Abstract
In this study, the 454 pyrosequencing platform was used for analyzing the comparative transcriptomic profiles of leaf and root tissues of 1-month-old cotton (Gossypium herbaceum) plants under drought stress. A total of 56,354 and 49,308 reads were generated from leaf and root tissues, respectively, and clustered into 6,313 and 5,858 unigenes. The differentially expressed unigenes that showed up-regulation (≥2-fold) or down-regulation (2≤-fold) were considered for further analysis. A total of 3,517 unigenes were differentially expressed in both tissues. The 1,528 genes specific to leaves and 1,128 specific to roots were obtained. The 28 biological pathways in two tissues were found to respond significantly to drought stress. A total of 289 in leaf and 277 in root unknown (novel) unigenes were found to be remarkably regulated by drought stress. Some key regulatory genes involved in abiotic stress such as WRKY, ERF, AP2 EREBP, MYB, and LEA were highly expressed in leaves. The genes RHD3, LBD, and transcription factor WRKY75, known for root development under various stress conditions, were expressed specifically in root. The genes related to chlorophyll a/b binding protein and photosystem-related proteins showed significant higher expression in roots and as compared to leaves. It can be concluded that cotton leaves are distinct from roots in terms of molecular mechanisms for responses to drought stress.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-014-0257-2) contains supplementary material, which is available to authorized users.
Keywords: G. herbaceum, Drought tolerance, Leaf, Root, Pyrosequencing, Reactive oxygen species
Introduction
Drought is one of the major constraints to cotton production worldwide. Cotton plants grown under unsuitable environmental conditions such as temperature extremes, water deficit, and salinity stress face reduced growth and productivity resulting from loss of cotton boll and altered fiber development. Choice of Gossypium herbaceum in these tracts was not preferential but obligatory due to harsh cotton growing conditions. In most regions of India, G. herbaceum is planted for its natural textile fibre. To cope with water stress, plants execute a number of physiological and metabolic responses (Arndt et al. 2001; Dias et al. 2003). The physiological and biological functions of plants are determined by expression pattern of genes which are influenced by several adverse environmental conditions (Shinozaki and Yamaguchi-Shinozaki 2007; Valliyodan and Nguyen 2006). Hence, an importance in stress-related studies is to detect the expression of unique genes or clusters of genes and to determine their expression pattern in specific plant tissue or organs which control the regulatory network. The plant leaves provide an adaptive mechanism for plants in water-deficit condition by reducing the leaf area, decreasing transpiration rate, and maintaining stomatal closure (Caldeira et al. 2014; Sanchez-Blanco et al. 2009). The small leaves or reducing leaf expansion is an important adaptive mechanism for drought tolerance (Caldeira et al. 2014). The proportion of leaf elongation and size has been attributed to root variability, root length and growth (Parent et al. 2010). The root architecture is determined by an endogenous genetic program as well as by external environmental factors (Ishida et al. 2008; Smith and Smet 2012). The few studies that have compared leaf and root tissue transcriptomes have highlighted the organ specificity of drought responses (Cohen et al. 2010; Libault et al. 2010; Milner et al. 2014; Narsai et al. 2010). Roots sense the edaphic water deficit, send chemical signals to the shoots, and maintenance of root growth despite reduced water availability can contribute to drought tolerance through water foraging (Lynch 1995). The comparative analysis of gene expression profiles in the roots of maize (Jansen et al. 2013; Yue et al. 2008), soybean (Libault et al. 2010; You et al. 2011), populous (Cohen et al. 2010) and Arabidopsis has revealed a considerable number of root-specific genes and these are important for abiotic or biotic tolerance (Smith and Smet 2012). The various research groups using transcriptome studies have revealed several downstream components involved in the complex network of root formation (Ishida et al. 2008; Milner et al. 2014; Sengupta and Reddy 2011). A