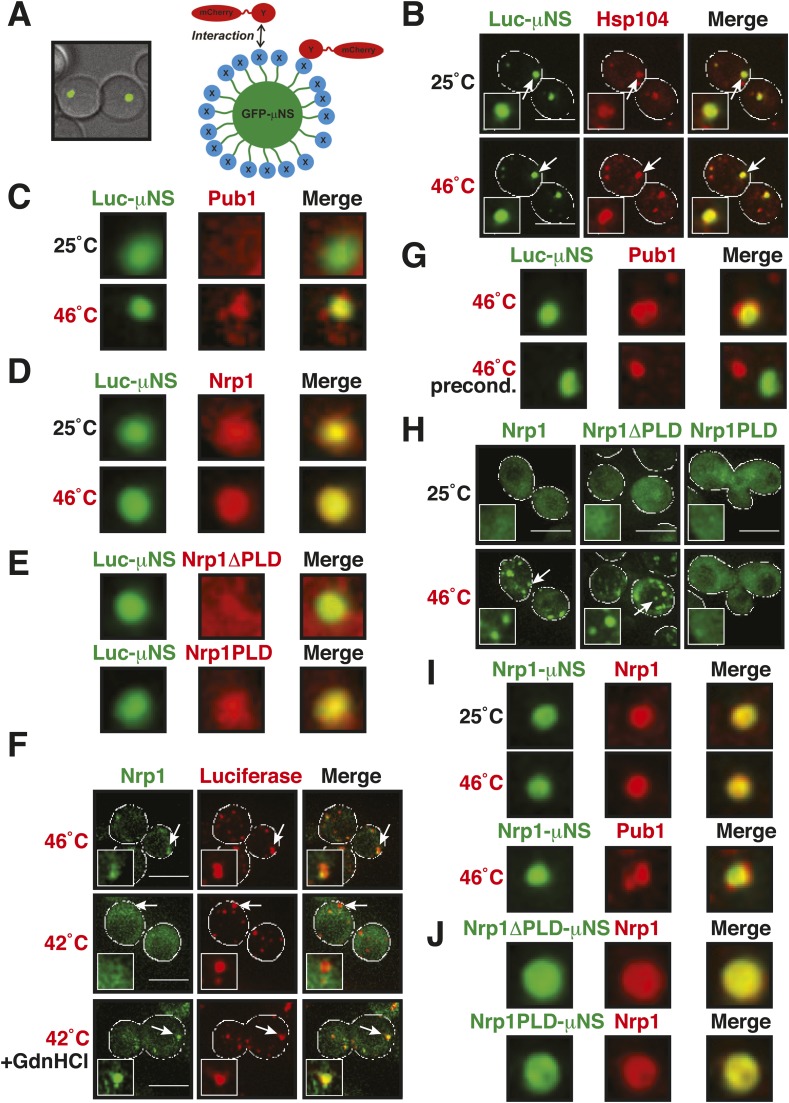
Figure 6. Stress granule assembly is redundant and highly adaptable.
