Abstract
The aim of the present study was to investigate the molecular mechanism of carbapenem resistance of three imipenem-resistant isolates of Myroides odoratimimus recovered from two livestock farms of cows and pigeons by rectal swab in Lebanon in January 2014. Investigation of imipenem resistance of these isolates using the modified Hodge test, the EDTA test, the modified CarbaNP test and the matrix-assisted laser desorption/ionization time-of-flight mass spectrometry Ultraflex assay showed a carbapenemase activity due to the presence of a chromosome-encoded β-lactamase MUS, verified by PCR. However amplification and sequencing of this chromosomal gene showed a novel variant of it designated MUS-2 by the curators of the Lahey database of β-lactamases (http://www.lahey.org/Studies/webt.asp). Cloning of the blaMUS-2 was performed, followed by protein expression in Escherichia coli TOP 10. Pulsed-field gel electrophoresis clearly showed that the three isolates belonged to the same clone. This study reports a novel variant of the chromosome-encoded blaMUS-1 associated with carbapenem resistance in Myroides odoratimimus and shows that animals may represent a reservoir of bacteria harbouring several variants of resistance genes.
Keywords: Animals, blaMUS-2, carbapenem-resistance, metallo-β-lactamase, Myroides odoratimimus
Introduction
Myroides species, previously named as Flavobacterium odoratum, are aerobic Gram-negative bacteria that can be found in environmental sources such as soil and water, but are not residents of the normal human microflora [1]. The genus Myroides comprises two species, Myroides odoratus and Myroides odoratimimus[2]. Taking into consideration their role as causative agents in human disease, both M. odoratimimus and M. odoratus are seen rarely as opportunistic pathogens, causing infections in severely immunocompromised patients and rarely, in immunocompetent hosts [1,3]. Myroides odoratimimus strains have been found to be responsible for urinary tract infections [4,5], cellulitis [6,7] in immunocompromised patients, and also for septic shock, pneumonia and soft-tissue infections [1,8] in immunocompetent hosts. In addition, three outbreaks of M. odoratimimus, two of urinary tract infections and the third of central venous catheter-associated bloodstream infections due to contaminated water, have been reported [2,4].
Antibiotic resistance patterns of Myroides strains show variable susceptibility to β-lactams [5], with a conserved decreased susceptibility to cephalosporins and carbapenems [9]. This decreased susceptibility is the result of the production of a chromosome-encoded β-lactamase MUS-1. This enzyme is a member of the subclass B1 of metallo-β-lactamases and is distantly related to other metalloenzymes, being most closely related to IND-1 from Chryseobacterium indologenes (42% amino acid identity). However, phylogenic analysis showed that MUS-1 belongs to the same phylogenic lineage of subclass B1 enzymes that groups the subclass B1 β-lactamases of Flavobacterium species [9]. Here we report a novel variant, MUS-2, of the chromosome-encoded β-lactamase MUS-1 from M. odoratimimus isolated from livestock animals in Lebanon.
Materials and methods
Bacterial isolates
Three imipenem-resistant M. odoratimimus strains (32, 35a and 104b) were isolated from two livestock farms of cows and pigeons by rectal swabs (32: farm 1, cow; 35a: farm 1, pigeon; 104b: farm 2, cow) in January 2014. These strains were collected during a study that aimed to detect carbapenemase-encoding genes in animals in Lebanon [10]. They were isolated on MacConkey agar supplemented by ertapenem (1 mg/L), from alive and non-sick animals without contact with any other animals. Strains were sub-cultured on trypticase soy agar plates at 37°C for 18–24 h and identification was confirmed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) (Microflex™; Bruker Daltonics, Bremen, Germany) with FlexControl software (Bruker Daltonics).