large number of genes encoding transcription factors (NLP, WRKY75, RAV and REM), osmoprotectants, ion transporters and heat shock proteins and pathways involved in hormone (ABA, ethylene and JA) biosynthesis and signal transduction are known to play an important role under drought stress (Narusaka et al. 2003; Ranjan et al. 2012b; Shinozaki and Yamaguchi-Shinozaki 2007; Tuteja and Sopory 2008). The genes involved in phenylpropanoid and flavonoid biosynthesis, pentose and glucuronate interconversions and starch and sucrose metabolism pathways are able to maintain adaptive condition during drought (Naika et al. 2013; Xiao et al. 2014). Cotton is an important source of natural fibre used in the textile industry and the productivity of the crop is adversely affected by drought stress (Trivedi et al. 2012; Zhu et al. 2013). Molecular processes in response to water-deficit stress have been studied at great length in cotton. Bowman et al. (2013) reported transcriptome analysis of upland cotton (G. hirsutum) root under drought stress. Chen et al. (2013) analyzed the drought-resistibility in two different cultivars of tetraploid upland cotton, using transcriptome profiling of leaf. Park et al. (2012) reported differentially expressed genes under water stress in root and leaf tissues in tetraploid cotton using cDNA-AFLP. Payton et al. (2011) investigated the transcriptional changes in the roots and leaves of diploid cotton under water-stress condition. The recent publication of the whole genome sequence of the cotton diploid relative Gossypium raimondii (Paterson et al. 2012) has expanded the use of NGS as a tool to study cotton development.
In our previous study we analyzed the drought-resistibility in different diploid cotton genotypes, and observed that the drought tolerance is due to several biochemical mechanisms working together (Ranjan et al. 2012a, b). In the present study, a genome-wide comparative analysis of genes in root and leaf tissue was performed to identify root and leaf specific genes and biological pathways that improve tolerance to drought stress. The comparative expression profiles of leaves and roots were useful in consolidating our knowledge of molecular mechanism of cotton pants in response to drought stress.
Methods
Drought stress treatment and plant growth
The genotypes of cotton (G. herbaceum), namely, RAHS-14 and GujCot-21 are drought sensitive and drought tolerant, respectively, were used for this study.
The seeds of both genotypes were sterilized and aseptically kept immersed in water for a day at 30 °C and then placed for germination in a moist petri dish under the following conditions: 28/25 °C as day and night temperature, 12 h of light and dark periods alternatively and relative humidity of 80 % in the first experiment. After 3 days, properly germinated seeds were transferred on paper rafts, which were submerged (3/4th) in Hoagland’s media containing different percentages (2, 4, 6 and 8 %) of mannitol and mannitol free Hoagland’s media was considered as control (Fig. 1a, b). Then seedlings were allowed to grow until they had shown healthy growth in control condition then root lengths of grown seedlings were compared. In second experiment, plants were grown in the earthen pot, drought stress was given to the plants by withholding watering till soil moisture reaches below 30 % in pots and drooping effects on plant leaves became prominent (Fig. 1c, e), whereas the control pots were irrigated daily (Fig. 1d, f).
Fig. 1.
Morphological changes in plant leaf due to drought stress and variation in root length development due to osmotic stress in a GujCot-21 b RAHS-14. GujCot-21 grown under c drought stress condition, d control condition and, e RAHS-14 grown under drought stress condition and f control condition
RNA isolation
The earthen pot grown plants were selected for the experiments after drought stress treatment. The first leaves of RAHS-14 and root of GujCot-21 plant were collected for RNA extraction. Total RNA of leaf and root tissues was extracted by Spectrum Plant Total RNA Kit® (Sigma-Aldrich) according to the manufacturer’s instructions. After DNase I treatment, RNA was purified by QIAquick PCR purification kit (Qiagen). Quality and quantity of the purified RNA were determined by measuring the absorbance at 260/280 nm (A260/A280). RNA integrity was further verified by 1.5 % agarose gel electrophoresis.
