Abstract
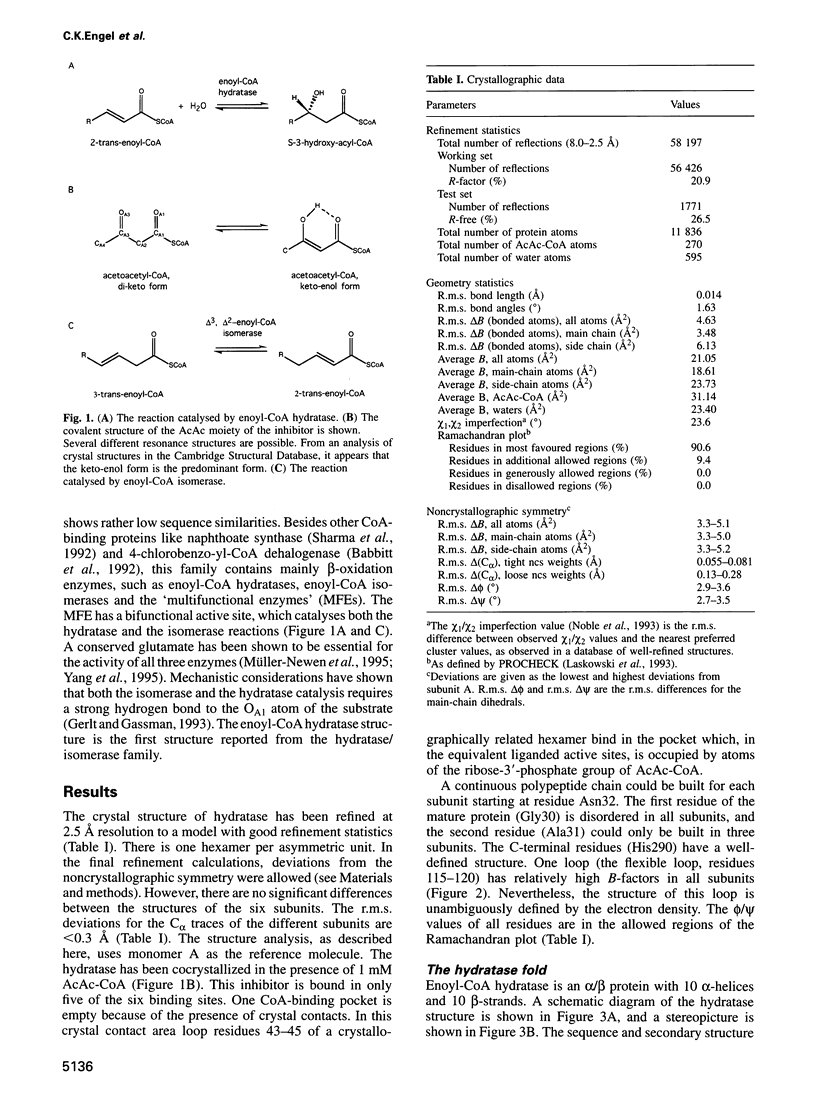
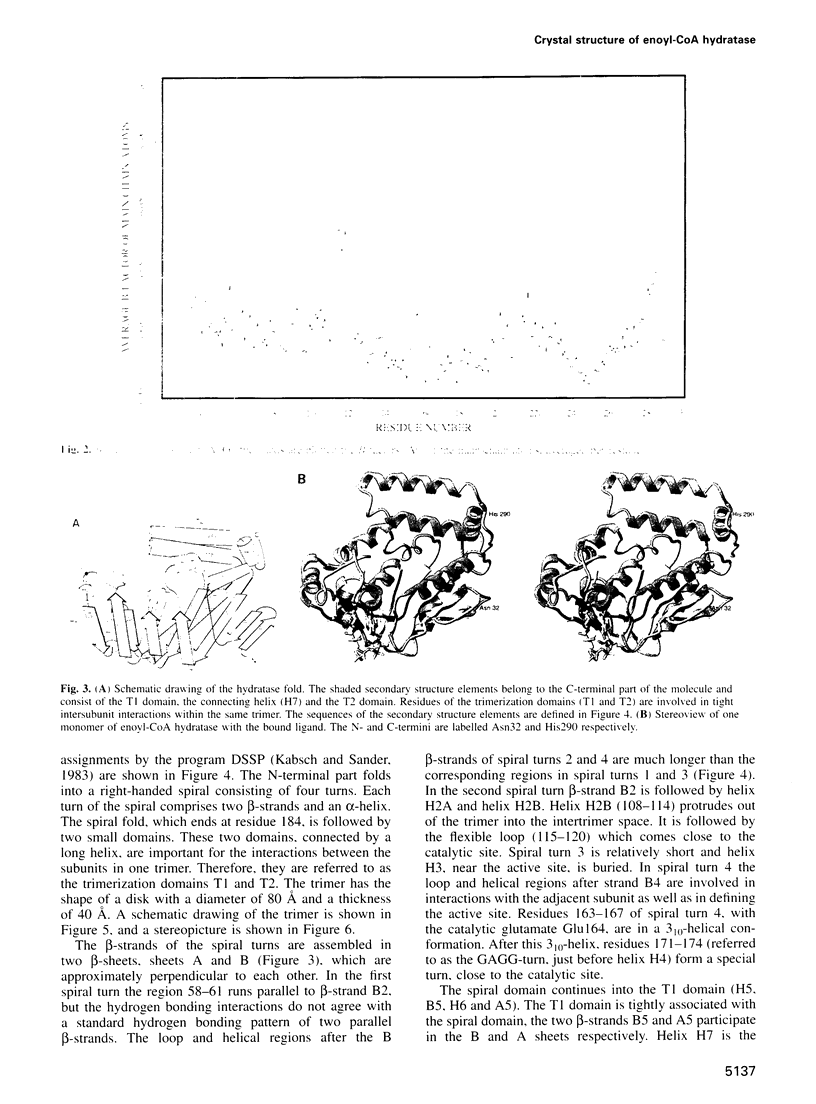
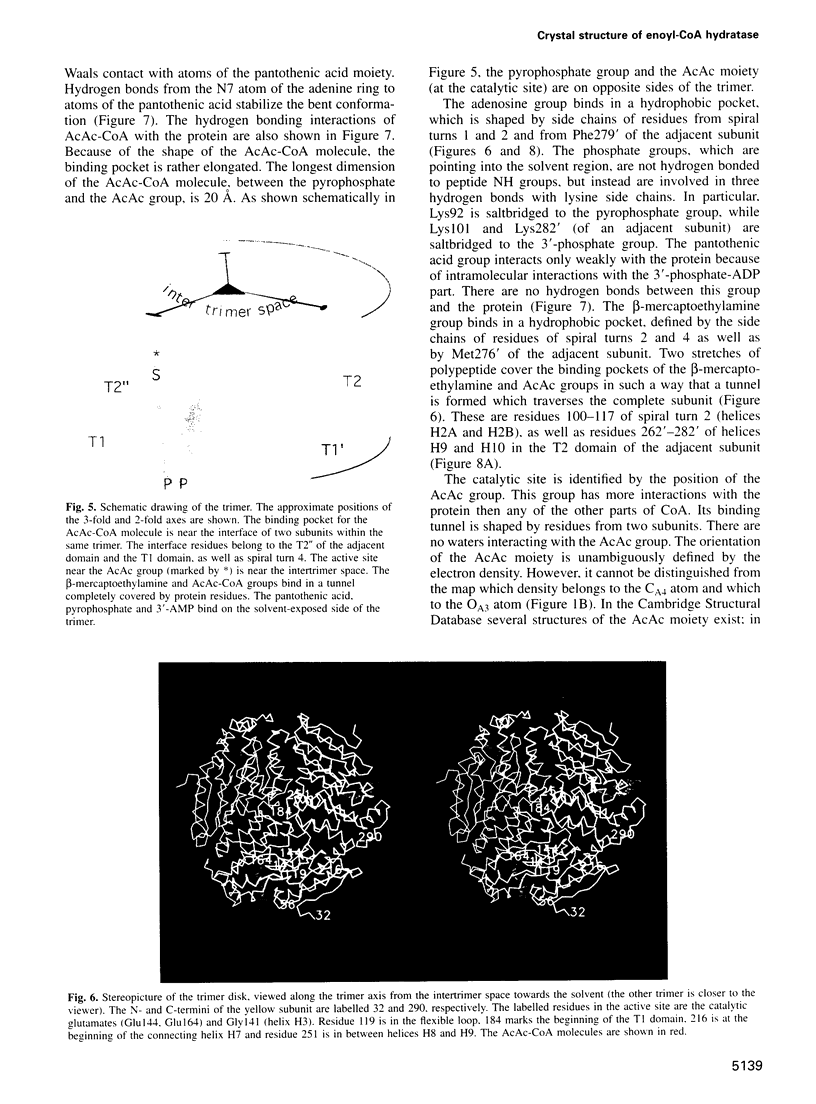
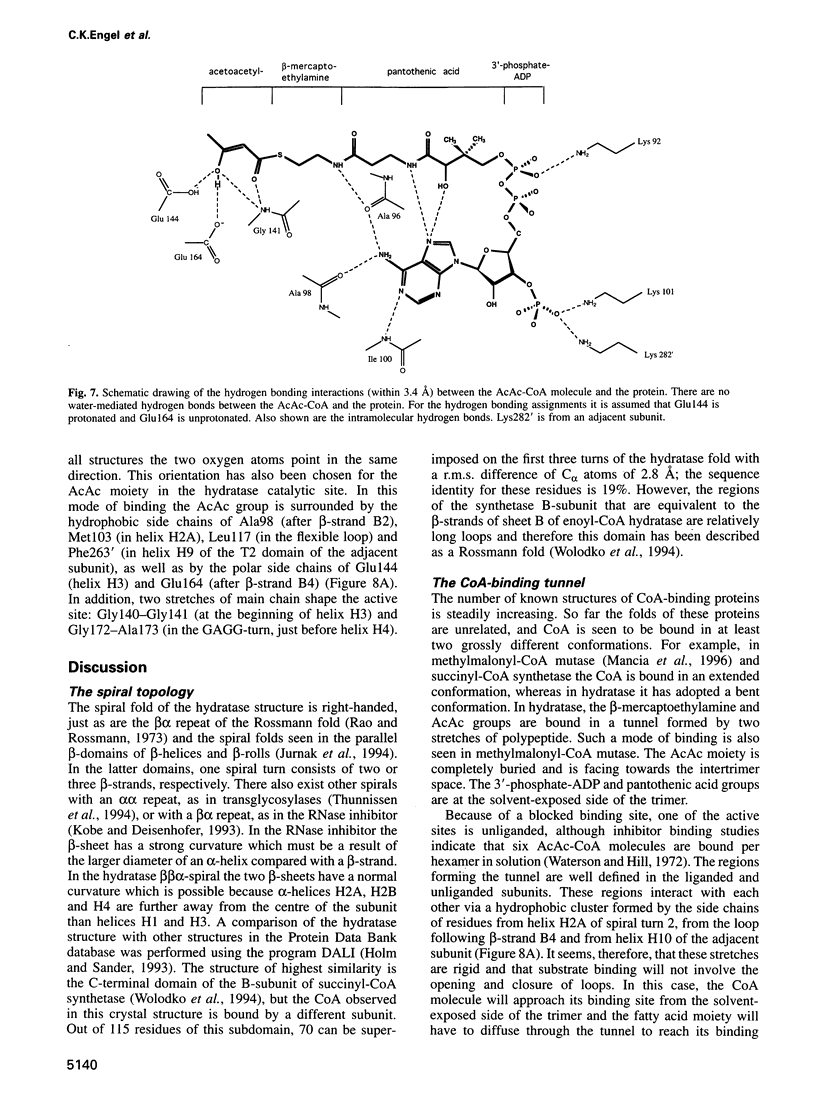
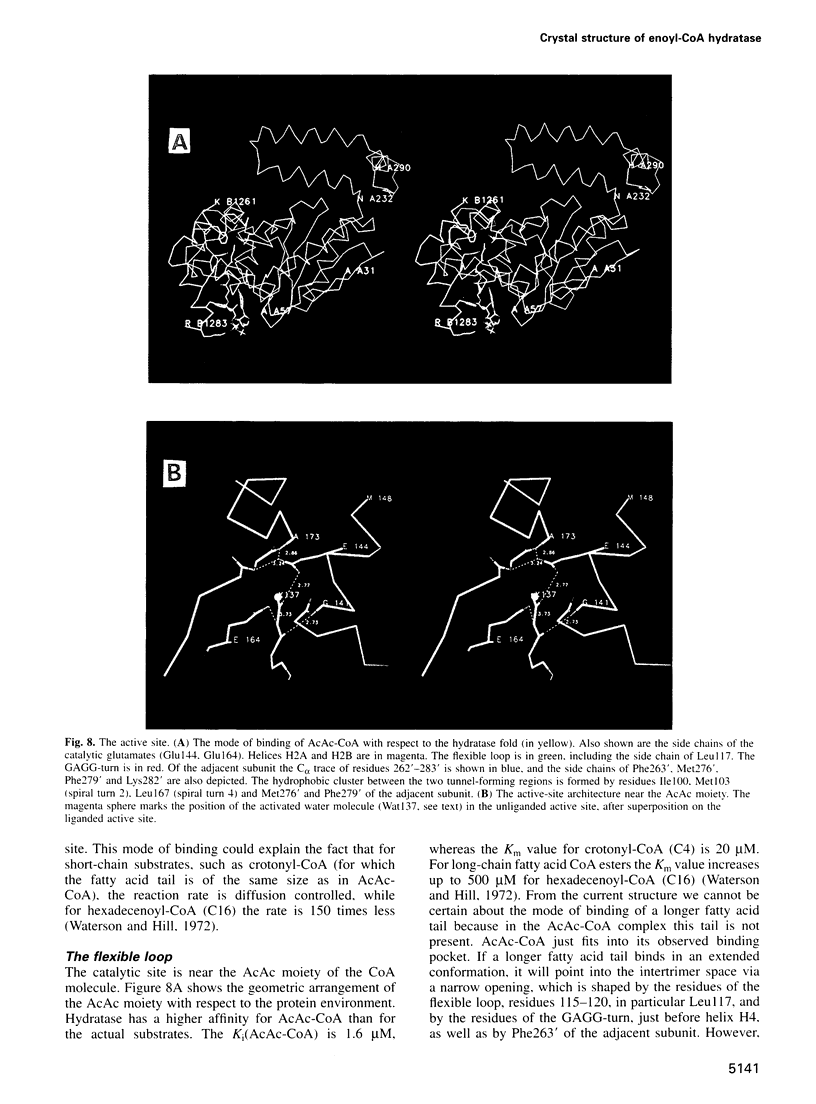
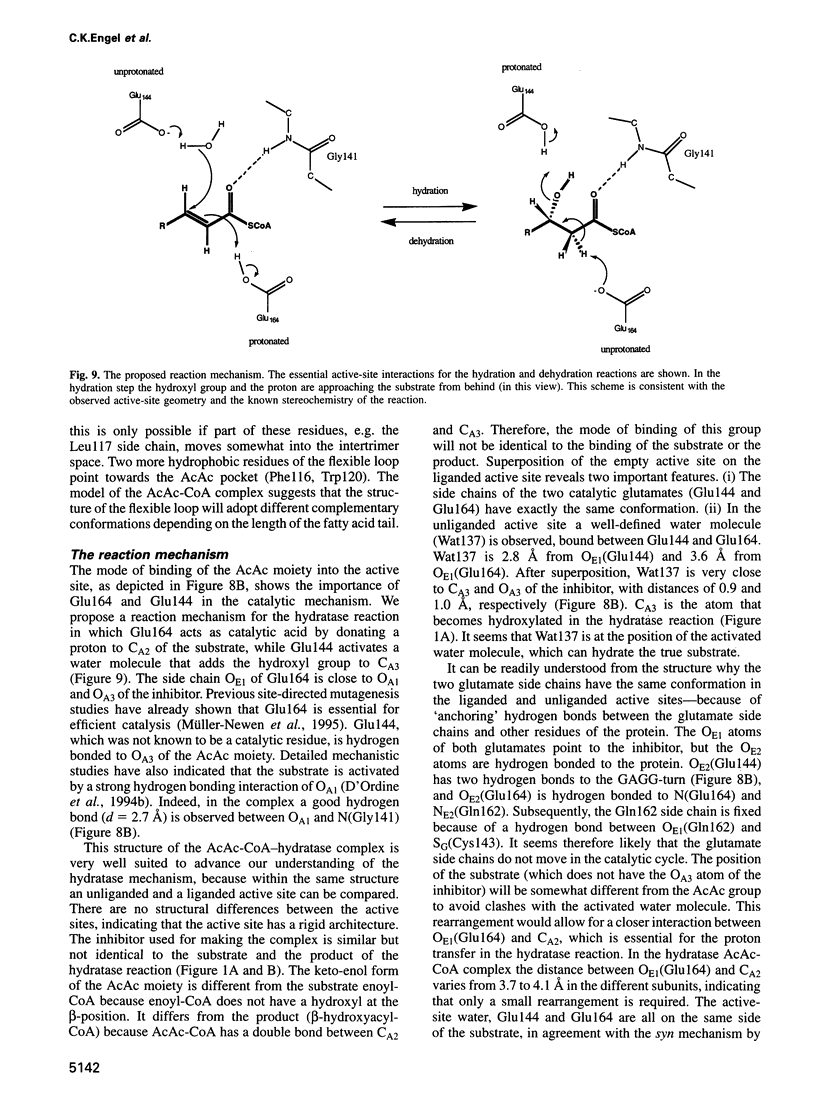
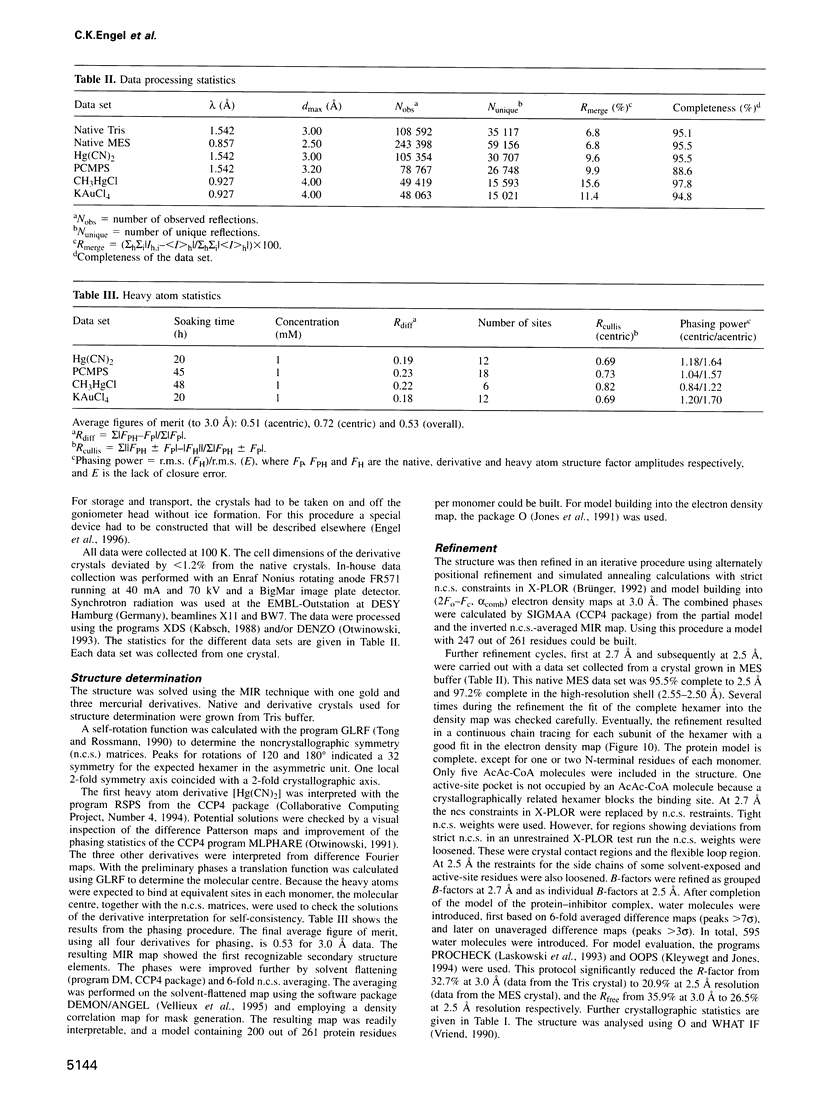
The crystal structure of rat liver mitochondrial enoyl-coenzyme A (CoA) hydratase complexed with the potent inhibitor acetoacetyl-CoA has been refined at 2.5 angstroms resolution. This enzyme catalyses the reversible addition of water to alpha,beta-unsaturated enoyl-CoA thioesters, with nearly diffusion-controlled reaction rates for the best substrates. Enoyl-CoA hydratase is a hexamer of six identical subunits of 161 kDa molecular mass for the complex. The hexamer is a dimer of trimers. The monomer is folded into a right-handed spiral of four turns, followed by two small domains which are involved in trimerization. Each turn of the spiral consists of two beta-strands and an alpha-helix. The mechanism for the hydratase/dehydratase reaction follows a syn-stereochemistry, a preference that is opposite to the nonenzymatic