Antibiotic susceptibility testing
Antibiotic susceptibility testing was determined on Müller–Hinton agar by standard disc diffusion method as recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST; www.eucast.org). Seventeen antibiotics were tested, including ticarcillin, ticarcillin-clavulanic acid, piperacillin, piperacillin-tazobactam, ceftazidime, cefotaxime, cefepime, aztreonam, amikacin, tobramycin, gentamicin, ciprofloxacin, rifampicin, ertapenem, meropenem, imipenem and colistin (Bio-Rad, Marnes-la-Coquette, France). The MIC for imipenem was determined using the Etest method (bioMérieux, La Balmes les Grottes, France) and the result was interpreted according to the EUCAST breakpoint for Enterobacteriaceae (susceptible if MIC ≤ 2 mg/L)
Phenotypic and molecular detection of carbapenemases
Isolates were screened for carbapenemase production using the modified Hodge test, the EDTA, the Modified CarbaNP test as previously described [11–13] and also by MALDI-TOF MS Ultraflex assay [14]. Screening for class A, B and D carbapenemases was carried out using PCR according to previous protocols and previously described primers including for blaVIM, blaIMP, blaNDM-1, blaKPC and all the blaOXA variants [15–20]. In addition, amplification of the chromosomal MUS gene was performed using primers designed in this study (MUS-F/R 5′-CGTTAGGCACACCAGAAGAA-3'/3′-TCCACACCATTCTGAGCGTA-5′). These primers were designed outside the region containing the blaMUS-1 located on the chromosome of a random M. odoratimimus genome deposited in GenBank. PCR products were purified and sequenced using the Big dye terminator chemistry on an ABI 3730 automated sequencer (Applied Biosystems, Foster City, CA, USA). The sequences obtained were analysed using BlastN and BlastP against the NCBI database (www.ncbi.nlm.nih.gov).
Cloning of the MUS-2 gene
The full length of the blaMUS-2 from M. odoratimimus was amplified using Herculase II Fusion Enzyme with dNTPs Combo (Agilent Technologies, Santa Clara, CA, USA) with primer MUS_F/MUS_R. The gene was then cloned into the high-copy-number plasmid pCR-BluntII-TOPO using the Zero Blunt TOPO PCR cloning kit (Invitrogen, Calrsbad, CA, USA). The resulting plasmid pTOPO-MUS-2 was transformed into TOP10 Electrocomp™ Escherichia coli. Transformants were selected on Luria–Bertani agar supplemented with 100 mg/L of kanamycin; the presence of the insert was confirmed by PCR. The antibiotic resistance profile of the TOP10 E. coli containing pTOPO-MUS vector was then determined.
Molecular strain typing
The three isolates were typed by the pulsed-field gel electrophoresis (PFGE) method as previously described [4]. The DNA was digested with SmaI restriction enzyme and the fragments of DNA were separated using a GenePath system (Bio-Rad). The running conditions were 6 V/cm at 14°C for 20 h.
Results
During a previous study that was aiming to detect carbapenemases encoding genes in animals in livestock farms in Lebanon [10], three isolates of M. odoratimimus were isolated and were studied separately. These isolates were obtained after culture of animal faeces in MacConkey agar supplemented with ertapenem (1 mg/L) and were identified by MALDI-TOF MS as M. odoratimimus (score: 2.439). The results of antibiotic susceptibility testing revealed that the isolates were resistant to almost all antibiotics, including β-lactams, aminoglycosides and colistin but remain susceptible to sulfamethoxazole-trimethroprim, rifampicin and fluoroquinolones (Table 1). In addition, all the isolates showed resistance to carbapenems with MIC for imipenem at 8 mg/L. The isolates were then analysed via the modified Hodge test, the MALDI-TOF MS Ultraflex assay, the EDTA-disc test and the modified CarbaNP test and they were all positive on the modified Hodge test, EDTA-disc and modified CarbaNP test positive. In addition, the MALDI-TOF MS Ultraflex assay was positive for all of them—suggesting a carbapenemase production, more specifically a metallo-β-lactamase as the EDTA test was positive. Screening for carbapenemase-encoding genes by PCR using carbapenemase gene primers showed a negative result for blaVIM, blaIMP, blaNDM-1, blaKPC and all the blaOXA variants. However the chromosomal MUS gene was amplified using primers MUS-F/R.
Table 1.
Antimicrobial susceptibility testing of the three Myroides isolates
| TIC | PRL | TIM | TPZ | CAZ | FEP | CTX | ETP | MEM | IPM | TOB | AMK | GEN | CIP | ATM | RA | SXT | CT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MO_32 | R | R | R | R | R | R | R | R | R | R | R | R | R | I | R | S | S | R |
| MO_35a | R | R | R | R | R | R | R | R | R | R | R | R | R | I | R | S | S | R |
| MO_104b | R | R | R | R | R | R | R | R | R | R | R | R | R | I | R | S | S | R |
MO, Myroides odoratimimus; TIC, ticarcillin; PRL, piperacillin; TIM, ticarcillin-clavulanic acid; TPZ, piperacillin-Tazobactam; CAZ, ceftazidime; FEP, cefepime; CTX, cefotaxime; ETP, ertapenem; MEM, meropenem; IPM, imipenem; TOB, tobramycin; AMK, amikacin; GEN, gentamicin; CIP, ciprofloxacin; ATM, aztreonam; RA, rifampicin; SXT, sulfamethoxazole-trimethroprim; CT, colistin; R, resistant; I, intermediate; S, susceptible.
