Abstract
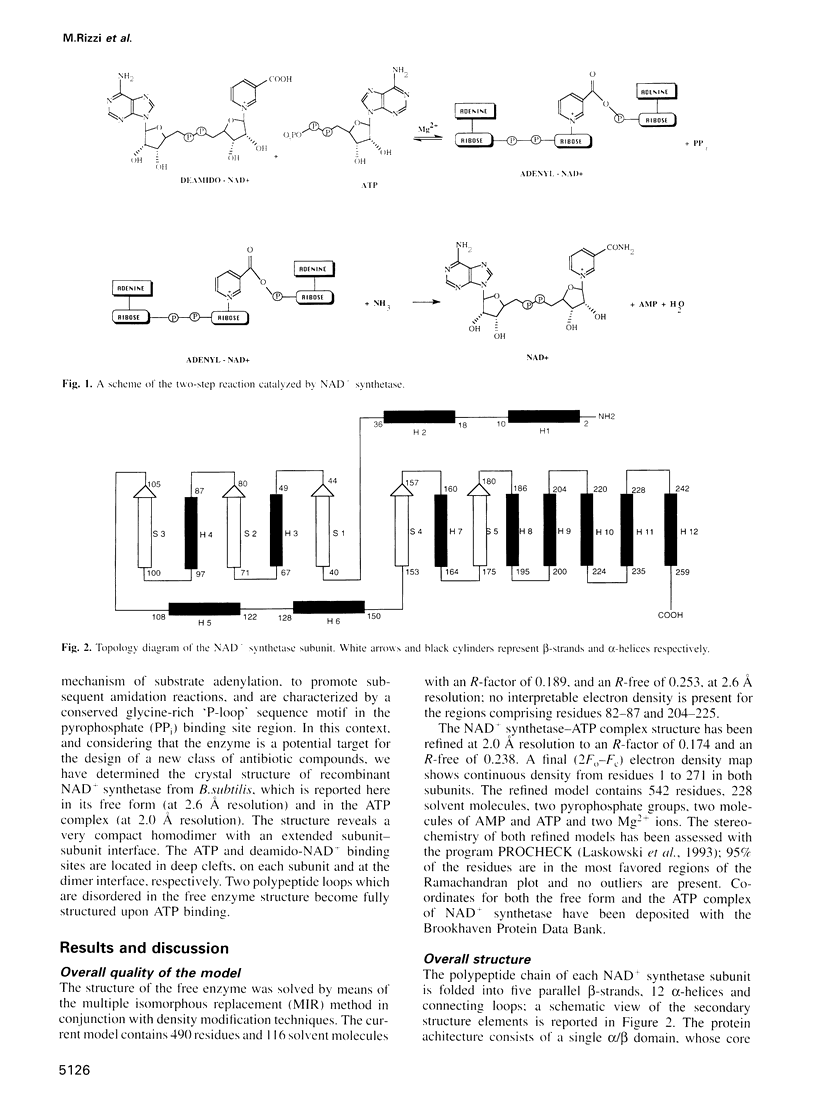
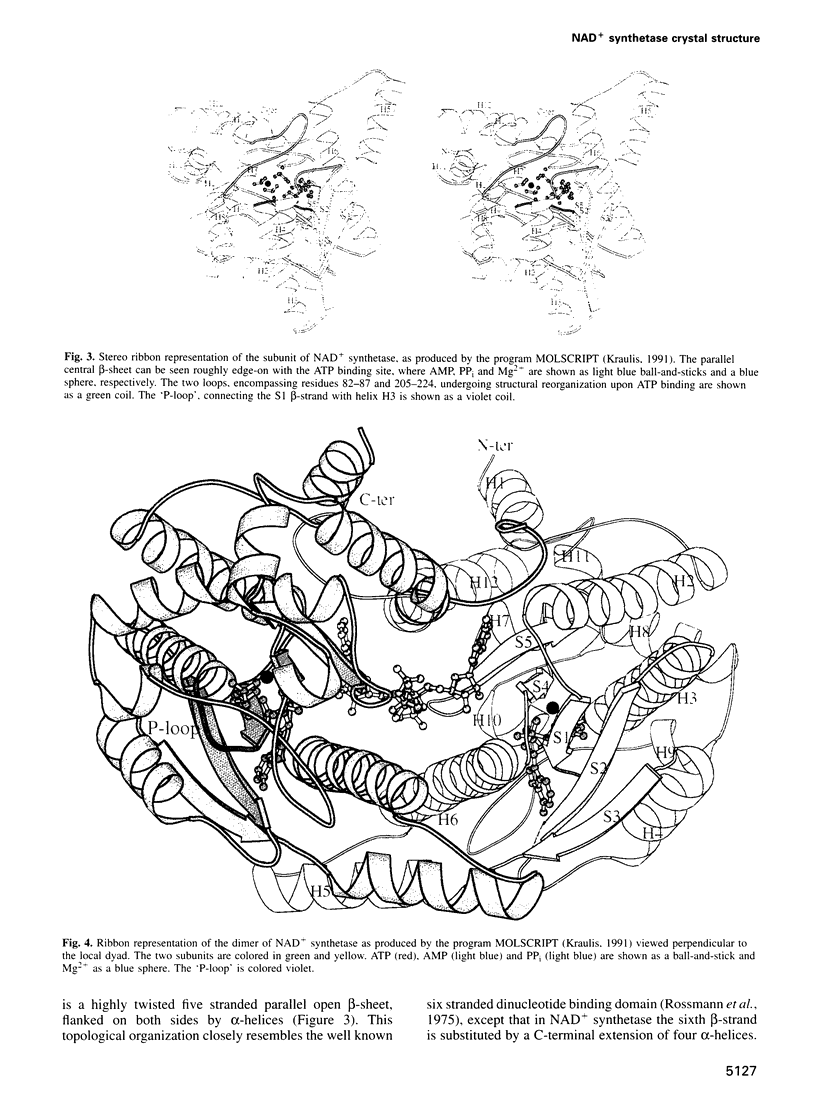
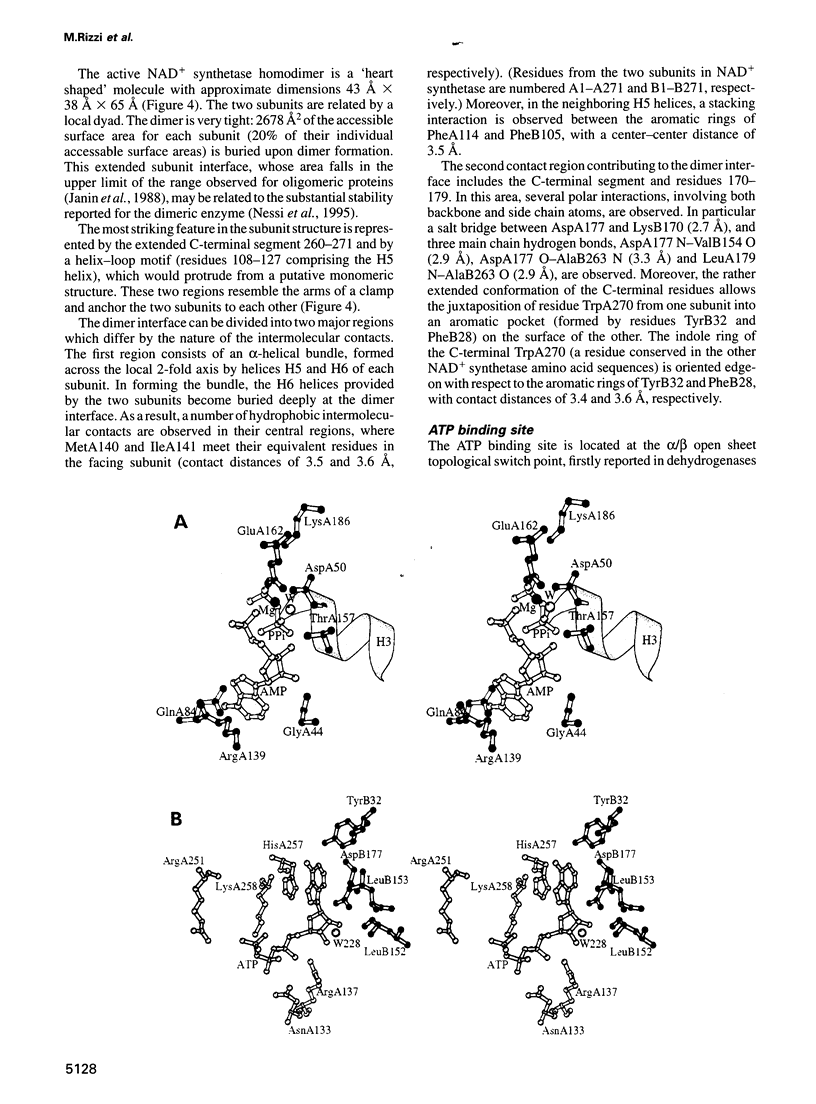
NAD+ synthetase catalyzes the last step in the biosynthesis of nicotinamide adenine dinucleotide. The three-dimensional structure of NH3-dependent NAD+ synthetase from Bacillus subtilis, in its free form and in complex with ATP, has been solved by X-ray crystallography (at 2.6 and 2.0 angstroms resolution, respectively) using a combination of multiple isomorphous replacement and density modification techniques. The enzyme consists of a tight homodimer with alpha/beta subunit topology. The catalytic site is located at the parallel beta-sheet topological switch point, where one AMP molecule, one pyrophosphate and one Mg2+ ion are observed. Residue Ser46, part of the neighboring 'P-loop', is hydrogen bonded to the pyrophosphate group, and may play a role in promoting the adenylation of deamido-NAD+ during the first step of the catalyzed reaction. The deamido-NAD+ binding site, located at the subunit interface, is occupied by one ATP molecule, pointing towards the catalytic center. A conserved structural fingerprint of the catalytic site, comprising Ser46, is very reminiscent of a related protein region observed in glutamine-dependent GMP synthetase, supporting the hypothesis that NAD+ synthetase belongs to the newly discovered family of 'N-type' ATP pyrophosphatases.
