Abstract
Although acquired mutations in KIT are commonly detected in various categories of mastocytosis, the methodologies applied to detect and quantify the mutant type and -burden in various tissues and cells are poorly defined. We here propose a consensus on methodologies used to detect KIT mutations in patients with mastocytosis at diagnosis and in the follow up with sufficient precision and sensitivity in daily practice. In addition, we provide recommendations for sampling and storage of diagnostic material as well as a robust diagnostic algorithm. Using highly-sensitive assays, KIT D816V can be detected in peripheral blood leukocytes in most patients with systemic mastocytosis (SM) which is a major step forward in screening and SM detection. In addition, the KIT D816V allele burden can be followed quantitatively during the natural course or during therapy in these patients. Our recommendations should greatly facilitate diagnostic and follow up investigations in SM in daily practice as well as in clinical trials. In addition, the new tools and algorithms proposed should lead to a more effective screen, early detection of SM, and help avoid unnecessary referrals.
Keywords: mast cell, mastocytosis, KIT, mutation, allele burden, allele specific PCR, classification, prognostication, monitoring of residual disease
Introduction
Mast cells (MCs) and their committed progenitors express KIT,{Krishnaswamy, 2006 #903} a transmembrane type III tyrosine kinase receptor also found in melanocytes, hematopoietic stem cells, germ cells and interstitial cells of Cajal.{Miettinen, 2005 #646} The ligand of KIT, stem cell factor (SCF), induces MC development from uncommitted and MC-committed hematopoietic precursor cells.{Ashman, 2000 #337} In the human system, MC-committed progenitors reside in the bone marrow (BM) and circulate in the peripheral blood (PB), whereas mature MCs usually are tissue-fixed non-circulating cells, found in virtually all vascularized organs.{Kirshenbaum, 1999 #549;Matarraz, 2008 #1493;Teodosio, 2010 #91}
Mastocytosis is a rare and heterogeneous disease characterized by the accumulation of clonal MCs in one or multiple organs. The disease is mostly acquired, although rare familial cases have been described.{Chang, 2001 #389} Mastocytosis can affect both children and adults.{Valent, 2012 #1559} In most pediatric patients, the disease affects only the skin, with different types of lesions.{Briley, 2008 #370} In these cases, the disease is termed cutaneous mastocytosis, CM. In CM the biopsy of lesional skin reveals MC infiltration, sometimes with atypical MCs forming aggregates in local areas.{Vano-Galvan, 2011 #1447} In children, the disease has a tendency to regress spontaneously,{Arock, 2010 #80} although some persist into adulthood.{Fried, 2013 #1466} By contrast, in most adult patients, the disease is systemic and chronic and affects almost invariably the BM. These cases are called systemic mastocytosis, SM. In most patients, indolent SM (ISM) is diagnosed and the skin is affected.{Arock, 2010 #80} More aggressive subtypes of SM are rare and include aggressive SM (ASM) and MC leukemia (MCL). Depending on data-source and center, 5 to 20% of all SM patients present with or develop an associated clonal hematological non-MC-lineage disease (SM-AHNMD).{Wang, 2013 #39} Symptoms in mastocytosis may occur from diverse effects of mediators that are released from neoplastic MCs, and, in case of ASM or MCL, from organ infiltration by MCs, with subsequent organomegaly and/or organ damage.{Metcalfe, 2008 #641}
Patients with mastocytosis are diagnosed and classified according to the proposal of the World Health Organization (WHO) which distinguishes 7 categories of mastocytosis: cutaneous mastocytosis (CM), indolent systemic mastocytosis (ISM), SM with an associated clonal hematopoietic non MC disease (SM-AHNMD), agressive systemic mastocytosis (ASM), MC leukemia (MCL), MC sarcoma (MCS) and extracutaneous mastocytoma.{Valent, 2001 #1646;Valent, 2001 #1490;Horny, 2008 #1449}. For some of these categories (CM, ISM, SM-AHNMD, ASM and MCL, subvariants have been identified, based on clinical and/or biological features.{Valent, 2001 #1646;Valent, 2001 #1490;Horny, 2008 #1449} For patients with SM, the WHO has provided one major diagnostic criterion (infiltrates of >15 aggregated MCs identified through tryptase immuno-histochemistry or other stains in sections obtained from BM and/or other extracutaneous organ(s)) and four minor SM diagnostic criteria based on i) the presence of atypical MCs in lesional tissues, ii) the presence of an activating point mutation in codon 816 in KIT in BM, PB, or any extracutaneous organ, iii) the abberant expression of CD2 and or CD25 by neoplastic MCs and iv) a persistently elevated (> 20 ng/mL) serum tryptase level .{Valent, 2007 #1598} The diagnosis SM can be established when at least the major and one minor or three minor criteria are found.{Valent, 2007 #1598}
In adult patients, an activating KIT D816V mutation (Nucleotide sequence 5′-3′: agagTcatc, position 2247), located in the phosphotransferase domain (PTD) of the receptor is found in >80% of all cases. The exact percentages vary depending on disease subtypes (e.g., ISM vs. ASM) and cell source (e.g., BM vs. PB).{Garcia-Montero, 2006 #1665;Sanchez-Munoz, 2011 #1494;Erben, 2014 #1659;Kristensen, 2014 #12} By contrast, a KIT mutation is found in skin biopsy samples in nearly 75% of affected children but only one third present with KIT D816V.{Bodemer, 2010 #96} Other KIT mutations found in childhood mastocytosis are mainly located in the extracellular domain of the receptor, the most frequently retrieved being a deletion at position 419 of the protein (Nucleotide sequence 5′-3′: acttac — aggctc, position 1255).{Bodemer, 2010 #96}
In most SM patients, particularly in ISM, the number of neoplastic MCs in the BM is low (low MC burden).{Tan, 2006 #143} Thus, the relatively low sensitivity of conventional PCR in combination with sequencing of PCR products may result in false-negative cases. Indeed, in these patients, the median level of KIT D816V mutant alleles (also referred as to “KIT D816V allele burden”), is often below 1%, as measured by recently developed allele-specific quantitative PCR assays, that are based on either DNA or on RNA/cDNA (ASO-qPCR), but have a similar (high) sensitivity.{Kristensen, 2012 #59;Erben, 2014 #1659;Hoermann, 2014 #4} In addition, although rarely seen, SM patients may present with KIT mutations other than D816V, e.g., D816H, D816Y, or even mutations outside the PTD of KIT.{Bibi, 2014 #5} The latter can only be detected by PCR assays that are not routinely used, e.g., RFLP or HPLC of PCR products, but not by ASO-qPCR.{Valent, 2005 #1621;Erben, 2014 #1659}
Recent reports using ASO-qPCR (on DNA or RNA/cDNA) have shown correlations between KIT D816V allele burden and clinical parameters such as disease subtype, prognosis and survival.{Escribano, 2009 #419;Erben, 2014 #1659;Hoermann, 2014 #4} Moreover, serial measurements of the KIT D816V allele burden by these ASO-qPCR techniques appear useful for monitoring of residual disease in aggressive subtypes during or after cytoreductive therapy or allogeneic stem cell transplantation.{Erben, 2014 #1659;Hoermann, 2014 #4}
The present article provides consensus recommendations from members of the European Competence Network on Mastocytosis (ECNM{Valent, 2004 #1616;Valent, 2012 #1548}) for standardized approaches and techniques to detect KIT mutations and to measure KIT mutant burden as part of the routine diagnostic work-up, prognostication and follow-up during or after treatment in patients with mastocytosis. Consensus recommendations have been developed and discussed among co-authoring members of the ECNM between September 2013 and September 2014. Consensus statements were discussed until complete consensus (all experts agreed) was reached (or not reached by September 2014) and only those consensus statements where complete consensus was reached have been included as such in the current article. Other points of discussions are labeled as open issues (under discussion or still under development) in our consensus document.
Short overview on the spectrum of KIT mutations and their clinical implications
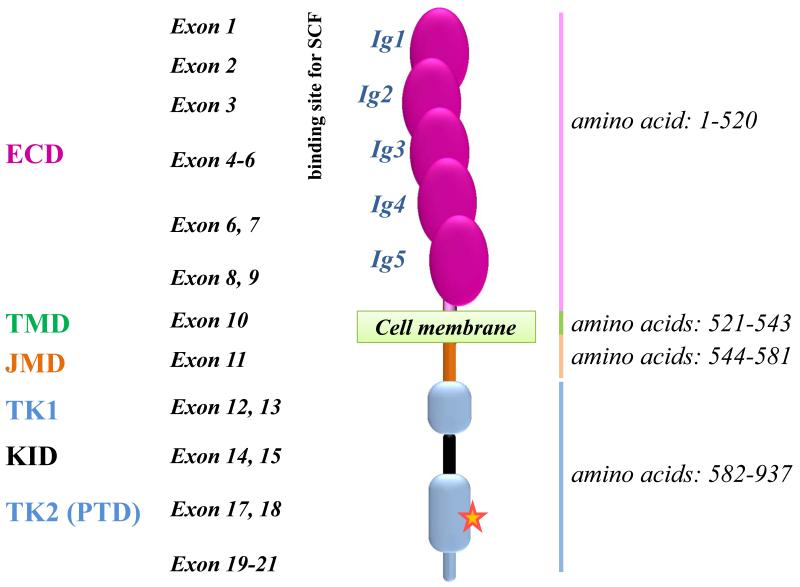
KIT is encoded by a 21 exon-containing gene located on human chromosome 4q12.{Giebel, 1992 #454} The resulting 976 amino acid-containing receptor has a molecular weight of 145 kDa.{Chabot, 1988 #1452} A schematic representation of wild type KIT (KIT WT) is shown in Figure 1. The receptor is composed of an extracellular domain (ECD), a transmembrane domain (TMD), a juxtamembrane domain (JMD) and a tyrosine kinase domain (TKD). The TKD contains an ATP-binding site and a phosphotransferase domain (PTD) (Figure 1). A few single nucleotide polymorphisms (SNPs) which might account for susceptibility to various diseases have been described in the KIT gene.{Cheng, 2013 #1677} The most common SNP found in the KIT gene in human is an ATG to CTG transition in codon 541 (exon 10) giving rise to a M541L KIT protein sequence variation, which seems to confer increased sensitivity to the KIT ligand (SCF), although the clinical impact of this SNP remains unclear.{Nagata, 1996 #1675}
Figure 1. Structure of normal KIT (KIT Wild-Type).
