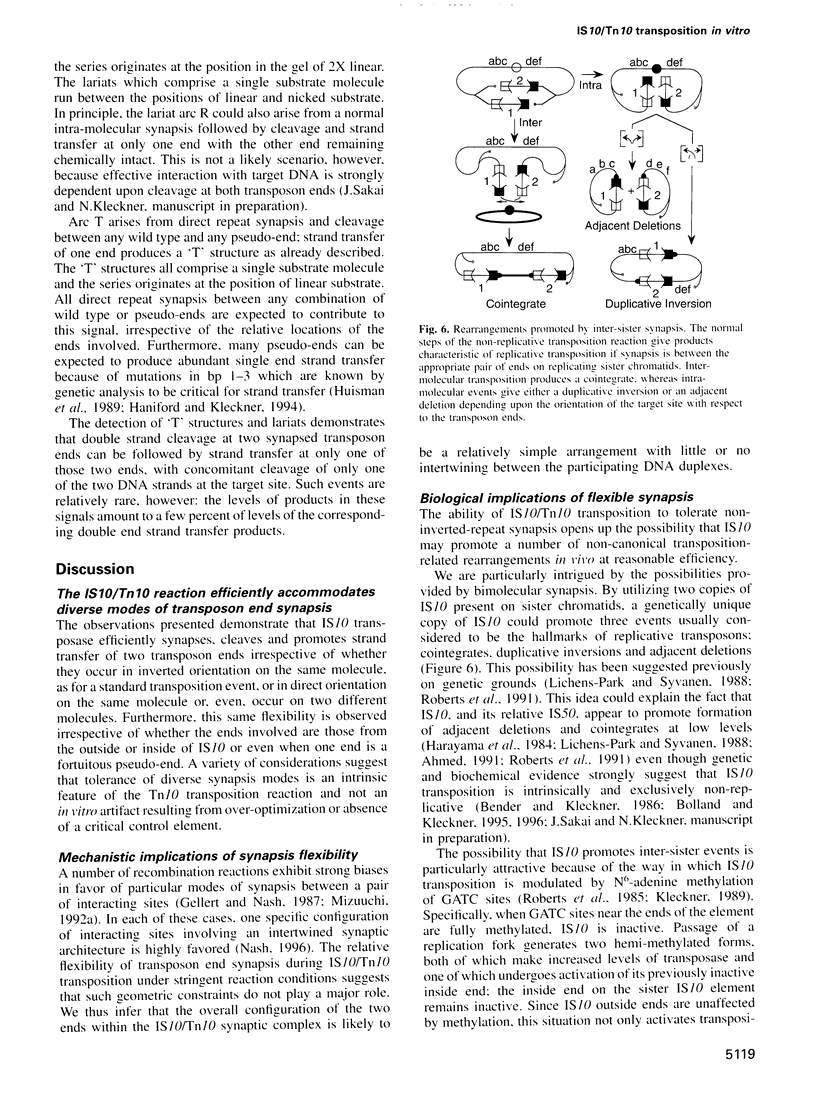
Abstract
Transposon Tn10 and its component insertion sequence IS10 move by non-replicative transposition. We have studied the array of reaction intermediates and products in a high efficiency in vitro IS10/Tn10 transposition reaction. Synapsis of two transposon ends, followed by cleavage and strand transfer, can occur very efficiently irrespective of the relative locations and orientations of the two ends. The two participating ends can occur in inverted or direct orientation on the same molecule or, most importantly, on two different molecules. This behavior contrasts sharply with that of Mu, in which transposition is strongly biased in favor of inverted repeat synapsis. Mechanistically, the absence of discrimination amongst various end configurations implies that the architecture within the IS10/Tn10 synaptic complex is relatively simple, i.e. lacking any significant intertwining of component DNA strands. Biologically these observations are important because they suggest that the IS10 insertion sequence module has considerable flexibility in the types of DNA rearrangements that it can promote. Most importantly, it now seems highly probable that a single non-replicative IS10 element can promote DNA rearrangements usually attributed to replicative transposition, i.e. adjacent deletions and cointegrates, by utilizing transposon ends on two sister chromosomes. Other events which probably also contribute to the diversity of IS10/Tn10-promoted rearrangements are discussed.