Transcriptome sequencing
Total RNA (3 μg) from leaf and root tissue were reverse transcribed using a T7-Oligo (dT) Promoter Primer in the first-strand cDNA synthesis (Affymetrix). The double strand cDNA was synthesized and enriched using in vitro transcription (IVT) reaction (Affymetrix). cRNA (3 μg) was reverse transcribed in the first-strand cDNA synthesis step by using random hexamer primer, followed RNase H-mediated second strand cDNA synthesis and purified in column (QIAquick PCR purification kit) and further used for GS-FLX pyrosequencing. These sequence data were deposited in NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov) with accession number SRX038499 (leaves) and SRX038496 (roots).
Assembly of transcriptome using CAP3
The analyses of sequences were conducted using custom Perl scripts and publicly available software, R package (www.R.project.org). The sequences for leaves and roots were tagged with the library name and pooled exemplars were queried against the NCBI nucleotide database (NR) and in Arabidopsis TAIR-10 version using BLASTN at e-value 10−10 and alignment length of more than 50 % of the query sequence for annotation (see additional information-1, Table 1). All the Gossypium ESTs available at NCBI were downloaded and pooled. The pooled exemplars were also queried against all public cotton EST database to identify new transcripts of Gossypium.
Table 1.
Assembly report of transcriptome sequencing data of G. herbaceum leaves and root under drought condition
| Parameters | Leaf | Root |
|---|---|---|
| (a) | ||
| Total reads | 56,354 | 49,308 |
| Total contigs (100 bp or greater) | 6,313 | 5,858 |
| Singleton | 20,780 | 30,776 |
| Average length of contigs (bp) | 481.7 bp | 532.9 bp |
| Supercontigs | 101,063 | 104,298 |
| TAIR | Cotton EST | NR | No-hits | ||||
|---|---|---|---|---|---|---|---|
| Contigs | Singleton | Contigs | Singleton | Contigs | Singleton | Contigs | |
| (b) | |||||||
| Leaf | 726 | 17,267 | 656 | 11,202 | 834 | 10,211 | 289 |
| Root | 682 | 16,806 | 488 | 12,383 | 713 | 9,885 | 275 |
(a) Total number of reads separated for both transcriptome libraries. Contigs generated by CAP3 assembly
(b) Contigs showing significant hits (e-value 10−10 and ≥50 % overlap) in the NCBI database. Contigs showing significant hits (e-value 10−10 and ≥50 % alignment of either the query or the subject) in the cotton EST database and TAIR-10
Differential expression analyses
The reads of both the tissues were tagged and pooled to create a dataset of 105,662 reads and used for digital profiling. These reads were assembled using the CAP3 programme at an overlap of 100 bp and 80 % identity. These reads assembled into 12,171 contigs which include only those contigs that have more than five reads. We calculated TPM value and R value using the R statistics for contigs and those with R value ≥3 and fold change ≥2 were considered significantly differentially expressed contigs. These filtered contigs were annotated using BLASTN against NCBI nucleotide (NT) database and BLASTX against NCBI non-redundant proteins (NR) (see additional information-2).
Heat map analysis
For visualization of the significant comparisons, heat maps of the significant genes after FDR adjustment were produced with the heat maps two package in R. Hierarchical clustering of individual sample with 1,000 bootstrap replications was performed with the R package pvclust and heat map were sorted accordingly. To visualize clusters of genes expression, we grouped the z-transformed expression ratios by using k-means in R.
GO analyses
The GO annotations for the sequences were derived using their UniProt annotation. The UniProt database was used as it has extensive GO mapping. The GO annotation for level 5 was extracted for each library and used for further analysis.
Biochemical pathway analysis
Pathway analysis was performed to find out the significant pathways of the differentially expressed genes in roots and leaves according to the KEGG databases. The two-way Fisher’s exact test and χ 2-test were used to classify the significant pathways, and the threshold of significance was defined by p < 0.05 and FDR <0.05. The FDR value was used to correct the ‘p value’ (Garcia-Alcalde et al. 2011).
MapMan analysis
To adapt MapMan software to cotton (G. herbaceum) the major MapMan BINs and their sub-BINS used for the classification of Arabidopsis genes were transferred to the G. herbaceum system. The classification based on the similarity to Arabidopsis proteins was then automatically checked against domains that have been assigned to a MapMan category (Usadel et al. 2006). Furthermore, cotton tentative consensus sequences that did not meet the prerequisites for an automatic draft assignment were classified manually according to their respective TIGR annotation (TIGR release 10). Finally, all draft assignments were corrected manually for potential mistakes.