The KIT gene, located on chromosome 4q12 in humans, contains 21 exons transcribed/translated into a transmembrane receptor tyrosine kinase (RTK) of 145 kDa and 976 amino acids. KIT structure is characterized by 5 Ig-like subunits in the extracellular domain (ECD) which contains a ligand binding site (SCF for KIT) and a dimerization site, and is linked to a cytoplasmic region by a single transmembrane helix. The cytoplasmic region of KIT consists of an autoinhibitory juxta-membrane domain (JMD) and a kinase domain (KD) arranged in a proximal (N−) and a distal (C−) lobe linked by a hinge region. The C-lobe of RTKs type III includes a large Kinase Insert Domain (KID) of ~ 60-100 residues. The red star represents the position 816 where an Asp to Val point mutation (KIT D816V) is found in > 80% of adult systemic mastocytosis patients. The receptor is presented under its monomeric form, whereas a dimer results from stem cell factor (SCF) ligation. ECD: Extracellular domain; JMD: Juxtamembrane domain; KID: Kinase insert domain; PTD: phosphotransférase domain; TK: tyrosine kinase; TMD: transmembrane domain.
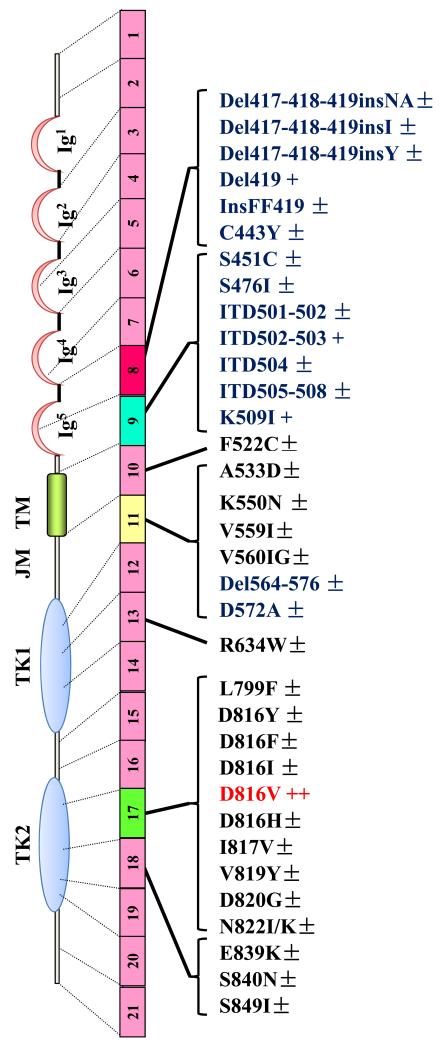
Activating mutations in KIT were first described in SM patients,{Longley, 1996 #612} and more recently in children with CM.{Bodemer, 2010 #96} In adult patients with ISM, ASM or SM-AHNMD, these mutations affect mainly the PTD of KIT, encoded by exon 17, usually at position 816 (most commonly KIT D816V),{Orfao, 2007 #129} whereas in children, ECD mutants are also found.{Bodemer, 2010 #96} In MCL, the presence of the KIT D816V mutation seems less frequent than in other categories of SM.{Georgin-Lavialle, 2013 #44} Finally, in the rare cases of familial mastocytosis, often presenting as pediatric CM, most cases exhibit no KIT mutations or uncommon germline mutations.{Zhang, 2006 #161;Wasag, 2011 #71;Hoffmann, 2008 #497;Wohrl, 2013 #1465;Pollard, 2014 #1674} However, even somatic KIT mutations have been described in familial mastocytosis.{Bodemer, 2010 #96;Broesby-Olsen, 2012 #56;Zanotti, 2013 #43} The Figure 2 presents all current KIT mutations retrieved in the literature for adult and children patient, together with their approximate frequencies.
Figure 2. Representation of the structure of KIT, illustrating the localization of the more frequently observed mutations in the KIT sequence in pediatric and adult patients with mastocytosis.
The receptor is presented under its monomeric form, whereas its wild-type counterpart dimerizes upon ligation with SCF before being activated in normal cells. In children,he KIT D816V PTD mutant (in red) is found in nearly 30% of the patients, whereas the ECD mutants (in blue) are found in nearly 40% of the affected children. In adults, depending of the categoty of mastocytosis, the KIT D81V mutant is found in at least 80% of all patients. The complete list of KIT mutants retrieved in the literature for mastocytosis is depicted here. In children, the structure of KIT is found WT in around 25% of the patients analyzed, whereas in adults, KIT is found WT in less than 20% of all patients analyzed so far. Some of the mutations (in black) are found only in a very few number of patients. Del: deletion; ECD: Extracellular domain; Ins: Insertion; ITD: Internal tandem duplication; JMD: Juxtamembrane domain; KI: Kinase insert; PTD: phosphotransferase domain; TMD: Transmembrane domain. ±: mutation found in less than 1% of the patients; +: mutation found in 1 to 5% of the patients; ++: mutation found in around 30% of pediatric patients and in > 80% of all adult patients.
SM-AHNMD (5 to 20% of all SM cases) is a special subvariant of the disease.{Wang, 2013 #39} In most patients, an associated myeloid neoplasm is diagnosed, such as chronic myelomonocytic leukemia (CMML), atypical chronic myeloid leukemia (aCML), myelodyspastic/myeloproliferative neoplasm unclassifiable (MDS/MPN), MDS, chronic eosinophilic leukemia (CEL), AML or infrequently BCR-ABL1-positive MPN or MPN-unclassifiable.{Sperr, 2002 #259;Stoecker, 2012 #1459;Wang, 2013 #39;Valent, 2013 #36;Piazza, #1492} By contrast, lymphoid variants of AHNMD (multiple myeloma, B-cell lymphoma, chronic lymphocytic leukemia) are rarely found.{Sperr, 2002 #259;Stellmacher, 2004 #1627;Horny, 2006 #1608} Interestingly, a recent study of 48 SM-AHNMD patients has shown that the KIT D816V mutation can be detected in almost all these individuals except those with SM with an associated CEL.{Sotlar, 2010 #1573} In most patients with SM-CMML, KIT D816V is detectable in AHNMD cells, suggesting the involvement of a common myelomono- and mastocytic precursor. In patients with SM-MPN and SM-AML, the KIT mutant D816V is also detectable, albeit at a lower frequency.{Fritsche-Polanz, 2010 #1661;Fritsche-Polanz, 2010 #1661} Finally, in patients with lymphoproliferative AHNMDs, the AHNMD cells are usually negative for KIT 816.{Garcia-Montero, 2006 #1665;Sotlar, 2010 #1573} Table 1 shows the frequency and distribution of the different KIT mutations in the diverse subtypes of mastocytosis based on current knowledge and recently published data.{Bibi, 2014 #5}
Table 1. Major characteristics of the various categories of mastocytosis, and distribution of KIT mutations along these categories.
| Categories of disease and their major characteristics | Relative frequency | Prognosis | Type and frequency of KIT mutations/presence of additional genetic lesions |
|---|---|---|---|
Pediatric mastocytosis
|
++ | Usually very good |
|
Indolent Systemic Mastocytosis (ISM)
|
++ | Very good to good |
|
Smoldering Systemic Mastocytosis
|
+ | Relatively good |
|
Aggressive Systemic Mastocytosis (ASM)
|
+/− | Poor |
|
Systemic mastocytosis with an Associated Hematological non MC disease (SM-AHNMD)
|
+ | Depending on the type of SM and on the prognosis of the AHNMD |
|
Mast Cell Leukemia (MCL)
|
+/− | Very poor |
|
Mast Cell Sarcoma (MCS)
|
+/−− | Very poor |
|
Familial mastocytosis
|
+/− | Usually very good |
|
B-findings: hypercellular BM, dysplasia, organomegaly without organ failure, and high serum tryptase levels.
C-findings: organ dysfunction, such as cytopenia (ANC<1×109/L, Hb<10 g/dL, or platelets<100×109/l), hepatomegaly with impaired liver function, palpable splenomegaly with signs of hypersplenism, malabsorption with significant hypoalbuminemia, significant weight loss >10% over the last 6months and/or large osteolyses.
AHNMD: Associated clonal hematologic non-mast cell lineage diseases; ASM: aggressive systemic mastocytosis; BM: bone marrow; CM: cutaneous mastocytosis; ECD: extracellular domain; ISM: indolent systemic mastocytosis; MCL: mast cell leukemia; MCS: mast cell sarcoma; MCs: mast cells; WT: wild type.
++: frequent; +: relatively frequent; +/−: rare; +/−− extremely rare.
For each SM patients, the precise knowledge of the structure of KIT in neoplastic cells is of great clinical significance since the A-loop mutations D816V/H/Y/N induce resistance to imatinib in vitro as well as ex vivo in patients’ cells,{Frost, 2002 #184;Lasota, 2007 #580} This resistance has been attributed to the fact that the KIT D816V mutant disrupts the structure of the receptor, leading permanently to an active conformation, which cannot be targeted by imatinib.{Foster, 2004 #1676;Laine, 2011 #70} This resistance of the most common KIT mutant against imatinib found in SM, has led to the use of non-targeted cytoreductive therapy, such as cladribine (2CdA),{Kluin-Nelemans, 2003 #564} or interferon-α,{Hauswirth, 2004 #490} and to clinical trials with novel tyrosine kinase inhibitors (TKIs) that overcome resistance in these patients, such as PKC412 (midostaurin) with encouraging results.{Gotlib, 2010 #1483;Gotlib, 2012 #1484}} By contrast, the rare SM patients presenting with a KIT mutant outside the PTD (e.g. JMD or ECD mutants), or with no KIT mutation may respond to imatinib. {Zhang, 2006 #161;Hoffmann, 2008 #497;Mital, 2011 #76;Alvarez-Twose, 2012 #1485}
Recommendations for cells and assays to be used for KIT mutation analysis
This section will first describe the differences in the management between pediatric and adult patients. We will then highlight the various samples and cell types to be used for detection of KIT mutations in different subtypes of mastocytosis, as well as the different techniques used for this purpose, their advantages and possible limitations. Finally, we will provide recommendations for the most appropriate procedures to be applied at time of diagnosis and during follow-up.
