Abstract
The relative degree to which stochastic and deterministic processes underpin community assembly is a central problem in ecology. Quantifying local-scale phylogenetic and functional beta diversity may shed new light on this problem. We used species distribution, soil, trait and phylogenetic data to quantify whether environmental distance, geographic distance or their combination are the strongest predictors of phylogenetic and functional beta diversity on local scales in a 20-ha tropical seasonal rainforest dynamics plot in southwest China. The patterns of phylogenetic and functional beta diversity were generally consistent. The phylogenetic and functional dissimilarity between subplots (10 × 10 m, 20 × 20 m, 50 × 50 m and 100 × 100 m) was often higher than that expected by chance. The turnover of lineages and species function within habitats was generally slower than that across habitats. Partitioning the variation in phylogenetic and functional beta diversity showed that environmental distance was generally a better predictor of beta diversity than geographic distance thereby lending relatively more support for deterministic environmental filtering over stochastic processes. Overall, our results highlight that deterministic processes play a stronger role than stochastic processes in structuring community composition in this diverse assemblage of tropical trees.
Community ecologists frequently debate the relative degree to which deterministic and stochastic processes underpin community assembly. Neutral models of community assembly, for example, emphasize the importance of dispersal limitation and demographic stochasticity1. Deterministic models of community assembly, on the other hand, emphasize the importance of ecological and evolutionary differentiation between species and non-random responses of species to their biotic and abiotic environments2. Although the details of neutral and niche theory may differ among studies, relatively few studies have examined their relative importance3.
Beta diversity represents the compositional differentiation between species assemblages. Recent conceptual advances have been made using patterns of beta diversity to disentangle the relative importance of stochastic and deterministic processes during community assembly4,5,6,7. Beta diversity along spatial and environmental gradients, in particular, has been a focal point in the neutrality versus determinism debate. Specifically, community ecologists have often focused on quantifying the degree to which beta diversity can be explained by the geographic or environmental distance (or their combination) between two assemblages. In other words, ecologists often seek to partition the variation in species beta diversity into its geographic and environmental components8,9,10. A neutral model, that assumes dispersal limitation and no consequential ecological interactions with the environment, predicts that geographic distance will be the best predictor of species beta diversity. A deterministic model, that assumes the importance of ecological interactions with the environment, predicts that environmental distance will be the best predictor of species beta diversity. Thus, partitioning beta diversity into environmental and geographic components provides a useful tool for disentangling the relative importance of deterministic or neutral processes in community assembly11.
While beta diversity approaches have been widespread, they have largely focused on the species turnover or dissimilarity between assemblages4,5,7,11. However, such an approach assumes that all species independent, ignoring the functional and phylogenetic differences between them12,13. Thus it is impossible to identify whether the species turnover from one assemblage to the next are ecologically and evolutionarily similar or dissimilar, which can provide very different insights about the ecological and evolutionary processes that structure communities14,15. For example, one could simultaneously detect a high species turnover and a low turnover of functional strategies or lineages in diverse tropical forests due to the presence of many con-generic species and many functionally similar species. Thus, it is increasingly acknowledged that an analysis of only species is not sufficient to understanding the community assembly processes13.
Given the potential limitations of species beta diversity measures13,15, ecologists have begun to develop and implement measures of phylogenetic and functional beta diversity in microbial communities16, tropical tree communities17,18,19,20, and temperate plant communities21,22. Some of these studies have demonstrated instances where measures of species beta diversity do not unveil the underlying patterns. A clear early example of this comes from Fukami et al.21 who found that experimental plant communities may functionally converge through time while maintaining divergent species compositions indicating the importance of functional determinism. Further, recent work by Graham & Fine15, Swenson et al.23,24 and Siefert et al.20 has shown that phylogenetic and functional beta diversity analyses reveal signals of deterministic assembly processes that are not evident from patterns of species beta diversity alone.
Variation partitioning of regional-scale plant phylogenetic and functional beta diversity has successfully dissected the relative contribution of deterministic and stochastic processes17,25,26. However, local-scale investigations integrating both phylogenetic and functional turnover have been rare24,27,28, despite the large number of papers on phylogenetic and functional alpha diversity in plant communities29,30,31.
Here we aim to provide a detailed analysis of the phylogenetic and functional beta diversity on local scales for tropical trees. Specifically, we utilized a large forest dynamics plot in tropical China. Species level functional trait data were collected and a molecular community phylogeny was generated for over 400 co-occurring species in this plot. The distribution, environment, trait and phylogenetic data were used to quantify whether environmental (soil nutrients) distance, geographic distance or their combination are stronger predictors of phylogenetic and functional beta diversity in tropical trees. Because many phylogenetic and functional patterns in tropical trees have been shown to be scale dependent30,32, all analyses were replicated across multiple spatial scales. Further, we compared the phylogenetic and functional beta diversity within and between habitats to determine whether a coarser scale categorical perspective of the environment can provide results comparable to those generated from fine scale continuous representations of the environment.