Results
Comparison of drought tolerance of GujCot-21 and RAHS-14
The GujCot-21 and RAHS-14 exhibited contrasting difference in their root growth under osmotic stress and control condition. GujCot-21 has longer root length as compared to RAHS-14 at 6 % mannitol concentration (Fig. 1a, b). In earthen pot experiments, RAHS-14 showed prominent effect of drought stress (Fig. 1e) as compared to control (Fig. 1d). However, GujCot-21 showed much better development, less wilting and higher biomass as compared to RAHS-14 (Fig. 1c). In addition, GujCot-21 exhibited better drought tolerance than RAHS-14 as observed by visual comparison of leaf senescence and wilting symptom in two genotypes (Fig. 1c, f.)
Overall features of the drought stress-responsive expression profiles in leaves and roots
In leaf tissue 1,528 genes were identified; out of these 289 genes showed no hits to any protein or nucleotide in the database (NR-non redundant and NT-nucleotide data base of NCBI) and the 26 genes were hypothetical or unknown proteins were obtained (see additional information 2). The majority of the annotated genes were from the photosynthesis pathway, with high expression in ribulose 1–5 bisphosphate carboxylase/oxygenase activase, photosystem II D and chlorophyll a/b binding proteins. The senescence-associated proteins were also observed in the differentially expressed genes. In root library 1,128 genes were identified and out of these 275 genes did not have any hit to any of the database (NR, UniProt, NT) and about 8 % genes matched with predicted/hypothetical proteins. The other genes related to drought stress that were upregulated in the root were ascorbate peroxidase, cysteine protease, delta-tonoplast intrinsic proteins, Lea proteins, etc. (see additional information 2).
Roots behave in a more focused response under drought stress compared to leaves
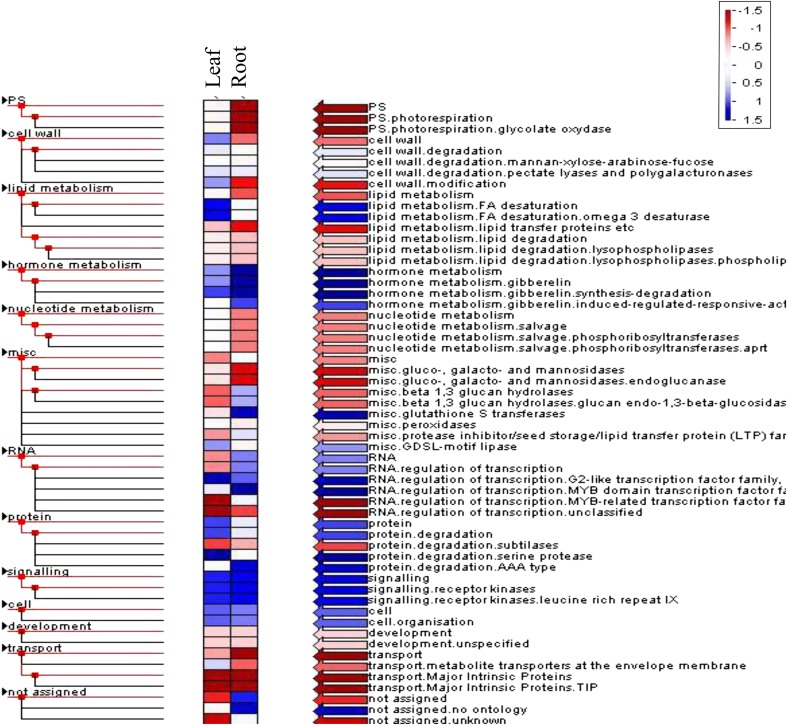
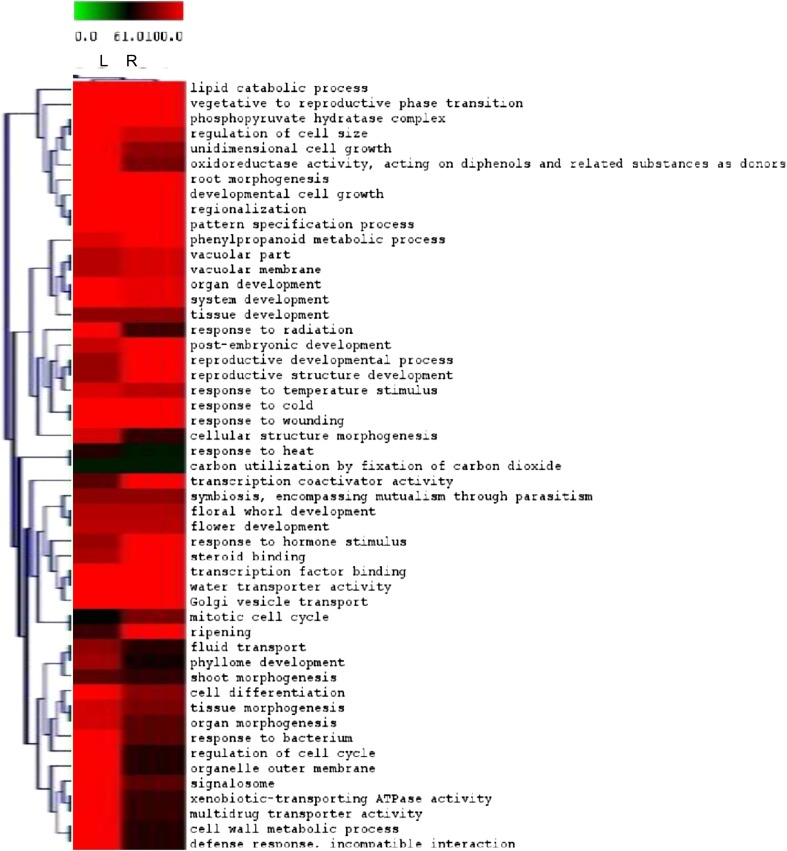