Pediatric mastocytosis
In childhood patients with CM, BM investigations are usually not required unless clinical findings and symptoms suggest the presence of a relevant (aggressive) systemic disease.{Fried, 2013 #1466} In the latter case, a thorough BM examination is recommended, including KIT mutation analysis.{Valent, 2007 #1598} In some patients in whom the diagnosis CM is in question because of atypical skin lesions and/or a questionable histology, lesional skin can be examined for the presence of KIT mutations.{Bodemer, 2010 #96} However this test is not regarded as standard in typical CM, which may change in the future, especially if it appears that the presence of a particular mutation is associated with a certain prognosis or with disease persistence into adulthood. In some of these cases, sequencing of the entire KIT coding region may be required and should be performed according to standard methodologies.{Bodemer, 2010 #96}
Adult patients
In SM, neoplastic MCs usually reside in the BM but are not found in the PB.{Krishnaswamy, 2006 #903} Therefore, it has been standard to examine BM cells for the presence of KIT D816V.{Valent, 2014 #1529} Until recently, the most frequently used assay for the detection of KIT mutations has been conventional sequencing of PCR-derived amplicons. However, the level of sensitivity of this technique is rather low (10-20%) which may lead to false-negative results, especially when the degree of BM infiltration is low.{Corless, 2006 #141} Consequently, considerable efforts have been made recently to improve assay sensitivities using different approaches, including PCR amplification followed by restriction digestion (PCR+RFLP),{Tan, 2006 #143} peptide nucleic acid (PNA)-mediated PCR clamping,{Sotlar, 2003 #1671} nested RT-PCR followed by denaturing high-performance liquid chromatography (DHPLC),{Erben, 2014 #1659} and ASO-qPCR assays.{Schumacher, 2008 #132;Kristensen, 2011 #77;Erben, 2014 #1659;Kristensen, 2014 #12} More sophisticated techniques, including enrichment of MCs and other cell types from BM samples via laser micro-dissection or fluorescence-activated cell sorting (FACS) followed by PNA-mediated PCR-clamping technique, are used for research purposes but are not routinely used for diagnostic algorithms in most centers.{Sotlar, 2000 #1673;Sotlar, 2002 #1672;Sotlar, 2003 #1671;Escribano, 2009 #419;Teodosio, 2012 #66}
Recommended tissues and cells for KIT mutation analysis
While it is generally recommended that fresh samples should be used whenever possible, storage of cells and isolation of total RNA or DNA from stored cells is also acceptable, provided that standard-methods are applied: cell suspensions should be stored in liquid nitrogen, extracted RNA should be stored at −70°C (or as cDNA at or below −20°C) and DNA at or below −20°C.
In suspected SM, the diagnostic standard is to examine unfractionated BM cells (after erythrocyte-lysis) or mononuclear marrow cells (MNC) for KIT mutations.{Valent, 2007 #1598} The marrow aspirate (1-2 mL) should be recovered in EDTA or in heparin. In certain circumstances, when no BM aspirate sample is available, KIT mutational analyses can be performed on cells detached from BM smears or from a paraffin-embedded biopsy sample, provided that the procedure used to determine the KIT mutational status exhibits sufficient sensitivity.{Sotlar, 2013 #1660} A practical issue here is that if marrow aspirate is not obtainable, the core biopsy preservative should be formalin, and not Zencker’s or Bouin’s which degrades DNA.
In typical ISM, MNCs from peripheral blood (PB) are frequently KIT D816V negative when investigated by low sensitivity techniques, such as sequencing of PCR-derived amplicons. However, recent studies using ASO-qPCR-assays have suggested the possibility of detecting the mutation in PB in nearly all adult patients with typical SM.{Kristensen, 2014 #12;Erben, 2014 #1659;Hoermann, 2014 #4} Thus, we recommend the use of such technique in PB samples as a screening test in cases with suspected SM.{Valent, 2014 #1529} However, the KIT D816V mutation may not be detected in all patients with SM by this approach, especially in the few cases where other mutations in KIT are present or the MC burden is extremely low. Therefore, we recommend testing of PB cells for initial screening, whereas a more detailed analysis must always include a thorough examination of the BM. Because of a variable burden of MCs it is recommended to either use a very sensitive technology for mutation-detection (e.g. ASO-qPCR),{Erben, 2014 #1659;Kristensen, 2014 #12} or to perform sequencing studies on MCs purified by laser microdissection or flow cytometry.{Sotlar, 2000 #1673;Garcia-Montero, 2006 #1665} However, most centers have focused on developing sensitive ASO-qPCR assays rather than methods to purify MCs. It is important to note that every center establishing these tests should focus carefully on the possibility of false positive results by examining an appropriate number of negative controls.{Mattocks, 2010 #1491}
Regarding adult patients with skin lesions (also referred as to “mastocytosis in the skin”, MIS), a skin biopsy can be performed, where the presence of KIT D816V is thus indicative for MIS.{Arock, 2010 #80} Nonetheless, this result is not diagnostic for systemic disease (SM) which still requires the demonstration of the presence of KIT D816V in the BM or PB.{Valent, 2014 #1529} In addition, other visceral organs, such as the spleen, liver, GI tract or lymph nodes, may be affected in SM.{Travis, 1988 #1470;Diebold, 1991 #1469;Metcalfe, 1991 #637;Horny, 1992 #281;Mican, 1995 #645;Horny, 1997 #272;Smith, 1999 #1468;Jensen, 2000 #527} However, we do not recommend routine screening of these organs for the presence of KIT mutations. Involvement of a single extramedullary (visceral) organ in SM in the absence of BM involvement is an extremely rare condition.{Parwaresch, 1985 #695;Horny, 1985 #292} In such exceptional cases, it has been demonstrated that KIT mutations may also be detected in the involved organs.{Jensen, 2000 #527;Wimazal, 2004 #1668}
Standard assays for detection of KIT D816V
Several assays for detection of the KIT D816V mutation have been reported. Based on literature-data, the most sensitive assays, briefly described below, are i) RT-PCR plus restriction fragment length polymorphism (RFLP), ii) nested RT-PCR followed by D-HPLC of PCR amplicons, iii) PNA-mediated PCR, and iv) ASO-qPCR on DNA or RNA/cDNA. These techniques should be regarded as standard, provided that they have sufficient specificity, sensitivity and precision. We thus recommend performing these techniques according to published protocols.{Fritsche-Polanz, 2001 #1448;Sotlar, 2003 #1671;Garcia-Montero, 2006 #1665;Bodemer, 2010 #96;Kristensen, 2011 #77;Kristensen, 2012 #59;Sotlar, 2013 #1660;Kristensen, 2014 #12;Erben, 2014 #1659} The advantages, disadvantages and sensitivity of the different tests recommended are summarized in Table 2.
Table 2. Summarized advantages, disadvantages and sensitivity of the different tests recommended to detect KIT mutations at position 816.
| Technique | RT-PCR plus restriction fragment length polymorphism (RFLP) | Nested RT-PCR followed by D-HPLC of PCR amplicons | PNA-mediated PCR | ASO-qPCR on DNA or RNA/cDNA |
|---|---|---|---|---|
| Advantages | -simple -fast -reliable -cost-saving |
-detects different KIT mutations at position 816 | -allows detection of KIT mutations at position 816 or at adjacent positions -recommended for formalin-fixed paraffin-embedded tissues |
-simple -fast -cost-saving -highly sensitive test -quantitative: allows the quantification of the KIT D816V allele burden in blood or BM at diagnosis or in the follow up |
| Disadvantages | -detects only KIT D816V mutant -not quantitative |
-relatively low sensitivity -not quantitative -time-consuming -needs special facilities (HPLC) |
-not quantitative -intermediate sensitivity |
-detects only KIT D816V mutant -needs standardization,validation and harmonization |
| Sensitivity | ± 0.05 % | 0.5-1.0 % | ± 0.1% | ± 0.01% |
ASO-qPCR: allele specific-quantitative PCR; BM: bone marrow; D-HPLC: denaturing high performance liquid chromatography; PNA-mediated PCR: peptide nucleic acid-mediated; PCR RT-PCR: reverse transcriptase-polymerase chain reaction.
