Abstract
Mutations in NYX and CACNA1F gene are responsible for the X-linked congenital stationary night blindness (CSNB). In this study, we described the clinical characters of the two Chinese families with X-linked CSNB and detected two novel mutations of c. 371_377delGCTACCT and c.214A>C in the NYX gene by direct sequencing. These two mutations would expand the mutation spectrum of NYX. Our study would be helpful for further studying molecular pathogenesis of CSNB.
Congenital stationary night blindness (CSNB) is a group of clinically and genetically heterogeneous retinal disorders characterized by night blindness, decreased visual acuity, and a reduced or absent b-wave in the electroretinogram (ERG)1. Other clinical features of CSNB may include variable degrees of myopia, a nearly normal fundus appearance, nystagmus and strabismus. Two subgroups of CSNB can be classified by ERG into the “complete form” (or type 1 CSNB), and “incomplete form” (or type 2 CSNB)2. The complete form is characterized by absence of rod b-wave and oscillatory potentials due to complete loss of the rod pathway function, whereas the incomplete form shows a reduced rod b-wave, cone a-wave, and 30-Hz flicker ERG response caused by impaired rod and cone pathway function3.
CSNB may be inherited as an autosomal dominant, autosomal recessive and X-linked inheritance mode. Up to date, mutations in 14 genes, including CACNA1F (MIM300110), NYX (MIM 300278), TRPM1 (MIM 603576), LRIT3 (OMIM, 615004), GRM6 (MIM 604096), GPR179 (MIM 614515), CABP4 (MIM 608965), GNAT1 (OMIM, 139330), PDE6B(OMIM, 163500) RHO(OMIM, 180380), GRK1 (OMIM, 180381), RDH5(OMIM, 136880), SAG(OMIM, 181031), and SLC24A1(OMIM, 603617) (Retnet: http://www.sph.uth.tmc.edu/retnet/), have been reported to be responsible for CSNB. These genes are involved either in different components of the phototransduction cascade or in signaling transduction from photoreceptors to adjacent bipolar cells4.
NYX and CACNA1F genes are two disease-causing genes for X-linked congenital stationary night blindness. Mutations in NYX gene could be responsible for 45% of the X-linked CSNB, and may lead to the complete form of CSNB (CSNB1A), whereas mutations in CACNA1F gene could explain 55% of the X-linked CSNB and result in the incomplete form of CSNB (CSNB2A)4.
Here we reported two Chinese families with X-linked CSNB. Molecular genetic analysis of the two candidate genes (NYX and CACNA1F) indicated that two novel mutations in the NYX were detected in these two families.
Materials and Methods
Patient Ascertainment
Two families with X-linked CSNB (CSNB-01, and CSNB-02) were recruited for this study (Fig. 1). All participants underwent an ophthalmologic and orthoptic examination, including best corrected visual acuities, visual field, anterior segment of the eyes, vitreous and fundus, measurement of the deviation angle in the cardinal eye positions, examination of ocular movement and binocular vision, and Electroretinography (ERG). ERG were recorded according to the standards of the International Society for Clinical Electrophysiology of Vision (http://www.iscev.org). Cycloplegic refraction were performed for children under the age of 14. The spherical equivalent of the refractive error is given in diopters (D). After informed consent, 7 affected and 13 unaffected individuals from the two unrelated families were taken 3 ml blood samples from their blood vessels and DNA was extracted from blood lymphocytes according to the standard methods of protocol (Roche Biochemical, Inc). This study obtained IRB approval from the Tianjin Eye Hospital and conformed to the tenets of the Declaration of Helsinki.
