Abstract
Isoniazid is used either alone or in combination with other drugs for the treatment of tuberculosis. It is also used for the prevention of tuberculosis. Chronic treatment of Isoniazid may cause severe liver damage leading to acute liver failure. The mechanism through which Isoniazid causes liver damage is investigated. Isoniazid treatment generates reactive oxygen species and induces apoptosis in Hep3B cells. It induces antioxidative and apoptotic genes leading to increase in mRNA expression and protein levels in Hep3B cells. Whole genome expression analysis of Hep3B cells treated with Isoniazid has resulted in differential expression of various genes playing prime role in regulation of apoptotic, antioxidative, DNA damage, cell signaling, cell proliferation and differentiation pathways. Isoniazid increased cytosolic Nrf2 protein level while decreased nuclear Nrf2 protein level. It also decreased ERK1 phosphorylation and treatment of Hep3B cells with ERK inhibitor followed by Isoniazid resulting in increased apoptosis in these cells. Two dimensional gel electrophoresis results have also shown differential expression of various protein species including heat shock proteins, proteins playing important role in oxidative stress, DNA damage, apoptosis, cell proliferation and differentiation. Results suggest that Isoniazid induces apoptosis through oxidative stress and also prevents Nrf2 translocation into the nucleus by reducing ERK1 phosphorylation thus preventing cytoprotective effect.
Keywords: Isoniazid, Oxidative stress, Differential expression, Phosphorylation, ERK inhibitor
Graphical abstract
Highlights
-
•
Isoniazid generates ROS and induces apoptosis in Hep3B cells.
-
•
Isoniazid resulted in differential expression of various genes playing prime role in various biological pathways.
-
•
Isoniazid decreased phosphorylation of ERK1 into phospho-ERK1 as a result of which Nrf2 phosphorylation was inhibited.
-
•
Nrf2 might not be able to translocate into the nucleus.
1. Introduction
Isoniazid is one of the cheapest and most effective drugs used for the treatment of tuberculosis since 1950. Isoniazid is metabolized in liver by various metabolizing enzymes such as N-acetyl transferase which acetylates it into N-acetylisoniazid. N-acetylisoniazid is then biotransformed into isonicotinic acid and monoacetylhydrazine. Monoacetylhydrazine is N-hydroxylated by the cytochrome P450 mixed oxidase system and causes hepatotoxicity via formation of a reactive intermediate metabolite. Antituberculosis drug-induced hepatotoxicity (ATDH) is a serious concern in tuberculosis treatment and management of hepatotoxicity. However, the mechanism of toxicity of ATDH is not yet known completely and further understanding of the mechanisms associated with ATDH is greatly required to facilitate a rational approach in the treatment.
In this study the role of Nrf2 (Nuclear factor, erythoid 2-like 2) is being investigated. Nrf2 induction occurs in the cell during oxidative stress which is caused by various environmental factors and chemicals including drugs [1–3]. These external stimuli induce generation of reactive oxygen species which in turn trigger Nrf2 activation and its translocation into the nucleus [4]. After its translocation into the nucleus, it binds to antioxidative response elements (ARE) sequences of various antioxidative and cytoprotective genes and then induces their transcription [5–7]. Protein products formed from these transcribed antioxidative and cytoprotective genes decrease reactive oxygen species and reduce the cell damages caused by them. Several reports have suggested a protective action for Nrf2-ARE signaling pathway against CYP2E1-dependent hepatic oxidative injury [8,9] indicating Nrf2-ARE pathway may exhibit a protective effect on INH and RFP-induced hepatotoxicity.
Extracellular signal regulated kinase 1 (ERK1) in the cytosol acts in signaling cascade and phosphorylate Nrf2 and induces its translocation into the nucleus [10]. ERK1 regulates various cellular processes like cell proliferation, cell differentiation and cell cycle progression in response to various extracellular signals [11,12]. In this study we hypothesized that Isoniazid inhibits Nrf2 phosphorylation by reducing ERK1 phosphorylation and interferes its translocation into the nucleus.
2. Results
2.1. Cell viability assay and IC50 determination
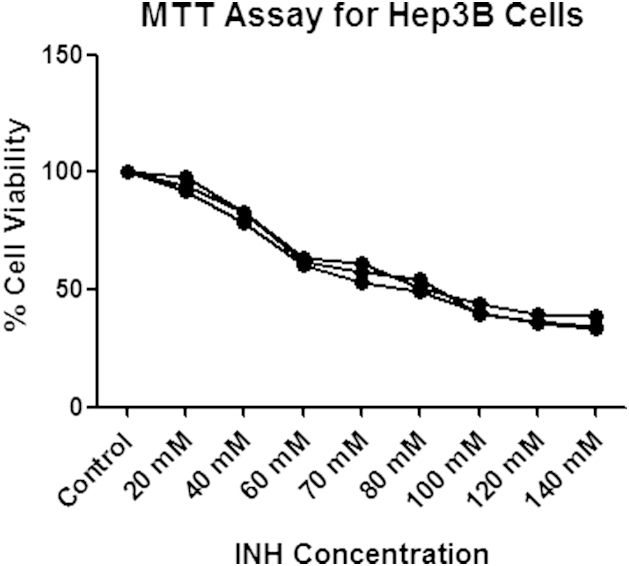
Hep3B cells were treated with 20, 40, 60, 80, 100, 120 and 140 mM Isoniazid concentrations while controls were treated with DMEM medium. Cell viability observed was 94.73% at 20 mM INH while at 140 mM of INH cell viability was 35.53% with controls having 100% cell viability. At 80 mM of INH 50% cell viability was observed. Therefore, IC50 for Hep3B cells for INH was calculated as 80 mM [Fig. 1].
Fig. 1.
Cell viability assay of Hep3B cells treated with different concentrations of Isoniazid for IC50 determination (INH-Isoniazid).
2.2. Reactive oxygen species estimation
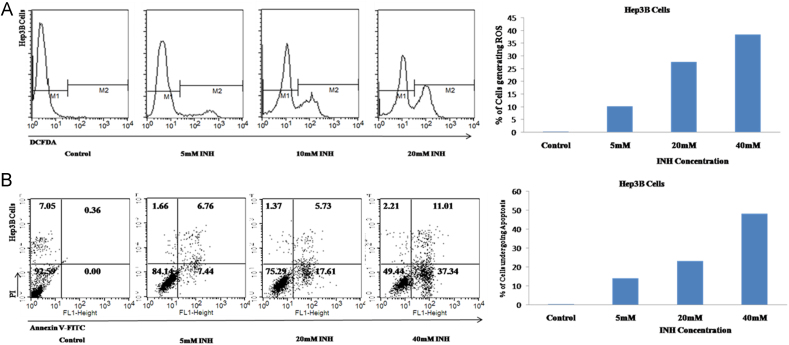
Isonaizid induced reactive oxygen species (ROS) generation in Hep3B cells in a dose dependent fashion. ROS generation was increased with increase in Isoniazid concentration in comparison to controls, where 0.36% cells generated ROS. When Hep3B cells were treated with 5, 10 and 20 mM INH, ROS was generated by 10.35%, 27.65% and 38.50% of the cells respectively [Fig. 2(A)].
Fig. 2.
Estimation of reactive oxygen species (ROS) generation and measurement of apoptosis induction by different concentrations of Isoniazid in Hep3B cells. (A) Flow cytometry results of Hep3B cells incubated with DCFDA following 24 h incubation of different concentrations of Isoniazid for estimation of reactive oxygen species generation (ROS). (B) Flow cytometry results of Hep3B following 24 h incubation of different concentrations of Isoniazid for measurement of apoptosis induction.
2.3. Apoptosis induction
In control Hep3B cell population, 0.36% of cells were in apoptosis while in cells treated with 5, 10 and 20 mM INH, percentage of cells undergoing apoptosis was found to be at 1.42, 23.34 and 48.35 respectively [Fig. 2(B)]. The result showed that Isoniazid induces apoptosis in Hep3B cells in a dose dependent manner.
2.4. Messenger RNA level of apoptotic and antioxidative genes
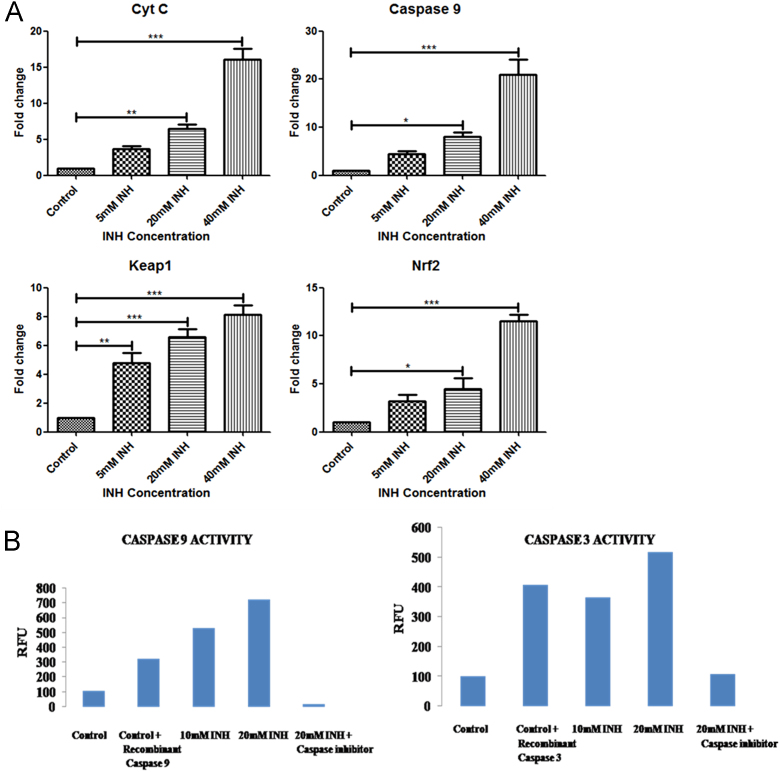
mRNA levels of apoptotic (Cyt C, Caspase 9) and antioxidative genes (Keap1, Nrf2) were observed to be significantly increased in a dose dependent fashion in Hep3B cells treated with different Isoniazid concentrations as compared to controls [Fig. 3(A)].