Full text
PDF
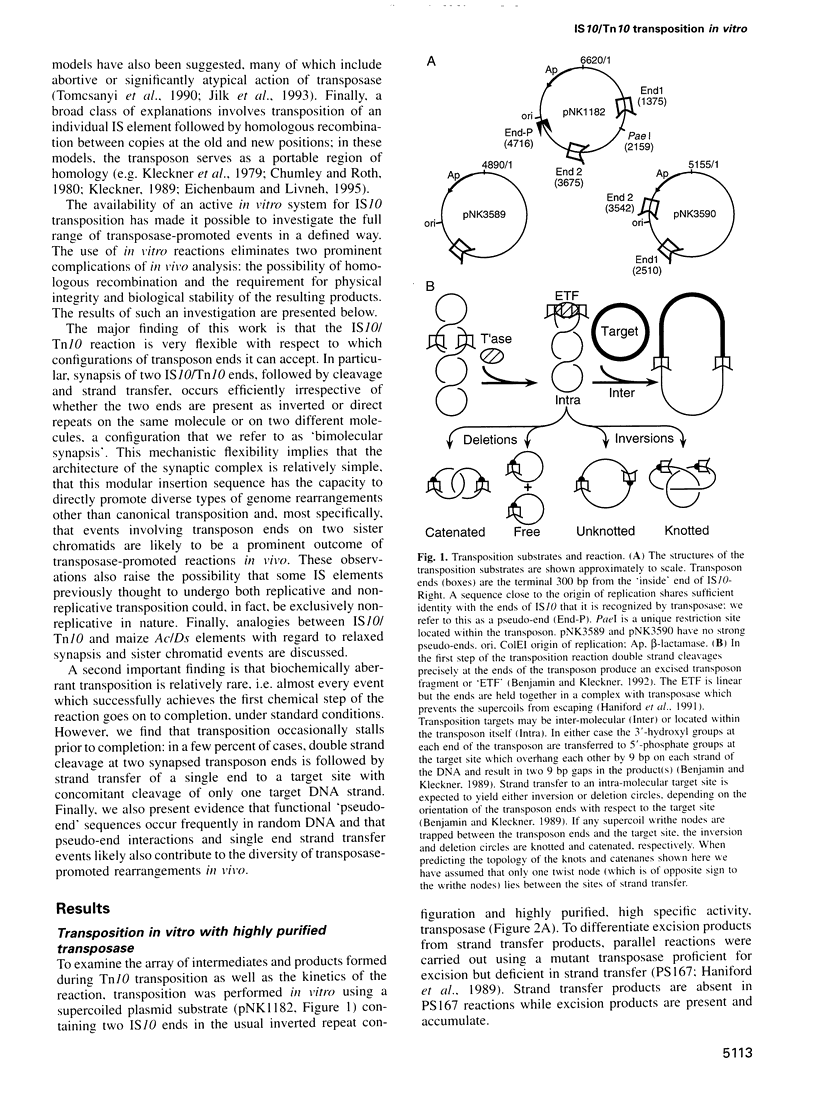

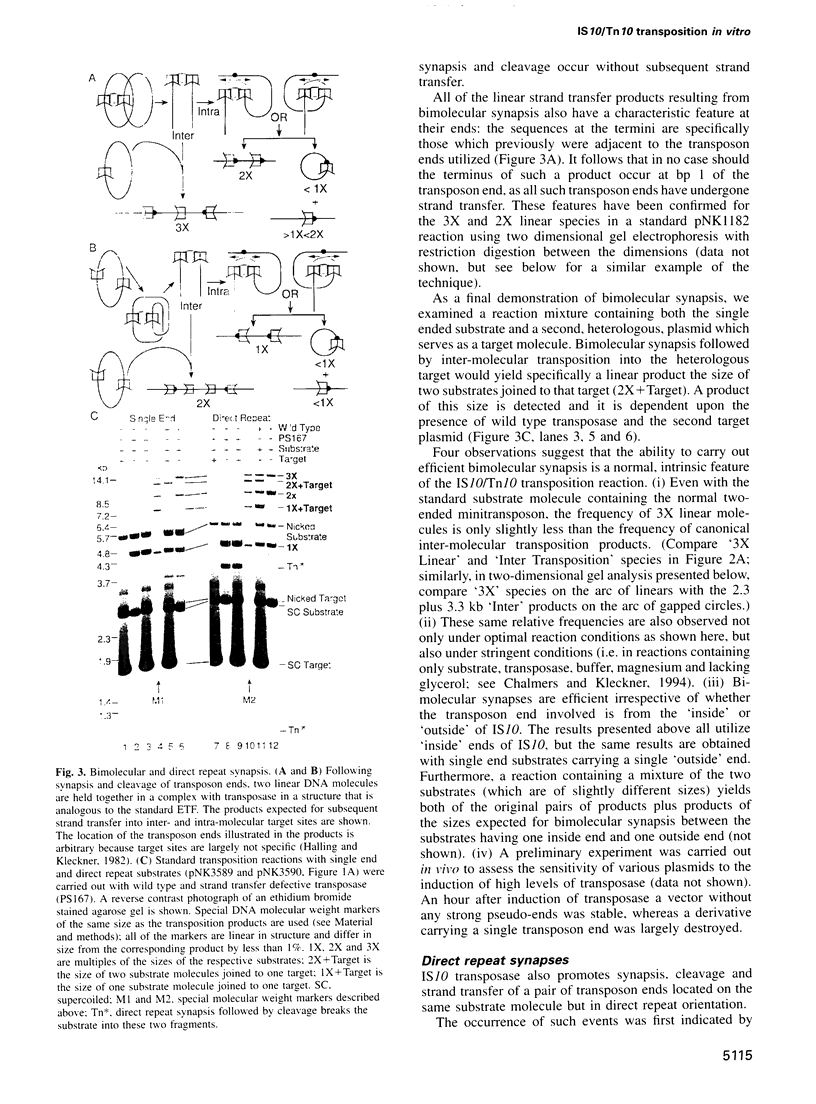

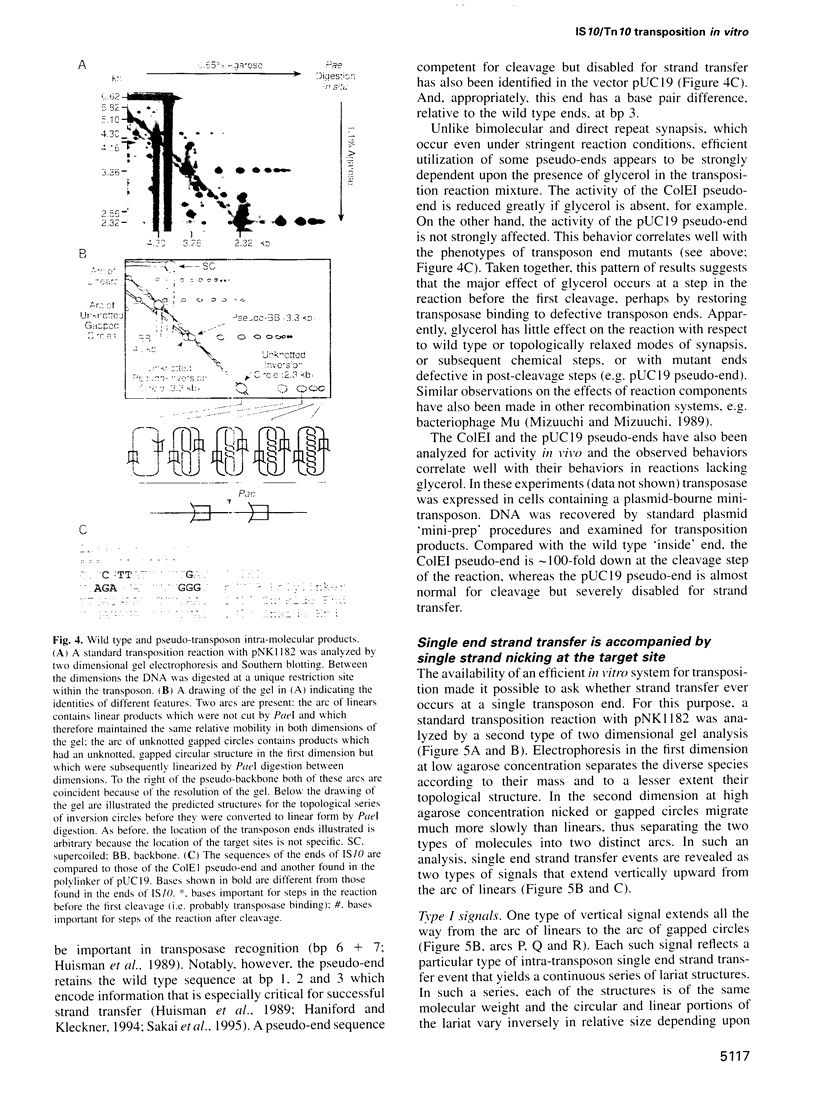




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arthur A., Sherratt D. Dissection of the transposition process: a transposon-encoded site-specific recombination system. Mol Gen Genet. 1979 Oct 1;175(3):267–274. doi: 10.1007/BF00397226. [DOI] [PubMed] [Google Scholar]
- Bender J., Kleckner N. Genetic evidence that Tn10 transposes by a nonreplicative mechanism. Cell. 1986 Jun 20;45(6):801–815. doi: 10.1016/0092-8674(86)90555-6. [DOI] [PubMed] [Google Scholar]
- Benjamin H. W., Kleckner N. Excision of Tn10 from the donor site during transposition occurs by flush double-strand cleavages at the transposon termini. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4648–4652. doi: 10.1073/pnas.89.10.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin H. W., Kleckner N. Intramolecular transposition by Tn10. Cell. 1989 Oct 20;59(2):373–383. doi: 10.1016/0092-8674(89)90298-5. [DOI] [PubMed] [Google Scholar]
- Berg D. E. Structural requirement for IS50-mediated gene transposition. Proc Natl Acad Sci U S A. 1983 Feb;80(3):792–796. doi: 10.1073/pnas.80.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biel S. W., Berg D. E. Mechanism of IS1 transposition in E. coli: choice between simple insertion and cointegration. Genetics. 1984 Oct;108(2):319–330. doi: 10.1093/genetics/108.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolland S., Kleckner N. The three chemical steps of Tn10/IS10 transposition involve repeated utilization of a single active site. Cell. 1996 Jan 26;84(2):223–233. doi: 10.1016/s0092-8674(00)80977-0. [DOI] [PubMed] [Google Scholar]
- Bolland S., Kleckner N. The two single-strand cleavages at each end of Tn10 occur in a specific order during transposition. Proc Natl Acad Sci U S A. 1995 Aug 15;92(17):7814–7818. doi: 10.1073/pnas.92.17.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers R. M., Kleckner N. Tn10/IS10 transposase purification, activation, and in vitro reaction. J Biol Chem. 1994 Mar 18;269(11):8029–8035. [PubMed] [Google Scholar]