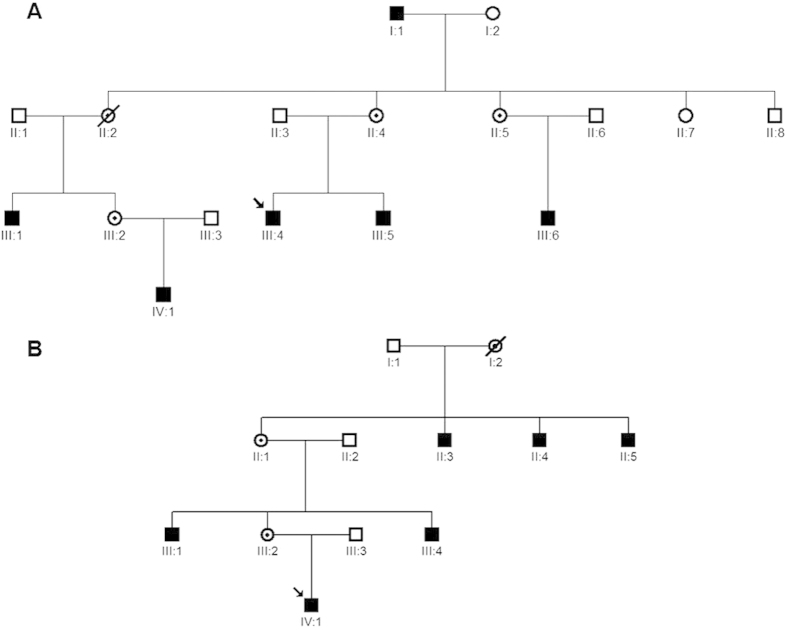
Figure 1. Two Chinese families with X-linked Congenital Stationary Night Blindness.
The family CSNB-01 consisted of 6 affected males (individuals I:1, III:1, III:4, III:5, III:6, IV:1) and 3 female carrier (II:4, II:5, III:2), in which the individual III:4 was the proband (A) The family CSNB-02 included 6 affected males (individuals II:3, II:4, II:5, III:1, III:4, IV:1) and 2 female carriers (II:1, III:2). The individual IV:1 was the proband. (B).
Mutation Analysis
Because NYX and CACNA1F are two candidate genes for X-linked congenital stationary night blindness, both of these two genes were directly sequenced in the CSNB-01 and CSNB-02 families. The coding regions and exon-intron boundaries of these two genes were amplified by PCR reaction using the primer pairs designed by the online software of the Primer 3. The exon 3 of NYX gene was split into five overlap fragments. The primer sequences were listed in the supplement Table. PCR were carried in 20 μL of standard PCR buffer containing 1.5 mM MgCl2, 0.2 mM of each dNTP, 0.5 μM of each primer, 1 U of Taq polymerase (Sangon, Shanghai, China), and 50 ng of DNA. The amplification program was an initial 2min denaturation at 98 °C, followed by 30 cycles of 30 s at 94 °C, 30 s at 55 °C, 1 min at 72 °C, and a final 7 min extension step at 72 °C. The PCR products were bi-directionally sequenced using the BigDye Terminator Cycle Sequencing V3.1 kit on an ABI PRISM 3130 Genetic Analyzer (Applied Biosystems) after purification with the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA). Sequencing results were assembled and analyzed with the Seqman program of DNASTAR software (DNASTAR Inc, Madison, WI). The reference cDNA sequences of the NYX and CACNA1F were obtained from Genebank (NM_022567 for NYX mRNA, and NM_001256789 for CACNA1F mRNA) and +1 corresponds to the A of the ATG translation initiation codon. Mutation naming followed the nomenclature recommended by the Human Genomic Variation Society (HGVS).
In silico analysis
Two online programs of Polymorphism Phenotype (PolyPhen) and Sorting Intolerant From Tolerant (SIFT) were used to predict the potential impact of an amino acid substitution on the function of the protein. The molecular model were built using Swiss-model5,6,7,8.
Results
Clinical features
In the family CSNB-01, all affected males complained about night blindness since their early childhood. Ophthalmologic examination showed that the patients had decreased visual acuity, variable degrees of myopia, latent nystagmus, and extropia combined with dissociated vertical deviation (DVD). Their detail clinical data were listed in the table 1. The proband (individual III:4, Fig. 1A) was a 24-year-old affected male and feel night blindness at the age of 4. Fundus examination revealed no obvious anomalies. Refraction examination showed a refractive error of −6.25D in his right eye and of −6.0D in his left eye. The best corrected visual acuity of his both eyes were 0.4. He had an extropia of about 15° at the primary eye position, and had fairly pronounced latent nystagmus combined with dissociated vertical deviation. The ERG showed absence of rod b-wave and oscillatory potentials under the scotopic condition, but a normal 30 HZ flicker response (Fig. 2). Thus, the clinical phenotype of the family CSNB-01 could be classified as the complete form of CSNB according to the ERG. The CSNB-02 family consisted of 6 affected males. The proband of the CSNB-02 family (individual IV:1, Fig. 1B) was a 7-year-old affected boy. He was diagnosed with CSNB by inquiring family history because his two uncles (individual III:1,III:4) were diagnosed with CSNB in other hospital. Refraction examination showed a refractive error of −9.0D sph in his right eye and of −10.0D sph in his left eye. The best corrected visual acuity of both eyes were 0.7. He had an intermittent extropia of 30 prism diopter (PD) at distance and 40 PD at near fixation. However, nystagmus and DVD were unremarkable. Fundus examination were high myopia changes but were otherwise normal. ERG examination was failed because he cannot cooperate very well. The CSNB form were also not able to established without ERG.