reaction. The active-site architecture agrees with this stereochemistry. It confirms the importance of Glu164 as the catalytic acid for providing the alpha-proton during the hydratase reaction. It also shows the importance of Glu144 as the catalytic base for the activation of a water molecule in the hydratase reaction. The comparison of an unliganded and a liganded active site within the same crystal form shows a water molecule in the unliganded subunit. This water molecule is bound between the two catalytic glutamates and could serve as the activated water during catalysis.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Babbitt P. C., Kenyon G. L., Martin B. M., Charest H., Slyvestre M., Scholten J. D., Chang K. H., Liang P. H., Dunaway-Mariano D. Ancestry of the 4-chlorobenzoate dehalogenase: analysis of amino acid sequence identities among families of acyl:adenyl ligases, enoyl-CoA hydratases/isomerases, and acyl-CoA thioesterases. Biochemistry. 1992 Jun 23;31(24):5594–5604. doi: 10.1021/bi00139a024. [DOI] [PubMed] [Google Scholar]
- Bahnson B. J., Anderson V. E. Crotonase-catalyzed beta-elimination is concerted: a double isotope effect study. Biochemistry. 1991 Jun 18;30(24):5894–5906. doi: 10.1021/bi00238a013. [DOI] [PubMed] [Google Scholar]
- D'Ordine R. L., Bahnson B. J., Tonge P. J., Anderson V. E. Enoyl-coenzyme A hydratase-catalyzed exchange of the alpha-protons of coenzyme A thiol esters: a model for an enolized intermediate in the enzyme-catalyzed elimination? Biochemistry. 1994 Dec 13;33(49):14733–14742. doi: 10.1021/bi00253a011. [DOI] [PubMed] [Google Scholar]
- D'Ordine R. L., Tonge P. J., Carey P. R., Anderson V. E. Electronic rearrangement induced by substrate analog binding to the enoyl-CoA hydratase active site: evidence for substrate activation. Biochemistry. 1994 Oct 25;33(42):12635–12643. doi: 10.1021/bi00208a014. [DOI] [PubMed] [Google Scholar]
- Engeland K., Kindl H. Purification and characterization of a plant peroxisomal delta 2, delta 3-enoyl-CoA isomerase acting on 3-cis-enoyl-CoA and 3-trans-enoyl-CoA. Eur J Biochem. 1991 Mar 28;196(3):699–705. doi: 10.1111/j.1432-1033.1991.tb15868.x. [DOI] [PubMed] [Google Scholar]