Full text
PDF
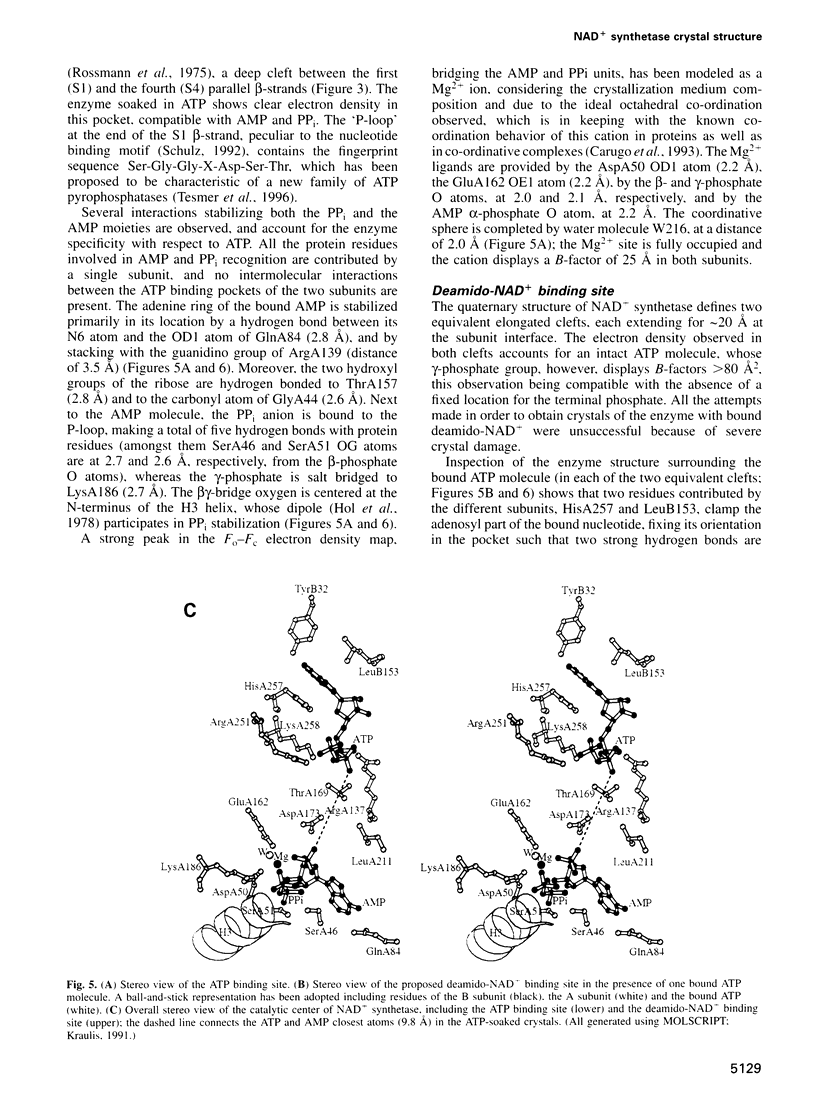



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Galizzi A. Mutant of Bacillus subtilis with a temperature-sensitive lesion in ribonucleic acid synthesis during germination. J Bacteriol. 1975 Oct;124(1):14–25. doi: 10.1128/jb.124.1.14-25.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramori T., Calogero S., Albertini A. M., Galizzi A. Functional analysis of the outB gene of Bacillus subtilis. J Gen Microbiol. 1993 Jan;139(1):31–37. doi: 10.1099/00221287-139-1-31. [DOI] [PubMed] [Google Scholar]
- Conti E., Franks N. P., Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996 Mar 15;4(3):287–298. doi: 10.1016/s0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- Delarue M. Aminoacyl-tRNA synthetases. Curr Opin Struct Biol. 1995 Feb;5(1):48–55. doi: 10.1016/0959-440x(95)80008-o. [DOI] [PubMed] [Google Scholar]
- Foster J. W., Moat A. G. Nicotinamide adenine dinucleotide biosynthesis and pyridine nucleotide cycle metabolism in microbial systems. Microbiol Rev. 1980 Mar;44(1):83–105. doi: 10.1128/mr.44.1.83-105.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galizzi A., Siccardi A. G., Mazza G., Canosi U., Polsinelli M. A recombination test to classify mutants of Bacillus subtilis of identical phenotype. Genet Res. 1976 Feb;27(1):47–58. doi: 10.1017/s0016672300016220. [DOI] [PubMed] [Google Scholar]