Fig. 3.
(A) Quantitative real time PCR analysis for relative mRNA expression of apoptotic and antioxidative genes in Hep3B cells following treatment with different concentrations (5, 20 and 40 mM) of Isoniazid. For normalization β-actin was used as housekeeping control. (B) Diagram showing Caspase 9 and Caspase 3 activities in Hep3B cells following treatment with different Isoniazid concentrations (10 mM and 20 mM).
2.5. Caspase activity assay
Activities of Caspase 9 and Caspase 3 were increased in Hep3B cells treated with 10 and 20 mM of Isoniazid respectively [Fig. 3(B)]. When caspase inhibitors were added to cell suspension before adding caspase substrate the activities of both Caspase 9 and Caspase 3 were observed to decrease in Hep3B cells.
2.6. Differential gene expression study
Following 20 mM INH treatment in Hep3B cells, whole genome mRNA expression was assessed by microarray analysis. A high statistically stringent criterion (p<0.05 and 2.2 fold change) for up-regulated genes filtered 39 genes (Table 2) while a high statistically stringent criterion (p<0.05 and 2.5 fold change) for down-regulated genes filtered 64 genes (Table 3). Few differentially expressed transcript cluster Ids have shown no sequence similarity with any known genes and have not been assigned any biological function yet.
Table 2.
List of differentially up-regulated genes identified with p-value cut-off 0.05 and fold change >2.2 following Isoniazid treatment to Hep3B cells at 20 mM concentration.
| S.No | Transcripts cluster Id | Gene symbol | Gene description | Regulation | Log FC ([Treat] vs [Control]) | FC ([Treat] vs [Control]) |
|---|---|---|---|---|---|---|
| Amino acid metabolism, developmental disorder, hereditary disorder | ||||||
| 1. | 7953211 | C12orf5 | Chromosome 12 open reading frame 5 | Up | 1.167182 | 2.245726 |
| 2. | 8153457 | EEF1D | Eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) | Up | 1.152429 | 2.2228777 |
| 3. | 8047097 | GLS | Glutaminase | Up | 1.271881 | 2.4147623 |
| 4. | 8121144 | MANEA | Mannosidase, endo-alpha | Up | 1.543634 | 2.915279 |
| 5. | 8110861 | MRPL36 | Mitochondrial ribosomal protein L36 | Up | 1.547498 | 2.9230971 |
| 6. | 8159687 | MRPL41 | Mitochondrial ribosomal protein L41 | Up | 1.248036 | 2.3751783 |
| 7. | 7985757 | MRPS11 | Mitochondrial ribosomal protein S11 | Up | 1.221251 | 2.331488 |
| 8. | 8156043 | PSAT1 | Phosphoserine aminotransferase 1 | Up | 1.228868 | 2.3438294 |
| 9. | 8006183 | SUZ12P | Suppressor of zeste 12 homolog pseudogene | Up | 1.619368 | 3.0724032 |
| 10. | 8165703 | UIMC1 | Ubiquitin interaction motif containing 1 | Up | 1.324133 | 2.5038245 |
| 11. | 7936346 | ZDHHC6 | Zinc finger, DHHC-type containing 6 | Up | 1.36511 | 2.5759594 |
| 12. | 8006377 | ZNF207 | ZInc finger protein 207 | Up | 1.273516 | 2.4175005 |
| 13. | 7995252 | ZNF720 | Zinc finger protein 720 | Up | 1.182164 | 2.2691684 |
| 14. | 7997904 | ZNF778 | Zinc finger protein 778 | Up | 1.935808 | 3.8259227 |
| Infectious disease, cell proliferation, gene expression | ||||||
| 15. | 8125766 | BAK1 | BCL2-antagonist/killer 1 | Up | 1.158309 | 2.2319558 |
| 16. | 8129562 | CTGF | Connective tissue growth factor | Up | 1.230378 | 2.3462842 |
| 17. | 8112914 | DHFR | Dihydrofolate reductase | Up | 1.695785 | 3.2395313 |
| 18. | 8154765 | DNAJA1 | DnaJ (Hsp40) homolog, subfamily A, member 1 | Up | 1.172203 | 2.2535555 |
| 19. | 7980535 | DYNLL1 | Dynein, light chain, LC8-type 1 | Up | 1.512186 | 2.8524194 |
| 20. | 7948839 | NXF1 | Nuclear RNA export factor 1 | Up | 1.880587 | 3.6822488 |
| 21. | 8147654 | POLR2K | Polymerase (RNA) II (DNA directed) polypeptide K, 7.0 kDa | Up | 1.241887 | 2.365076 |
| 22. | 8060758 | PRNP | Prion protein | Up | 1.313068 | 2.4846935 |
| 23. | 7914791 | SFPQ | Splicing factor proline/glutamine-rich (polypyrimidine tract binding protein associated) | Up | 1.217865 | 2.3260224 |
| 24. | 8165674 | SH3KBP1 | SH3-domain kinase binding protein 1 | Up | 1.336754 | 2.5258236 |
| 25. | 8150698 | SNAI2 | Snail homolog 2 (Drosophila) | Up | 1.375106 | 2.5938694 |
| 26. | 7992789 | TNFRSF12A | Tumor necrosis factor receptor superfamily, member 12A | Up | 1.205973 | 2.3069274 |
| RNA processing, DNA damage | ||||||
| 27. | 7898375 | RNU1A|RNU1A3|RNU1C1|RNU1C2|RNU1G2|RNU1F1|RNU1G1|RNU1G3|GSTM2 | RNA, U1A small nuclear|RNA, U1A3 small nuclear|RNA, U1C1 small nuclear|RNA, U1C2 small nuclear|RNA, U1G2 small nuclear|RNA, U1F1 small nuclear|RNA, U1G1 small nuclear|RNA, U1G3 small nuclear|glutathione S-transferase mu 2 (muscle) | Up | 1.19027 | 2.2819536 |
| 28. | 7911329 | SEPT14|C20orf69|DKFZP434B2016 | Septin 14|chromosome 20 open reading frame 69|similar to hypothetical protein LOC284701 | Up | 1.383161 | 2.6083918 |
| 29. | 7942783 | C11orf67|INTS4 | Chromosome 11 open reading frame 67|integrator complex subunit 4 | Up | 1.189677 | 2.2810163 |
| 30. | 7949916 | CHKA | Choline kinase alpha | Up | 1.142337 | 2.207383 |
| 31. | 8005553 | SNORD3A|SNORD3B-1|SNORD3B-2|SNORD3C|SNORD3D | Small nucleolar RNA, C/D box 3A|small nucleolar RNA, C/D box 3B-1|Usmall nucleolar RNA, C/D box 3B-2|small nucleolar RNA, C/D box 3C|small nucleolar RNA, C/D box 3D | Up | 2.225389 | 4.676368 |
| 32. | 8024485 | GADD45B | Growth arrest and DNA-damage-inducible, beta | Up | 1.182639 | 2.2699163 |
| 33. | 8031646 | LOC100288114|MGC9913 | Hypothetical protein LOC100288114|hypothetical protein MGC9913 | Up | 1.318619 | 2.494272 |
| 34. | 8035779 | ZNF626|ZNF93|ZNF253|LOC100128975 | Zinc finger protein 626|zinc finger protein 93|zinc finger protein 253| similar to zinc finger protein 626 | Up | 1.64639 | 3.1304922 |
| 35. | 8044961 | RNU4ATAC | RNA, U4atac small nuclear (U12-dependent splicing) | Up | 1.146462 | 2.213704 |
| 36. | 8137008 | C7orf11 | Chromosome 7 open reading frame 11 | Up | 1.158495 | 2.2322443 |
| 37. | 8145795 | LOC100293539 | Similar to ribosomal protein 10 | Up | 1.159232 | 2.2333846 |
| 38. | 8148000 | C8orf85 | Chromosome 8 open reading frame 85 | Up | 1.320166 | 2.4969485 |
| 39. | 8161575 | CBWD5|CBWD3|CBWD1|CBWD6|CBWD7|CBWD2|LOC653510 | COBW domain containing 5|COBW domain containing 3| COBW domain containing 1 | Up | 1.209761 | 2.3129926 |
Table 3.
List of differentially down-regulated genes identified with p-value cut-off 0.05 and fold change >2.5 following Isoniazid treatment to Hep3B cells at 20 mM concentration.