Results
Phylogenetic and functional beta diversity
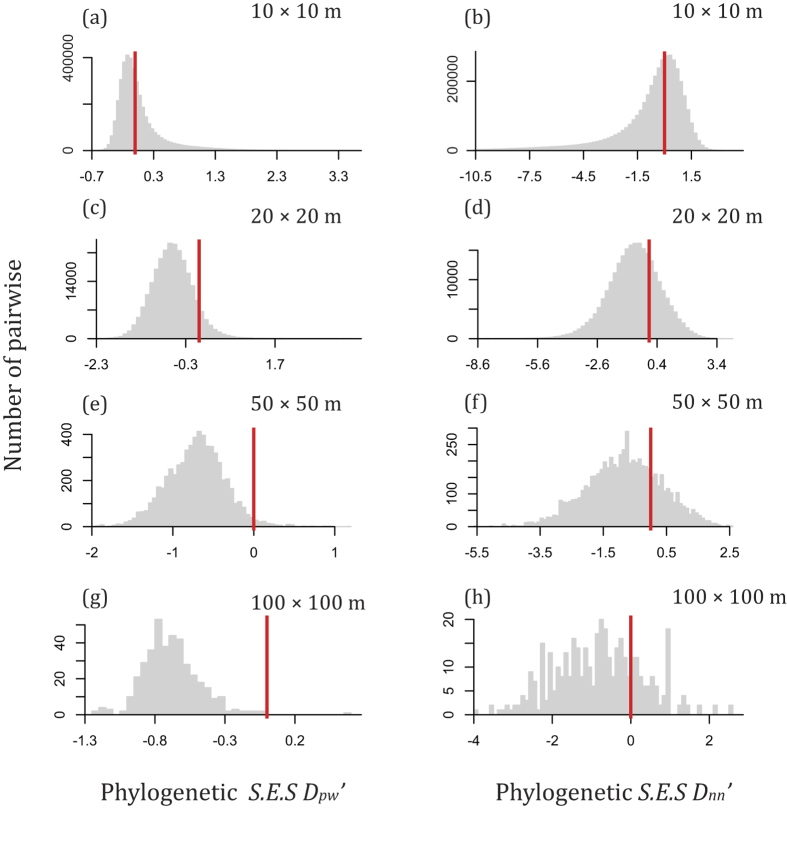
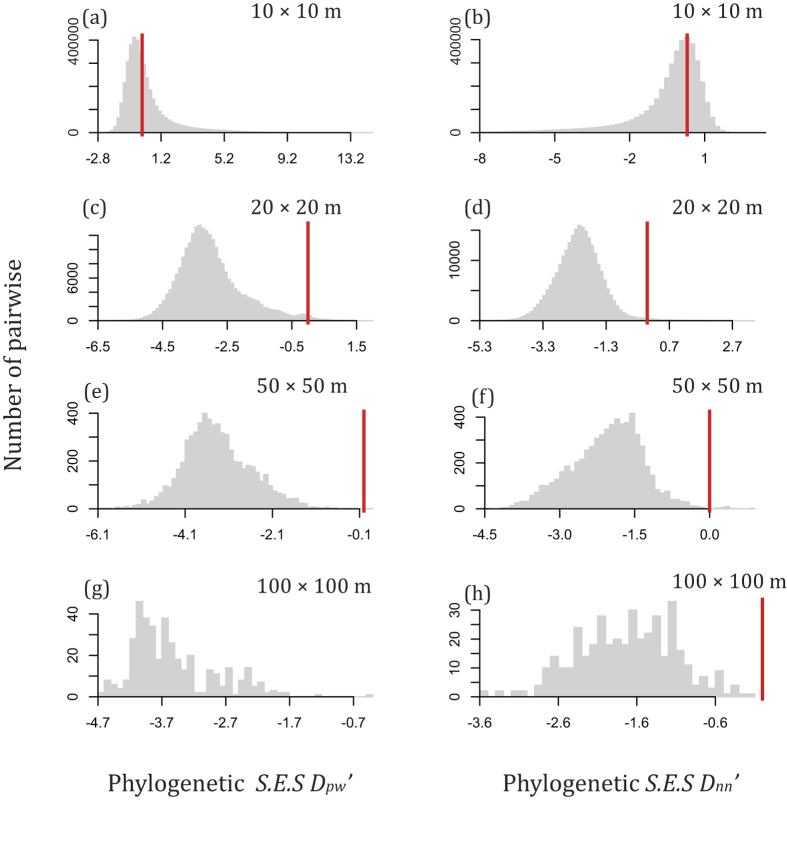
In general, the values of S.E.S. Dpw and S.E.S. Dnn deviated from zero across spatial scales (Figs 1 and 2). In most cases, the values of S.E.S. Dpw and S.E.S. Dnn were less than zero (Figs 1 and 2), indicating that the phylogenetic and functional dissimilarity between subplots was often higher than expected. However, there were also some cases of positive S.E.S. Dpw and S.E.S. Dnn, indicating that lower phylogenetic and functional turnover than expected also occurred. High phylogenetic and functional turnover between pairs of subplots was observed using indices based on species abundances and based on species occurrences (Figs S3 and S4). These patterns of non-random phylogenetic and functional beta diversity are consistent with habitat specialization being an important process in community assembly. We also found that the proportion of negative S.E.S. Dpwvalues was generally higher than that of negative S.E.S. Dnnvalues across spatial scales (Figs 1 and 2).
Figure 1.
The distribution of standard effective size for abundance-weighted mean pairwise phylogenetic distance (S.E.S. Dpw’) and standard effective size for abundance-weighted mean nearest taxon phylogenetic distance (S.E.S. Dnn’) across scales. Bars to the left of the red zero line indicates phylogenetic turnover is faster than expected. The proportions of S.E.S values below zero were as follows: (a) 57.16%, (b) 57.35%, (c) 91.18%, (d) 71.96%, (e) 97.80%, (f) 75.81%, (g) 99.75%, (h) 76%.
Figure 2.
The distribution of standard effective size for abundance-weighted mean pairwise functional distance (S.E.S. Dpw’) and standard effective size for abundance-weighted mean nearest taxon functional distance (S.E.S. Dnn’) across scales. Bars to the left of the red zero line indicates functional turnover is faster than expected. The proportions of S.E.S values below zero were as follows: (a) 58.35%, (b) 61.26%, (c) 94.7%, (d) 94.84%, (e) 99.14%, (f) 95.91%, (g) 100%, (h) 98.25%.