Table 1. Clinical features of individuals in Two Chinese families with X-linked CSNB.
| Family | ID | Patient/carrier | Age | Mutation | Status | CN,XT,DVD | BCVA OD;OS | Refraction OD;OS |
|---|---|---|---|---|---|---|---|---|
| CSNB-01 | I:1 | patient | 78 | c. 371_377del | Hemizygous | CN,XT,DVD | 0.4; 0.4 | −6.75D; −6.75D |
| CSNB-01 | II:4 | carrier | 53 | c. 371_377del | Heterozygous | none | 0.8; 0.8 | −1.00D; −1.00D |
| CSNB-01 | II:5 | carrier | 47 | c. 371_377del | Heterozygous | none | 1.0; 1.0 | −0.50D; −0.75D |
| CSNB-01 | III:1 | patient | 33 | c. 371_377del | Hemizygous | CN,XT,DVD | 0.2; 0.2 | −9.25D; −8.75D |
| CSNB-01 | III:2 | carrier | 31 | c. 371_377del | Heterozygous | none | 0.8; 0.8 | −1.00D; −1.00D |
| CSNB-01 | III:4 | patient | 24 | c. 371_377del | Hemizygous | CN,XT,DVD | 0.4; 0.4 | −6.25D; −6.00D |
| CSNB-01 | III:5 | patient | 22 | c. 371_377del | Hemizygous | CN,XT,DVD | 0.1; 0.1 | −9.75D; −9.50D |
| CSNB-01 | III:6 | patient | 22 | c. 371_377del | Hemizygous | CN,XT,DVD | 0.3; 0.3 | −5.00D; −4.50D |
| CSNB-01 | IV:1 | patient | 6 | c. 371_377del | Hemizygous | CN,XT,DVD | 0.4; 0.4 | −3.75D; −3.00D |
| CSNB-02 | III:1 | patient | 35 | c.214A>C | Hemizygous | XT | 0.6; 0.6 | −9.25D; −9.75D |
| CSNB-02 | III:2 | carrier | 32 | c.214A>C | Heterozygous | none | 1.0; 1.0 | −1.50D; −2.75D |
| CSNB-02 | IV:1 | patient | 7 | c.214A>C | Hemizygous | XT | 0.7; 0.7 | −9.00D; −10.00D |
CN: congenital nystagmus.
XT: extropia.
DVD: dissociated vertical deviation.
BCVA: Best corrected Visual acuity.
OD: Right eye.
OS: Left eye.
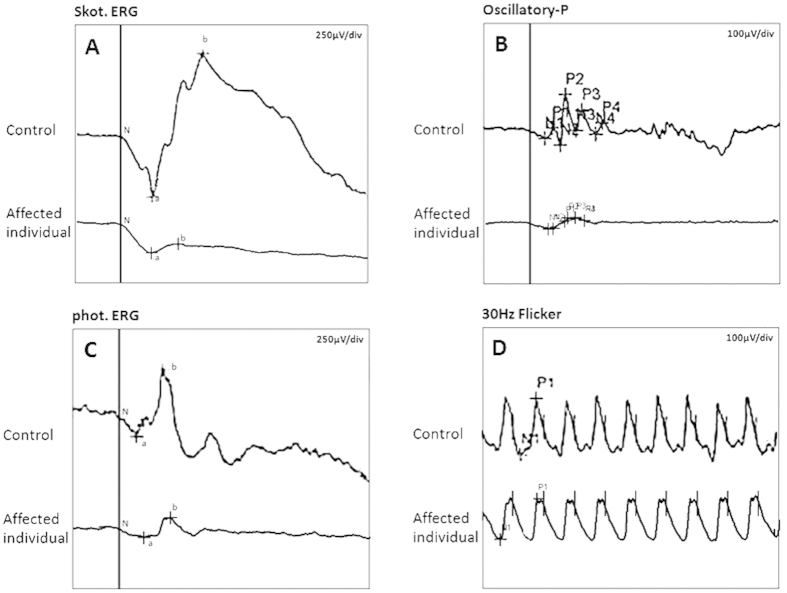
Figure 2. ERG recordings for the right eye of the proband and a normal control (individual III:3) in the family CSNB-01.