- Euler-Bertram S., Stoffel W. Purification and characterization of bovine liver 3-cis-2-trans-enoyl-CoA isomerase. Biol Chem Hoppe Seyler. 1990 Jul;371(7):603–610. doi: 10.1515/bchm3.1990.371.2.603. [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993 Sep 5;233(1):123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Jurnak F., Yoder M. D., Pickersgill R., Jenkins J. Parallel beta-domains: a new fold in protein structures. Curr Opin Struct Biol. 1994 Dec;4(6):802–806. doi: 10.1016/0959-440x(94)90259-3. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kleywegt G. J., Jones T. A. Efficient rebuilding of protein structures. Acta Crystallogr D Biol Crystallogr. 1996 Jul 1;52(Pt 4):829–832. doi: 10.1107/S0907444996001783. [DOI] [PubMed] [Google Scholar]
- Kobe B., Deisenhofer J. Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature. 1993 Dec 23;366(6457):751–756. doi: 10.1038/366751a0. [DOI] [PubMed] [Google Scholar]
- Mancia F., Keep N. H., Nakagawa A., Leadlay P. F., McSweeney S., Rasmussen B., Bösecke P., Diat O., Evans P. R. How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 A resolution. Structure. 1996 Mar 15;4(3):339–350. doi: 10.1016/s0969-2126(96)00037-8. [DOI] [PubMed] [Google Scholar]