Phylogenetic and functional beta diversity across habitats
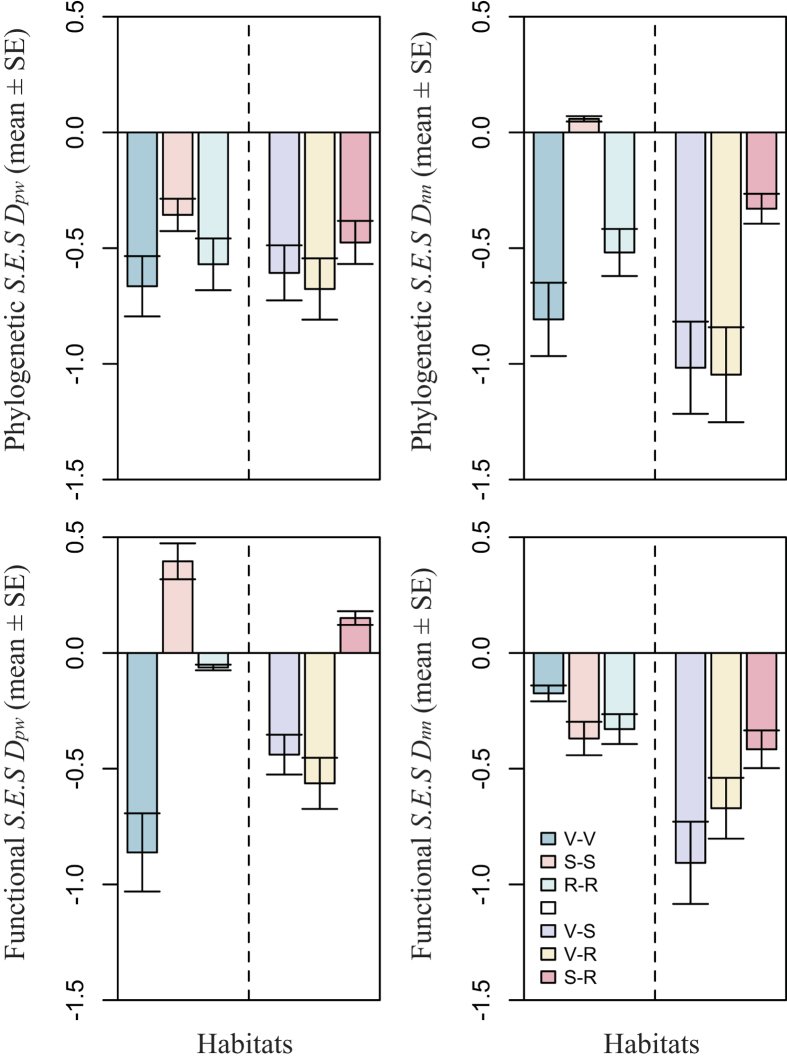
In general, phylogenetic and functional turnover was higher than expected by chance both within or across habitat types as indicated by predominantly negative S.E.S. Dpw and S.E.S. Dnn values (Fig. 3). However, we found that the phylogenetic and functional turnover within habitat types was significantly lower than that between habitat types (Table S5).
Figure 3. Bar charts illustrating phylogenetic and functional dissimilarity between subplot (20 m × 20 m) pairs both within habitats and across habitats.
V: Valley; S: Slope; R: Ridge. The same letters represent phylogenetic and functional dissimilarity within habitats (i.e. V-V, S-S, R-R) and the different letters represent that across habitats (i.e. V-S, V-R, S-R).
Relationships between phylogenetic and functional dissimilarity and geographic and environmental distance
The phylogenetic and functional dissimilarity between subplots was significantly correlated to geographic and environmental distance, except at the scale of 10 × 10 m (Tables 1, 2, S3 and S4). The explanatory power of geographic and environmental distance increased with spatial scales. The multiple regressions on distance matrices (MRM) results demonstrated that only a small proportion of variation in phylogenetic or functional dissimilarity using both Dpw and Dnn could be explained by geographic distance together with environmental distance on the finest scale (10 × 10 m), but about 40% of the variation in phylogenetic or functional turnover (Dnn) at the scale of 100 × 100 m can be explained by combination of environmental and geographic distance (Tables 1 and 2). Environmental distance explained much more variation than geographic distance using both the Dpw and Dnn metrics (Tables 1 and 2). Lastly, the amount of variation in Dpw explained by environmental distance was less than that explained for Dnn by environmental distance (Tables 1 and 2).
Table 1. The variation of phylogenetic dissimilarity explained by geographic and environmental distances across scales using multiple regressions on distance matrices.
| Scale (m2) | Response distance matrix |
Explanatory distance matrices |
||
|---|---|---|---|---|
| Combination of geographic and environmental distance | Geographic distance | Environmental distance | ||
| 10 × 10 | Dpw’ | 0.004* | 0.003 | 0.001 |
| Dnn’ | 0.009* | 0.009 | 0.000 | |
| 20 × 20 | Dpw’ | 0.139*** | 0.005*** | 0.139*** |
| Dnn’ | 0.207*** | 0.027*** | 0.197*** | |
| 50 × 50 | Dpw’ | 0.199*** | 0.012*** | 0.199*** |
| Dnn’ | 0.279*** | 0.072*** | 0.246*** | |
| 100 × 100 | Dpw’ | 0.244*** | 0.018*** | 0.235*** |
| Dnn’ | 0.410*** | 0.177*** | 0.385*** | |
***P < 0.001, **P < 0.01, *P < 0.05.
“Combination of geographic and environmental distance” represents the whole variation explained by the geographic and environmental components; “Geographic distance” represents the pure variation explained by the spatial component; “Environmental distance” represents the pure variation explained by the soil nutrients component.