Comparing with the normal control, the affected individual had abnormal ERG responses showing absence of b-wave under scotopic condition (A) loss of the wavelets of the oscillatory potentials (B) a normal photopic ERG (C) and 30 Hz flicker response (D).
Identification of Mutations
After sequence analysis to the NYX and CACNA1F genes, a 7-base pair deletion of c. 371_377delGCTACCT(p.Y125TfsX138) was detected in exon 3 of NYX gene in all affected males and female carriers in the family CSNB-01, while in the same exon, a missense mutation of c.214A>C(p.N72H) was identified in the family CSNB-02 (Fig. 3). Multiple sequence alignment of the NYX protein shows that N72 is conserved among Homo sapiens, Pongo abelii, Rattus norvegicus, Mus musculus, Bos taurus, Gallus gallus, Xenopus, and Danio rerio (Fig. 4 ). A PISC score of 1.0 and a SIFT score of 0.02 were yielded after using the POLYPHEN 2 (http://coot.embl.de/PolyPhen/) and SIFT (http://sift.bii.a-star.edu.sg/) program to predict the functional and structural changes of the amino acid substitution, which means that the substitution of Asparagine (Asn, N) at codon 72 by Histidine would be deleterious to the structure and function of the NYX protein. The above two mutations in NYX were absent in 100 normal controls after sequencing of the NYX.
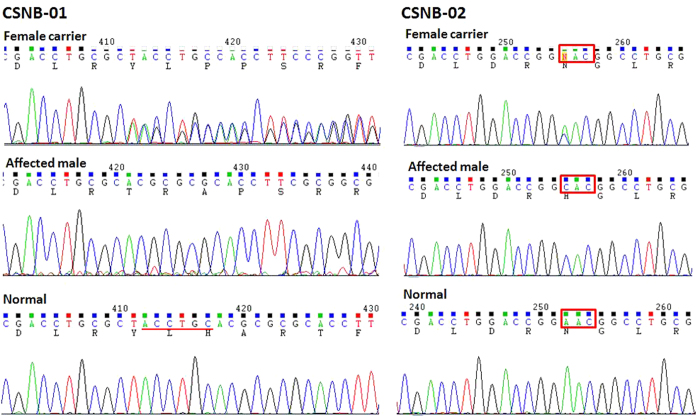
Figure 3. Sequence analysis of NYX gene for two families with X-linked CSNB.
Sequencing chromatograms from a female carrier (top), an affected male (middle) and a normal individual (bottom) in each family. Mutations in NYX were identified in each of two families: c. 371_377delGCTACCT in the family CSNB-01 and c.214A>C in the family CSNB-02.
Figure 4. Multiple sequence alignment of the NYX protein.
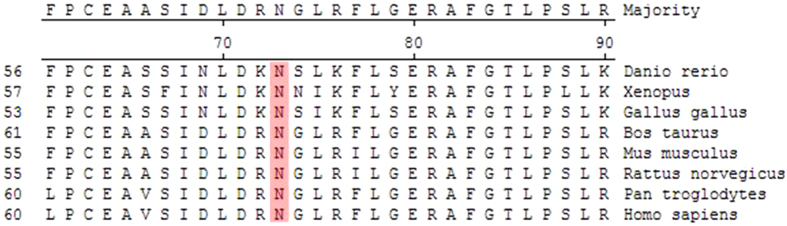
Multiple alignment of amino acids around p.N72H (denoted by the black framework) revealed evolutionary conservation of the Asparagine among Homo sapiens, Pongo abelii, Rattus norvegicus, Mus musculus, Box Taurus, Gallus gallus, Xenopus, and Danio rerio.