- Minami-Ishii N., Taketani S., Osumi T., Hashimoto T. Molecular cloning and sequence analysis of the cDNA for rat mitochondrial enoyl-CoA hydratase. Structural and evolutionary relationships linked to the bifunctional enzyme of the peroxisomal beta-oxidation system. Eur J Biochem. 1989 Oct 20;185(1):73–78. doi: 10.1111/j.1432-1033.1989.tb15083.x. [DOI] [PubMed] [Google Scholar]
- Mohrig J. R., Moerke K. A., Cloutier D. L., Lane B. D., Person E. C., Onasch T. B. Importance of historical contingency in the stereochemistry of hydratase-dehydratase enzymes. Science. 1995 Jul 28;269(5223):527–529. doi: 10.1126/science.7624773. [DOI] [PubMed] [Google Scholar]
- Müller-Newen G., Janssen U., Stoffel W. Enoyl-CoA hydratase and isomerase form a superfamily with a common active-site glutamate residue. Eur J Biochem. 1995 Feb 15;228(1):68–73. doi: 10.1111/j.1432-1033.1995.tb20230.x. [DOI] [PubMed] [Google Scholar]
- Noble M. E., Zeelen J. P., Wierenga R. K., Mainfroid V., Goraj K., Gohimont A. C., Martial J. A. Structure of triosephosphate isomerase from Escherichia coli determined at 2.6 A resolution. Acta Crystallogr D Biol Crystallogr. 1993 Jul 1;49(Pt 4):403–417. doi: 10.1107/S0907444993002628. [DOI] [PubMed] [Google Scholar]