Table 2. The variation of functional dissimilarity explained by geographic and environmental distances across scales using multiple regressions on distance matrices.
| Scale (m2) | Response distance matrix |
Explanatory distance matrices |
||
|---|---|---|---|---|
| Combination of geographic and environmental distance | Geographic distance | Environmental distance | ||
| 10 × 10 | Dpw’ | 0.006 | 0.002 | 0.003 |
| Dnn’ | 0.012* | 0.011 | 0.001 | |
| 20 × 20 | Dpw’ | 0.172*** | 0.006*** | 0.166*** |
| Dnn’ | 0.224*** | 0.040*** | 0.184*** | |
| 50 × 50 | Dpw’ | 0.201*** | 0.007*** | 0.201*** |
| Dnn’ | 0.271*** | 0.074*** | 0.236*** | |
| 100 × 100 | Dpw’ | 0.202*** | 0.026*** | 0.200*** |
| Dnn’ | 0.401*** | 0.121*** | 0.396*** | |
***P < 0.001, **P < 0.01, *P < 0.05.
“Combination of geographic and environmental distance” represents the whole variation explained by the spatial and environmental components; “Geographic distance” represents the pure variation explained by the spatial component; “Environmental distance” represents the pure variation explained by the environmental component.
Discussion
The present study focused on quantifying the phylogenetic and functional beta diversity and partitioning their variation along environmental and spatial gradients at multiple spatial scales in a Chinese tropical forest. We found that the amount of variation in phylogenetic and functional beta diversity explained by environmental distance was consistently greater than that explained by geographic distance. Our results provided strong support for a deterministic model underlying the beta diversity of trees in our forest.
The first goal of this study was to quantify the degree to which the observed phylogenetic or functional beta diversity differed from that expected given the observed species beta diversity. This part of the study was accomplished by performing null model analyses across four spatial scales. A neutral model would predict that the observed phylogenetic and functional beta diversity should not deviate from that expected given the observed species beta diversity15,23. Our results show that the observed phylogenetic and functional beta diversity was generally greater than that expected given the species turnover as indicated by the negative S.E.S. Dpw and S.E.S. Dnn in most cases (Figs 1 and 2). This result indicates that niche-based deterministic processes are relatively more important in structuring the tree community studied than neutral processes15,20,24,33.
The second goal of this study was to partition the amount of variation in phylogenetic and functional beta diversity with respect to environmental and geographic distance. The MRM analysis showed the phylogenetic and functional dissimilarity between subplots were significantly correlated with both geographic and environmental distance except the scale of 10 × 10 m (Tables 1 and 2). The pure spatial component as the explanatory variable is often interpreted as evidence of dispersal limitation and the potential importance of neutrality17,26. The explanatory power of geographic distance can be explained by not only dispersal limitation but also the spatially structured environmental variables34,35,36 not included in our analyses. However, the explanatory power of environmental distance was generally greater than geographic distance across spatial scales (Tables 1 and 2). This is consistent with the rapid turnover in lineages and functions through space in our forest plot which has rugged terrain with rapid changes in topography and the abiotic environment over short spatial distances (Figs 1 and 2). Thus, we can conclude that environmental filtering plays a more important role in driving the phylogenetic and functional turnover in our forest than neutral processes. Our results also support previous findings that phylogenetic or functional turnover is significantly related to environmental gradients from local to regional scales26.
On very fine spatial scales 100 m2 little variation in phylogenetic or functional beta diversity could be predicted by the geographic distance or environmental dissimilarity based upon the soil variables measured. This result is most likely driven by one to three possible, not necessarily mutually exclusive, possibilities. First, random dispersal on this local scale and a lack of strong interactions between species and their biotic and abiotic environments could result in a spatially heterogenous (i.e. non-autocorrelated) turnover. Second, unmeasured environmental variables that are not spatially autocorrelated (e.g. vertical light gradients or fine scale soil nutrient gradients) may be important on this local scale. Third, biotic interactions promote the consistent dissimilarity between species within subplots (i.e. high phylogenetic or functional alpha diversity) in similar environments thereby reducing the overall phylogenetic or functional turnover on very local scales. It is most likely that all three of these possibilities are important in the forest plot and future more detailed analyses on very fine spatial scales are required.
Habitat specialization is often thought to influence taxonomic, phylogenetic and functional turnover in tropical forests5,17,24. Given this expectation we further examined the phylogenetic and functional beta diversity within and across the three different broad categorical habitat types in the plot. If these habitats filter lineages and functions, we would expect that the phylogenetic and functional beta diversity within habitats would be lower than expected by positive S.E.S. Dpw and S.E.S. Dnn. Further, we might expect this to result in faster phylogenetic and functional turnover than expected across habitats as indicated by negative S.E.S. Dpw and S.E.S. Dnn. However, we found S.E.S. Dpw and S.E.S. Dnn were generally negative whether within or across habitats indicating a higher than expected turnover (Fig. 3). Taken together with the evidence of how variance in phylogenetic and functional beta diversity is partitioned along continuous soil nutrient axes in this forest, it suggests that local scale environmental heterogeneity within habitats plays a predominant role in structuring the phylogenetic and functional turnover in this forest.
Our phylogenetic and functional beta diversity results were generally consistent, though there were slight differences that are likely explained by the fact that most functional traits had weak, albeit significant, phylogenetic signal37. As many researchers have argued, phylogenetic similarity is not always a good predictor of ecological similarity38, and thus only using phylogenetic information may be misleading when coexisting species simultaneously converge and diverge in function13. Here we calculated the phylogenetic and functional dissimilarity by using the algorithms of pairwise distance (Dpw) and nearest neighbor distances (Dnn). The Dpw and Dnn are ‘basal’ and ‘terminal’ metrics of phylogenetic or functional beta diversity18. Although there were subtle differences between Dpw and Dnn, our results based on these two metrics were generally consistent (Figs 1 and 2). However, S.E.S. Dpwseems to reflect stronger phylogenetic and functional dissimilarity across four spatial scales than did Dnn (Figs 1 and 2). The generally negative S.E.S Dpw and S.E.S Dnn across spatial scales indicate that phylogenetic and functional turnover was faster than that expected by chance given the species turnover in both of distantly related species (basal turnover) and species within terminal clades (terminal turnover). However, less of the variation in the S.E.S. Dpw values could be explained by environmental distance, geographic distance, and their combination than Dnn (Tables 1 and 2). This result was also consistent with the studies of phylogenetic dissimilarity of 96 tree communities in India18 and in the Gutianshan forest dynamics plot in China39.