Sequencing of the chromosomal MUS gene of the three isolates and nucleotide alignment between the blaMUS-1 and the new blaMUS sequence showed some differences. The blaMUS-2 differed from blaMUS-1 due to substitution in nucleotides at positions 69 (A→G), 525 (A→G), 171 (C→T), 177 (C→T), 397 (C→T), 671 (C→T), 179 (T→C), 514 (T→C) and 675 (C→G), resulting in a different protein MUS-2 that differ from MUS-1 protein due to substitution of amino acid at position 60 (Valine→Alanine), 133 (Histidine→Tyrosine) and 224 (Threonine→Isoleucine) (Fig. 1). The percentage of homology between the two amino acid sequences was 98.78.
Fig. 1.
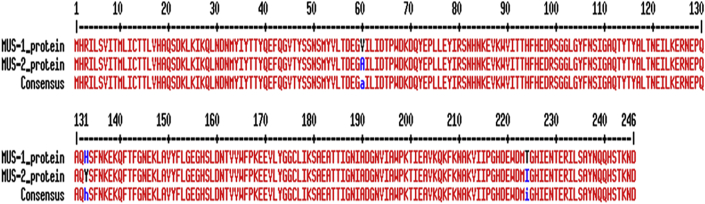
Amino acid sequence alignment between the MUS-1 and MUS-2 proteins.
The new enzyme variant was designated MUS-2 by the curators of the Lahey database of β-lactamases (http://www.lahey.org/Studies/webt.asp) and deposited in GenBank under accession number KP658209.
Cloning of the BlaMUS-2 successfully gave plasmids harbouring this gene; these plasmids were transformed into TOP10 Electrocomp™ E. coli. The presence of the insert was verified by PCR. MIC values of β-lactams for the E. coli TOP10 strain, which harbours recombinant plasmid pTOPO-MUS-2, showed that the bacteria was resistant to amoxicillin and ticarcillin and had a reduced susceptibility to piperacillin, in addition it showed an increased resistance to extended-spectrum cephalosporins and carbapenems by at least four-fold of MIC (Table 2).
Table 2.
MICs of β-lactams for Myroides odoratimimus 35a, TOP10 Escherichia coli and TOP10 E. coli pTOPO-MUS-2
| Substratea | MIC (μg/mL) |
||
|---|---|---|---|
| M. odoratimimus 35a | TOP10 E. coli pTOPO-MUS-2 | TOP10 E. coli | |
| Amoxicillin | 32 | 128 | 2 |
| Amoxicillin-CLA | 32 | 128 | 2 |
| Ticarcillin | 256 | 256 | 2 |
| Piperacillin | 128 | 8 | 2 |
| Piperacillin-TZB | 24 | 8 | 1 |
| Cephalotin | 128 | 8 | 2 |
| Cefoxitin | 32 | 4 | 2 |
| Moxalactam | 32 | 0.064 | 0.19 |
| Cefotaxime | 32 | 0.094 | 0.047 |
| Cefuroxime | 128 | 6 | 1 |
| Ceftazidime | 128 | 0.38 | 0.064 |
| Cefepime | 32 | 0.064 | 0.064 |
| Cefpirone | 32 | 0.064 | 0.064 |
| Aztreonam | 64 | 0.064 | 0.064 |
| Imipenem | 8 | 0.38 | 0.064 |
| Meropenem | 4 | 0.38 | 0.064 |
CLA, clavulanic acid at 2 μg/mL; TZB, tazobactam at 4 μg/mL.
Finally, PFGE analysis showed that the three strains of M. odoratimimus were genetically clonal since their PFGE profiles were similar (Fig. 2).
Fig. 2.
Pulsed-field gel electrophoresis of SmaI DNA restriction digests of the three Myroides odoratimimus isolates.