Among the cotton genes, 6,313 and 5,858 from leaves and roots, respectively, were either twofold up- or down-regulated with adjusted p ≤ 0.05. Together, 12,171 genes were either significantly up- or down-regulated under drought stress (Table 1). Thus 3,588 genes were differentially regulated in both leaves (1,528) and roots (1,128). However, various tissues behaved differently during the stress response. In terms of the number of genes, there appeared to be a balance between leaves and roots, while in terms of induction high number of genes was primarily observed in the root (see additional information 2). To have a general view of the whole picture, we grouped and classified them into Gene Ontology terms then biological processes were further examined. There were five significantly represented GOs in leaves while roots showed eight indicating the earlier response of roots. It is likely because of the faster dehydration on root represented by shorter root length (Table 2; Fig. 1e); in other words, roots are more water stressed than leaves. In the field, root also is the earlier organ under drought because plants uptake water through their roots. When soils become dry, roots sense this water deficit and produce chemical signals to transport to leaves via the xylems resulting in physiological changes in leaves (Arndt et al. 2001; Sengupta and Reddy 2011; Tuteja and Sopory 2008). Further we analyzed these genes by MapMan, This finding suggested that, the leaves differentially express genes in a more random/scattered manner that lead to less statically significant results. Compared to that, roots seemed to behave in a more focused response. Unlike the responses of the leaves, root tissue showed notable expression of stress related and protein signaling pathway (Fig. 2). The results suggest that, under drought stress the leaves try to maintain normal functional condition and re-direct to the whole system on stress response, whereas, root tissues became more active. The heat map analysis showed that the photosynthesis and related processes were among enhanced processes in the roots, while it was repressed in leaves, In particular, the simultaneous reduction in leaves and induction in root of photosynthesis as well as secondary metabolism suggests the possibility of biological processes functioning in an opposite and tissue-specific manner (Fig. 3).
Table 2.