- Eichenbaum Z., Livneh Z. Intermolecular transposition of IS10 causes coupled homologous recombination at the transposition site. Genetics. 1995 Jul;140(3):861–874. doi: 10.1093/genetics/140.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English J. J., Harrison K., Jones JDG. Aberrant Transpositions of Maize Double Ds-Like Elements Usually Involve Ds Ends on Sister Chromatids. Plant Cell. 1995 Aug;7(8):1235–1247. doi: 10.1105/tpc.7.8.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English J., Harrison K., Jones J. D. A genetic analysis of DNA sequence requirements for Dissociation state I activity in tobacco. Plant Cell. 1993 May;5(5):501–514. doi: 10.1105/tpc.5.5.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Nash H. Communication between segments of DNA during site-specific recombination. 1987 Jan 29-Feb 4Nature. 325(6103):401–404. doi: 10.1038/325401a0. [DOI] [PubMed] [Google Scholar]
- Halling S. M., Kleckner N. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell. 1982 Jan;28(1):155–163. doi: 10.1016/0092-8674(82)90385-3. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Benjamin H. W., Kleckner N. Kinetic and structural analysis of a cleaved donor intermediate and a strand transfer intermediate in Tn10 transposition. Cell. 1991 Jan 11;64(1):171–179. doi: 10.1016/0092-8674(91)90218-n. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Chelouche A. R., Kleckner N. A specific class of IS10 transposase mutants are blocked for target site interactions and promote formation of an excised transposon fragment. Cell. 1989 Oct 20;59(2):385–394. doi: 10.1016/0092-8674(89)90299-7. [DOI] [PubMed] [Google Scholar]
- Haniford D., Kleckner N. Tn 10 transposition in vivo: temporal separation of cleavages at the two transposon ends and roles of terminal basepairs subsequent to interaction of ends. EMBO J. 1994 Jul 15;13(14):3401–3411. doi: 10.1002/j.1460-2075.1994.tb06643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Oguchi T., Iino T. Does Tn10 transpose via the cointegrate molecule? Mol Gen Genet. 1984;194(3):444–450. doi: 10.1007/BF00425556. [DOI] [PubMed] [Google Scholar]