Discussion
NYX gene is located on chromosome Xp11.3 and consists of 3 exons spanning approximately 28 kb. It is mainly expressed in the kidney and retina. Within the retina, NYX is expressed in the inner segment of photoreceptors, outer and inner nuclear layers and the ganglion cell layer9. Nyctalopin is the encoded protein of the NYX gene, and consists of 481 amino acids. Structurally, nyctalopin belongs to a member of the leucine-rich repeat (LRR) superfamilies, containing 11 consecutive LRRs flanked by N- and C-terminal cysteine-rich LRRs, as well as an N-terminal ER signal peptide and a C-terminal glycosylphosphatidylinositol (GPI) membrane anchor10. LRRs are short-sequence motifs presenting in a number of proteins and are believed to be involved in the protein–protein interactions, cell adhesion and axon guidance11,12,13. Up to date, more than 50 mutations in NYX have been reported to be associated with congenital stationary night blindness. These mutations include missense and nonsense, splicing site mutations, as well as insertions and deletions. However, the exact molecular pathogenesis of CSNB caused by NYX mutations remains to be elucidated. It was suggested that nyctalopin might play an important role in keeping localization of TRPM1 in the dendritic tips of ON bipolar cells14,15. As an accessory subunit of the transient receptor potential (TRP) channel, nyctalopin might interact directly with TRPM1, or together with LRIT3 (another LRR family member), to keep the signaling channel proteins in the right place. The dislocation of TRPM1 would lead to abnormal function of retinal depolarizing bipolar cells (DBCs) and be predicated to result in abnormal signal transmission from photoreceptors. Mutations in these three interacted proteins (Nyctalopin, LRIT3 and TRPM1) have been documented to be associated with the complete form CSNB.
The mutation of c. 371_377delGCTACCT(p.Y125TfsX138) is located in exon 3 of the NYX gene. The 7 base pair of GCTACCT deletion would result in a frameshift change of NYX gene and produce a truncated proteins with initial 125 amino acids similar to the wild type NYX protein and additional 12 aberrant amino acids followed by a premature stop codon. Thus, the NMD surveillance mechanism could be activated by a PTC-containing mRNA. The abnormal mRNA with PTC would be degraded under the NMD surveillance mechanism or produce a defective truncated protein through escaping the NMD surveillance16,17.
Mutation of c.214A>C (p.N72H) in the family CSNB-02 is a novel mutation and not detected in 100 normal controls. N72 is located in the first repeat of leucine-rich repeat (LRR) in the nyctalopin, and is highly conserved among human and the lower animals. Substitution of Asparagine by Histidine would be probably damaging to the protein structure and/or the protein function, predicted by either POLYPHEN 2 or SIFT program. In addition, Asparagine 72 forms the inter-side chain hydrogen bonds with Cysteine 48, Leucine 53, Leucine 69 and Aspartate 70. Substitution of Asparagine by Histidine would break hydrogen bonds with Leu 53 and Leu 69, and forms a new hydrogen bond with Alanine 51 in the Swiss-model (Fig. 5), which could be expected to change the protein structure. However, changing Asparagine to Histidine were not able to disrupt the concave exterior surface of the secondary molecular structure in the constructed model as to be expected (Fig. 5). Previous study documented that different constructs containing NYX mutations could not produce any effect on the cellular localization in comparison to wild type constructs18, and the mutated variants in the LRR might cause CSNB through disrupting the protein-protein interactions11,19. The detailed molecular mechanism should be elucidated further by in vitro and in vivo experiments.
Figure 5. Structural model of nyctalopin built by the Swiss model.
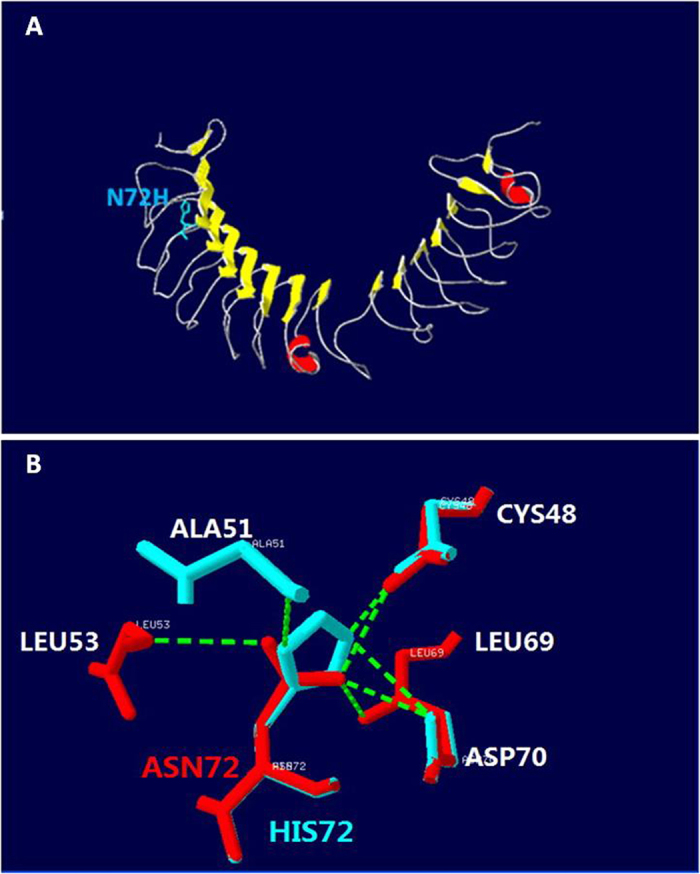
(A) 3-Dimensional structure of nyctalopin. Yellow arrows show beta-strands. Red parts show α-helix. The side chains of N72 are marked in blue. (B) Detail of the effect of the N72H mutation showing loss of hydrogen bonds. The normal and mutated nyctalopin structures are shown in red and blue, respectively. Hydrogen bonds are shown by green dashed solid lines.