Discussion
To date, infections caused by M. odoratimimus in immunocompromised and immunocompetent hosts had been rarely reported [1,2,4–8]. The fact that this bacterium might cause infection in immunocompetent hosts is a real health problem because it is resistant to β-lactams including carbapenems, aminoglycoside but also to colistin. Biochemical detection of β-lactamases in clinical strains of Myroides spp. (formerly F. odoratum) was performed in 1985 by Sato et al. [21], but the molecular characteristics of β-lactamase were not reported. We would have to wait until 2002 when Mammeri et al. biochemically and genetically characterized β-lactamases expressed in M. odoratimimus and M. odoratus[9]. In fact, M. odoratimimus species produce chromosomal MUS-1 β-lactamase, which belongs to the subclass B1 of Ambler classification, and their antibiotic resistance patterns show a constant decreased susceptibility to imipenem [9].The β-lactamase produced by the three M. odoratimimus isolates in our study shared 98.78% amino acid identity with the known MUS-1 protein. At the present time, each bacterial species of the Flavobacteriaceae family that has been investigated for β-lactamase characterization produces a subclass B1 metalloenzyme [9]. Our study confirms a previous study performed on M. odoratimimus species; however, here we report a new variant of this enzyme in Lebanon. PFGE analysis showed that these isolates belong to the same clone because they had the same PFGE profile. This confirms that contamination between farms has occurred and precautions should be established to limit the emergence of this clone in livestock farms because that could be a source of human infection.
Taking into consideration the biochemical criteria established by Rasmussen and Bush to classify metallo-β-lactamases [22], MUS-1 and most probably MUS-2 belong to functional subgroup 3a, as their catalytic efficiencies for penicillins are at least 60% of that for imipenem [9]. However, these data are discrepant with those from Rasmussen and Bush [22], who mentioned that a metallo-β-lactamase produced by F. odoratum strain belongs to subgroup 3b, which groups true carbapenem-hydrolysing β-lactamases. Regarding this conflict, it is very difficult to estimate the exact role of either MUS-1 or MUS-2 in the intrinsic β-lactam resistance of M. odoratimimus because the metalloenzymes expressed in E. coli give much lower levels of resistance to β-lactams than those seen in the original producers as seen in the TOP10 E. coli after cloning.
Finally, it remains unknown if these bacteria could be a source of such a variety of metalloenzymes. The most serious finding is that animals could be a reservoir for such genes and are a source of new and/or emerging multidrug-resistant bacteria and due to possible links with animals, humans may have high risk factors for colonization/infection with such bacteria.
In conclusion, we report a novel variant of the chromosome-encoded β-lactamase MUS-1 that has been designated MUS-2 from M. odoratimimus isolates from livestock animals in Lebanon.
GenBank accession number
The full sequence of the BlaMUS-2 described in this study is deposited in the GenBank database with accession no. KP658209.
Funding
This work was supported by AZM research centre for biotechnology and its application, Lebanese University, the National Council for Scientific Research, Lebanon, the IHU Méditerranée Infection and the French CNRS.
Transparency declarations
None to declare; all authors have read and approved the manuscript.
Conflict of Interest
None declared.
Acknowledgements
The authors thank Linda Hadjadj, Saiid Azza, Taha Abdo and Maryam Yehya for technical assistance.
References
- 1.Maraki S., Sarchianaki E., Barbagadakis S. Myroides odoratimimus soft tissue infection in an immunocompetent child following a pig bite: case report and literature review. Braz J Infect Dis. 2012;16:390–392. doi: 10.1016/j.bjid.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Ktari S., Mnif B., Koubaa M. Nosocomial outbreak of Myroides odoratimimus urinary tract infection in a Tunisian hospital. J Hosp Infect. 2012;80:77–81. doi: 10.1016/j.jhin.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Schröttner P., Rudolph W.W., Eing B.R., Bertram S., Gunzer F. Comparison of VITEK2, MALDI-TOF MS, and 16S rDNA sequencing for identification of Myroides odoratus and Myroides odoratimimus. Diagn Microbiol Infect Dis. 2014;79:155–159. doi: 10.1016/j.diagmicrobio.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Yagci A., Cerikcioglu N., Kaufmann M.E. Molecular typing of Myroides odoratimimus (Flavobacterium odoratum) urinary tract infections in a Turkish hospital. Eur J Clin Microbiol Infect Dis. 2000;19:731–732. doi: 10.1007/s100960070001. [DOI] [PubMed] [Google Scholar]