| S.No. | Transcripts Cluster Id | Gene Symbol | Gene Description | Regulation | Log FC ([Treat] vs [Control]) | FC ([Treat] vs [Control]) |
|---|---|---|---|---|---|---|
| Cell death and survival, cellular development, nervous system development and function | ||||||
| 1. | 8158671 | ASS1 | Argininosuccinate synthetase 1 | Down | −1.58433 | −2.99868 |
| 2. | 8077441 | BHLHE40 | Basic helix-loop-helix family, member e40 | Down | −2.2831 | −4.86722 |
| 3. | 8060850 | BMP2 | Bone morphogenetic protein 2 | Down | −1.528 | −2.88387 |
| 4. | 7928308 | DDIT4 | DNA-damage-inducible transcript 4 | Down | −2.79568 | −6.9436 |
| 5. | 8019392 | FASN | Fatty acid synthase | Down | −1.66354 | −3.16794 |
| 6. | 8151457 | HEY1 | Hairy/enhancer-of-split related with YRPW motif 1 | Down | −1.47183 | −2.77374 |
| 7. | 8061564 | ID1 | Inhibitor of DNA binding 1, dominant negative helix-loop-helix protein | Down | −1.56954 | −2.96809 |
| 8. | 8174444 | IRS4 | Insulin receptor substrate 4 | Down | −2.78425 | −6.88877 |
| 9. | 7956301 | LRP1 | Low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | Down | −1.41548 | −2.66749 |
| 10. | 8153002 | NDRG1 | N-myc downstream regulated 1 | Down | −1.79439 | −3.46868 |
| 11. | 8008627 | NOG | Noggin | Down | −1.38934 | −2.6196 |
| 12. | 8046408 | PDK1 | Pyruvate dehydrogenase kinase, isozyme 1 | Down | −1.74592 | −3.35409 |
| 13. | 8008263 | PDK2 | Pyruvate dehydrogenase kinase, isozyme 2 | Down | −1.47028 | −2.77077 |
| 14. | 8119161 | PIM1 | Pim-1 oncogene | Down | −1.46096 | −2.75292 |
| 15. | 8145977 | PLEKHA2 | Pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 | Down | −1.45269 | −2.73718 |
| 16. | 8166469 | SAT1 | Spermidine/spermine N1-acetyltransferase 1 | Down | −1.41249 | −2.66197 |
| Developmental disorder, hereditary disorder, neurological disease | ||||||
| 17. | 8080168 | ACY1 | Aminoacylase 1 | Down | −1.74711 | −3.35686 |
| 18. | 7935403 | ARHGAP19 | Rho GTPase activating protein 19 | Down | −1.3689 | −2.58274 |
| 19. | 7967072 | COQ5 | Coenzyme Q5 homolog, methyltransferase (S. cerevisiae) | Down | −1.48488 | −2.79895 |
| 20. | 7923596 | ETNK2 | Ethanolamine kinase 2 | Down | −1.67516 | −3.19356 |
| 21. | 8046524 | HOXD13 | Homeobox D13 | Down | −1.71756 | −3.2888 |
| 22. | 7915827 | LRRC41 | Leucine rich repeat containing 41 | Down | −1.34957 | −2.54836 |
| 23. | 8148917 | MFSD3 | Major facilitator superfamily domain containing 3 | Down | −1.96708 | −3.90977 |
| 24. | 7984922 | MPI | Mannose phosphate isomerase | Down | −1.40341 | −2.64525 |
| 25. | 8085431 | NUP210 | Nucleoporin 210 kDa | Down | −1.51115 | −2.85037 |
| 26. | 8165711 | PLCXD1 | Phosphatidylinositol-specific phospholipase C, X domain containing 1 | Down | −1.6955 | −3.2389 |
| 27. | 8088664 | SUCLG2 | Succinate-CoA ligase, GDP-forming, beta subunit | Down | −1.63177 | −3.09893 |
| 28. | 7980720 | TTC7B | Tetratricopeptide repeat domain 7B | Down | −1.33148 | −2.5166 |
| 29. | 8169598 | ZCCHC12 | Zinc finger, CCHC domain containing 12 | Down | −1.93981 | −3.83656 |
| 30. | 8166500 | ZFX | Zinc finger protein, X-linked | Down | −1.35301 | −2.55444 |
| 31. | 8055377 | ZRANB3 | Zinc finger, RAN-binding domain containing 3 | Down | −1.63365 | −3.10297 |
| Amino acid metabolism, post-translational modification, small molecule biochemistry | ||||||
| 32. | 8082444 | ACAD9 | Acyl-Coenzyme A dehydrogenase family, member 9 | Down | −1.43318 | −2.70041 |
| 33. | 7901110 | AKR1A1 | Aldo-keto reductase family 1, member A1 (aldehyde reductase) | Down | −1.68036 | −3.2050 |
| 34. | 8013660 | ALDOC | Aldolase C, fructose-bisphosphate | Down | −1.74079 | −3.34219 |
| 35. | 7981427 | CKB | Creatine kinase, brain | Down | −1.39591 | −2.63155 |
| 36. | 8036602 | ECH1 | Enoyl Coenzyme A hydratase 1, peroxisomal | Down | −1.54757 | −2.92324 |
| 37. | 7953532 | ENO2 | Enolase 2 (gamma, neuronal) | Down | −1.34501 | −2.54032 |
| 38. | 8136293 | EXOC4 | Exocyst complex component 4 | Down | −1.3335 | −2.52013 |
| 39. | 8159373 | FAM69B | Family with sequence similarity 69, member B | Down | −1.40775 | −2.65322 |
| 40. | 8154951 | GLUL | Glutamate-ammonia ligase (glutamine synthetase) | Down | −1.8001 | −3.48245 |
| 41. | 8095303 | LPHN3 | Latrophilin 3 | Down | −1.49011 | −2.8091 |
| 42. | 7934278 | P4HA1 | Prolyl 4-hydroxylase, alpha polypeptide I | Down | −1.81848 | −3.52709 |
| 43. | 8027330 | PCGF6 | Polycomb group ring finger 6 | Down | −1.7239 | −3.30328 |
| 44. | 8091283 | PLOD2 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | Down | −1.40192 | −2.64253 |
| 45. | 7944382 | VPS11 | Vacuolar protein sorting 11 homolog | Down | −1.41151 | −2.66016 |
| Connective tissue disorders, developmental disorder, hereditary disorder | ||||||
| 46. | 8035494 | FKBP8 | FK506 binding protein 8, 38 kDa | Down | −1.33595 | −2.52441 |
| 47. | 8101324 | HNRNPD | Heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37 kDa) | Down | −1.32701 | −2.50882 |
| 48. | 7937335 | IFITM1 | Interferon induced transmembrane protein 1 (9–27) | Down | −1.74811 | −3.35919 |
| 49. | 7969574 | KRT18 | Keratin 18 | Down | −1.52955 | −2.88695 |
| 50. | 7938777 | LDHA | Lactate dehydrogenase A | Down | −1.42929 | −2.69313 |
| 51. | 8167656 | MAGED1 | Melanoma antigen family D, 1 | Down | −1.46751 | −2.76543 |
| 52. | 8168215 | MED12 | Mediator complex subunit 12 | Down | −1.60053 | −3.03255 |
| 53. | 8013120 | PEMT | Phosphatidylethanolamine N-methyltransferase | Down | −1.54938 | −2.92691 |
| 54. | 8069057 | PFKL | Phosphofructokinase, liver | Down | −1.44216 | −2.71727 |
| 55. | 8156905 | TMEFF1 | Transmembrane protein with EGF-like and two follistatin-like domains 1 | Down | −1.38198 | −2.60625 |
| 56. | 7950307 | UCP2 | Uncoupling protein 2 (mitochondrial, proton carrier) | Down | −1.41558 | −2.66768 |
| 57. | 7993872 | UQCRC2 | Ubiquinol-cytochrome c reductase core protein II | Down | −1.33492 | −2.52262 |
| Cell-to-cell signaling and interaction, embryonic development, tissue development | ||||||
| 58. | 8170393 | CXorf40A | Chromosome X open reading frame 40A | Down | −1.43056 | −2.69551 |
| 59. | 8123739 | NRN1 | Neuritin 1 | Down | −1.64898 | −3.13611 |
| Cellular development, embryonic development, nervous system development and function | ||||||
| 60. | 7920664 | THBS3 | Thrombospondin 3 | Down | −1.3366 | −2.52556 |
| 61. | 7934906 | ACTA2 | Actin, alpha 2, smooth muscle, aorta | Down | −1.65103 | −3.14058 |
| 62. | 7955317 | ACCN2 | Amiloride-sensitive cation channel 2, neuronal | Down | −1.62671 | −3.08809 |
| 63. | 8098604 | ANKRD37|UFSP2 | Ankyrin repeat domain 37|UFM1-specific peptidase 2 | Down | −1.98257 | −3.95196 |
| 64. | 8178609 | VARS|VARS2 | Valyl-tRNA synthetase|valyl-tRNA synthetase 2, mitochondrial (putative) | Down | −1.36886 | −2.58267 |
2.7. Affected pathways
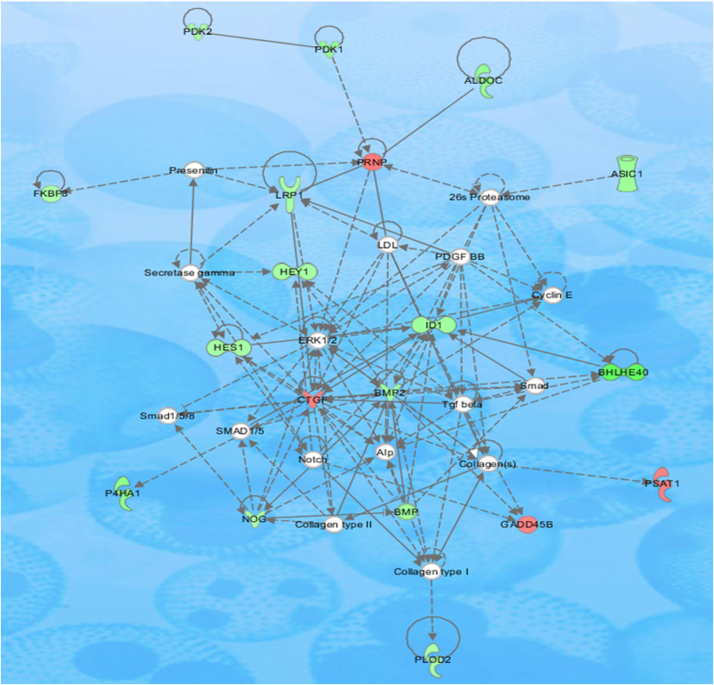
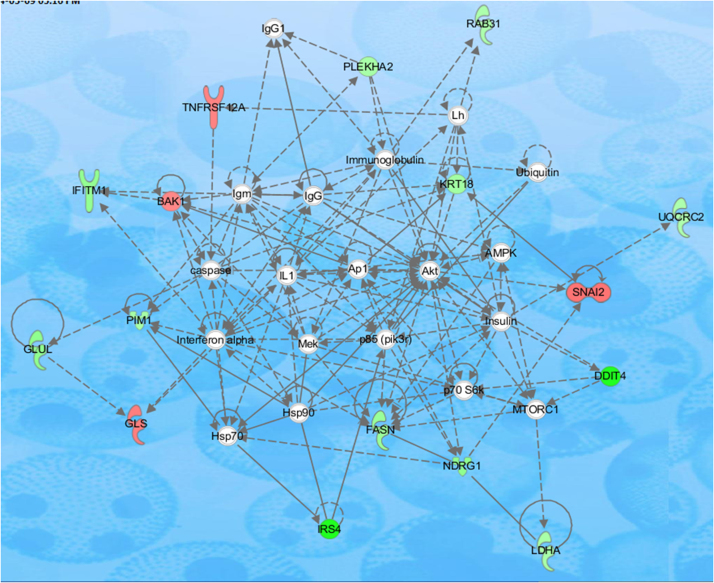
Differentially expressed genes were clustered by IPA (Ingunity Pathway Analysis, Qiagen) software in which genes were classified based upon functional category and pathways. Genes belonging to same mechanisms were joined together to form a single pathway. Major pathways that showed signs of Isoniazid induced perturbations included development function, cellular movement, post-translational modifications, energy production, cell to cell signaling, cell death, survival, cell growth and proliferation pathways [Figs. 4 and 5].