- Huisman O., Errada P. R., Signon L., Kleckner N. Mutational analysis of IS10's outside end. EMBO J. 1989 Jul;8(7):2101–2109. doi: 10.1002/j.1460-2075.1989.tb03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilk R. A., Makris J. C., Borchardt L., Reznikoff W. S. Implications of Tn5-associated adjacent deletions. J Bacteriol. 1993 Mar;175(5):1264–1271. doi: 10.1128/jb.175.5.1264-1271.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Reichardt K., Botstein D. Inversions and deletions of the Salmonella chromosome generated by the translocatable tetracycline resistance element Tn10. J Mol Biol. 1979 Jan 5;127(1):89–115. doi: 10.1016/0022-2836(79)90461-3. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Kunze R. The maize transposable element activator (Ac). Curr Top Microbiol Immunol. 1996;204:161–194. doi: 10.1007/978-3-642-79795-8_8. [DOI] [PubMed] [Google Scholar]
- Lane D., Cavaillé J., Chandler M. Induction of the SOS response by IS1 transposase. J Mol Biol. 1994 Sep 30;242(4):339–350. doi: 10.1006/jmbi.1994.1585. [DOI] [PubMed] [Google Scholar]
- Lichens-Park A., Syvanen M. Cointegrate formation by IS50 requires multiple donor molecules. Mol Gen Genet. 1988 Feb;211(2):244–251. doi: 10.1007/BF00330600. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K. Polynucleotidyl transfer reactions in transpositional DNA recombination. J Biol Chem. 1992 Oct 25;267(30):21273–21276. [PubMed] [Google Scholar]
- Mizuuchi K. Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- Mizuuchi M., Mizuuchi K. Efficient Mu transposition requires interaction of transposase with a DNA sequence at the Mu operator: implications for regulation. Cell. 1989 Jul 28;58(2):399–408. doi: 10.1016/0092-8674(89)90854-4. [DOI] [PubMed] [Google Scholar]