RT-PCR and RFLP
The sensitivity of this technique allows the detection of one KIT D816V+ cell among 2000 KIT WT cells (0.05%).{Valent, 2007 #1598} However, the technique has limitations since it does not allow the detection of other mutations in codon 816, and it is not quantitative.{Valent, 2007 #1598}
Nested RT-PCR followed by D-HPLC of PCR amplicons
The major interest of this technique is that it allows detection of all mutations in the amplified region and would be perfect for simultaneous detection of KIT D816V or of other mutations in codon 816 in the same reaction.{Erben, 2014 #1659} The technique has been optimized by the authors to detect mutant KIT D816V down to 0.5–1 % of mutant in normal cells based on cell line and RNA dilutions.{Erben, 2014 #1659}
Peptide nucleic acid-mediated (PNA)-mediated PCR
Three interesting features of this technique are that i) it allows detection of the mutated KIT D816V allele in a 1000-fold excess of WT background,{Sotlar, 2003 #1671} ii) it allows the identification of other mutations in codon 816 or in adjacent codons,{Sotlar, 2013 #1660} iii) it is the recommended method to be applied on formalin-fixed paraffin-embedded BM trephine biopsies.{Sotlar, 2003 #1671;Garcia-Montero, 2006 #1665;Sotlar, 2013 #1660}
Allele specific PCR (ASO-PCR) and ASO-quantitative PCR (ASO-qPCR)
ASO-PCR is a simple, fast and reliable method to detect specifically the KIT D816V mutant in various tissues.{Schumacher, 2008 #132} Interestingly, the quantitative variant of the method (ASO-qPCR) has the advantage to allow simultaneously detection and allele burden measurement of KIT D816V in different tissues and cells (PB, BM and organ biopsies) at low cost (reagents <US $50).{Kristensen, 2011 #77;Kristensen, 2012 #59;Kristensen, 2014 #12} As an example, Kristensen et al. presented recently an assay that detects less than 0.01% KIT D816V mutation-positive cells.{Kristensen, 2011 #77} In their study, the authors analyzed a total of 61 samples derived from 31 cases diagnosed with various SM subtypes and were able to detect the mutation in 95% of BM and 81% of PB samples, respectively.{Kristensen, 2011 #77} The Danish group further confirmed the sensitivity and reliability of their assay on lesional skin biopsies with positive results in all out of 29 adult SM patients.{Kristensen, 2013 #27} In another study conducted on 25 ISM patients, the same authors showed that the method detects the KIT mutant in circulating PB cells even if no circulating MCs are detectable.{Kristensen, 2012 #59} From these data, the group proposed adopting mutation analysis of PB as a “routine” diagnostic screen test in mastocytosis.{Kristensen, 2014 #12} The sensitivity and specificity of the ASO-qPCR analysis of PB cells was also examined. For this purpose, the authors analyzed PB samples in a total of 83 SM patients and 4 CM patients as well as 10 patients with a provisional diagnosis of MIS.{Kristensen, 2014 #12} In this study, KIT D816V was detected in 94% of all cases with SM. Interestingly, all 10 patients with MIS tested positive in this study, demonstrating the high sensitivity of the test in suspected SM.{Kristensen, 2014 #12} Based on this study and similar data from other centers,{Hoermann, 2014 #4;Erben, 2014 #1659} the ECNM suggests that PB should be used for the initial screening of KIT mutations in suspected SM patients in daily practice.{Valent, 2014 #1529} However, an issue is that in case of very low numbers of clonal MCs, the KIT D816V mutation may not be detectable in PB. Therefore, BM cells need to be investigated in all cases with high suspicion of SM. Finally, concerning the ASO-qPCR, multi-center standardization projects (ring trials) aimed at inter-center validation as well as harmonization will be required in the future.
Usefulness of allelic burden measurements of KIT D816V in PB and BM
Very recent reports have shown that allele burden measurement of KIT D816V in BM and PB using sensitive ASO-qPCR on DNA or on RNA/cDNA are not only useful for diagnostic purposes but also for prognostication, follow-up of the disease as well as for quantification of response to treatment such as chemotherapy or allogeneic SCT.{Erben, 2014 #1659;Hoermann, 2014 #4}
Indeed, Hoermann and colleagues quantified the KIT D816V allele burden from genomic DNA derived from BM aspirates and PB by ASO-qPCR in a cohort of 105 patients with mastocytosis, including cases with CM (n=12), MIS (n=3), ISM (n=67), SSM (n=5), ASM (n=7), SM-AHNMD (n=10) and MCL (n=1).{Hoermann, 2014 #4} Interestingly, the authors reported significant differences in the median allele burden between disease subgroups, with an increased burden found in advanced disease: CM (0.042%), ISM (0.285%), SSM (5.991%), ASM (9.346%), and SM-AHNMD (3.761%). Confirming previous data,{Kristensen, 2013 #35} the authors also reported a good correlation between the KIT D816V allele burden and the serum tryptase levels. However, no significant correlation was found between the allele burden and the MC infiltration in the BM, which may be explained by the fact that non-MC-lineage cells in the BM also harbor KIT D816V, but at levels that may greatly vary from patient to patient.{Hoermann, 2014 #4} Moreover, the authors were able to demonstrate that the KIT D816V allele burden was of prognostic significance. In fact, patients with ISM showed a stable allele burden during the follow-up, whereas in case of progression, a marked increase in KIT D816V was found.{Hoermann, 2014 #4} Finally, the authors were able to show that the KIT D816V allele burden decreases during cytoreductive therapy.{Hoermann, 2014 #4}
In a similar study, Erben and colleagues used a KIT D816V ASO-qPCR on RNA/cDNA to diagnose mastocytosis and to monitor the allele burden as marker of residual disease.{Erben, 2014 #1659} The SM patients analyzed (n=147) were classified as follows: ISM (n=63), SSM (n=8), SM-AHNMD (n=16) and ASM/MCL±AHNMD, (n=60). In 62 patients, samples were collected from BM and PB (ISM, n=28; SSM, n=4; SM-AHNMD, n=5; ASM/MCL, n=25). Thus, overall, 210 samples were analyzed.{Erben, 2014 #1659} The authors observed that when compared to nested RT-PCR followed by HPLC and sequencing, their ASO-qPCR was more sensitive, detecting 0.01-0.1% of mutated cells. Independent of disease subtype, they identified the KIT D816V mutation in 97% of BM samples and in 78% of all PB samples tested.{Erben, 2014 #1659} In addition, they showed a strong correlation between the KIT D816V allele burden, disease subtype (e.g., indolent vs. advanced SM) and survival.{Erben, 2014 #1659} In terms of monitoring residual disease, the analysis was performed on 4 ASM/MCL±AHNMD patients treated either with cladribine (1 patient), aggressive chemotherapy (1 patient) or allogeneic SCT (2 patients). The authors reported a significant reduction of the KIT D816V allele burden in the BM and PB during treatment.{Erben, 2014 #1659}
Thus, it appears that ASO-qPCR assays on DNA or on RNA/cDNA are highly sensitive methods for the quantification of the KIT D816V burden, and possibly preferable to nested PCR plus RFLP. Consequently, the ECNM recommends the use of the KIT D816V ASO-qPCR assays for the analysis of patients with suspected SM in PB and BM samples in routine screening tests.{Valent, 2007 #1598} However, a few issues remain to be solved for the future. Indeed, neither of the ASO-qPCR assays can detect other mutations at codon 816 of KIT (such as D816Y or D816H) or at positions outside the kinase domain.{Erben, 2014 #1659;Kristensen, 2011 #77}
Guidelines to search for and monitor KIT D816V+ SM
RT-PCR+RFLP, PNA-mediated PCR, and ASO-qPCR are fast, reliable, sensitive, and cost-effective methods for routine screening of KIT D816V in all SM subtypes. When applied on BM cells, all these assays have demonstrated their ability to detect the KIT D816V mutation in > 80% of SM patients, a sensitivity considered sufficient in daily practice.{Valent, 2007 #1598} In addition, it has emerged that PB is sufficient for mutation analysis in many patients when the most sensitive assays are applied.{Erben, 2014 #1659;Hoermann, 2014 #4;Valent, 2014 #1529} Thus, we propose to use KIT D816V ASO-qPCR assays in PB in combination with measurement of serum tryptase and other relevant parameters, such as specific clinical symptoms indicating potential organ involvement, for the routine screening of adult patients with suspected mastocytosis, and these recommendations are summarized in a algorithm for the diagnostic of adult patients with suspected mastocytosis (Figure 3). In addition, Table 3 describes the usefulness of KIT D816V ASO-qPCR assays, together with several other clinical, biological, molecular and radiographic explorations, in the diagnosis and monitoring of the disease. However, the applied methodology should be validated carefully with respect to the expected diagnostic yield which can be reduced dramatically by even minor reductions in sensitivity.{Kristensen, 2014 #12} Thus, KIT mutational analysis in BM samples may be always performed following a positive test on PB samples, and should be considered in cases with high suspicion of SM even when no KIT mutation is detectable in the PB. In addition, the frequency of measurement of the allele burden should be adapted to the individual situation in each patient. Thus, in ISM patients with low MC burden and stable clinical course, the KIT D816V allele burden should be measured at diagnosis, but should not necessarily be repeated, unless signs of disease progression occur. In patients with more aggressive forms enrolled in clinical trials with cytoreductive therapies, the KIT D816V allele burden should be measured repeatedly before and during therapy.
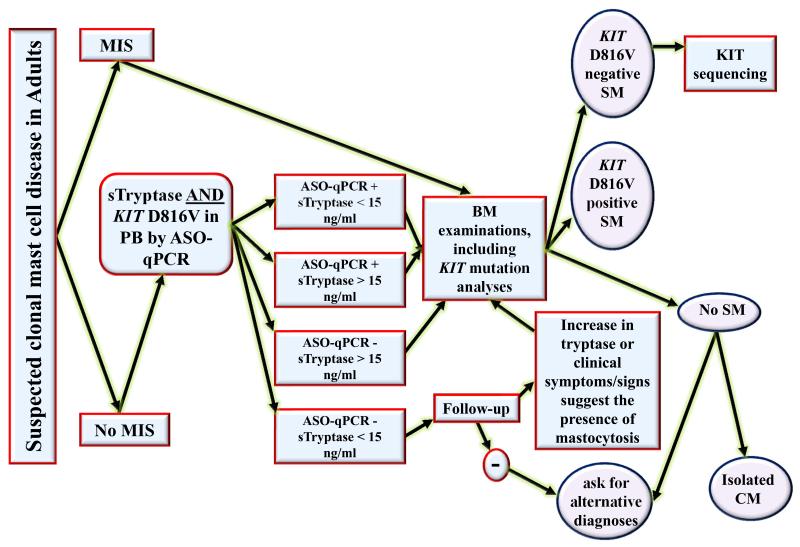
Figure 3. Proposed algorithm for the use of KIT D816V allele-specific quantitative PCR or RT-PCR (ASO-qPCR) in peripheral blood (PB), together with serum tryptase measurement, for the screening of adult patients with suspected mastocytosis.