- Palosaari P. M., Kilponen J. M., Sormunen R. T., Hassinen I. E., Hiltunen J. K. Delta 3,delta 2-enoyl-CoA isomerases. Characterization of the mitochondrial isoenzyme in the rat. J Biol Chem. 1990 Feb 25;265(6):3347–3353. [PubMed] [Google Scholar]
- Palosaari P. M., Vihinen M., Mäntsälä P. I., Alexson S. E., Pihlajaniemi T., Hiltunen J. K. Amino acid sequence similarities of the mitochondrial short chain delta 3, delta 2-enoyl-CoA isomerase and peroxisomal multifunctional delta 3, delta 2-enoyl-CoA isomerase, 2-enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase enzyme in rat liver. The proposed occurrence of isomerization and hydration in the same catalytic domain of the multifunctional enzyme. J Biol Chem. 1991 Jun 15;266(17):10750–10753. [PubMed] [Google Scholar]
- Rao S. T., Rossmann M. G. Comparison of super-secondary structures in proteins. J Mol Biol. 1973 May 15;76(2):241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- Sharma V., Suvarna K., Meganathan R., Hudspeth M. E. Menaquinone (vitamin K2) biosynthesis: nucleotide sequence and expression of the menB gene from Escherichia coli. J Bacteriol. 