- Morisato D., Kleckner N. Tn10 transposition and circle formation in vitro. Cell. 1987 Oct 9;51(1):101–111. doi: 10.1016/0092-8674(87)90014-6. [DOI] [PubMed] [Google Scholar]
- Polard P., Prère M. F., Fayet O., Chandler M. Transposase-induced excision and circularization of the bacterial insertion sequence IS911. EMBO J. 1992 Dec;11(13):5079–5090. doi: 10.1002/j.1460-2075.1992.tb05615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polard P., Seroude L., Fayet O., Prère M. F., Chandler M. One-ended insertion of IS911. J Bacteriol. 1994 Feb;176(4):1192–1196. doi: 10.1128/jb.176.4.1192-1196.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D., Hoopes B. C., McClure W. R., Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell. 1985 Nov;43(1):117–130. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- Ross D. G., Swan J., Kleckner N. Physical structures of Tn10-promoted deletions and inversions: role of 1400 bp inverted repetitions. Cell. 1979 Apr;16(4):721–731. doi: 10.1016/0092-8674(79)90088-6. [DOI] [PubMed] [Google Scholar]
- Sakai J., Chalmers R. M., Kleckner N. Identification and characterization of a pre-cleavage synaptic complex that is an early intermediate in Tn10 transposition. EMBO J. 1995 Sep 1;14(17):4374–4383. doi: 10.1002/j.1460-2075.1995.tb00112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomcsanyi T., Berg C. M., Phadnis S. H., Berg D. E. Intramolecular transposition by a synthetic IS50 (Tn5) derivative. J Bacteriol. 1990 Nov;172(11):6348–6354. doi: 10.1128/jb.172.11.6348-6354.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Derbyshire K. M., Hughson F. M., Grindley N. D. Replicative and conservative transpositional recombination of insertion sequences. Cold Spring Harb Symp Quant Biol. 1984;49:251–260. doi: 10.1101/sqb.1984.049.01.029. [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Schaus N. A., Grindley N. D. Insertion sequence duplication in transpositional recombination. Science. 1983 Nov 18;222(4625):755–765. doi: 10.1126/science.6314502. [DOI] [PubMed] [Google Scholar]