In case of suspected mastocytosis in an adult patient, the first step is the search for clear signs of mastocytosis in the skin (MIS). If MIS is present, we recommend that a BM examination should be performed directly. If there is no MIS, we recommend to measure serum tryptase level, together with a KIT D816V ASO-qPCR using PB cells. If the serum tryptase level is > 15 ng/mL and/or the KIT D816V ASO-qPCR detects the mutation, a bone marrow examination is recommended, including a KIT D816V ASO-qPCR. If the BM studies are compatible with the diagnosis SM, and the KIT D816V ASO-qPCR is positive, then the final diagnosis is KIT D816V+ SM. If the BM studies are compatible with the diagnosis SM, and the KIT D816V ASO-q(RT)PCR is negative, sequencing of the entire KIT gene should be considered on a BM sample, in order to determine the KIT structure in this D816V-negative patient. In MIS-negative patients who have a serum tryptase level < 15 ng/mL and who are negative for the search of the KIT D816V mutant by ASO-qPCR on PB, we recommend to follow up the patients and to perform a BM investigation only if there is an increase in tryptase or clinical symptoms/signs suggesting the presence of mastocytosis. Otherwise, alternative explanations and differential diagnoses should be considered. ASO-qPCR: allele specific-quantitative PCR on RNA or DNA; BM: bone marrow; CM: cutaneous mastocytosis; MIS: “mastocytosis in the skin”; PB: peripheral blood; SM: systemic mastocytosis; sTryptase: serum total tryptase level.
Table 3. Recommended use of major clinical, biological and molecular parameters/explorations for the diagnosis and the follow up of adult patients with SM.
| Parameters/explorations | Diagnosis | Follow up in patients with stable SM | Follow up in case of (suspected) progression in SM | Follow up in during cytoreductive/targeted therapy |
|---|---|---|---|---|
|
| ||||
| Clinical parameters | ||||
| -Careful dermatological examination (including Darier’s sign) | + | + a | + a | + b |
| - Palpation for hepatomegaly, splenomegaly and/or lymphadenopathy | + | + c | + | + b |
| - Neuropshycological evaluation | + | + c | + | + |
|
| ||||
| Biological parameters | ||||
| - Serum tryptase level | + | + c | + | + d |
| -CBC and differential | + | + c | + | + |
| - BM biopsy with KIT (CD117) and tryptase staining by IHC | + | − | + | + |
| -Cytological examination of BM smears | + | − | + | + |
| - CD2/CD25 on BM MCs by IHC or flow cytometry | + | − | + | + |
| - CD30 on BM MCs by IHC or flow cytometry | + | − | + | + |
| -Serum LDH | + | + c | + | + |
| - Serum albumin | + | + c | + | + |
| -Serum calcium | + | + c | + | + |
| - Total IgE | + | |||
| -Serum alkaline phosphatase | + | + c | + | + |
| -Liver function tests | + | + c | + | + |
| -Serum β2-microglobulin | + | + c | + | + |
| -Urinary histamine metabolites | + | − | − | − |
| -Urinary PGD2 | + | − | − | − |
|
| ||||
| Molecular parameters | ||||
| -ASO-qPCR for KIT D816V mutation in PB | + | + c | + | + d |
| -ASO-qPCR for KIT D816V mutation in BM | + (see Figure 3) | − | + | + d |
| -KIT sequencing | + (see Figure 3) | − | − | − |
| -Testing for additional genetic lesions (NSG, myeloid gene panels) | +(if ASM/MCL or SM-AHNMD) | − | + | + |
|
| ||||
| Radiographic explorations | + | − | + | + |
| -Computed tomography | + | − | + | + |
| -Ultrasonography | + | − | + | + |
| -Osteodensitometry (T Score by Dexa-Scan) | + | + | + | + |
ASM: aggressive systemic mastocytosis; ASO-qPCR: allele specific-quantitative PCR; BM: bone marrow; CBC: complete blood count, IHC: immunohistochemistry; MCL: mast cell leukemia; MCs: mast cells; MRD: minimum residual disease; NSG: nest generation sequencing; PGD2: prostaglandin D2; SM: systemic mastocytosis; SM-AHNMD: systemic mastocytosis with associated hematological non MC disease.
If skin lesions are evolutive
Lesions may regress upon treatment
At regular intervals
Useful to appreciate the efficacy of the treatment and to evaluate the level of MRD
Recommendations for SM cases with a “negative” KIT D816V test result
In a small percentage of cases (<5-10%) no mutation is detectable in codon 816 of KIT,{Garcia-Montero, 2006 #1665;Sanchez-Munoz, 2011 #1494} which may have several explanations: i) patients are in fact KIT D816V+ but the (very) low MC burden leads to a false negative result because the sensitivity of the applied assay is too low, ii) patients indeed only bear wild type KIT or iii) patients are positive for other mutations at codon 816 (D816H, D816Y, others) or in other regions of KIT which are not detectable by the ASO-qPCR assays for KIT D816V. In scenario i) a KIT mutation may only be detected in highly enriched (sorted or microdissected) MCs or by using one of the highly sensitive techniques presented above.{Sotlar, 2003 #1671;Garcia-Montero, 2006 #1665;Kristensen, 2011 #77;Erben, 2014 #1659;Hoermann, 2014 #4} However, enrichment is usually not required for decision-making in daily clinical practice as demonstration or exclusion of KIT D816V has no therapeutic consequence.
By contrast, in patients with ASM or MCL (huge MC-infiltrates), a negative screen-result is a diagnostic challenge. In such cases, certain KIT mutations in the JMD, TMD or ECD domains or even KIT wild-type may be excellent candidates for targeted treatment with imatinib,{Frost, 2002 #184;Akin, 2004 #315;Yang, 2010 #1451;Mital, 2011 #76;Georgin-Lavialle, 2012 #63;Alvarez-Twose, 2012 #1485} whereas KIT D816V (and other mutations at position 816) are imatinib-resistant,{Valent, 2010 #81;Mital, 2011 #76;Georgin-Lavialle, 2012 #63;Ashman, 2013 #1471} leading to the use of other TKIs with a broader activity, such as PKC412 (midostaurin),{Gotlib, 2010 #1483;Gotlib, 2012 #1484;Gleixner, 2013 #38} or of non-targeted therapies such as Interferon-alpha, cladribine or allogeneic SCT.{Kluin-Nelemans, 1992 #563;Kluin-Nelemans, 2003 #564;Ustun, 2014 #1498} Therefore, a negative result should be confirmed, preferably in a reference laboratory, by using the most sensitive technique available today, i.e. ASO-qPCR. This is then followed by examination of KIT for other mutations in codon 816, which requires amplification of the codon 17 and sequencing of the resulting amplicons or preferably PNA-mediated PCR.{Garcia-Montero, 2006 #1665} Finally, if no mutation is found at codon 816, sequencing of the whole KIT coding sequences should be considered.{Bodemer, 2010 #96}
Concluding remarks and future perspectives
Highly sensitive and specific PCR-based assays for the detection of KIT D816V are now available. These methods have become standard in the routine screening of patients with suspected SM. Furthermore, quantification of the KIT mutational load may be rapidly adopted as gold standard for assessing the disease burden in advanced SM, thereby improving prognostication and monitoring of the disease course prior, during and after cytoreductive therapies, including KIT-targeting drugs and allogeneic SCT. However, although more than 80% of all SM patients harbor KIT D816V, SM is a heterogeneous disease regarding mutational patterns, course and aggressiveness. It has also been shown that additional genetic lesions such as mutations in TET2, SRSF2, ASXL1, SRSF2 or RUNX1 are frequently identified in patients with advanced SM, preferably in ASM or MCL with AHNMD.{Bibi, 2014 #5;Soucie, 2014 #1487;Schwaab, 2013 #1542} These lesions might decisively contribute to the clinical heterogeneity and the aggressiveness of SM and may represent new therapeutic targets. Thus, methods such as myeloid gene mutation panels, e.g. 20-50 genes being evaluated in parallel, or next generation sequencing (NGS), applied on the whole genome, will allow the mapping of additional functionally important molecular lesions, which may complement or even replace present techniques used for KIT mutation analyses as basis to improve our knowledge about the clinical course of patients with mastocytosis.
Acknowledgments
The authors were unable to cite all the original papers due to reference limits and apologize for not citing all relevant original papers. M. Arock is supported by Fondation de France; P. Dubreuil is supported by La Ligue Nationale Contre le Cancer (équipe labellisée) and INCa; A. Garcia-Montero and A. Orfao are Supported by grants from the Instituto de Salud Carlos III, Ministry of Economy and Competitivity, Madrid, Spain (grant numbers RD12/0036/0048 and PI11/02399, FEDER) and from Fundacion Ramon Areces, Madrid, Spain (grant number CIVP16A1806); D.D. Metcalfe is supported in part by the Division of Intramural Research, NIAID; P. Valent is supported by Austrian Science Funds (FWF) Project SFB F4611 and SFB F4704-B20.
Footnotes
Conflict-of-interest disclosures:
C. Akin is a consultant in a Novartis trial. O. Hermine and P. Dubreuil receive research funding and honorarium from AB Science N.C.P. Cross receives honoraria and research funding from Novartis. K. Hartmann is a consultant in a Novartis trial and received research grants from Novartis. J. Gotlib is a consultant in a Novartis trial and receives research funding from Novartis. P. Valent receives honoraria and a research grant from Novartis and is Consultant in a Global Novartis trial. The other authors declare no conflict of interest.