Fig. 4.
Cardiovascular system development and function, connective tissue development and function, embryonic development pathway affected by Isoniazid treatment in Hep3B cells. In the shown pathway genes represented by red color were upregulated while genes respresented by green color were downregulated by Isoniazid treatment to Hep3B cells.
Fig. 5.
Amino acid metabolism, post-translational modification, small molecule biochemistry pathway affected by Isoniazid treatment in Hep3B cells. In the shown pathway genes represented by red color were upregulated while genes respresented by green color were downregulated by Isoniazid treatment to Hep3B cells.
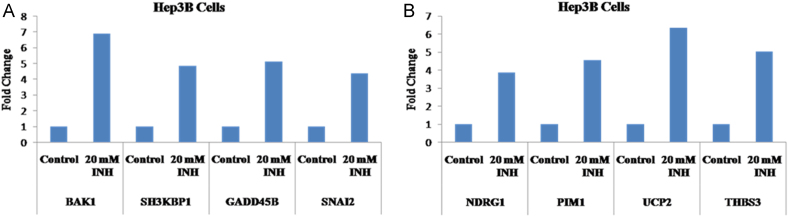
2.8. Validation of microarray results by quantitative real time PCR analysis
Differential expression of four up-regulated genes (Bak1, SH3KBP1, GADD45B and SNAI2) and four down-regulated genes (NDRG1, PIM1, UCP2 and THBS3) as observed in microarray analysis were further validated by quantitative real time PCR analysis [Fig. 6(A) and (B)]. The up regulated genes Bak1, SH3KBP1, GADD45B and SNAI2 were observed to be up-regulated by 6.91, 4.87, 5.13 and 4.39 folds respectively, following 20 mM INH treatment to Hp3B cells. Similarly the down-regulated genes NDRG1, PIM1, UCP2 and THBS3 were down regulated by 3.87, 4.56, 6.34 and 5.02 fold respectively, following 20 mM INH treatment to Hep3B cells.
Fig. 6.
Bar graph presents the fold change in selected genes with quantative real time-PCR. The genes were selected from those that showed differential expression either (A) at p<0.05 and up regulated >2.2 fold or (B) at p<0.05 and down regulated >2.5 fold in microarray analysis after 20 mM Isoniazid treatment to Hep3B cells.
2.9. Expression of apoptotic genes at protein level
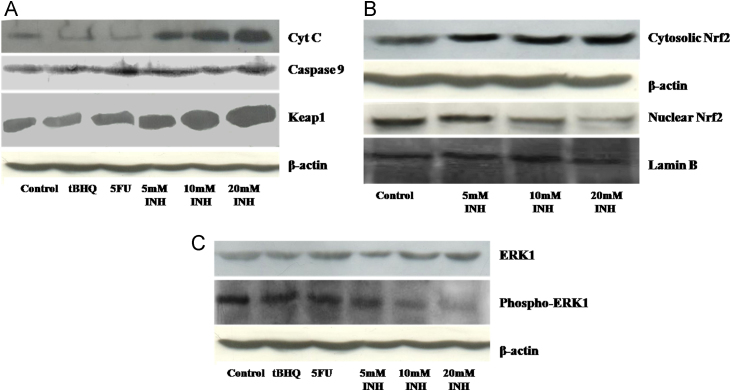
Isoniazid induced oxidative stress in Hep3B cells was confirmed by Cyt C release from mitochondrial inner membrane. Cytochrome C released from mitochondrial inner membrane was observed by Western blot analysis and this increase was proportional to Isoniazid concentrations. Cyt C binds apoptotic protease activating factor 1 (Apaf 1) which in turn binds to Procaspase 9 and converts it into Caspase 9. Caspase 9 protein level in Isoniazid treated Hep3B cells, was also observed to increase in a dose dependent fashion. Protein amount of both Cyt C and Caspase 9 was increased, from 5 mM to 10 and 20 mM of INH in comparison to control Hep3B cells [Fig. 7(A)]. Keap1 protein was also observed to increase in Hep3B cells treated with Isoniazid and this increase in Keap1 protein was also proportional to Isoniazid concentrations [Fig. 7(A)]. Densitometric results of bands of Cytochrome C, Caspase 9 and Keap1 are shown in Fig. S1.
Fig. 7.
Effect of different concentrations (5 , 10 , and 20 mM) of Isoniazid on protein levels of various proteins, (A) Western blot result of Cyt C, Caspase 9 and Keap1 following treatment with Isoniazid to Hep3B cells. (B) Western blot result of Cytosolic and Nuclear Nrf2 protein level following treatment with Isoniazid. (C) Western blot result of ERK1 and Phospho-ERK1 following treatment with Isoniazid to Hep3B cells. All the experiments were performed in triplicate independently and image is representative image of one of them. For cytosolic fraction β-actin was used as housekeeping and for nuclear fraction Lamin B was used as housekeeping (5FU- 5-fluorouracil, tBHQ- tertiary butylhydroquinone, INH- Isoniazid).
2.10. Cytosolic and nuclear Nrf2 protein level
Cytosolic Nrf2 protein level was observed to be increased in Hep3B cells treated with Isoniazid while the nuclear Nrf2 protein was observed to decrease [Fig. 7(B)]. This change in cytosolic and nuclear Nrf2 protein levels was proportional to Isoniazid concentrations. Protein amount of cytosolic Nrf2 was increased, from 5 mM to 10 and 20 mM of INH, as compared to control Hep3B cells while protein amount of nuclear Nrf2 was decreased, from 5 mM to 10 and 20 mM of Isoniazid, as compared to control Hep3B cells. Densitometric results of bands of Cytosolic Nrf2 and Nuclear Nrf2 are shown in Fig. S2.
2.11. Phosphorylation of ERK1 into Phospho-ERK1
Phosphorylation of extra-cellular regulated kinase 1 (ERK1) protein into phospho-ERK1 was found to be decreased in the cytosol of Hep3B cells following Isoniazid treatment as compared to control Hep3B cells [Fig. 7(C)]. Phosphorylation of ERK1 in Hep3B cells was decreased when we increased INH concentrations from 5 mM to 10 and 20 mM. Densitometric results of bands of ERK1 and phospho-ERK1 are shown in Fig. S3.
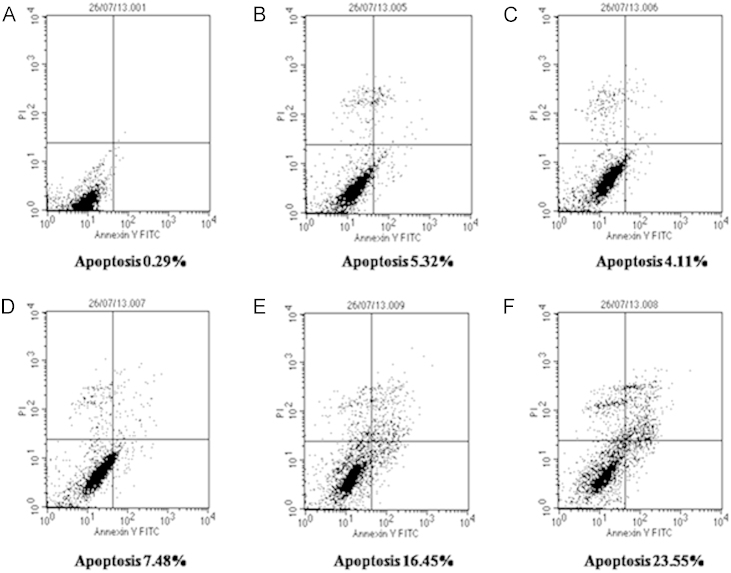
2.12. ERK1 inhibitor and apoptosis
When Hep3B cells were treated with ERK inhibitor, it induced apoptosis. 50 µM of ERK inhibitor induced apoptosis in 5.32% of Hep3B cells [Fig. 8(B)] while cells treated with 10 mM and 20 mM of INH induced apoptosis in 4.11% and 7.48% of Hep3B cells [Fig. 8(C) and (D)] respectively compared to untreated Hep3B cells [Fig. 8(A)]. When Hep3B cells were pre-treated with 50 µM ERK inhibitor followed by 10 mM and 20 mM of INH, percent of Hep3B cells undergoing apoptosis was increased and it was 16.45 and 23.55 respectively compared to untreated Hep3B cells [Fig. 8(E) and (F)].
Fig. 8.
Measurement of apoptosis induced by different concentrations of Isoniazid and combination of Isoniazid and ERK inhibitor in Hep3B cells by Annexin V FITC method. (A) Untreated Hep3B cells, (B) Hep3B cells treated with 50 µM ERK inhibitor, (C) Hep3B cells treated with 10 mM INH, (D) Hep3B cells treated with 20 mM INH, (E) Hep3B cells treated with combination of 50 µM ERK inhibitor and 10 mM INH, (F) Hep3B cells treated with combination of 50 µM ERK inhibitor and 20 mM INH. Experiments were performed in triplicate independently.
2.13. Two dimensional gel electrophoresis
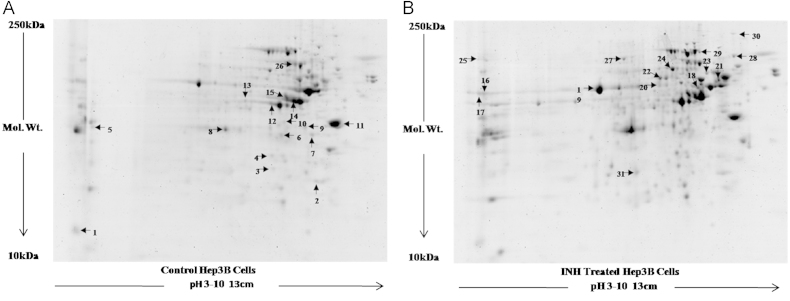
After analyzing two dimensional gels, 30 protein spots, which were differentially regulated, were selected and cut [Fig. 9]. These spots were either up-regulated by >1.5 times or down-regulated by <0.65 times. We observed that out of 30, 15 proteins were up-regulated and 15 proteins were down-regulated (Table 4). These proteins were identified by MALDI TOF/TOF analysis. These differentially expressed proteins were found to have role in oxidative stress, apoptosis, cell proliferation, t-RNA export, protein synthesis, development and cytoskeleton formation.