Although more than 50 mutations in NYX have been reported and could be responsible for 45% of the X-linked CSNB, only three NYX mutations, including p.Arg94Pro20, p.Leu184Arg20 and p.Thr258Pro21, have been reported in the Chinese patients with CSNB. Mutations of p.Arg94Pro and p.Leu184Arg are located in the second and the third LRR motif respectively, whereas mutation of p.Thr258Pro located in the ninth LRR motif. To our knowledge, mutation of N72H in the first LRR motif have not been reported before. With exception of the CSNB, NYX mutations could be responsible for some Chinese high myopia patients without CSNB22,23.
The forms of CSNB1 and CSNB2 can be differentiated by ERG. However, the correlation of the genotype with the clinical phenotypes “complete” and “incomplete” cannot be established very well24. Previous study suggested that patients with CSNB1 mainly had rod-related problems, and patients with CSNB2 had both rod- and cone-related problems, so that the visual acuity on average was better in CSNB1 than in CSNB2. However, the expressivity may be variable within the CSNB1 patients due to the clinical and genetic heterogeneity. Some CSNB1 patients may have dramatically decreased visual acuity, whereas a few patients might have good visual acuity even better than 0.8, and experience only high refractive error and nyctalopia25,26. In our patients, the patients carrying NYX mutation of c.371_377del GCTACCT (p.Y125TfsX138) had reduced visual acuity ≤0.4, myopia of the spherical equivalent range from −3.0D to −10.0D, dissociated vertical deviation, extropia and nystagmus, whereas the patient carrying NYX mutation of c.214A>C (p.N72H) had only high myopia of −10.0D and intermittent extropia. Although we cannot establish the correlation of the NYX mutations with the clinical phenotype because we don’t have enough patients to be observed, we think that variable expressivity might be related to other genetic or environmental factors rather than to the different mutant types in the same gene.
In summary, we report the clinical characterization of two Chinese families with X-linked CSNB and identified two novel mutations of c. 371_377delGCTACCT and c.214A>C in the NYX gene. These two mutations would expand the mutation spectrum of NYX and help to study molecular pathogenesis of CSNB.
Additional Information
How to cite this article: Dai, S. et al. Two Novel NYX Gene Mutations in the Chinese Families with X-linked Congenital Stationary Night Blindness. Sci. Rep. 5, 12679; doi: 10.1038/srep12679 (2015).
Acknowledgments
The authors thank all participants for their enthusiastic participation. This research is partially supported by the National Natural Science Foundation of China (Grant No. 30940081 and No. 81170884), Technology Foundation of Tianjin Health Bureau (Grant No.09KR09), The Key Scientific and Technological Project of Tianjin Health Bureau.
Footnotes
Author Contributions D.S. and M.Y. finished most part of the experiment and wrote the manuscript. K.W., L.W. and did some experiment including DNA sequencing and analyzed some data. R.H. and P.H. evaluated clinical characters for some patients. N.L. was in charge of the project design and revised the manuscript.