1992 Aug;174(15):5057–5062. doi: 10.1128/jb.174.15.5057-5062.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thunnissen A. M., Dijkstra A. J., Kalk K. H., Rozeboom H. J., Engel H., Keck W., Dijkstra B. W. Doughnut-shaped structure of a bacterial muramidase revealed by X-ray crystallography. Nature. 1994 Feb 24;367(6465):750–753. doi: 10.1038/367750a0. [DOI] [PubMed] [Google Scholar]
- Tong L. A., Rossmann M. G. The locked rotation function. Acta Crystallogr A. 1990 Oct 1;46(Pt 10):783–792. doi: 10.1107/s0108767390005530. [DOI] [PubMed] [Google Scholar]
- Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990 Mar;8(1):52-6, 29. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- Waterson R. M., Hill R. L. Enoyl coenzyme A hydratase (crotonase). Catalytic properties of crotonase and its possible regulatory role in fatty acid oxidation. J Biol Chem. 1972 Aug 25;247(16):5258–5265. [PubMed] [Google Scholar]
- Willadsen P., Eggerer H. Substrate stereochemistry of the enoyl-CoA hydratase reaction. Eur J Biochem. 1975 May;54(1):247–252. doi: 10.1111/j.1432-1033.1975.tb04134.x. [DOI] [PubMed] [Google Scholar]
- Wolodko W. T., Fraser M. E., James M. N., Bridger W. A. The crystal structure of succinyl-CoA synthetase from Escherichia coli at 2.5-A resolution. J Biol Chem. 1994 Apr 8;269(14):10883–10890. doi: 10.2210/pdb1scu/pdb. [DOI] [PubMed] [Google Scholar]
- Yang S. Y., He X. Y., Schulz H. Glutamate 139 of the large alpha-subunit is the catalytic base in the dehydration of both D- and L-3-hydroxyacyl-coenzyme A but not in the isomerization of delta 3, delta 2-enoyl-coenzyme A catalyzed by the multienzyme complex of fatty acid oxidation from Escherichia coli. Biochemistry. 1995 May 16;34(19):6441–6447. doi: 10.1021/bi00019a025. [DOI] [PubMed] [Google Scholar]
- Zeelen J. P., Hiltunen J. K., Ceska T. A., Wierenga R. K. Crystallization experiments with 2-enoyl-CoA hydratase, using an automated 'fast-screening' crystallization protocol. Acta Crystallogr D Biol Crystallogr. 1994 Jul 1;50(Pt 4):443–447. doi: 10.1107/S0907444994001277. [DOI] [PubMed] [Google Scholar]









