–789. doi: 10.1038/344787a0. [DOI] [PubMed] [Google Scholar]
- Perret V., Garcia A., Puglisi J., Grosjean H., Ebel J. P., Florentz C., Giegé R. Conformation in solution of yeast tRNA(Asp) transcripts deprived of modified nucleotides. Biochimie. 1990 Oct;72(10):735–743. doi: 10.1016/0300-9084(90)90158-d. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Pütz J., Florentz C., Giegé R. Influence of tRNA tertiary structure and stability on aminoacylation by yeast aspartyl-tRNA synthetase. Nucleic Acids Res. 1993 Jan 11;21(1):41–49. doi: 10.1093/nar/21.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pütz J., Florentz C., Benseler F., Giegé R. A single methyl group prevents the mischarging of a tRNA. Nat Struct Biol. 1994 Sep;1(9):580–582. doi: 10.1038/nsb0994-580. [DOI] [PubMed] [Google Scholar]
- Pütz J., Puglisi J. D., Florentz C., Giegé R. Additive, cooperative and anti-cooperative effects between identity nucleotides of a tRNA. EMBO J. 1993 Jul;12(7):2949–2957. doi: 10.1002/j.1460-2075.1993.tb05957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pütz J., Puglisi J. D., Florentz C., Giegé R. Identity elements for specific aminoacylation of yeast tRNA(Asp) by cognate aspartyl-tRNA synthetase. Science. 1991 Jun 21;252(5013):1696–1699. doi: 10.1126/science.2047878. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Steitz T. A. Structural basis of anticodon loop recognition by glutaminyl-tRNA synthetase. Nature. 1991 Jul 18;352(6332):213–218. doi: 10.1038/352213a0. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Saks M. E., Sampson J. R., Abelson J. N. The transfer RNA identity problem: a search for rules. Science. 1994 Jan 14;263(5144):191–197. doi: 10.1126/science.7506844. [DOI] [PubMed] [Google Scholar]
- Shimizu M., Asahara H., Tamura K., Hasegawa T., Himeno H. The role of anticodon bases and the discriminator nucleotide in the recognition of some E. coli tRNAs by their aminoacyl-tRNA synthetases. J Mol Evol. 1992 Nov;35(5):436–443. doi: 10.1007/BF00171822. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Steegborn C., Hübel F., Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1996 Jan 1;24(1):68–72. doi: 10.1093/nar/24.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Himeno H., Asahara H., Hasegawa T., Shimizu M. In vitro study of E.coli tRNA(Arg) and tRNA(Lys) identity elements. Nucleic Acids Res. 1992 May 11;20(9):2335–2339. doi: 10.1093/nar/20.9.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990 Aug 3;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Weissenbach J., Martin R., Dirheimer G. The primary structure of tRNAIIAgr from brewers' yeast. 2. Partial digestion with ribonuclease T1 and derivation of the complete sequence. Eur J Biochem. 1975 Aug 15;56(2):527–532. doi: 10.1111/j.1432-1033.1975.tb02258.x. [DOI] [PubMed] [Google Scholar]
- Wyatt J. R., Chastain M., Puglisi J. D. Synthesis and purification of large amounts of RNA oligonucleotides. Biotechniques. 1991 Dec;11(6):764–769. [PubMed] [Google Scholar]