Methods
Study site
This study was conducted in the 20-ha Xishuangbanna forest dynamics plot (FDP) located in the Yunnan Province, southwest China (21°36′N, 101°34′E) (Fig. S1). This forest is characterized as a seasonal tropical rainforest and dominated by large individuals of Parashorea chinensis (Dipterocarpaceae). There are 111,177 stems and 468 species of trees in this plot. The climate is strongly seasonal with distinct alterations between the dry season (from May to October) and the wet season (from November to April). The annual mean precipitation is 1493 mm, of which 1256 mm (84%) occurs in the wet season37. Elevation within the FDP ranges from 708.2 m to 869.1 m. The Xishuangbanna FDP was established in 2007 where all freestanding woody stems ≥1 cm diameter at 130 cm from the ground DBH (Diameter at Breast Height) were measured, mapped, tagged and identified to species between November 2006 and April 2007. A detailed description of the climate, geology and flora of Xishuangbanna can be found in Cao et al.40.
Functional traits
Trait data for the tree species in the plot were collected using standardized protocols41 with the exception of leaf chlorophyll content and stem specific resistance. Leaf area, specific leaf area (SLA), leaf thickness, leaf dry matter content (LDMC) and leaf chlorophyll content are regarded as key traits related to resource allocation strategies41,42. Maximum tree height is directly related to the adult light niche43. Wood density represents a tradeoff between volumetric growth and mechanical vulnerability on the tissue and organismal scale and it is often the trait most closely related to hydrological niche and demographic rates44. Stem specific resistance has been shown to be strongly correlated with wood density and was therefore used as a substitute45. Seed mass represents a trade-off between producing many small seeds per unit energy versus producing a few large seeds per unit energy46. In total, we measured eight functional traits that are believed to represent fundamental functional trade-offs in leaves, wood and seeds among tree species47. We used the mean values of traits for each tree species in this study. Detailed information regarding the collection and measurement of functional traits is provided in Yang et al.37.
Soil nutrients
Soil samples were collected and analyzed for the Xishuangbanna FDP following the standardized protocols described in John et al.48. Specifically, soil was sampled using a regular grid of 30 × 30 m in the 20-ha plot. The 252 grid intersections were basal collection points, together with each base point, three additional sampling points were located at random combination of 2 and 5 m, 2 and 15 m or 5 and 15 m along a random compass bearing away from the associated base point. At each sample point, we removed the litter and humus layer, then collected 500 g topsoil from depth of 0–10 cm. A total of 756 soil samples were taken. Fresh soil samples were placed into pre-labeled plastic bags and shipped to the Biogeochemistry Laboratory at the Xishuangbanna Tropical Botanical Garden. In the laboratory, one sub-sample was used for measuring pH values as immediately as possible using a potentiometer in fresh soil after water extraction (soil : water was 1 : 2.5). The other sub-samples were air-dried, smashed, sieved using 1 mm and 0.15 mm mesh and stored in plastic bags for soil bulk density, total C, total N, total P, total K, available N, extractable P and extractable K analysis. Soil sample data were subjected to variogram modeling, which was then used in block kriging to estimate mean soil nutrient concentrations in each subplot48,49. Finally, we obtained the spatial maps of average soil water content and 9 soil nutrients (pH, total N, total P, total K, available N, extractable P, extractable K, total C, bulk density) in the plot. Using the soil maps we characterized the environment of each subplot at each spatial scale. Additional detailed information regarding the measurement of soil variables can be found in Hu49.
Phylogenetic tree reconstruction
We reconstructed a community phylogeny representing 428 of the species in the Xishuangbanna FDP (Fig. S2). A DNA supermatrix was generated from three chloroplast sequence regions – rbcL, matK, trnH-psbA and nuclear ribosomal internal transcribed spacer (ITS)50. The DNA supermatrix was then analyzed using RAxML via the CIPRES supercomputer cluster to infer a maximum likelihood (ML) phylogeny using the APG III phylogenetic tree as a constraint or guide tree as described in Kress et al.50. Finally, we implemented non-parametric rate smoothing (NPRS) in the software r8s51. Detailed information regarding the phylogenetic tree reconstruction is provided in Yang et al.37.
Phylogenetic and functional beta diversity
We sequentially divided the 20-ha plot into 2000 10 × 10 m, 500 20 × 20 m, 80 50 × 50 m and 20 100 × 100 m subplots. The beta diversity analyses were performed between subplots using the above multiple spatial scales. To eliminate trait redundancy, we performed a principle components analysis (PCA) on the functional trait data (Table S1). We utilized the first three principle components, that explain over 70% of the variation in the trait data, to construct a Euclidean trait distance matrix. An Unweighted Pair Group Method with Arithmetic Mean (UPGMA) hierarchical clustering was then applied to this matrix to produce a trait dendrogram52. This dendrogram represented the trait similarity between taxa23.