- 5.Holmes B., Snell J.J., Lapage S.P. Flavobacterium odoratum: a species resistant to a wide range of antimicrobial agents. J Clin Pathol. 1979;32:73–77. doi: 10.1136/jcp.32.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachmeyer C., Entressengle H., Khosrotehrani K. Cellulitis due to Myroides odoratimimus in a patient with alcoholic cirrhosis. Clin Exp Dermatol. 2008;33:97–98. doi: 10.1111/j.1365-2230.2007.02590.x. [DOI] [PubMed] [Google Scholar]
- 7.Bachman K.H., Sewell D.L., Strausbaugh L.J. Recurrent cellulitis and bacteremia caused by Flavobacterium odoratum. Clin Infect Dis. 1996;22:1112–1113. doi: 10.1093/clinids/22.6.1112. [DOI] [PubMed] [Google Scholar]
- 8.Benedetti P., Rassu M., Pavan G., Sefton A., Pellizzer G. Septic shock, pneumonia, and soft tissue infection due to Myroides odoratimimus: report of a case and review of Myroides infections. Infection. 2011;39:161–165. doi: 10.1007/s15010-010-0077-1. [DOI] [PubMed] [Google Scholar]
- 9.Mammeri H., Bellais S., Nordmann P. Chromosome-encoded β-lactamases TUS-1 and MUS-1 from Myroides odoratus and Myroides odoratimimus (formerly Flavobacterium odoratum), new members of the lineage of molecular subclass B1 metalloenzymes. Antimicrob Agents Chemother. 2002;46:3561–3567. doi: 10.1128/AAC.46.11.3561-3567.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al Bayssari C., Dabboussi F., Hamze M., Rolain J.M. Emergence of carbapenemase-producing Pseudomonas aeruginosa and Acinetobacter baumannii in livestock animals in Lebanon. J Antimicrob Chemother. 2015;70:950–951. doi: 10.1093/jac/dku469. [DOI] [PubMed] [Google Scholar]
- 11.Lee K., Lim Y.S., Yong D., Yum J.H., Chong Y. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-β-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2003;41:4623–4629. doi: 10.1128/JCM.41.10.4623-4629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dortet L., Poirel L., Errera C., Nordmann P. CarbAcineto NP test for rapid detection of carbapenemase-producing Acinetobacter spp. J Clin Microbiol. 2014;52:2359–2364. doi: 10.1128/JCM.00594-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dortet L., Poirel L., Nordmann P. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob Agents Chemother. 2012;56:6437–6440. doi: 10.1128/AAC.01395-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kempf M., Bakour S., Flaudrops C. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS One. 2012;7:e31676. doi: 10.1371/journal.pone.0031676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mesli E., Berrazeg M., Drissi M., Bekkhoucha S.N., Rolain J.M. Prevalence of carbapenemase-encoding genes including New Delhi metallo-β-lactamase in Acinetobacter species, Algeria. Int J Infect Dis. 2013;17:e739–e743. doi: 10.1016/j.ijid.2013.02.024. [DOI] [PubMed] [Google Scholar]
- 16.Poirel L., Naas T., Nordmann P. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob Agents Chemother. 2010;54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz M., Marti S., Fernandez-Cuenca F., Pascual A., Vila J. High prevalence of carbapenem-hydrolysing oxacillinases in epidemiologically related and unrelated Acinetobacter baumannii clinical isolates in Spain. Clin Microbiol Infect. 2007;13:1192–1198. doi: 10.1111/j.1469-0691.2007.01825.x. [DOI] [PubMed] [Google Scholar]
- 18.El-Ageery S.M., Al-Hazmi S.S. Microbiological and molecular detection of VIM-1 metallo β lactamase-producing Acinetobacter baumannii. Eur Rev Med Pharmacol Sci. 2014;18:965–970. [PubMed] [Google Scholar]
- 19.Kusradze I., Diene S.M., Goderdzishvili M., Rolain J.M. Molecular detection of OXA carbapenemase genes in multidrug-resistant Acinetobacter baumannii isolates from Iraq and Georgia. Int J Antimicrob Agents. 2011;38:164–168. doi: 10.1016/j.ijantimicag.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 20.Amudhan S.M., Sekar U., Arunagiri K., Sekar B. OXA β-lactamase-mediated carbapenem resistance in Acinetobacter baumannii. Indian J Med Microbiol. 2011;29:269–274. doi: 10.4103/0255-0857.83911. [DOI] [PubMed] [Google Scholar]
- 21.Sato K., Fujii T., Okamoto R., Inoue M., Mitsuhashi S. Biochemical properties of β-lactamase produced by Flavobacterium odoratum. Antimicrob Agents Chemother. 1985;27:612–614. doi: 10.1128/aac.27.4.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasmussen B.A., Bush K. Carbapenem-hydrolyzing β-lactamases. Antimicrob Agents Chemother. 1997;41:223–232. doi: 10.1128/aac.41.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]