References
- 1.Krishnaswamy G, Ajitawi O, Chi DS. The human mast cell: an overview. Methods Mol Biol. 2006;315:13–34. doi: 10.1385/1-59259-967-2:013. [DOI] [PubMed] [Google Scholar]
- 2.Miettinen M, Lasota J. KIT (CD117): a review on expression in normal and neoplastic tissues, and mutations and their clinicopathologic correlation. Appl Immunohistochem Mol Morphol. 2005;13:205–220. doi: 10.1097/01.pai.0000173054.83414.22. [DOI] [PubMed] [Google Scholar]
- 3.Ashman LK, Ferrao P, Cole SR, Cambareri AC. Effects of mutant c-kit in early myeloid cells. Leuk Lymphoma. 2000;37:233–243. doi: 10.3109/10428190009057652. [DOI] [PubMed] [Google Scholar]
- 4.Kirshenbaum AS, Goff JP, Semere T, Foster B, Scott LM, Metcalfe DD. Demonstration that human mast cells arise from a progenitor cell population that is CD34(+), c-kit(+), and expresses aminopeptidase N (CD13) Blood. 1999;94:2333–2342. [PubMed] [Google Scholar]
- 5.Matarraz S, Lopez A, Barrena S, Fernandez C, Jensen E, Flores J, et al. The immunophenotype of different immature, myeloid and B-cell lineage-committed CD34+ hematopoietic cells allows discrimination between normal/reactive and myelodysplastic syndrome precursors. Leukemia. 2008;22:1175–1183. doi: 10.1038/leu.2008.49. [DOI] [PubMed] [Google Scholar]
- 6.Teodosio C, Garcia-Montero AC, Jara-Acevedo M, Sanchez-Munoz L, Alvarez-Twose I, Nunez R, et al. Mast cells from different molecular and prognostic subtypes of systemic mastocytosis display distinct immunophenotypes. J Allergy Clin Immunol. 2010;125:719–726. 726 e711–726 e714. doi: 10.1016/j.jaci.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 7.Chang A, Tung RC, Schlesinger T, Bergfeld WF, Dijkstra J, Kahn TA. Familial cutaneous mastocytosis. Pediatr Dermatol. 2001;18:271–276. doi: 10.1046/j.1525-1470.2001.01939.x. [DOI] [PubMed] [Google Scholar]
- 8.Valent P, Akin C, Arock M, Brockow K, Butterfield JH, Carter MC, et al. Definitions, criteria and global classification of mast cell disorders with special reference to mast cell activation syndromes: a consensus proposal. Int Arch Allergy Immunol. 2012;157:215–225. doi: 10.1159/000328760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briley LD, Phillips CM. Cutaneous mastocytosis: a review focusing on the pediatric population. Clin Pediatr (Phila) 2008;47:757–761. doi: 10.1177/0009922808318344. [DOI] [PubMed] [Google Scholar]
- 10.Vano-Galvan S, Alvarez-Twose I, De las Heras E, Morgado JM, Matito A, Sanchez-Munoz L, et al. Dermoscopic features of skin lesions in patients with mastocytosis. Arch Dermatol. 2011;147:932–940. doi: 10.1001/archdermatol.2011.190. [DOI] [PubMed] [Google Scholar]
- 11.Arock M, Valent P. Pathogenesis, classification and treatment of mastocytosis: state of the art in 2010 and future perspectives. Expert Rev Hematol. 2010;3:497–516. doi: 10.1586/ehm.10.42. [DOI] [PubMed] [Google Scholar]
- 12.Fried AJ, Akin C. Primary mast cell disorders in children. Curr Allergy Asthma Rep. 2013;13:693–701. doi: 10.1007/s11882-013-0392-6. [DOI] [PubMed] [Google Scholar]
- 13.Wang SA, Hutchinson L, Tang G, Chen SS, Miron PM, Huh YO, et al. Systemic mastocytosis with associated clonal hematological non-mast cell lineage disease: clinical significance and comparison of chomosomal abnormalities in SM and AHNMD components. Am J Hematol. 2013;88:219–224. doi: 10.1002/ajh.23380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metcalfe DD. Mast cells and mastocytosis. Blood. 2008;112:946–956. doi: 10.1182/blood-2007-11-078097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valent P, Horny HP, Escribano L, Longley BJ, Li CY, Schwartz LB, et al. Diagnostic criteria and classification of mastocytosis: a consensus proposal. Leuk Res. 2001;25:603–625. doi: 10.1016/s0145-2126(01)00038-8. [DOI] [PubMed] [Google Scholar]
- 16.Valent P, Horny H-P, Li CY. Mastocytosis. In: Jaffe ES, Harris NL, Stein H, Vardiman JW, editors. World Health Organization (WHO) Classification of Tumours. Vol. 12001. IARC Press; Lyon, France: pp. 291–302. [Google Scholar]
- 17.Horny HP, Akin C, Metcalfe DD, Escribano L, Bennett JM, Valent P, Bain BJ. Mastocytosis (mast cell disease) In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, editors. World Health Organization (WHO) Classification of Tumours. IARC Press; Lyon, France: 2008. pp. 54–63. [Google Scholar]
- 18.Valent P, Akin C, Escribano L, Fodinger M, Hartmann K, Brockow K, et al. Standards and standardization in mastocytosis: consensus statements on diagnostics, treatment recommendations and response criteria. Eur J Clin Invest. 2007;37:435–453. doi: 10.1111/j.1365-2362.2007.01807.x. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Montero AC, Jara-Acevedo M, Teodosio C, Sanchez ML, Nunez R, Prados A, et al. KIT mutation in mast cells and other bone marrow hematopoietic cell lineages in systemic mast cell disorders: a prospective study of the Spanish Network on Mastocytosis (REMA) in a series of 113 patients. Blood. 2006;108:2366–2372. doi: 10.1182/blood-2006-04-015545. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez-Munoz L, Alvarez-Twose I, Garcia-Montero AC, Teodosio C, Jara-Acevedo M, Pedreira CE, et al. Evaluation of the WHO criteria for the classification of patients with mastocytosis. Mod Pathol. 2011;24:1157–1168. doi: 10.1038/modpathol.2011.84. [DOI] [PubMed] [Google Scholar]
- 21.Erben P, Schwaab J, Metzgeroth G, Horny HP, Jawhar M, Sotlar K, et al. The KIT D816V expressed allele burden for diagnosis and disease monitoring of systemic mastocytosis. Ann Hematol. 2014;93:81–88. doi: 10.1007/s00277-013-1964-1. [DOI] [PubMed] [Google Scholar]
- 22.Kristensen T, Vestergaard H, Bindslev-Jensen C, Moller MB, Broesby-Olsen S. Sensitive KIT D816V mutation analysis of blood as a diagnostic test in mastocytosis. Am J Hematol. 2014;89:493–498. doi: 10.1002/ajh.23672. [DOI] [PubMed] [Google Scholar]
- 23.Bodemer C, Hermine O, Palmerini F, Yang Y, Grandpeix-Guyodo C, Leventhal PS, et al. Pediatric mastocytosis is a clonal disease associated with D816V and other activating c-KIT mutations. J Invest Dermatol. 2010;130:804–815. doi: 10.1038/jid.2009.281. [DOI] [PubMed] [Google Scholar]
- 24.Tan A, Westerman D, McArthur GA, Lynch K, Waring P, Dobrovic A. Sensitive detection of KIT D816V in patients with mastocytosis. Clin Chem. 2006;52:2250–2257. doi: 10.1373/clinchem.2006.068205. [DOI] [PubMed] [Google Scholar]
- 25.Kristensen T, Broesby-Olsen S, Vestergaard H, Bindslev-Jensen C, Moller MB. Circulating KIT D816V mutation-positive non-mast cells in peripheral blood are characteristic of indolent systemic mastocytosis. Eur J Haematol. 2012;89:42–46. doi: 10.1111/j.1600-0609.2012.01789.x. [DOI] [PubMed] [Google Scholar]
- 26.Hoermann G, Gleixner KV, Dinu GE, Kundi M, Greiner G, Wimazal F, et al. The KIT D816V allele burden predicts survival in patients with mastocytosis and correlates with the WHO type of the disease. Allergy. 2014;69:810–813. doi: 10.1111/all.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bibi S, Langenfeld F, Jeanningros S, Brenet F, Soucie E, Hermine O, et al. Molecular defects in mastocytosis: KIT and beyond KIT. Immunol Allergy Clin North Am. 2014;34:239–262. doi: 10.1016/j.iac.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Valent P, Akin C, Sperr WR, Mayerhofer M, Fodinger M, Fritsche-Polanz R, et al. Mastocytosis: pathology, genetics, and current options for therapy. Leuk Lymphoma. 2005;46:35–48. doi: 10.1080/10428190400010775. [DOI] [PubMed] [Google Scholar]
- 29.Escribano L, Alvarez-Twose I, Sanchez-Munoz L, Garcia-Montero A, Nunez R, Almeida J, et al. Prognosis in adult indolent systemic mastocytosis: a long-term study of the Spanish Network on Mastocytosis in a series of 145 patients. J Allergy Clin Immunol. 2009;124:514–521. doi: 10.1016/j.jaci.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Valent P, Arock M, Bischoff SC, Buhring HJ, Brockow K, Escribano L, et al. The European Competence Network on Mastocytosis (ECNM) Wien Klin Wochenschr. 2004;116:647–651. doi: 10.1007/s00508-004-0253-3. [DOI] [PubMed] [Google Scholar]
- 31.Valent P, Arock M, Bonadonna P, Brockow K, Broesby-Olsen S, Escribano L, et al. European Competence Network on Mastocytosis (ECNM): 10-year jubilee, update, and future perspectives. Wien Klin Wochenschr. 2012;124:807–814. doi: 10.1007/s00508-012-0293-z. [DOI] [PubMed] [Google Scholar]
- 32.Giebel LB, Strunk KM, Holmes SA, Spritz RA. Organization and nucleotide sequence of the human KIT (mast/stem cell growth factor receptor) proto-oncogene. Oncogene. 1992;7:2207–2217. [PubMed] [Google Scholar]