Based on the molecular phylogenetic tree and trait dendrogram, we calculated the phylogenetic and functional dissimilarity between each pair of subplots using the mean pairwise phylogenetic or trait distance (Dpw) and the mean nearest neighbor phylogenetic or trait distance (Dnn)53,54. We also calculated abundance-weighted mean pairwise phylogenetic or trait distance (Dpw’) and abundance-weighted mean nearest neighbor phylogenetic or trait distance (Dnn’). Dpw, Dnn, Dpw’ and Dnn’ are calculated as follows:
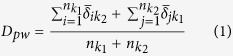 |
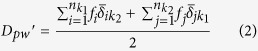 |
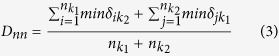 |
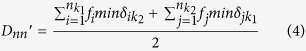 |
Where 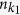 represents the number of species in community k1;
represents the number of species in community k1; 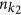 represents the number of species in community k2;
represents the number of species in community k2; 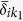 is the mean pairwise phylogenetic or trait distance between species i in community k1 to all species in community k2 and
is the mean pairwise phylogenetic or trait distance between species i in community k1 to all species in community k2 and 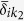 is the mean pairwise phylogenetic or trait distance between species j in community k2 to all species in community k1; where min
is the mean pairwise phylogenetic or trait distance between species j in community k2 to all species in community k1; where min 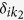 is the nearest neighbor phylogenetic or trait distance between species i in community k1 to all species in community k2 and min
is the nearest neighbor phylogenetic or trait distance between species i in community k1 to all species in community k2 and min 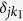 is the nearest neighbor phylogenetic or trait distance between species j in community k2 to all species in community k1, where fi and fj are the relative abundance of species i in community k1 and species j in community k2. Dpw and Dpw’ are ‘basal’ metrics of phylogenetic or trait beta diversity, while Dnn and Dnn’ are ‘terminal’ metric of phylogenetic or trait beta diversity18.
is the nearest neighbor phylogenetic or trait distance between species j in community k2 to all species in community k1, where fi and fj are the relative abundance of species i in community k1 and species j in community k2. Dpw and Dpw’ are ‘basal’ metrics of phylogenetic or trait beta diversity, while Dnn and Dnn’ are ‘terminal’ metric of phylogenetic or trait beta diversity18.
Although there are many methods for calculating phylogenetic and functional beta diversity33, the advantage of Dpw and Dnn is that they represent the two main mathematically independent classes of phylogenetic and functional dissimilarity metrics while avoiding the redundancy of calculating other nearly identical metrics17,18.
We addressed whether the observed phylogenetic or functional dissimilarity deviated from a random expectation by comparing the observed values with 999 null values generated by a null model. The null model randomly shuffled the names of species across the tips of the phylogenetic tree or trait dendrogram 999 times. This approach randomized the phylogenetic relatedness or trait similarity of species to one another, while maintained the observed community data matrix. Thus, species occupancy frequencies, abundances and spatial distributions (e.g. any patterns of intraspecific aggregation or dispersal limitation) were fixed in each randomization.
A standardized effect size of Dpw (S.E.S. Dpw) and the standardized effect size of Dnn (S.E.S. Dnn) were quantified using the null distribution18,20,24,39. Specifically, the mean of the null distribution was subtracted from the observed value and divided by the standard deviation of the null distribution. This value was then multiplied by negative one to stick with convention. Thus, negative S.E.S.Dpw or S.E.S.Dnn values indicated a higher observed Dpw or Dnn than expected and positive S.E.S.Dpw or S.E.S.Dnn values indicated a lower observed Dpw or Dnn than expected. We calculated the S.E.S.Dpw and S.E.S.Dnn for each pair of subplots at each spatial scale.
Lastly, we were also interested in whether the phylogenetic and functional beta diversity between subplots in the same habitat type was lower than that between habitat types. To quantify this, the 20-ha plot was divided into three habitat types using the topographic variables including slope, elevation and convexity (Table S2). The three habitat types, valley, slope and ridge, were assigned to each 20 × 20 m subplot in the FDP. The spatial distribution of three habitat types is given in Figure S1. We calculated phylogenetic and functional S.E.S. Dpw and S.E.S Dnn, for each pair of subplots within the same habitats and that across the different habitats, and then use Student’s t test to compare the means of S.E.S. Dpw or S.E.S Dnn between within habitats and across habitats.
Variation partitioning of phylogeneitc and functional beta diversity
We utilized MRM55,56 to relate phylogenetic or functional dissimilarity (Dpw or Dnn) with geographic distance and environmental distance. MRM is similar to a partial Mantel’s test and can be used to examine the correlation between the dependent distance matrix and the independent explanatory distance matrices. Next we used variation partitioning to determine the explanatory power of independent effects and joint effects of the explanatory factors8. We constructed the geographic distance matrix by calculating the geographic distance between each pair of subplots using the coordinates in the center of each subplot. A PCA was conducted on all soil nutrient variables and an environmental distance matrix was then constructed by the Euclidean distance between each pair of subplots based on the first three principle components. The above analyses were performed using the R packages “ecodist”, “vegan” and “picante”57,58,59,60.
Additional Information
How to cite this article: Yang, J. et al. Local-scale Partitioning of Functional and Phylogenetic Beta Diversity in a Tropical Tree Assemblage. Sci. Rep. 5, 12731; doi: 10.1038/srep12731 (2015).
Supplementary Material
Acknowledgments
This study was supported by the National Key Basic Research Program of China (Grant No. 2014CB954100), National Natural Science Foundation of China (31400362, 31370445 and 31000201), the West Light Foundation of Chinese Academy of Sciences and the Applied Fundamental Research Foundation of Yunnan Province (2014GA003). NGS was supported by two US-China Dimensions of Biodiversity grants (DEB-1046113; DEB-1241136). We thank David L. Erickson and W. John Kress for advice and assistance with DNA barcoding analyses. We thank all the people who have contributed to the establishment of the 20-ha Xishuangbanna tropical seasonal rainforest dynamics plot. Logistical support was provided by Xishuangbanna Station of Tropical Rainforest Ecosystem Studies (National Forest Ecosystem Research Station at Xishuangbanna), Chinese Academy of Sciences.