Fig. 9.
Two dimensional gel electrophoresis of proteins isolated from Hep3B cells following 20 mM INH treatment. 2D gel images of Control and 20 mM INH treated Hep3B cells showing differential expression of various proteins spots. All the experiments were performed in triplicate independently and image is representative image of one of them.
Table 4.
List of proteins identified using MALDI-TOF from 2D Gels from NCBInr.
| Spot ID | Identified protein | Accession number | Molecular mass (Da) | Sequence coverage (%) | Score | Fold difference | Up or down |
|---|---|---|---|---|---|---|---|
| 1. | 60 kDa heat shock protein | gi|31542947 | 61,187 | 18 | 284 | 0.65 | Down |
| 2. | Beta actin like protein | gi|63055057 | 42,318 | 2 | 82 | 1.94 | UP |
| 3. | Heterogenous nuclear ribonucleoprotein H | gi|5031753 | 49,484 | 2 | 117 | 1.69 | Up |
| 4. | BiP Protein | gi|6470150 | 71,002 | 7 | 991 | 1.52 | Up |
| 5. | Heterogenous nuclear ribonucleoprotein A2 | gi|4504447 | 36,041 | 10 | 363 | 2.38 | Up |
| 6. | Inorganic pyrophosphatase | gi|11056044 | 33,095 | 2 | 64 | 0.27 | Down |
| 7. | Cofilin 1 | gi|5031635 | 18,719 | 5 | 127 | 0.42 | Down |
| 8. | Peroxiredoxin 4 | gi|5453549 | 30,749 | 4 | 242 | 2.33 | Up |
| 9. | Enolase 1 | gi|62896593 | 47,453 | 16 | 843 | 0.48 | Down |
| 10. | Chain A Horf6A Novel Human Peroxidase enzyme | gi|331884 | 25,011 | 9 | 468 | 1.53 | Up |
| 11. | KRT73 Protein | gi|80475848 | 42,270 | 2 | 59 | 0.56 | Down |
| 12. | Alpha globin | gi|28549 | 13,631 | 1 | 71 | 0.56 | Down |
| 13. | Beta globin chain | gi|66473265 | 11,537 | 2 | 60 | 0.46 | Down |
| 14. | Pote Ankyrin domain family member E | gi|578804361 | 77,074 | 6 | 326 | 0.54 | Down |
| 15. | CKB | gi|49457530 | 42,933 | 2 | 137 | 0.46 | Down |
| 16. | EEF1A1 protein | gi|48735185 | 50,437 | 3 | 111 | 1.79 | Up |
| 17. | Eukaryotic translation initiation | gi|4503513 | 45,035 | 8 | 342 | 1.80 | Up |
| 18. | Heterogenous nuclear ribonucleoprotein F | gi|4826760 | 45,985 | 3 | 67 | 0.48 | Down |
| 19. | Endoplasmin precursor | gi|4507677 | 92,696 | 6 | 155 | 1.80 | Up |
| 20. | B23 nucleophosmin | gi|82567 | 31,080 | 9 | 348 | 1.82 | Up |
| 21. | ATP synthase subunit beta | gi|32189394 | 56,525 | 10 | 681 | 0.46 | Down |
| 22. | Transformation upregulated nuclear protein | gi|460789 | 51,325 | 1 | 70 | 1.82 | Up |
| 23. | Alpha tubulin | gi|37492 | 37,492 | 1 | 77 | 0.53 | Down |
| 24. | Chain A Tapasin ERP57 | gi|220702 | 54,541 | 7 | 337 | 2.03 | Up |
| 25. | non-POU domain containing octamer protein binding isoform 1 | gi|34932414 | 54,311 | 5 | 200 | 3.7 | Up |
| 26. | Chaperonin (Hsp60) | gi|306890 | 61,157 | 2 | 76 | 0.51 | Down |
| 27. | Stress induced phophoprotein 1 | gi|5803181 | 63,227 | 3 | 142 | 2.34 | Up |
| 28. | Rho GDP-dissociation inhibitor 1 | gi|4757768 | 23,250 | 6 | 221 | 0.62 | Down |
| 29. | Triose phosphate isomerase | gi|136066 | 26,894 | 3 | 134 | 0.46 | Down |
| 30. | Cyclophilin | gi|181250 | 22,654 | 7 | 281 | 3.06 | Up |
3. Discussion
Drug induced liver injury during the treatment of tuberculosis infection, is a major challenge to modern hepatology [15,16]. Therefore, the assessment and monitoring of the hepatotoxicity of anti-tuberculosis drugs for prevention of liver injury are great concerns during disease treatment. The exact mechanism of anti-tuberculosis drugs induced hepatotoxicity is not clearly understood yet. Some authors point to oxidative stress in hepatocytes by anti-tuberculosis drugs [17,18]. Isoniazid is the most effective agent against tuberculosis and is used both for the treatment and the prophylaxis of this disease [19]. Toxic metabolites may also play an important role in the development of anti-TB drug induced hepatotoxicity (ATDH) [20].
During oxidative stress, ROS is generated and it attacks the mitochondrial membrane resulting in Cyt C release. This released Cyt C binds to Apoptotic protease activating factor 1 (Apaf-1) which in turn binds to pro-caspase-9 to form apoptosome. The apoptosome cleaves pro-caspase-9 to its active form caspase-9, which in turn activates the effector caspase-3 [21].
In Hep3B cells, twenty-four hours after treatment of 20 mM Isoniazid led to significant increase in number of annexin V FITC positive cells demonstrating apoptotis. ROS generation and apoptosis were found to be directly proportional to Isoniazid concentrations in Hep3B cells in this study. Apoptosis can be induced through two distinct pathways; one involves the ligation of the TNF/Fas-receptor with its ligand and followed by caspase-8 activation, leading to either direct activation of caspase-3, or through merger with the mitochondrial pathway via cleavage of the Bcl-2 family member, Bid. The other pathway is the mitochondria mediated caspase-9 activation pathway. Both pathways converge in caspase-3, culminating to cell death. Subsequent accumulation of cytochrome c in the cytosol has been shown to be responsible for activation of downstream proteolytic caspase and DNA fragmenting enzymes responsible for apoptotic cell death, and subsequent release of cytochrome c to peripheral circulation [22,23].
In the present study, it was observed that relative mRNA expressions of apoptotic genes (Cyt C and Caspase 9) as well as antioxidative genes (Keap1 and Nrf2) were increased in Hep3B cells following INH treatment. Further it was confirmed by activity assay of Caspase 9 and its down stream effector Caspase 3 that apoptosis in Hep3B cells is occurring through intrinsic pathway and not through extrinsic pathway following INH treatment. In Hep3B cells, administration of 10 and 20 mM of INH induced activation of caspase 9 and caspase 3 and it increased from 10 to 20 mM of INH. When cell suspensions of Hep3B cells incubated with 20 mM of INH for twenty-four hours were treated with caspase inhibitor following administration of caspase substrate, activities of both caspases were observed to be decreased.
Further, whole genome expression analysis was performed in Hep3B cells treated with 20 mM of Isoniazid to identify differentially expressed genes and their involvement in different pathways by microarray study. High throughput expression profiling made it possible for identification of various genes whose expression has changed following INH treatment. Their differential expression indicates hindrance of various roles in diverse pathways. Hep3B cells treated with Isoniazid resulted in differential regulation of 103 genes of which 39 were up-regulated and 64 were down-regulated. Most of the genes were engaged in apoptosis, oxidative stress, cell proliferation, post-translational modification, cell signaling, cellular movement and developmental disorder. In addition, validation of few important differentially expressed genes in microarray findings using quantitative real time PCR [24] substantiates these results. As a result, identification of genes with similar expression changes, at such a high stringency indicates important biological functions under these circumstances.
Several genes were found up regulated. For example following five genes: C12orf5, GLS, GADD45B, DDIT4 and GLUL were involved in oxidative stress and DNA damage. Genes like BAK1, SH3KBP1, SNAI2, TNFRSF12A and MAGED1 were involved in programmed cell death and DHFR, CTGF, ASS1, ID1, NDRG1, PIM1, ARHGAP19, PFKL, THBS3 and ACCN2 were found to play role in the cell proliferation. Similarly seven genes i.e. UIMC1, BMP2, IRS4, NOG, EXOC4, IFITM1 and NRN1 were found to be engaged in cell signaling pathways. One of the identified genes was Chromosome 12 open reading frame 5 (C12orf5) that increases NADPH production to help limit reactive oxygen species (ROS) and lowers cell death [25]. In BCL2-antagonist/killer 1 (BAK1), SH3-domain kinase binding protein 1 (SH3KBP1), Snail homolog 2 (SNAI2), Tumor necrosis factor receptor superfamily (TNFRSF12A), member 12A and Melanoma antigen family D 1 (MAGED1) genes BAK1, SH3KBP1, SNAI2 and TNFRSF12A were up-regulated while MAGED1 was down-regulated following Isoniazid treatment. BAK is a fundamental outer mitochondrial membrane protein and a key cell death initiator [26]. It further confirms that INH induced apoptosis through intrinsic pathway.
Dihydrofolate reductase and Connective tissue growth factor (DHFR and CTGF) were up-regulated while Argininosuccinate synthetase1(ASS1), Inhibitor of DNA binding 1 (ID1), N-myc downstream regulated 1(NDRG1), Pim-1 oncogene (PIM1), Rho GTPase activating protein 19(ARHGAP19), Phosphofructokinase (PFKL) liver, Thrombospondin 3 (THBS3), and Amiloride-sensitive cation channel 2 neuronal (ACCN2) genes were down-regulated following Isoniazid treatment. All the above genes play an important role in cell proliferation. Out of the 10 genes having role in cell proliferation, 8 were down-regulated following Isoniazid treatment indicating that INH prevents cell proliferation.