- 33.Chabot B, Stephenson DA, Chapman VM, Besmer P, Bernstein A. The proto-oncogene c-kit encoding a transmembrane tyrosine kinase receptor maps to the mouse W locus. Nature. 1988;335:88–89. doi: 10.1038/335088a0. [DOI] [PubMed] [Google Scholar]
- 34.Cheng P, Chen H, Liu SR, Pu XY, A ZC. SNPs in KIT and KITLG genes may be associated with oligospermia in Chinese population. Biomarkers. 2013;18:650–654. doi: 10.3109/1354750X.2013.838307. [DOI] [PubMed] [Google Scholar]
- 35.Nagata H, Worobec AS, Metcalfe DD. Identification of a polymorphism in the transmembrane domain of the protooncogene c-kit in healthy subjects. Exp Clin Immunogenet. 1996;13:210–214. [PubMed] [Google Scholar]
- 36.Longley BJ, Tyrrell L, Lu SZ, Ma YS, Langley K, Ding TG, et al. Somatic c-KIT activating mutation in urticaria pigmentosa and aggressive mastocytosis: establishment of clonality in a human mast cell neoplasm. Nat Genet. 1996;12:312–314. doi: 10.1038/ng0396-312. [DOI] [PubMed] [Google Scholar]
- 37.Orfao A, Garcia-Montero AC, Sanchez L, Escribano L. Recent advances in the understanding of mastocytosis: the role of KIT mutations. Br J Haematol. 2007;138:12–30. doi: 10.1111/j.1365-2141.2007.06619.x. [DOI] [PubMed] [Google Scholar]
- 38.Georgin-Lavialle S, Lhermitte L, Dubreuil P, Chandesris MO, Hermine O, Damaj G. Mast cell leukemia. Blood. 2013;121:1285–1295. doi: 10.1182/blood-2012-07-442400. [DOI] [PubMed] [Google Scholar]
- 39.Zhang LY, Smith ML, Schultheis B, Fitzgibbon J, Lister TA, Melo JV, et al. A novel K509I mutation of KIT identified in familial mastocytosis-in vitro and in vivo responsiveness to imatinib therapy. Leuk Res. 2006;30:373–378. doi: 10.1016/j.leukres.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 40.Wasag B, Niedoszytko M, Piskorz A, Lange M, Renke J, Jassem E, et al. Novel, activating KIT-N822I mutation in familial cutaneous mastocytosis. Exp Hematol. 2011;39:859–865. e852. doi: 10.1016/j.exphem.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Hoffmann KM, Moser A, Lohse P, Winkler A, Binder B, Sovinz P, et al. Successful treatment of progressive cutaneous mastocytosis with imatinib in a 2-year-old boy carrying a somatic KIT mutation. Blood. 2008;112:1655–1657. doi: 10.1182/blood-2008-03-147785. [DOI] [PubMed] [Google Scholar]
- 42.Wohrl S, Moritz KB, Bracher A, Fischer G, Stingl G, Loewe R. A c-kit mutation in exon 18 in familial mastocytosis. J Invest Dermatol. 2013;133:839–841. doi: 10.1038/jid.2012.394. [DOI] [PubMed] [Google Scholar]
- 43.Pollard WL, Beachkofsky TM, Kobayashi TT. Novel R634W c-kit Mutation Identified in Familial Mastocytosis. Pediatr Dermatol. 2014 doi: 10.1111/pde.12381. [DOI] [PubMed] [Google Scholar]
- 44.Broesby-Olsen S, Kristensen TK, Moller MB, Bindslev-Jensen C, Vestergaard H. Adult-onset systemic mastocytosis in monozygotic twins with KIT D816V and JAK2 V617F mutations. J Allergy Clin Immunol. 2012;130:806–808. doi: 10.1016/j.jaci.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 45.Zanotti R, Simioni L, Garcia-Montero AC, Perbellini O, Bonadonna P, Caruso B, et al. Somatic D816V KIT mutation in a case of adult-onset familial mastocytosis. J Allergy Clin Immunol. 2013;131:605–607. doi: 10.1016/j.jaci.2012.11.040. [DOI] [PubMed] [Google Scholar]
- 46.Sperr WR, Horny HP, Valent P. Spectrum of associated clonal hematologic non-mast cell lineage disorders occurring in patients with systemic mastocytosis. Int Arch Allergy Immunol. 2002;127:140–142. doi: 10.1159/000048186. [DOI] [PubMed] [Google Scholar]
- 47.Stoecker MM, Wang E. Systemic mastocytosis with associated clonal hematologic nonmast cell lineage disease: a clinicopathologic review. Arch Pathol Lab Med. 2012;136:832–838. doi: 10.5858/arpa.2011-0325-RS. [DOI] [PubMed] [Google Scholar]
- 48.Valent P. Mastocytosis: a paradigmatic example of a rare disease with complex biology and pathology. Am J Cancer Res. 2013;3:159–172. [PMC free article] [PubMed] [Google Scholar]
- 49.Piazza R, Redaelli S, Pirola A, Valletta S, Magistroni V, Cross NCP, et al. Recurrent KIT D816V mutation in atypical chronic myeloid leukemia. 19th Congress of the European Hematology Association; Milan, Italy. 2014 12-15 june 2014.2014. [Google Scholar]
- 50.Stellmacher F, Sotlar K, Balleisen L, Valent P, Horny HP. Bone marrow mastocytosis associated with IgM kappa plasma cell myeloma. Leuk Lymphoma. 2004;45:801–805. doi: 10.1080/10428190310001615693. [DOI] [PubMed] [Google Scholar]
- 51.Horny HP, Sotlar K, Stellmacher F, Valent P, Grabbe J. An unusual case of systemic mastocytosis associated with chronic lymphocytic leukaemia (SM-CLL) J Clin Pathol. 2006;59:264–268. doi: 10.1136/jcp.2005.026989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sotlar K, Colak S, Bache A, Berezowska S, Krokowski M, Bultmann B, et al. Variable presence of KITD816V in clonal haematological non-mast cell lineage diseases associated with systemic mastocytosis (SM-AHNMD) J Pathol. 2010;220:586–595. doi: 10.1002/path.2677. [DOI] [PubMed] [Google Scholar]
- 53.Fritsche-Polanz R, Fritz M, Huber A, Sotlar K, Sperr WR, Mannhalter C, et al. High frequency of concomitant mastocytosis in patients with acute myeloid leukemia exhibiting the transforming KIT mutation D816V. Mol Oncol. 2010;4:335–346. doi: 10.1016/j.molonc.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frost MJ, Ferrao PT, Hughes TP, Ashman LK. Juxtamembrane mutant V560GKit is more sensitive to Imatinib (STI571) compared with wild-type c-kit whereas the kinase domain mutant D816VKit is resistant. Mol Cancer Ther. 2002;1:1115–1124. [PubMed] [Google Scholar]
- 55.Lasota J. Not all c-kit mutations can be corrected by imatinib. Lab Invest. 2007;87:317. [PubMed] [Google Scholar]
- 56.Foster R, Griffith R, Ferrao P, Ashman L. Molecular basis of the constitutive activity and STI571 resistance of Asp816Val mutant KIT receptor tyrosine kinase. J Mol Graph Model. 2004;23:139–152. doi: 10.1016/j.jmgm.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Laine E, Chauvot de Beauchene I, Perahia D, Auclair C, Tchertanov L. Mutation D816V alters the internal structure and dynamics of c-KIT receptor cytoplasmic region: implications for dimerization and activation mechanisms. PLoS Comput Biol. 2011;7:e1002068. doi: 10.1371/journal.pcbi.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kluin-Nelemans HC, Oldhoff JM, Van Doormaal JJ, Van ’t Wout JW, Verhoef G, Gerrits WB, et al. Cladribine therapy for systemic mastocytosis. Blood. 2003;102:4270–4276. doi: 10.1182/blood-2003-05-1699. [DOI] [PubMed] [Google Scholar]
- 59.Hauswirth AW, Simonitsch-Klupp I, Uffmann M, Koller E, Sperr WR, Lechner K, et al. Response to therapy with interferon alpha-2b and prednisolone in aggressive systemic mastocytosis: report of five cases and review of the literature. Leuk Res. 2004;28:249–257. doi: 10.1016/s0145-2126(03)00259-5. [DOI] [PubMed] [Google Scholar]
- 60.Gotlib J, DeAngelo DJ, George TI, Corless CL, Linder A, Langford C, et al. KIT Inhibitor Midostaurin Exhibits a High Rate of Clinically Meaningful and Durable Responses in Advanced Systemic Mastocytosis: Report of a Fully Accrued Phase II Trial. ASH Annual Meeting Abstracts. 2010;116 : 316- [Google Scholar]
- 61.Gotlib J, Kluin-Nelemans HC, George TI, Akin C, Sotlar K, Hermine O, et al. KIT Inhibitor Midostaurin in Patients with Advanced Systemic Mastocytosis: Results of a Planned Interim Analysis of the Global CPKC412D2201 Trial. ASH Annual Meeting Abstracts. 2012;120 : 799- [Google Scholar]
- 62.Mital A, Piskorz A, Lewandowski K, Wasag B, Limon J, Hellmann A. A case of mast cell leukaemia with exon 9 KIT mutation and good response to imatinib. Eur J Haematol. 2011;86:531–535. doi: 10.1111/j.1600-0609.2011.01598.x. [DOI] [PubMed] [Google Scholar]
- 63.Alvarez-Twose I, Gonzalez P, Morgado JM, Jara-Acevedo M, Sanchez-Munoz L, Matito A, et al. Complete response after imatinib mesylate therapy in a patient with well-differentiated systemic mastocytosis. J Clin Oncol. 2012;30:e126–129. doi: 10.1200/JCO.2011.38.9973. [DOI] [PubMed] [Google Scholar]
- 64.Valent P, Escribano L, Broesby-Olsen S, Hartmann K, Grattan C, Brockow K, et al. Proposed diagnostic algorithm for patients with suspected mastocytosis: a proposal of the European Competence Network on Mastocytosis. Allergy. 2014;69:1267–1274. doi: 10.1111/all.12436. [DOI] [PubMed] [Google Scholar]