- Hol W. G., van Duijnen P. T., Berendsen H. J. The alpha-helix dipole and the properties of proteins. Nature. 1978 Jun 8;273(5662):443–446. doi: 10.1038/273443a0. [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993 Sep 5;233(1):123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Janin J., Miller S., Chothia C. Surface, subunit interfaces and interior of oligomeric proteins. J Mol Biol. 1988 Nov 5;204(1):155–164. doi: 10.1016/0022-2836(88)90606-7. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Matthews B. W. Solvent content of protein crystals. J Mol Biol. 1968 Apr 28;33(2):491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- McElroy W. D., DeLuca M., Travis J. Molecular uniformity in biological catalyses. The enzymes concerned with firefly luciferin, amino acid, and fatty acid utilization are compared. Science. 1967 Jul 14;157(3785):150–160. doi: 10.1126/science.157.3785.150. [DOI] [PubMed] [Google Scholar]
- Nessi C., Albertini A. M., Speranza M. L., Galizzi A. The outB gene of Bacillus subtilis codes for NAD synthetase. J Biol Chem. 1995 Mar 17;270(11):6181–6185. doi: 10.1074/jbc.270.11.6181. [DOI] [PubMed] [Google Scholar]
- Perona J. J., Rould M. A., Steitz T. A. Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. Biochemistry. 1993 Aug 31;32(34):8758–8771. doi: 10.1021/bi00085a006. [DOI] [PubMed] [Google Scholar]
- Spencer R. L., Preiss J. Biosynthesis of diphosphopyridine nucleotide. The purification and the properties of diphospyridine nucleotide synthetase from Escherichia coli b. J Biol Chem. 1967 Feb 10;242(3):385–392. [PubMed] [Google Scholar]
- Tesmer J. J., Klem T. J., Deras M. L., Davisson V. J., Smith J. L. The crystal structure of GMP synthetase reveals a novel catalytic triad and is a structural paradigm for two enzyme families. Nat Struct Biol. 1996 Jan;3(1):74–86. doi: 10.1038/nsb0196-74. [DOI] [PubMed] [Google Scholar]
- Tong L. A., Rossmann M. G. The locked rotation function. Acta Crystallogr A. 1990 Oct 1;46(Pt 10):783–792. doi: 10.1107/s0108767390005530. [DOI] [PubMed] [Google Scholar]
- Willison J. C., Tissot G. The Escherichia coli efg gene and the Rhodobacter capsulatus adgA gene code for NH3-dependent NAD synthetase. J Bacteriol. 1994 Jun;176(11):3400–3402. doi: 10.1128/jb.176.11.3400-3402.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalkin H. The amidotransferases. Adv Enzymol Relat Areas Mol Biol. 1993;66:203–309. doi: 10.1002/9780470123126.ch5. [DOI] [PubMed] [Google Scholar]