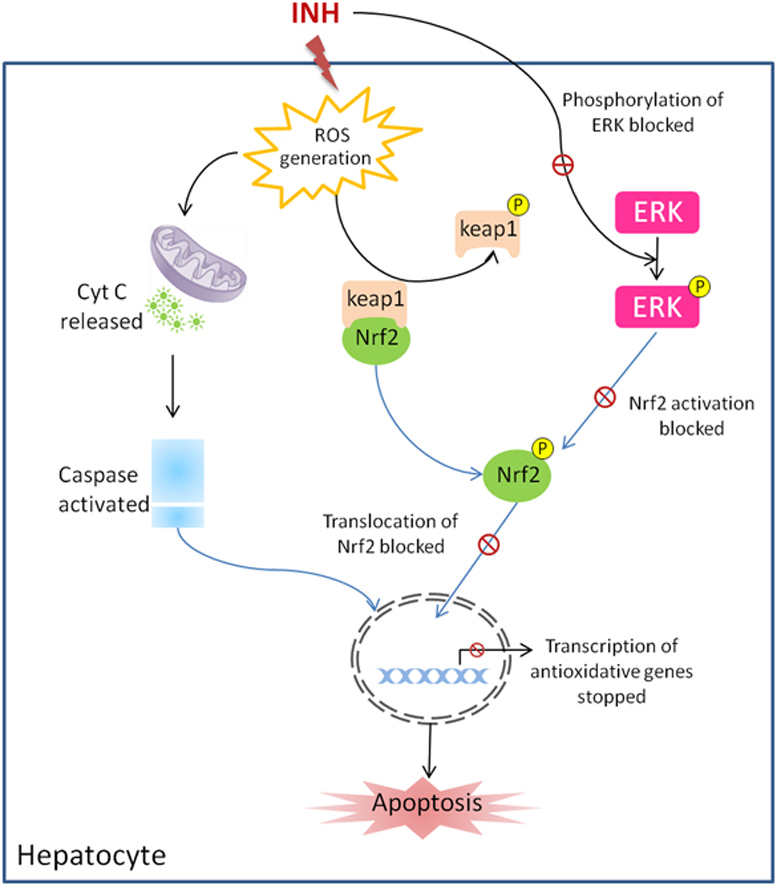
At the basal level, Nrf2 retained in the cytosol by binding its inhibitor Keap1 or INrf2 [(Kelch like ECH-associated protein 1 or inhibitor of Nrf2)] [27]. The xenobiotics and antioxidants lead to dissociation of Nrf2 from INrf2 resulting in stabilization and nuclear translocation of Nrf2. Nrf2 attaches to ARE and induces the expression of defensive genes.
The Hep3B cells treated with 5, 10 and 20 mM of INH showed an increased level of cytosolic Nrf2 protein level while a decreased level of nuclear Nrf2 protein level. ERK2/ERK1 (also known as p42/p44MAPK, in that order, and authoritatively named MAPK 1 and 3) are activated through a sequential phosphorylation cascade that amplifies and transduces signals from the cell membrane to the nucleus.
Phospho-ERK activates Nrf2 by phosphorylating it [10] and Nrf2 translocates into the nucleus by Karyopherin β1[28]. This study has shown that Isoniazid decreases ERK phosphorylation following INH treatment to Hep3B cells and interferes with the activation and translocation of Nrf2 into the nucleus. Isoniazid does not change ERK level, but decreases activation of existing ERK protein as evident from decreased amount of phosphorylated ERK1. As a result, possibly due to the inhibition of Nrf2 translocation, Nrf2 may not be able to bind ARE sequences of the anti-oxidative and cytoprotective genes and activate them.
In 2D gel electrophoresis results of Hep3B cells treated with INH, we have selected only those protein spots which were either up-regulated by >1.5 fold or down-regulated by <0.65 fold. It was observed that total 30 proteins were differentially expressed under the criteria selected by us. Out of these 30 differentially expressed proteins, 15 proteins were detected to be up-regulated whereas 15 proteins were detected to be down-regulated. Thirteen differentially expressed proteins were found to be engaged in oxidative stress and DNA damage response. These proteins were Chaperonin Hsp60, Stress induced phosphoprotein1, 60 kDa Heat shock protein, Creatine kinase B, Cofilin, Inorganic pyrophoaphatase, Chain A Horf6A Novel Human Peroxidase enzyme, Alpha globin, Beta globin, Rho protein GDP dissociation inhibitor, Endoplasmin precursor, Transformation upregulated nuclear protein and Chain A Tapasin Erp57. Creatine kinase B (CKB), a cytosolic isoform of creatine kinase, demonstrates up-regulated expression in a variety of cancers. CKB knockdown reduced glucose consumption and lactate production, and raised ROS production and oxygen consumption. This recommended that CKB knockdown reduced cytosolic glycolysis and resulted in a tumor suppressive metabolic state in Skov3 cells [29]. Cofilin1 (CFL-1) over-expression enhances radiosensitivity and is associated with reduced DNA repair capacity [30].
Five differentially expressed proteins observed from the 2D results in Hep3B cells treated with INH had role in apoptosis pathway. These proteins were BiP protein, Peroxiredoxin 4, Nucleophosmin/B23, EF1A1 and Eukaryotin translation initiator. The endoplasmic reticulum is the place where folding occurs for newly synthesized proteins that are destined for the cell surface. Additionally, it is the primary cellular storage site for calcium. Calcium homeostasis is needed for proper polypeptide folding and secretion of selective proteins as well as for intracellular signaling events that occur inside the cell. BiP/GRP78 is a lumenal stress protein of the endoplasmic reticulum (ER) that interacts with polypeptide folding intermediates transiting the secretory compartment [31]. Enzymes CK2, PKC, PLK1, CDKs, and PKA could phosphorylate B23 at higher number of sites. B23 phosphorylation is associated to cellular processes such as cell survival, apoptosis, cell proliferation, and reply to DNA damage stimulus [32]. eEF1A1 expression has clear effects on the proliferation enhancement and apoptosis inhibition of Jurkat cells, which may be intervened by the up-regulation of PI3K/Akt/NF-κB and PI3K/Akt/ mTOR signaling pathway [33].
Six differentially expressed proteins observed were found to have role in Cell proliferation and differentiation. These were Cyclophilin, Heterogenous nuclear ribonucleoprotein A2, Heterogenous nuclear ribonucleoprotein F, Heterogenous nuclear ribonucleoprotein H, Non-POU containing octamer binding protein and POTE Ankyrin domain family member E. CypB induced by hypoxia stimulates the survival of HCC via a positive feedback loop with HIF-1α, demonstrating that CypB is a novel candidate target for developing chemotherapeutic agents against HCC and colon cancer [34]. Three differentially expressed proteins (KRT73 protein, Alpha tubulin and Beta actin like protein) were found to have role in maintaining cytoskeleton of the cell while three differentially expressed proteins (Enolase, ATP synthase subunit beta and Triose phosphate isomerase) were seen to play role in ATP synthesis pathway and maintaining energy reservoir of the cell.
4. Conclusions
Isoniazid generated reactive oxygen species and induced apoptosis in Hep3B cells by increasing mRNA and protein level of various genes playing important roles in apoptosis, oxidative stress, cell signaling, cell proliferation and differentiation pathways. It also decreased the phosphorylation of ERK1 as a result of which it might not able to activate Nrf2 and its translocation into the nucleus and could not activate ARE to protect the Hep3B cells undergoing apoptosis following Isoniazid treatment.
5. Materials and method
5.1. Chemicals and biochemicals
All the chemicals were purchased from Sigma Aldrich. DMEM (Invitrogen), Whole genome expression kit (Ambion), cDNA synthesis kit (Applied Biosystem), SYBR Green (Roche), Annexin V FITC kit (Calbiochem), IPG strip, IPG buffer (GE Healthcare), Primary antibodies Caspase 9, Cyt C, Nrf2, ERK1, Phospho-ERK1, β-actin, Lamin B (all from Santacruz Biotechnology, USA), Keap1 (Thermo Scientific, USA) and Secondary antibodies (Santacruz Biotechnology, USA) were purchased. ERK1 inhibitor was purchased from Calbiochem.
5.2. Cell culture
Hep3B cells were obtained from our institutional cell repository. These cells were maintained at sub-confluence in 95% air and 5% CO2 in a humidified atmosphere at 37 °C. DMEM (Dulbecco's modified eagles medium) was used for cell cultivation. Cells were supplemented with 10% FBS, penicillin (100 units/ml) (both from GIBCO, Invitrogen). Hep3B cells were sub-cultured by discarding culture media and subsequently washing them with PBS, then adding Trypsin–EDTA (0.25% Trypsin and 0.53 mM EDTA) and incubating the cells for 2–5 min at 37 °C in CO2 incubator. Now cells were aspirated by gentle pipetting. Fresh growth media was added to it and appropriate aliquots of cell suspension were added to new culture vessel.
5.3. Cell viability assay
In order to calculate 50% growth inhibitory concentration (IC50) of Isoniazid, Hep3B cells were seeded and grown for 24 h. Now, cells were treated with different concentrations of Isoniazid. 24 h later media was removed and cells were incubated in MTT for 4 h. Then MTT was removed and DMSO was added to each well and cells were incubated for 45 min in dark. Then OD was taken at 570 nm on Elisa Reader (BIO-TEK).
5.4. Estimation of reactive oxygen species generation (ROS)
Intracellular ROS generated in Hep3B cells following INH treatment were analyzed using the fluorescent probe 2',7'-dichlorofluorescin diacetate(DCFDA), a non-fluorescent compound under normal condition, which is converted into highly fluorescent dichlorofluorescein (DCF) by ROS. Hep3B cells were seeded in T25 flask and grown for next 24 h. After 24 h, these cells were washed with 1× PBS pH 7.4 and treated with different concentrations of INH for 24 h. Then, after 24 h cells were scraped in the media. Now cells were taken in 15 ml centrifuge tube and centrifuged at 1200 RPM for 5 min at 4 °C. Supernatant was removed and cells were washed with 1× PBS pH 7.4. These cells were then re-suspended in 500 µL of PBS. Subsequently, 10 µL of 1 mM DCFDA was added to these cells for reactive oxygen species (ROS) estimation. Samples were incubated for 20–30 min at 37 °C and fluorescence was monitored on a fluorescence activated cell sorting (FACS) flow cytometer (Beckman Coulter, USA) with excitation wavelength at 488 nm and emission wavelength at 530 nm [13].