- 65.Corless CL, Harrell P, Lacouture M, Bainbridge T, Le C, Gatter K, et al. Allele-specific polymerase chain reaction for the imatinib-resistant KIT D816V and D816F mutations in mastocytosis and acute myelogenous leukemia. J Mol Diagn. 2006;8:604–612. doi: 10.2353/jmoldx.2006.060089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sotlar K, Escribano L, Landt O, Mohrle S, Herrero S, Torrelo A, et al. One-step detection of c-kit point mutations using peptide nucleic acid-mediated polymerase chain reaction clamping and hybridization probes. Am J Pathol. 2003;162:737–746. doi: 10.1016/S0002-9440(10)63870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schumacher JA, Elenitoba-Johnson KS, Lim MS. Detection of the c-kit D816V mutation in systemic mastocytosis by allele-specific PCR. J Clin Pathol. 2008;61:109–114. doi: 10.1136/jcp.2007.047928. [DOI] [PubMed] [Google Scholar]
- 68.Kristensen T, Vestergaard H, Moller MB. Improved detection of the KIT D816V mutation in patients with systemic mastocytosis using a quantitative and highly sensitive real-time qPCR assay. J Mol Diagn. 2011;13:180–188. doi: 10.1016/j.jmoldx.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sotlar K, Marafioti T, Griesser H, Theil J, Aepinus C, Jaussi R, et al. Detection of c-kit mutation Asp 816 to Val in microdissected bone marrow infiltrates in a case of systemic mastocytosis associated with chronic myelomonocytic leukaemia. Mol Pathol. 2000;53:188–193. doi: 10.1136/mp.53.4.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sotlar K, Fridrich C, Mall A, Jaussi R, Bultmann B, Valent P, et al. Detection of c-kit point mutation Asp-816 --> Val in microdissected pooled single mast cells and leukemic cells in a patient with systemic mastocytosis and concomitant chronic myelomonocytic leukemia. Leuk Res. 2002;26:979–984. doi: 10.1016/s0145-2126(02)00041-3. [DOI] [PubMed] [Google Scholar]
- 71.Teodosio C, Garcia-Montero AC, Jara-Acevedo M, Alvarez-Twose I, Sanchez-Munoz L, Almeida J, et al. An immature immunophenotype of bone marrow mast cells predicts for multilineage D816V KIT mutation in systemic mastocytosis. Leukemia. 2012;26:951–958. doi: 10.1038/leu.2011.293. [DOI] [PubMed] [Google Scholar]
- 72.Sotlar K. c-kit mutational analysis in paraffin material. Methods Mol Biol. 2013;999:59–78. doi: 10.1007/978-1-62703-357-2_4. [DOI] [PubMed] [Google Scholar]
- 73.Mattocks CJ, Morris MA, Matthijs G, Swinnen E, Corveleyn A, Dequeker E, et al. A standardized framework for the validation and verification of clinical molecular genetic tests. Eur J Hum Genet. 2010;18:1276–1288. doi: 10.1038/ejhg.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Travis WD, Li CY. Pathology of the lymph node and spleen in systemic mast cell disease. Mod Pathol. 1988;1:4–14. [PubMed] [Google Scholar]
- 75.Diebold J, Riviere O, Gosselin B, Janin-Mercier A, Canelhas A, Le Tourneau A, et al. Different patterns of spleen involvement in systemic and malignant mastocytosis. A histological and immunohistochemical study of three cases. Virchows Arch A Pathol Anat Histopathol. 1991;419:273–280. doi: 10.1007/BF01606518. [DOI] [PubMed] [Google Scholar]
- 76.Metcalfe DD. The liver, spleen, and lymph nodes in mastocytosis. J Invest Dermatol. 1991;96:45S–46S. [PubMed] [Google Scholar]
- 77.Horny HP, Ruck MT, Kaiserling E. Spleen findings in generalized mastocytosis. A clinicopathologic study. Cancer. 1992;70:459–468. doi: 10.1002/1097-0142(19920715)70:2<459::aid-cncr2820700214>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 78.Mican JM, Di Bisceglie AM, Fong TL, Travis WD, Kleiner DE, Baker B, et al. Hepatic involvement in mastocytosis: clinicopathologic correlations in 41 cases. Hepatology. 1995;22:1163–1170. doi: 10.1016/0270-9139(95)90625-8. [DOI] [PubMed] [Google Scholar]
- 79.Horny HP, Ruck P, Krober S, Kaiserling E. Systemic mast cell disease (mastocytosis). General aspects and histopathological diagnosis. Histol Histopathol. 1997;12:1081–1089. [PubMed] [Google Scholar]
- 80.Smith JD, Lazarchick J. Systemic mast cell disease with marrow and splenic involvement associated with chronic myelomonocytic leukemia. Leuk Lymphoma. 1999;32:391–394. doi: 10.3109/10428199909167403. [DOI] [PubMed] [Google Scholar]
- 81.Jensen RT. Gastrointestinal abnormalities and involvement in systemic mastocytosis. Hematol Oncol Clin North Am. 2000;14:579–623. doi: 10.1016/s0889-8588(05)70298-7. [DOI] [PubMed] [Google Scholar]
- 82.Parwaresch MR, Horny HP, Lennert K. Tissue mast cells in health and disease. Pathol Res Pract. 1985;179:439–461. doi: 10.1016/s0344-0338(85)80184-9. [DOI] [PubMed] [Google Scholar]
- 83.Horny HP, Parwaresch MR, Lennert K. Bone marrow findings in systemic mastocytosis. Hum Pathol. 1985;16:808–814. doi: 10.1016/s0046-8177(85)80252-5. [DOI] [PubMed] [Google Scholar]
- 84.Wimazal F, Schwarzmeier J, Sotlar K, Simonitsch I, Sperr WR, Fritsche-Polanz R, et al. Splenic mastocytosis: report of two cases and detection of the transforming somatic C-KIT mutation D816V. Leuk Lymphoma. 2004;45:723–729. doi: 10.1080/1042819032000140979. [DOI] [PubMed] [Google Scholar]
- 85.Fritsche-Polanz R, Jordan JH, Feix A, Sperr WR, Sunder-Plassmann G, Valent P, et al. Mutation analysis of C-KIT in patients with myelodysplastic syndromes without mastocytosis and cases of systemic mastocytosis. Br J Haematol. 2001;113:357–364. doi: 10.1046/j.1365-2141.2001.02783.x. [DOI] [PubMed] [Google Scholar]
- 86.Kristensen T, Broesby-Olsen S, Vestergaard H, Bindslev-Jensen C, Moller MB. KIT D816V mutation-positive cell fractions in lesional skin biopsies from adults with systemic mastocytosis. Dermatology. 2013;226:233–237. doi: 10.1159/000349986. [DOI] [PubMed] [Google Scholar]
- 87.Kristensen T, Broesby-Olsen S, Vestergaard H, Bindslev-Jensen C, Moller MB. Serum tryptase correlates with the KIT D816V mutation burden in adults with indolent systemic mastocytosis. Eur J Haematol. 2013;91:106–111. doi: 10.1111/ejh.12128. [DOI] [PubMed] [Google Scholar]
- 88.Akin C, Fumo G, Yavuz AS, Lipsky PE, Neckers L, Metcalfe DD. A novel form of mastocytosis associated with a transmembrane c-kit mutation and response to imatinib. Blood. 2004;103:3222–3225. doi: 10.1182/blood-2003-11-3816. [DOI] [PubMed] [Google Scholar]
- 89.Yang Y, Letard S, Borge L, Chaix A, Hanssens K, Lopez S, et al. Pediatric mastocytosis-associated KIT extracellular domain mutations exhibit different functional and signaling properties compared with KIT-phosphotransferase domain mutations. Blood. 2010;116:1114–1123. doi: 10.1182/blood-2009-06-226027. [DOI] [PubMed] [Google Scholar]
- 90.Georgin-Lavialle S, Lhermitte L, Suarez F, Yang Y, Letard S, Hanssens K, et al. Mast cell leukemia: identification of a new c-Kit mutation, dup(501-502), and response to masitinib, a c-Kit tyrosine kinase inhibitor. Eur J Haematol. 2012;89:47–52. doi: 10.1111/j.1600-0609.2012.01761.x. [DOI] [PubMed] [Google Scholar]
- 91.Soucie E, Brenet F, Dubreuil P. Molecular basis of mast cell disease. Mol Immunol. 2014 doi: 10.1016/j.molimm.2014.03.013. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 92.Schwaab J, Schnittger S, Sotlar K, Walz C, Fabarius A, Pfirrmann M, et al. Comprehensive mutational profiling in advanced systemic mastocytosis. Blood. 2013;122:2460–2466. doi: 10.1182/blood-2013-04-496448. [DOI] [PubMed] [Google Scholar]
- 93.Hussein K, Horny HP, Busche G, Gorner M, Gohring G, Kreipe H, et al. Systemic mastocytosis (SM) with associated BCR-ABL-positive myelogenous leukaemia (SM-AHNMD): evidence that mast cells do not belong to the leukaemic clone. Leukemia. 2011;25:1050–1053. doi: 10.1038/leu.2011.41. [DOI] [PubMed] [Google Scholar]
- 94.Joris M, Georgin-Lavialle S, Chandesris MO, Lhermitte L, Claisse JF, Canioni D, et al. Mast Cell Leukaemia: c-KIT Mutations Are Not Always Positive. Case Rep Hematol. 2012;2012:517546. doi: 10.1155/2012/517546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ryan RJ, Akin C, Castells M, Wills M, Selig MK, Nielsen GP, et al. Mast cell sarcoma: a rare and potentially under-recognized diagnostic entity with specific therapeutic implications. Mod Pathol. 2013;26:533–543. doi: 10.1038/modpathol.2012.199. [DOI] [PubMed] [Google Scholar]
- 96.Kim YS, Wu H, Pawlowska AB, Bautista-Quach MA, Huang Q, Gaal K, et al. Pediatric mast cell sarcoma of temporal bone with novel L799F (2395 C>T) KIT mutation, mimicking histiocytic neoplasm. Am J Surg Pathol. 2013;37:453–458. doi: 10.1097/PAS.0b013e31828446d6. [DOI] [PubMed] [Google Scholar]
- 97.Georgin-Lavialle S, Aguilar C, Guieze R, Lhermitte L, Bruneau J, Fraitag S, et al. Mast cell sarcoma: a rare and aggressive entity--report of two cases and review of the literature. J Clin Oncol. 2013;31:e90–97. doi: 10.1200/JCO.2012.41.9549. [DOI] [PubMed] [Google Scholar]