Footnotes
Author Contributions J.Y., N.G.S., L.L. and M.C. designed the study, J.Y., N.G.S. and L.L. performed analyses, J.Y., G.Z., X.C., J.W.F.S., L.S. and J.L. collected data, J.Y. wrote the first draft of the manuscript, and J.Y., N.G.S. and L.L. contributed substantially to revisions.
References
- Hubbell S. P. The unified neutral theory of biodiversity and biogeography (Princeton University Press, 2001). [DOI] [PubMed] [Google Scholar]
- Tilman D. Resource competition and community structure. (Princeton University Press, 1982). [PubMed] [Google Scholar]
- Chase J. M. & Myers J. A. Disentangling the importance of ecological niches from stochastic processes across scales. Phil. Trans. R. Soc. B. 366, 2351–2363 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomisto H., Ruokolainen K. & Yli-Halla M. Dispersal, environment, and floristic variation of western Amazonian forests. Science, 299, 241–244 (2003). [DOI] [PubMed] [Google Scholar]
- Anderson M. J. et al. Navigating the multiple meanings of β diversity: a roadmap for the practicing ecologist. Ecol. Lett. 14, 19–28 (2011). [DOI] [PubMed] [Google Scholar]
- Laliberté E., Paquette A., Legendre P. & Bouchard A. Assessing the scale-specific importance of niches and other spatial processes on beta diversity: a case study from a temperate forest. Oecologia, 159, 377–388 (2009). [DOI] [PubMed] [Google Scholar]
- Kraft N. J. B. et al. Disentangling the drivers of β diversity along latitudinal and elevational gradients. Science, 333, 1755–1758 (2011). [DOI] [PubMed] [Google Scholar]
- Legendre P., Borcard D. & Peres-Neto P. R. Analyzing beta diversity: partitioning the spatial variation of community composition data. Ecol. Monogr. 75, 435–450 (2005). [Google Scholar]
- Legendre P. et al. Partitioning beta diversity in a subtropical broad-leaved forest of China. Ecology, 90, 663–674 (2009). [DOI] [PubMed] [Google Scholar]
- De Cáceres M. et al. The variation of tree beta diversity across a global network of forest plots. Global. Ecol. Biogeogr. 21, 1191–1202 (2012). [Google Scholar]
- Myers J. A. et al. Beta-diversity in temperate and tropical forests reflects dissimilar mechanisms of community assembly. Ecol. Lett. 16, 151–157 (2013). [DOI] [PubMed] [Google Scholar]
- Chave J., Chust G. & Thébaud C. The importance of phylogenetic structure in biodiversity studies. In Scaling Biodiversity (Institute Editions, Santa Fe, 2007). [Google Scholar]
- Swenson N. G. The assembly of tropical tree communities - the advances and shortcomings of phylogenetic and functional trait analyses. Ecography, 36, 264–276 (2013). [Google Scholar]
- Hardy O. J. & Senterre B. Characterizing the phylogenetic structure of communities by an additive partitioning of phylogenetic diversity. J. Ecol. 95, 493–506 (2007). [Google Scholar]
- Graham C. H. & Fine P. V. A. Phylogenetic beta diversity: Linking ecological and evolutionary processes across space in time. Ecol. Lett. 11, 1265–1277 (2008). [DOI] [PubMed] [Google Scholar]
- Martin A. P. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microb. 68, 3673–3682 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine P. V. A. & Kembel S. W. Phylogenetic community structure and phylogenetic turnover across space and edaphic gradients in western Amazonian tree communities. Ecography. 34, 552–565 (2011). [Google Scholar]
- Swenson N. G. Phylogenetic beta diversity metrics, trait evolution and inferring the functional beta diversity of communities. PLoS One, 6, e21264 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy O. J., Couteron P., Munoz F., Ramesh B. R. & Pélissier R. Phylogenetic turnover in tropical tree communities: impact of environmental filtering, biogeography and mesoclimatic niche conservatism. Global. Ecol. Biogeogr. 21, 1007–1016 (2012). [Google Scholar]
- Siefert A., Ravenscroft C., Weiser M. D. & Swenson N. G. Functional beta diversity patterns reveal deterministic community assembly processes in eastern North American trees. Global. Ecol. Biogeogr. 22, 682–691 (2013). [Google Scholar]
- Fukami T., Bezemer T. M., Mortimer S. R. & van der Putten W. H. Species divergence and trait convergence in experimental plant community assembly. Ecol. Lett. 8, 1283–1290 (2005). [Google Scholar]
- Ricotta C. & Burrascano S. Testing for differences in beta diversity with asymmetric dissimilarities. Ecol. Indic. 9, 719–724 (2009). [Google Scholar]
- Swenson N. G., Anglada-Cordero P. & Barone J. A. Deterministic tropical tree community turnover: Evidence from patterns of functional beta diversity along an elevational gradient. Proc. R. Soc. B. 278, 877–884 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson N. G. et al. Phylogenetic and functional alpha and beta diversity in temperate and tropical tree communities. Ecology, 93, S112–S125 (2012a). [Google Scholar]
- Qian H., Swenson N. G. & Zhang J. Phylogenetic beta diversity of angiosperms in North America. Global. Ecol. Biogeogr. 22, 1152–1161 (2013). [Google Scholar]
- Zhang J. et al. Phylogenetic beta diversity in tropical forests: implications for the roles of geographical and environmental distance. J. Syst. Evol. 51, 71–85 (2013). [Google Scholar]
- Swenson N. G. et al. Temporal turnover in the composition of tropical tree communities: functional determinism and phylogenetic stochasticity. Ecology. 93, 490–499 (2012b). [DOI] [PubMed] [Google Scholar]