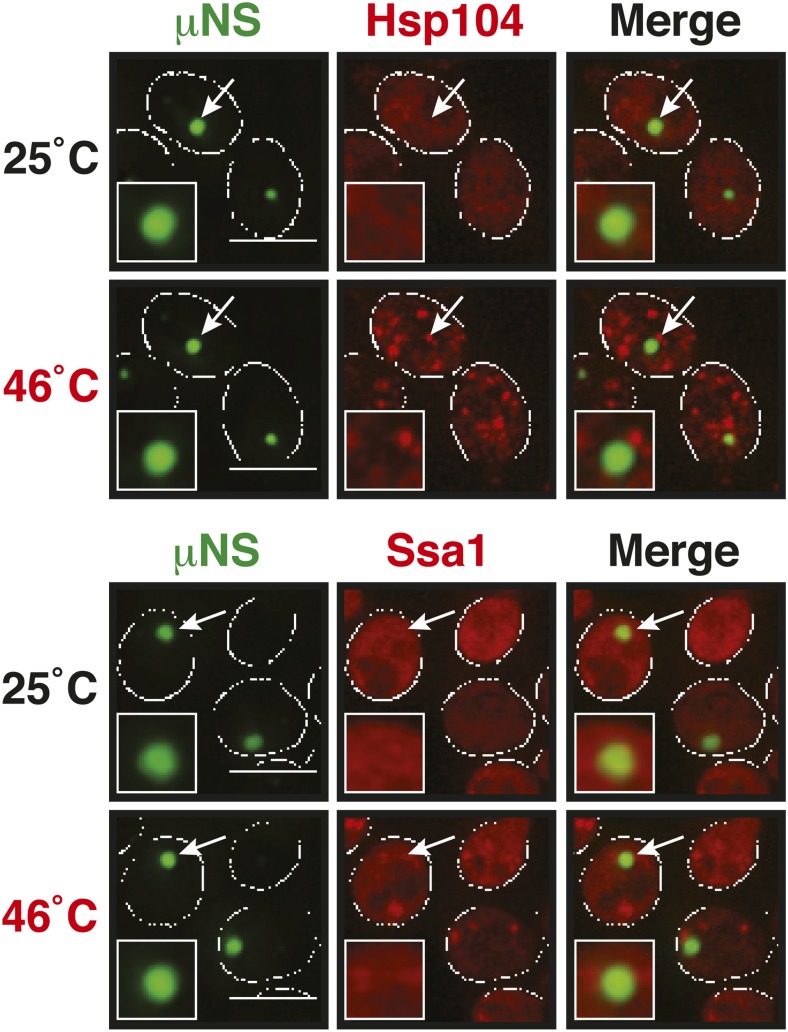
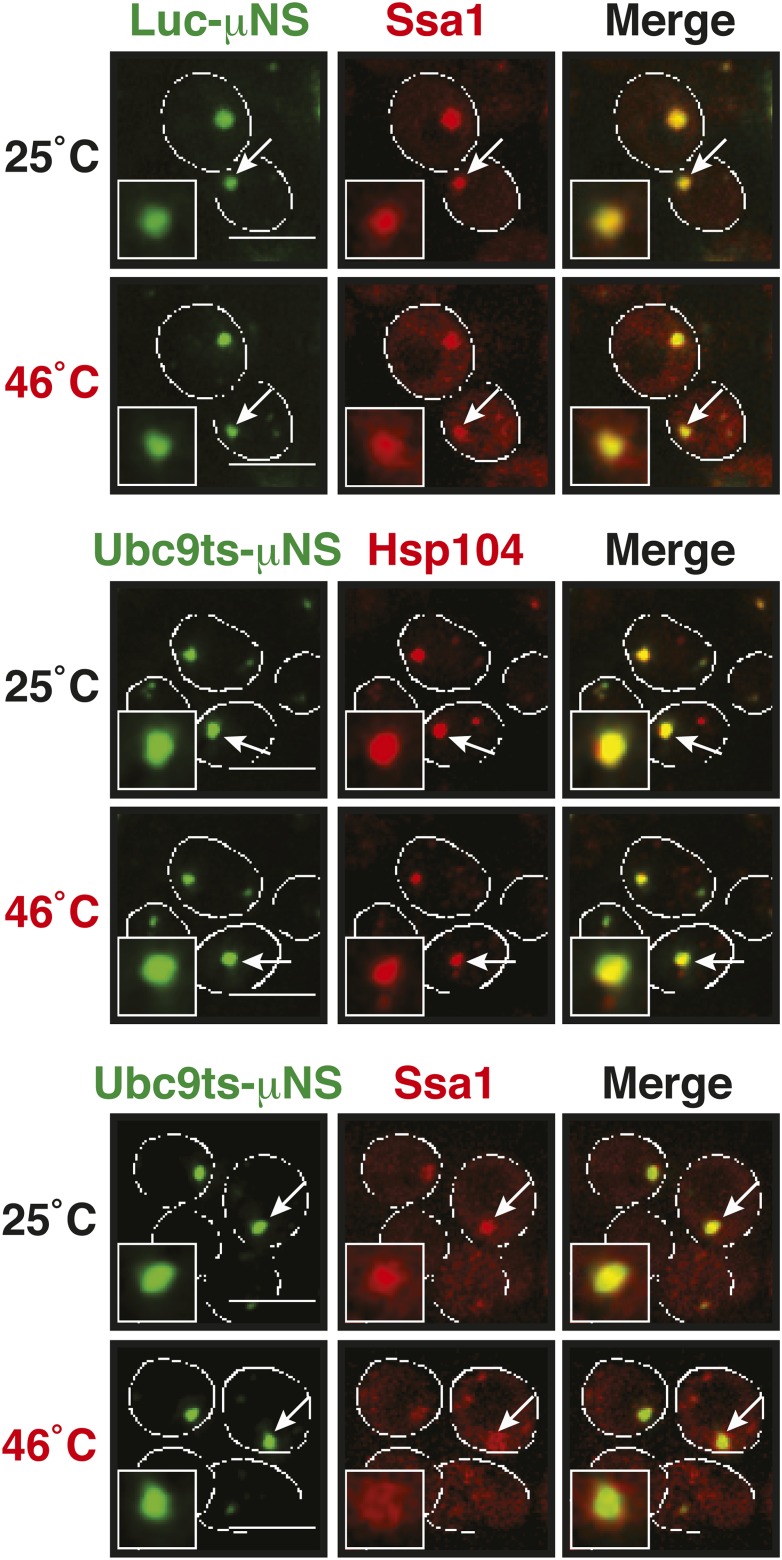
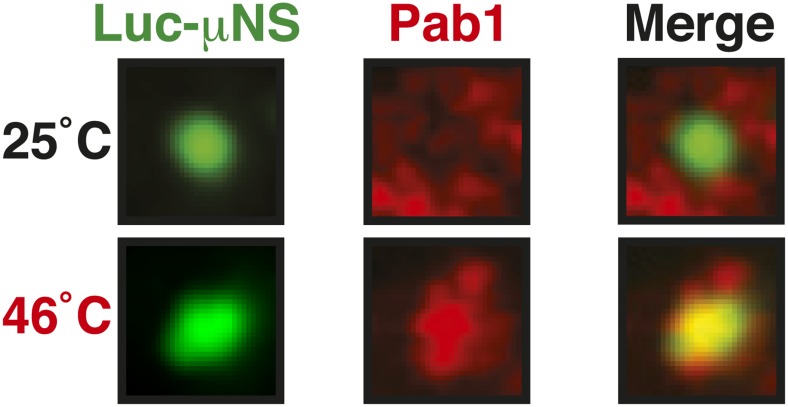
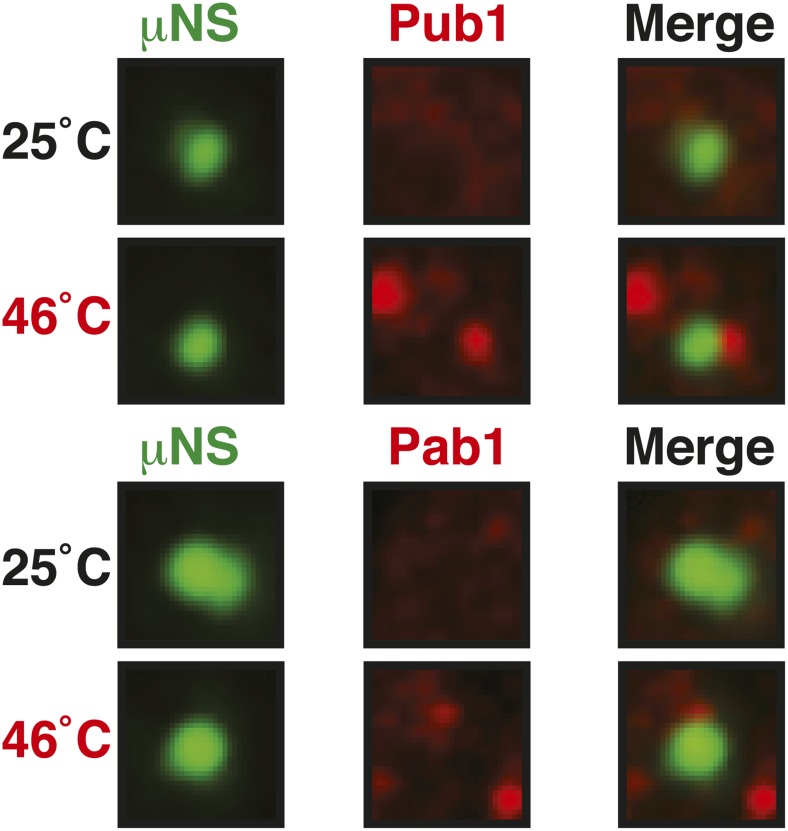
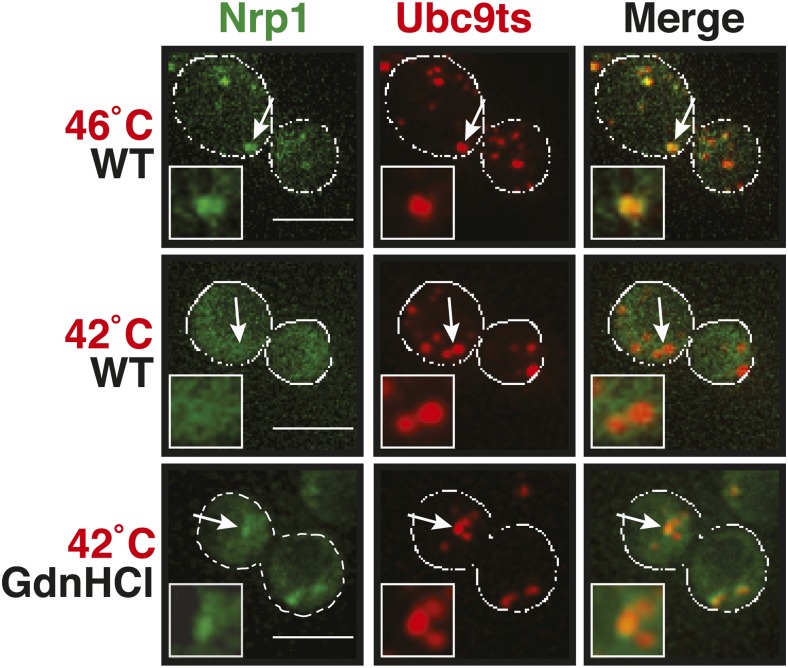
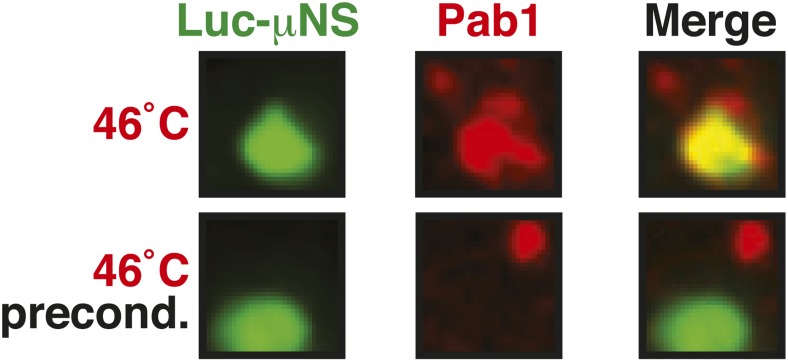
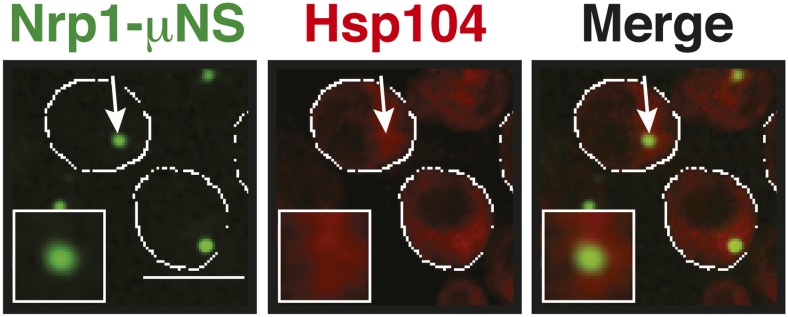
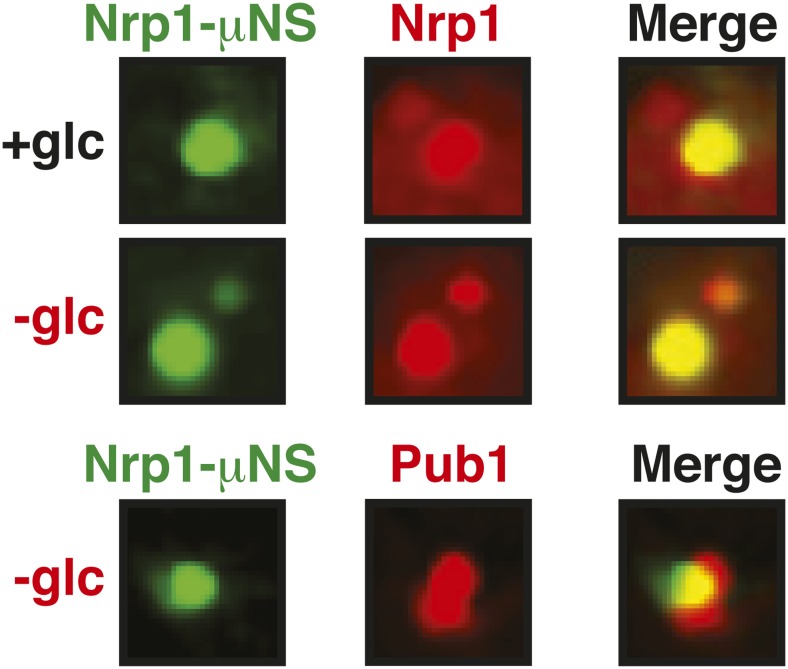
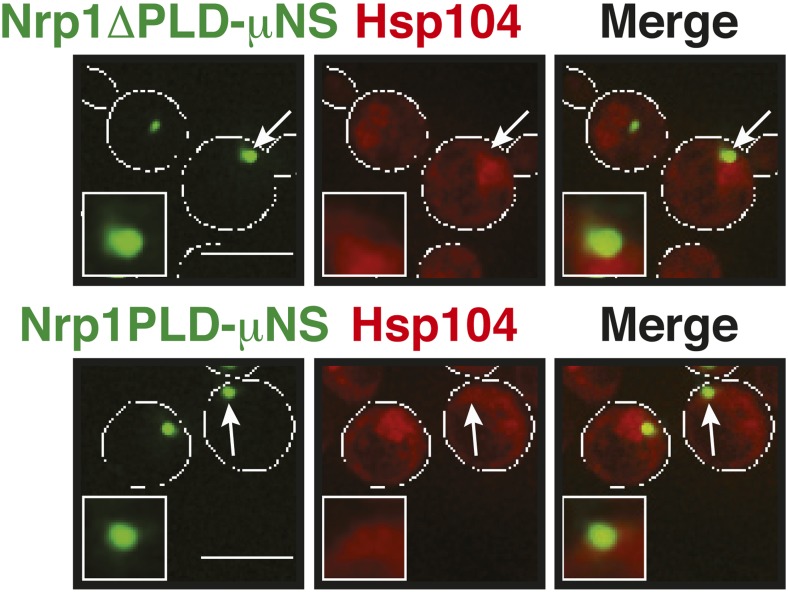
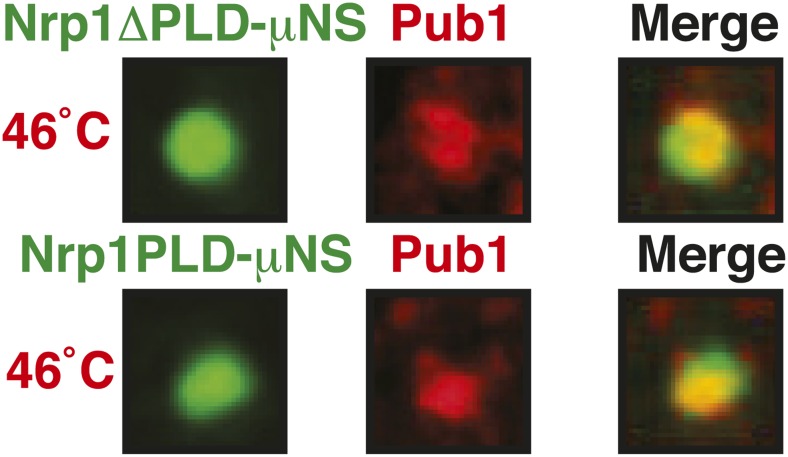
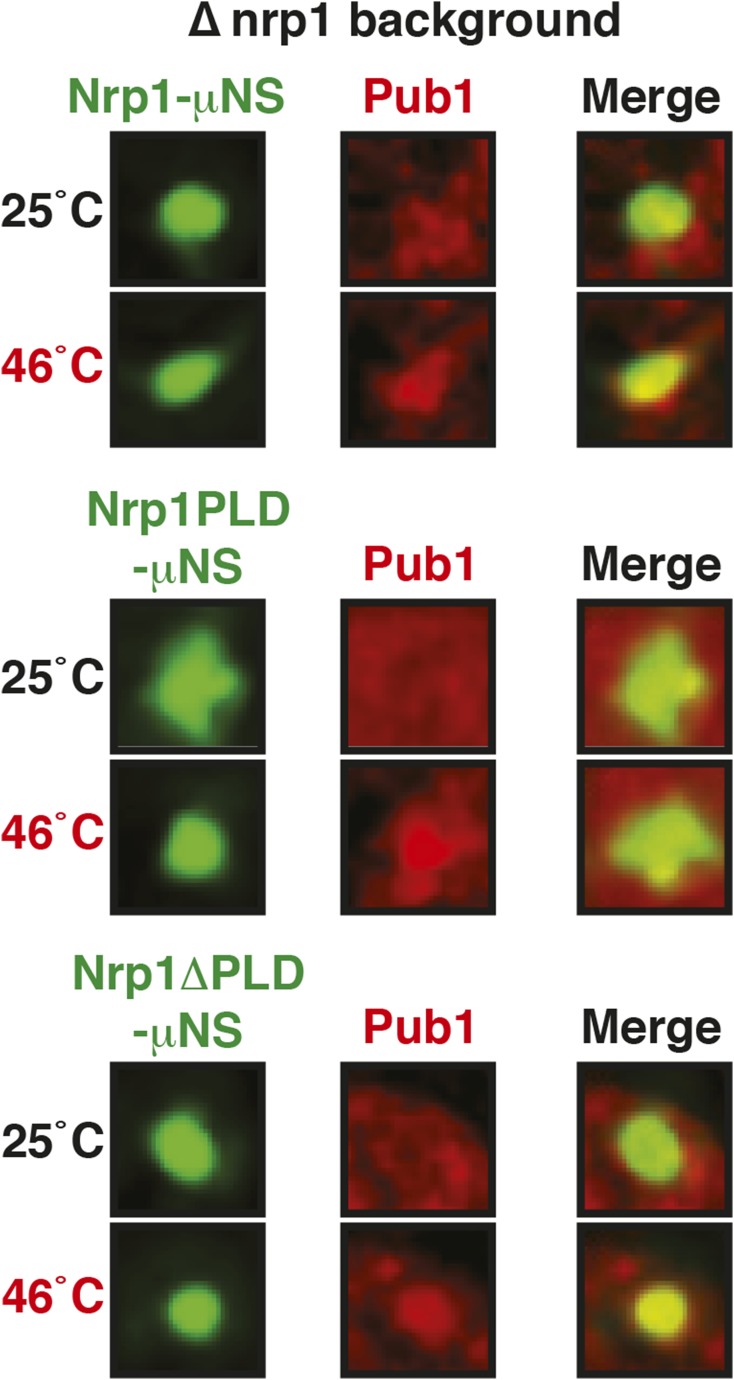
(A) Using a genetically encoded particle to study dynamic interactions in living yeast cells. Left: a fragment of the viral capsid protein μNS comprising the 250 C-terminal amino acids forms self-assembling particles in yeast cells. The particles were visualized using an N-terminal sfGFP tag. Right: interaction assay. Fusion of a protein X to sfGFP-tagged μNS particles allows interaction studies with a mCherry-tagged protein Y. (B) μNS particles carrying mutant luciferase on the surface interact with endogenous mCherry-tagged Hsp104. Cells were observed before and after a 10-min heat shock at 46°C. White lines indicate the cell boundaries. Scale bars: 5 µm. Also see related Figure 6—figure supplements 1, 2. (C) Same as (B), except that cells were used in which mCherry-tagged Pub1 was expressed from the endogenous locus. Only one representative μNS particle is shown at high magnification. Note that Pub1 only interacts with luciferase in cells exposed to robust heat stress. Also see related Figure 6—figure supplements 3–5. (D) Same as (C), except that mCherry-tagged Nrp1 was mildly overexpressed from a plasmid carrying an ADH1 promoter. Note that Nrp1 interacts with luciferase already in unstressed cells, and that the amount of Nrp1 accumulating on the particle is strongly increased upon heat stress. (E) Same as (D), except that the prion-like domain (PLD) of Nrp1 (Nrp1PLD) or a deletion mutant lacking the PLD (Nrp1ΔPLD) was observed at 25˚C. (F) The cellular chaperone machinery prevents interactions between misfolded proteins and stress granule components. Cells expressing Nrp1-GFP from the endogenous locus and mCherry-tagged mutated luciferase from a plasmid were exposed to a 10-min heat shock at 42°C or 46°C. The cells in the bottom panel were exposed to 3 mM guanidinium hydrochloride (GdnHCl) to inhibit Hsp104. Also see related Figure 6—figure supplement 6. (G) Same as (C), except that the temperature was increased slowly from 25°C to 46°C (preconditioning). Note that preconditioning prevents co-assembly of stress granules and misfolded proteins. Also see related Figure 6—figure supplement 7. (H) PLDs mediate interactions only when present in high local concentrations. Yeast cells were transformed with plasmids for the expression of GFP-tagged wild-type Nrp1 or deletion mutants lacking the RNA-binding domain (RBD) (Nrp1PLD) or PLD domain (Nrp1ΔPLD). The resulting cells were exposed to heat shock. (I) Upon heat shock, stress granules form on µNS particles that present Nrp1 on the surface. Same conditions as (C) and (D). Also see related Figure 6—figure supplements 8–9. (J) Same as (I), except that mutants lacking the RBD (Nrp1PLD) or PLD domain (Nrp1ΔPLD) were presented on the particle. Note that both mutants are able to recruit full-length Nrp1 at 25˚C. Also see related Figure 6—figure supplement 10–12.