5.5. Apoptosis by Annexin V FITC
7×105 Hep3B cells were seeded in T25 flasks and grown for 24 h. Then these cells were treated with different concentrations of Isoniazid for 24 h. FITC Annexin V apoptosis detetion kit I (BD Bioscience) was used for flow cytometric analysis of apoptotic cells as per the manufacturer's protocol. Cells were harvested, collected in 15 ml falcon tube and centrifuged. Now cells were washed with PBS. Annexin V FITC and PI were added to 1x binding buffer as manufacturer's protocol. Cells were incubated with annexin V FITC for 15 min in dark at room temperature. After 15 min, PI was added to cell suspension and incubated for 10 min. Subsequently, apoptosis was analyzed in a Flow Cytometer (FACS Calibur, Becton Dickinson, USA). Following the same procedure apoptosis induction was measured in Hep3B cells treated with ERK inhibitor followed by 10 and 20 mM of Isoniazid.
5.6. cDNA synthesis and qRT-PCR analysis
Hep3B cells were seeded and treated with 5, 20 and 40 mM of Isoniazid. Total RNA was isolated with Trizol (Invitrogen) manually. 5 µg of total RNA was used for cDNA synthesis which was performed using cDNA synthesis kit (High Capacity cDNA Reverse Transcription Kit, Applied Biosystems, USA) according to manufacturer's protocol. Program for cDNA synthesis was incubation steps at 25 °C for 10 min, 37 °C for 120 min, 85 °C 5 min and 4 °C forever. Expression of CytC, Caspase 9, Nrf2, Keap1 and PKCδ genes was examined using qRT-PCR on LightCycler480 PCR system (Roche Diagnostics, Germany). The reaction mixture contained 50 ng of analyzed cDNA. The amplification of each sample was performed in triplicate in three independent experiments using a Light Cycler 480 SYBR Green I master (Roche Diagnostics, Germany) after which Cp values were averaged. For each primer pair, a melting curve analysis was performed according to instrument. The program in brief was an initial incubation of 50 °C for 2 min hold (UDG incubation) and 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s, 58 °C for 30 s and final extension at 72 °C for 20 s. Differential expression was calculated by 2−ΔΔCT method. β-actin was used as internal control and used to normalize ratios between samples [14]. Primer sequences for this study are given in Table 1.
Table 1.
Primer sequences used for real time PCR for Hep3B cell lines.
| S.No. | Primers | Sequence |
|---|---|---|
| 1 | Actin FP | 5′-ACTACCTCATGAAGATCCTC-3′ |
| Actin RP | 5′-CTAGAAGCATTTGCGGTCGACGATGG-3′ | |
| 2 | CytC FP | 5′-GGACCTCTTTACCTTCTCCCC-3′ |
| CytC RP | 5′-GTTGGGACATACATCAGCAGC-3′ | |
| 3 | Caspase9 FP | 5′-GCGACCTGACTGCCAAGAAA-3′ |
| Caspase9 FP | 5′-TCACAATCTTCTCGACCGACA-3′ | |
| 4 | Nrf2 FP | 5′-TCAGCGACGGAAAGAGTATGA-3′ |
| Nrf2 RP | 5′-CCACTGGTTTCTGACTGGATG-3′ | |
| 5 | Keap1 FP | 5′-CTGGAGGATCATACCAAGCAG-3′ |
| Keap1 RP | 5′-GAACATGGCCTTGAAGACAGG-3′ | |
| 6 | BAK1 FP | 5′-GTTTTCCGCAGCTACGTTTTT-3′ |
| BAK1 RP | 5′-GCAGAGGTAAGGTGACCATCTC-3′ | |
| 7 | SH3KBP1 FP | 5′-CATCGACGTAGGCTGGTGG-3′ |
| SH3KBP1 RP | 5′-CCTTCCTTTTCAAAGTCCGGTG-3′ | |
| 8 | GADD45B FP | 5′-TGCTGTGACAACGACATCAAC-3′ |
| GADD45B RP | 5′-GTGAGGGTTCGTGACCAGG-3′ | |
| 9 | SNAI2 FP | 5′-TGTGACAAGGAATATGTGAGCC-3′ |
| SNAI2 RP | 5′-TGAGCCCTCAGATTTGACCTG-3′ | |
| 10 | NDRG1 FP | 5′-CTCCTGCAAGAGTTTGATGTCC-3′ |
| NDRG1 RP | 5′-TCATGCCGATGTCATGGTAGG-3′ | |
| 11 | PIM1 FP | 5′-GAGAAGGACCGGATTTCCGAC-3′ |
| PIM1 RP | 5′-CAGTCCAGGAGCCTAATGACG-3′ | |
| 12 | UCP2 FP | 5′-GGAGGTGGTCGGAGATACCAA-3′ |
| UCP2 RP | 5′-ACAATGGCATTACGAGCAACAT-3′ | |
| 13. | THBS3 FP | 5′-ATGGAGACGCAGGAACTTCG-3′ |
| THBS3 RP | 5′-AGCTACCATCTGCCGAGACT-3′ |
5.7. Caspase activity assay
Hep3B cells were seeded and grown for 24 h. After that cells were treated with 10 and 20 mM of Isoniazid for next 24 h. Then Caspase 9 and Caspase 3 activity were measured using Caspase 3 and Caspase 9 activity assay kit (Calbiochem, USA) according to manufacturer's protocol. Caspase activities were measured by detection with a fluorescence spectrophotometer (Varian, Cary Eclipse) and the fluorescence was measured at an excitation/emission wavelength of 400/505 nm.
5.8. Whole gene expression studies
Whole genome expression studies were performed by Microarray analysis. Total RNA from untreated and INH treated (20 mM) Hep3B cells was isolated manually by TriZol (Invitrogen). RNA quality and quantity were checked by formaldehyde gel electrophoresis and spectrophotometer respectively. RNA samples with approximately 2:1 ratio of 28 S: 18 S rRNA and 260/280 values ≥1.8 were used for gene expression analysis. First strand was synthesized from total RNA and then second strand was synthesized. Then cRNA was synthesized by in vitro transcription and purified. Further sense strand cDNA was synthesized from cRNA and purified. Now cDNA was fragmented, labeled and loaded on Human Gene 1.0 ST Array Chip and sealed with white tough tag. For hybridization, Chip was transferred to the Hybridization Chamber (Affymetrix Gene Chip Hybridization Oven 645) and incubated for 16 h at 45 °C with 60 rpm speed. After hybridization, Human Gene 1.0 ST Array Chip was washed in wash chamber (Affymetrix Gene Chip Fluidisc Station 450) to remove the unbounded fragmented cDNA. After washing, array was scanned (Affymetrix Gene Chip Scanner) and Microarray Data was analyzed by Gene Spring software and pathway analysis was performed by Ingunity Pathway Analysis (IPA) software from Qiagen. Eight differentially regulated genes were validated by quantitative real time PCR.
5.9. Western blot analysis
Cytosolic and nuclear proteins were isolated from Hep3B cells and liver tissue by ProteoJET cytosolic and nuclear protein extraction kit (Fermentas); according to manufacturer's protocol proteins were quantified using Bradford reagent. 50 µg protein from each sample was separated on 15% SDS-PAGE and transferred on to a nitrocellulose membrane using a semi-dry electroblotting apparatus (GE Healthcare, UK). Transfer was examined by Ponceau S stain and washed with TDW until the stain disappeared. Membrane was blocked overnight in 5% Non-Fat dried milk at 4 °C. Blocking membrane was washed with 0.1% PBST and probed with 1° antibodies. After 1° antibody incubation, further washing was done in 0.1% PBST. Membrane was incubated in HRP conjugated 2° antibody and washed again. Enhanced chemi-luminescent detection reagent was used to develop the blots. Blots were further used for densitometric analysis (Image J) and normalization. Experiments were carried out in triplicates and values were averaged.
5.10. 2D gel electrophoresis
Hep3B cells were incubated with INH for 24 h. Now whole cell lysates were prepared (7 M Urea, 2 M Thiourea ) and total protein concentration was estimated by Bradford assay. Six gels (three control and three treatment gels) were run independently in a pH range of 3–10 (13 cm long IPG strip). IEF was performed with 400 µg of protein samples on immobilized pH gradient IPG strips (GE Healthcare) in IEF machine. Proteins were separated in the second dimension by 12% SDS-PAGE, and protein spots were visualized by coomassie brilliant blue (CBB) staining. All the gels were scanned with an Image scanner (GE Healthcare) and quantitatively analyzed using Image Master 2D Platinum 7.0 software (GE Healthcare). For each protein spot, from both control and treatment gels, fold volume change of each spot was calculated by dividing % volume of treatment spot with control spot. We have selected the spots whose fold volume change was >1.5 were up-regulated, whereas <0.65 were down-regulated. Gel to gel and staining techniques variations were overcome by comparing each protein spot to a number of other protein spots in the same gel whose expression did not change under given experimental conditions.
5.11. Protein identification by MALDI-TOF
The differentially appearing protein spots were carefully excised with a cut pipette tip, collected in eppendorf tubes, and destained by washing in 50 mM ammonium bicarbonate–ACN solution followed by two alternative washing steps with 50% ACN and 50 mM ammonium bicarbonate (ABC). The gel pieces were dehydrated at room temperature with 100% ACN and covered with 10 µl of trypsin (working solution: ng/µL prepared from 100 ng/µL stock solution in 50 mM ammonium bicarbonate) overnight at 37 °C. The spots were grinded and the peptides were extracted in 10 µL of 70% ACN. The eluate was dried and stored at −80 °C. The peptides were resuspended in 5 µl of 20% acetonitrile (ACN) and 0.1% trifluoro acetic acid (TFA) and sonicated for 3 min before processing for MS. Thirty measurements were performed using MALDI-TOF/TOF tandem mass spectrometer (AB4800) and protein pilot software (Applied Biosystems, Foster City, CA, USA). After acquisition of spectra, peptide masses for each protein spot were analyzed by 4000 Series Explorer Software Version 3.5.3 (AB SCIEX) and a peak list of mono-isotopic masses was generated prior to MS/MS data search. The tandem mass spectra were stored in a combined Mascot Generic Format (MGF) file using “Peak to Mascot” tool, searched against NCBI-nr database (June 2014) through Mascot search engine version 2.4.01 (http://www.matrixscience.com). Search was performed on the following parameters; taxonomy, all; single missed cleavage; precursor mass tolerance 20 ppm; fragment ion tolerance 0.05 Da; carbamido methylation of cysteines as fixed and oxidation of methionine as variable modification. Decoy parameter was checked to minimize the false discovery rate. ~70% protein spots were identified.