References
- Carr R. E. Congenital stationary nightblindness. Trans Am Ophthalmol Soc 72, 448–487 (1974). [PMC free article] [PubMed] [Google Scholar]
- van Genderen M. M. et al. Mutations in TRPM1 are a common cause of complete congenital stationary night blindness. Am J Hum Genet 85, 730–736 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audo I., Robson A. G., Holder G. E. & Moore A. T. The negative ERG: clinical phenotypes and disease mechanisms of inner retinal dysfunction. Surv Ophthalmol 53, 16–40 (2008). [DOI] [PubMed] [Google Scholar]
- Zeitz C. Molecular genetics and protein function involved in nocturnal vision. Expert Rev. Ophthalmol. 2, 467–485 (2007). [Google Scholar]
- Arnold K., Bordoli L., Kopp J. & Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006). [DOI] [PubMed] [Google Scholar]
- Guex N., Peitsch M. C. & Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30 Suppl 1, S162–S173 (2009). [DOI] [PubMed] [Google Scholar]
- Biasini M. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42, W252–W258 (1093). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer F., Arnold K., Kunzli M., Bordoli L. & Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37, D387–D392 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bech-Hansen N. T. et al. Mutations in NYX, encoding the leucine-rich proteoglycan nyctalopin, cause X-linked complete congenital stationary night blindness. Nat Genet 26, 319–323 (2000). [DOI] [PubMed] [Google Scholar]
- Pusch C. M. et al. The complete form of X-linked congenital stationary night blindness is caused by mutations in a gene encoding a leucine-rich repeat protein. Nat Genet 26, 324–327 (2000). [DOI] [PubMed] [Google Scholar]
- Kobe B. & Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci 19, 415–421 (1994). [DOI] [PubMed] [Google Scholar]
- Krantz D. E. & Zipursky S. L. Drosophila chaoptin, a member of the leucine-rich repeat family, is a photoreceptor cell-specific adhesion molecule. Embo J 9, 1969–1977 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nose A., Takeichi M. & Goodman C. S. Ectopic expression of connectin reveals a repulsive function during growth cone guidance and synapse formation. Neuron 13, 525–539 (1994). [DOI] [PubMed] [Google Scholar]
- Zeitz C. et al. Whole-exome sequencing identifies LRIT3 mutations as a cause of autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet 92, 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearring J. N. et al. A role for nyctalopin, a small leucine-rich repeat protein, in localizing the TRP melastatin 1 channel to retinal depolarizing bipolar cell dendrites. J Neurosci 31, 10060–10066 (1523). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook J. A., Neu-Yilik G., Hentze M. W. & Kulozik A. E. Nonsense-mediated decay approaches the clinic. Nat Genet 36, 801–808 (2004). [DOI] [PubMed] [Google Scholar]
- Tazi J., Bakkour N. & Stamm S. Alternative splicing and disease. Biochim Biophys Acta. 1792, 14–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitz C. et al. NYX (nyctalopin on chromosome X), the gene mutated in congenital stationary night blindness, encodes a cell surface protein. Invest Ophthalmol Vis Sci 44, 4184–4191 (2003). [DOI] [PubMed] [Google Scholar]
- Zeitz C. et al. Novel mutations in CACNA1F and NYX in Dutch families with X-linked congenital stationary night blindness. Mol Vis 11, 179–183 (2005). [PubMed] [Google Scholar]
- Xiao X. et al. CSNB1 in Chinese families associated with novel mutations in NYX. J Hum Genet 51, 634–640 (2006). [DOI] [PubMed] [Google Scholar]
- Sui R., Li F., Zhao J. & Jiang R. Clinical and genetic characterization of a Chinese family with CSNB1. Adv Exp Med Biol 613, 245–252 (2008). [DOI] [PubMed] [Google Scholar]
- Zhang Q. et al. Mutations in NYX of individuals with high myopia, but without night blindness. Mol Vis 13, 330–336 (2007). [PMC free article] [PubMed] [Google Scholar]
- Yip S. P. et al. A novel missense mutation in the NYX gene associated with high myopia. Ophthalmic Physiol Opt 33, 346–353.. [DOI] [PubMed] [Google Scholar]
- Allen L. E. et al. Genotype-phenotype correlation in British families with X linked congenital stationary night blindness. Br J Ophthalmol 87, 1413–1420 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijveld M. M. et al. Genotype and phenotype of 101 dutch patients with congenital stationary night blindness. Ophthalmology 120, 2072–2081. [DOI] [PubMed] [Google Scholar]
- Boycott K. M., Pearce W. G. & Bech-Hansen N. T. Clinical variability among patients with incomplete X-linked congenital stationary night blindness and a founder mutation in CACNA1F. Can J Ophthalmol 35, 204–213 (2000). [DOI] [PubMed] [Google Scholar]