Commonly regulated GO terms in leaf and root
| Differentially regulated GO terms in leaf and root | p value | ||
|---|---|---|---|
| GO annotation | GO terms | Leaf | Root |
| GO:0006950 | Response to stress | 0.0006 | 0.0058 |
| GO:0009725 | Response to hormone stimulus | 0.0002 | 0.0049 |
| GO:0009644 | Response to high light intensity | 0.0021 | 0.0009 |
| GO:0009408 | Response to heat | 0.0047 | 0.0012 |
| GO:0009266 | Response to temperature stimulus | 0 | 0 |
| GO:0009808 | Lignin metabolic process | 0 | 0.0017 |
| GO:0042221 | Response to chemical stimulus | 0 | 0 |
| GO:0006720 | Isoprenoid metabolic process | 0.029 | 0.0341 |
| GO:0009719 | Response to endogenous stimulus | 0.0358 | 0.0296 |
| GO:0009725 | Response to hormone stimulus | 0.0361 | 0.0122 |
| GO:0008610 | Response to lipid metabolism | 0.0359 | 0.00165 |
| GO:0009628 | Response to abiotic stimulus | 0.0053 | 0.002 |
| GO:0050896 | Response to stimulus | 0.0017 | 0.0014 |
| GO:0051716 | Cellular response to stimulus | 0.0021 | 0.0032 |
| GO:0032268 | Regulation of cellular protein metabolic process | 0.0029 | 0.0065 |
| GO:0009314 | Response to radiation | 0.0024 | 0.0067 |
| GO:0042221 | Response to chemical stimulus | 0.00032 | 0.0009 |
| GO:0042254 | Ribosome biogenesis | 0.004 | 0.001 |
| GO:0050896 | Response to stimulus | 0 | 0.008 |
| GO:0034470 | ncRNA processing | 0 | 0.002 |
| GO:0043412 | Biopolymer modification | 0 | 0 |
| GO:0043687 | Post-translational protein modification | 0 | 0 |
| GO:0006464 | Protein modification process | 0.0138 | 0.002 |
| GO:0006793 | Phosphorus metabolic process | 0 | 0 |
| GO:0006796 | Phosphate metabolic process | 0.0165 | 0.0114 |
| GO:0008152 | Metabolic process | 0.0113 | 0 |
| GO:0016072 | rRNA metabolic process | 0.0117 | 0 |
| GO:0006261 | DNA-dependent DNA replication | 0.0022 | 0 |
| GO:0006364 | rRNA processing | 0.013 | 0.0008 |
| GO:0006952 | Defense response | 0 | 0 |
| GO:0051704 | Multi-organism process | 0.0055 | 0 |
| GO:0007166 | Cell surface receptor linked signal transduction | 0.0054 | 0 |
| GO:0008283 | Cell proliferation | 0.0023 | 0.0062 |
Fig. 2.
MapMan analysis of differentially expressed genes in leaf and root
Fig. 3.
Heat maps of expression profiles of top ranked differentially expressed genes in leaf and root
Differentially regulated cellular metabolism in leaves and roots
To identify the stress affected biochemical network of cells, we retrieved Arabidopsis homologs of cotton genes from PLEXdb (http://www.plexdb.org/) and calculated the effected pathway using KOBAS. Table 3 shows the 28 biological pathways were commonly affected in roots and leaves and the majority of affected pathways were observed in leaves. Some pathways were significantly affected in both leaves and roots; they’re involved in energy production, biosynthesis of phytohormones, amino acids, pigments and many others. Not only did each tissue differently regulate genes within the shared pathway, they also possessed tissue specific reactions. Specifically, leaves showed strong induction of choline and trehalose biosyntheses when cellulose and initial step of fatty acid biosynthesis were negatively regulated only in root (Table 3).
Table 3.