5.12. Statistical analysis
GraphPad Prism 5 was used for analyzing mice serum parameters and quantitative real time PCR data. Image J was used for densitometric calculation of Western blotting bands. All the experiments were performed in triplicate independently. ANOVA method was used for analyzing 2D gel spots. All experiments were performed in triplicate independently and each image is representative of one of those images.
Conflict of interests
There are no competing interests between authors.
Authors' contributions
Ajeet Kumar Verma: Experiment planning, and execution, conception and design, acquisition of data, analysis and interpretation of data, drafting and revising manuscript critically.
Arti Yadav: Experiment planning, and execution, analysis and interpretation of data, drafting and revising manuscript critically.
Jayant Dewangan: Experiment planning, and execution, conception and design.
Sarvendra Vikram Singh: Conception and design, acquisition of data.
Manisha Mishra: Acquisition of data.
Pradhyumna Kumar Singh: Analysis and interpretation of data.
Srikanta Kumar Rath: Conception and design, acquisition of data, drafting and revising manuscript critically, and final approval of the version to be published.
Acknowledgments
This study was supported by Council of Scientific and Industrial Research (CSIR) as CSIR-JRF, CSIR-SRF and CSIR network project BSC0103.
Authors want to acknowledge Mr. A.L. Vishwakarma for support in flow cytometric studies. CSIR-CDRI communication number for this article is 9027.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2015.06.020
Contributor Information
Ajeet Kumar Verma, Email: ajeetsonicdri@gmail.com.
Arti Yadav, Email: artiyadav.srm@gmail.com.
Jayant Dewangan, Email: jayantdewangan@gmail.com.
Sarvendra Vikram Singh, Email: sarvendravs@gmail.com.
Manisha Mishra, Email: manisha.nbri@gmail.com.
Pradhyumna Kumar Singh, Email: pradhyumnasingh@hotmail.com.
Srikanta Kumar Rath, Email: skrath@cdri.res.in.
Supplementary material
Supplementary material
Refrences
- 1.Itoh K., Ishii T. Regulatory mechanisms of cellular response to oxidative stress. Free Radic. Res. 1999;31(4):319–324. doi: 10.1080/10715769900300881. [DOI] [PubMed] [Google Scholar]
- 2.Kong A.N., Owuor E. Induction of xenobiotic enzymes by the MAP kinase pathway and the antioxidant or electrophile response element (ARE/EpRE) Drug Metab. Rev. 2001;33(3–4):255–271. doi: 10.1081/dmr-120000652. [DOI] [PubMed] [Google Scholar]
- 3.Owuor E.D., Kong A.N. Antioxidants and oxidants regulated signal transduction pathways. Biochem. Pharmacol. 2002;64(5–6):765–770. doi: 10.1016/s0006-2952(02)01137-1. [DOI] [PubMed] [Google Scholar]
- 4.Hsieh T.C., Lu X. Induction of quinone reductase NQO1 by resveratrol in human K562 cells involves the antioxidant response element ARE and is accompanied by nuclear translocation of transcription factor Nrf2. Med. Chem. 2006;2(3):275–285. doi: 10.2174/157340606776930709. [DOI] [PubMed] [Google Scholar]
- 5.Galloway D.C., Blake D.G. Regulation of human gamma-glutamylcysteine synthetase: co-ordinate induction of the catalytic and regulatory subunits in HepG2 cells. Biochem. J. 1997;328:99–104. doi: 10.1042/bj3280099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nioi P., McMahon M. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem. J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wasserman W.W., Fahl W.E. Functional antioxidant responsive elements. Proc. Natl. Acad. Sci. USA. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cederbaum A. Nrf2 and antioxidant defense against CYP2E1 toxicity. Expert Opin. Drug Metab. Toxicol. 2009;5:1223–1244. doi: 10.1517/17425250903143769. [DOI] [PubMed] [Google Scholar]
- 9.Liu J., Wu Q. New insights into generalized hepatoprotective effects of oleanolic acid: key roles of metallothionein and Nrf2 induction. Biochem. Pharmacol. 2008;76:922–928. doi: 10.1016/j.bcp.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 10.Tan Y., Ichikawa T. Diabetic downregulation of Nrf2 activity via ERK contributes to oxidative stress-induced insulin resistance in cardiac cells in vitro and in vivo. Diabetes. 2011;60:625–633. doi: 10.2337/db10-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonni A., Brunet A. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 12.Huang C., Jacobson K. MAP kinases and cell migration. J. Cell Sci. 2004;117:4619–4628. doi: 10.1242/jcs.01481. [DOI] [PubMed] [Google Scholar]
- 13.LeBel C.P., Ischiropoulos H. Evaluation of the probe 2′,7′-dichlorofluorescein as an indicator of reactive oxygen species formation and oxidative stress. Chem. Res. Toxicol. 1992;5:227–231. doi: 10.1021/tx00026a012. [DOI] [PubMed] [Google Scholar]
- 14.Livak K.J., Schmitten T.D. Analysis of relative geneexpression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Holt M.P., Ju. C. Mechanisms of drug-induced liver injury. AAPS J. 2006;8:E48–E54. doi: 10.1208/aapsj080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bjornsson E. Drug-induced liver injury: Hy's rule recisited. Clin. Pharmacol. Ther. 2006;79:521–528. doi: 10.1016/j.clpt.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 17.Chowdhury A., Santra A. Induction of oxidative stress in antitubercular drug-induced hepatotoxicity. Indian J. Gastroenterol. 2001;20:97–100. [PubMed] [Google Scholar]
- 18.Bhadauria S., Mishra R. Isoniazid-induced apoptosis in HepG2 cells: generation of oxidative stress and Bcl-2 down-regulation. Toxicol. Mech. Methods. 2010;20(5):242–251. doi: 10.3109/15376511003793325. [DOI] [PubMed] [Google Scholar]
- 19.Sahbazian B., Weis S.E. Treatment of active tuberculosis: challenges and prospects. Clin. Chest Med. 2005;26:273–282. doi: 10.1016/j.ccm.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 20.Schaberg T., Rebhan K. Risk factors for side-effects of isoniazid, rifampin and PZA in patients hospitalized for pulmonary tuberculosis. Eur. Respir. J. 1996;9:2026–2030. doi: 10.1183/09031936.96.09102026. [DOI] [PubMed] [Google Scholar]
- 21.McDonnell M.A., Wang D. Caspase-9 is activated in a cytochrome c-independent manner early during TNF-induced apoptosis in murine cells. Cell Death Differ. 2003;10:1005–1015. doi: 10.1038/sj.cdd.4401271. [DOI] [PubMed] [Google Scholar]
- 22.Green D., Reed J.C. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi Y., Mori M. Dynamic movement of cytochrome c from mitochondria into cytosol and peripheral circulation in massive hepatic cell injury. Pediatr. Int. 2004;46:685–692. doi: 10.1111/j.1442-200x.2004.01980.x. [DOI] [PubMed] [Google Scholar]
- 24.Chuaqui R.F., Bonner R.F. Post-analysis follow-up and validation of microarray experiments. Nat. Genet. 2002;32:509–514. doi: 10.1038/ng1034. [DOI] [PubMed] [Google Scholar]
- 25.Cheung E.C., Ludwig R.L. Mitochondrial localization of TIGAR under hypoxia stimulates HK2 and lowers ROS and cell death. Proc. Natl. Acad. Sci. USA. 2012;109(50):20491–20496. doi: 10.1073/pnas.1206530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams J., Cory. S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:324–337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhakshinamoorthy S., Jaiswal A.K. Functional characterization and role of INrf2 in antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene. 2001;20(29):3906–3917. doi: 10.1038/sj.onc.1204506. [DOI] [PubMed] [Google Scholar]
- 28.Theodore M., Kawai Y. Multiple nuclear localization signals function in the nuclear import of the transcription factor Nrf2. J. Biol. Chem. 2008;283(14):8984–8994. doi: 10.1074/jbc.M709040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X.H., Chen X.J. Knockdown of creatine kinase B inhibits ovarian cancer progression by decreasing glycolysis. Int. J. Biochem. Cell Biol. 2013;45(5):979–986. doi: 10.1016/j.biocel.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Leu J.D., Chiu Y.W. Enhanced cellular radiosensitivity induced by cofilin-1 over-expression is associated with reduced DNA repair capacity. Int. J. Radiat. Biol. 2013;89(6):433–444. doi: 10.3109/09553002.2013.767992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris J.A., Dorner A.J. Immunoglobulin binding protein (BiP) function is required to protect cells from endoplasmic reticulum stress but is not required for the secretion of selective proteins. J. Biol. Chem. 1997;272(7):4327–4334. doi: 10.1074/jbc.272.7.4327. [DOI] [PubMed] [Google Scholar]
- 32.Ramos-Echazábal G., Chinea G. In silico studies of potential phosphoresidues in the human nucleophosmin/B23: its kinases and related biological processes. J. Cell Biochem. 2012;113(7):2364–2374. doi: 10.1002/jcb.24108. [DOI] [PubMed] [Google Scholar]
- 33.Huang Y., Hu J.D. Effects of eEF1A1 re-expression on proliferation and apoptosis of Jurkat cells with knocked down eEF1A1 gene and its mechanisms. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2013;21(2):279–284. doi: 10.7534/j.issn.1009-2137.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Kim Y., Jang M. Role of cyclophilin B in tumorigenesis and cisplatin resistance in hepatocellular carcinoma in humans. Hepatology. 2011;54(5):1661–1678. doi: 10.1002/hep.24539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material