- Bernard-Verdier M., Flores O., Navas M. & Garnier E. Partitioning phylogenetic and functional diversity into alpha and beta components along an environmental gradient in a Mediterranean rangeland. J. Veg. Sci. 24, 877–889 (2013). [Google Scholar]
- Kraft N. J. B., Valencia R. & Ackerly D. D. Functional traits and niche-based tree community assembly in an Amazonian forest. Science. 322, 580–582 (2008). [DOI] [PubMed] [Google Scholar]
- Kraft N. J. B. & Ackerly D. D. Functional trait and phylogenetic tests of community assembly across spatial scales in an Amazonian forest. Ecol. Monog. 80, 401–422 (2010). [Google Scholar]
- Baraloto C. et al. Using functional traits and phylogenetic trees to examine the assembly of tropical tree communities. J. Ecol. 100, 690–701 (2012). [Google Scholar]
- Swenson N. G., Enquist B. J., Pither J., Thompson J. & Zimmerman J. K. The problem and promise of scale dependency in community phylogenetics. Ecology. 87, 2418–2424 (2006). [DOI] [PubMed] [Google Scholar]
- Leprieur F. et al. Quantifying phylogenetic beta diversity: distinguishing between ‘True’ turnover of lineages and phylogenetic diversity gradients. PLoS One, 7, e42760 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. W. & Lundholm J. T. Variation partitioning as a tool to distinguish between niche and neutral processes. Ecography. 33, 648–655 (2010). [Google Scholar]
- Gilbert B. & Bennett J. R. Partitioning variation in ecological communities: do the numbers add up? J. App. Ecol. 47, 1071–1082 (2010). [Google Scholar]
- Legendre P. Spatial autocorrelation: trouble or new paradigm? Ecology. 74, 1659–1673 (1993). [Google Scholar]
- Yang J. et al. Functional and phylogenetic assembly in a Chinese tropical tree community across size class, spatial scales and habitats. Funct. Ecol. 28, 520–529 (2014). [Google Scholar]
- Losos J. B. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett. 11, 995–1003 (2008). [DOI] [PubMed] [Google Scholar]
- Feng G. et al. Comparison of phylobetadiversity indices based on community data from Gutianshan forest plot. Chin. Sci. Bull. 57, 623–630 (2012). [Google Scholar]
- Cao M. et al. Xishuangbanna Tropical Seasonal Rainforest Dynamics Plot: Tree Distribution Maps, Diameter Tables and Species Documentation. (Yunnan Science and Technology Press, 2008). [Google Scholar]
- Cornelissen J. H. C. et al. A Handbook of protocols for standardised and easy measurement of plant functional traits worldwide. Aust. J. Bot. 51, 335–380 (2003). [Google Scholar]
- Pérez-Harguindeguy N. et al. New handbook for standardised measurement of plant functional traits worldwide. Aust. J. Bot. 61, 167–234 (2013). [Google Scholar]
- Kohyama T., Suzuki E., Partomihardjo T., Yamada T. & Kubo T. Tree species differentiation in growth, recruitment and allometry in relation to maximum height in a Bornean mixed dipterocarp forest. J. Ecol. 91, 797–806 (2003). [Google Scholar]
- Chave J. et al. Towards a worldwide wood economics spectrum. Ecol. Lett. 12, 351–336 (2009). [DOI] [PubMed] [Google Scholar]
- Isik F. & Li B. Rapid assessment of wood density of live tree using the Resistograph for selection in tree improvement programs. Can. J. Forest. Res. 33, 2426–2435 (2003). [Google Scholar]
- Moles A. T. & Westoby M. Seed size and plant strategy across the whole life cycle. Oikos, 113, 91–105 (2006). [Google Scholar]
- Westoby M. A leaf-height-seed (LHS) plant ecology strategy scheme. Plant. Soil. 199, 213–227 (1998). [Google Scholar]
- John R. et al. Soil nutrients influence spatial distributions of tropical tree species. Pro. Natl. Acad. Sci. 104, 864–869 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y. H. A study on the habitat variation and distribution patterns of tree species in a 20-ha forest dynamics plot at Bubeng, Xishuangbanna, SW China, PhD Thesis. Chinese Academy of Sciences (2010).
- Kress W. J. et al. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Pro. Natl. Acad. Sci. 106, 18621–18626 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson M. J. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 19, 301–302 (2003). [DOI] [PubMed] [Google Scholar]
- Swenson N. G. Functional and phylogenetic ecology in R. (Springer, 2014). [Google Scholar]
- Rao C. R. Diversity and dissimilarity coefficients: A unified approach. Theor. Popul. Biol. 21, 24–43 (1982). [Google Scholar]
- Ricotta C. & Burrascano S. Beta diversity for functional ecology. Preslia. 80, 61–71 (2008). [Google Scholar]
- Legendre P., Lapointe F. & Casgrain P. Modeling brain evolution from behavior: A permutational regression approach. Evolution. 48, 1487–1499 (1994). [DOI] [PubMed] [Google Scholar]
- Lichstein J. W. Multiple regression on distance matrices: a multivariate spatial analysis tool. Plant. Ecol. 188, 117–131 (2007). [Google Scholar]
- R Development Core Team R: a language and environment for statistical computing. (2012) Available at: http://www.R-project.org/. (Accessed: 6th June 2012).
- Goslee S. C. & Urban D. L. The ecodist package for dissimilarity-based analysis of ecological data. J. Stat. Softw. 22, 1–19 (2007). [Google Scholar]
- Oksanen J. et al. Vegan: Community ecology package. (2012) Available at: http://CRAN.R-project.org/package=vegan. (Accessed: 6th October 2012).
- Kembel S. W., Ackerly D. D., Blomberg S. P., Cornwell W. K. & Cowan P. D. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 26, 1463–1464 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.