Different biochemical pathways significantly affected under drought stress in leaf and root tissue
| Biological pathways | p value | |
|---|---|---|
| Leaf | Root | |
| Beta alanine metabolism | 0.022 | 0.033 |
| Histidine metabolism | 0.043 | 0.026 |
| Terpenoid backbone biosynthesis | 0.026 | 0.017 |
| Fatty acid metabolism | 0.041 | 0.027 |
| Flavonoid biosynthesis | 0.042 | 0.024 |
| Glycolysis/gluconeogenesis | 0.036 | 0.034 |
| Spliceosomes | 0.0003 | 0.048 |
| Ubiquitin mediated proteolysis | 0.029 | 0.0036 |
| Mevalonate pathways | 0.028 | 0.00001 |
| Tryptophan biosynthesis | 0.044 | 8.63E-06 |
| Carotenoid biosynthesis | 6.35E-06 | 0.000045 |
| Starch degradation | 0.026 | 0.00137 |
| IAA biosynthesis | 0.046 | 0.000039 |
| Ascorbate glutathione cycle | 0.018 | 0.0019 |
| Carotenoid biosynthesis | 0.023 | 0.024 |
| Sterol biosynthesis | 0.048 | 0.034 |
| Glycine degradation | 0.030 | 0.041 |
| Cysteine biosynthesis | 0.024 | 0.040 |
| Triacylglycerol degradation | 0.007 | 0.041 |
| Chlorophyll biosynthesis | 0.043 | 0.033 |
| Lignin biosynthesis | 0.033 | 0.036 |
| Choline biosynthesis | 0.011 | 0.014 |
| Trehalose biosynthesis | 0.022 | 0.008 |
| Asparagine degradation I | 0.024 | 0.046 |
| Calvin cycle | 0.00042 | 6.02E-07 |
| Fatty acid elongation saturated | 0.041 | 1.75E-06 |
| Threonine degradation | 0.0003 | 0.00012 |
| Cellulose biosynthesis | 0.024 | 2.43E-07 |
Unique and shared transcription factors in roots and leaves in response to drought stress
TFs are the main components to understand the complexity of expression of stress induced genes in their signaling network as suggested in various studies (Agarwal et al. 2006a, b; Ranjan et al. 2012b). In order to further investigate on these molecules, we determined the number of unique and shared TF’s expressed in leaves and roots using conserved domain database in NCBI. Among 29 groups that are affected by drought, three and six were uniquely expressed in leaves and roots, respectively. MybDNA-binding bHLH, HSF and WRKY were the most strongly affected groups even though the expressions were different in both the tissue. Transcription factors, such as WRKY 75, RAV, REM, CAMTA, SBP, NLP and G2 like, Orphan, TCP were uniquely expressed in root and leaf tissue, respectively (Table 4).
Table 4.
Differentially and uniquely expressed transcription factors in leaf and root under drought
| TFs | Differentially expressed | Uniquely expressed | ||
|---|---|---|---|---|
| Leaf | Root | Leaf | Root | |
| ARF | 9 | 4 | – | – |
| bHLH | 16 | 11 | – | – |
| bZIP | 3 | 5 | – | – |
| BZR | 6 | 2 | – | – |
| C2C2-like | 7 | 9 | – | – |
| C2C2-YABBY | 3 | 6 | – | – |
| C2C2 | 1 | 7 | – | – |
| C3H | 5 | 4 | – | – |
| CCAAT-HAP5 | 6 | 8 | – | – |
| EIL | 2 | 2 | – | – |
| G2-like | – | – | 1 | – |
| GRAS | 6 | 4 | – | – |
| GRF | 8 | 4 | – | – |
| Homeobox | 3 | 7 | – | – |
| HSF | 11 | 8 | – | – |
| MAKS | 7 | 4 | ||
| MYB | 19 | 13 | ||
| NAC | 5 | 2 | ||
| NLP | – | – | – | 4 |
| Orphan | – | – | 2 | – |
| SBP | – | – | – | 3 |
| TCP | 4 | 5 | – | – |
| WRKY | 12 | 9 | – | – |
| CAMTA | – | – | – | 7 |
| REM | – | – | – | 6 |
| TCP | – | – | 8 | – |
| RAV | – | – | – | 4 |
| WRKY75 | – | – | – | 8 |
Drought enhanced the expression of photosynthesis gene in root
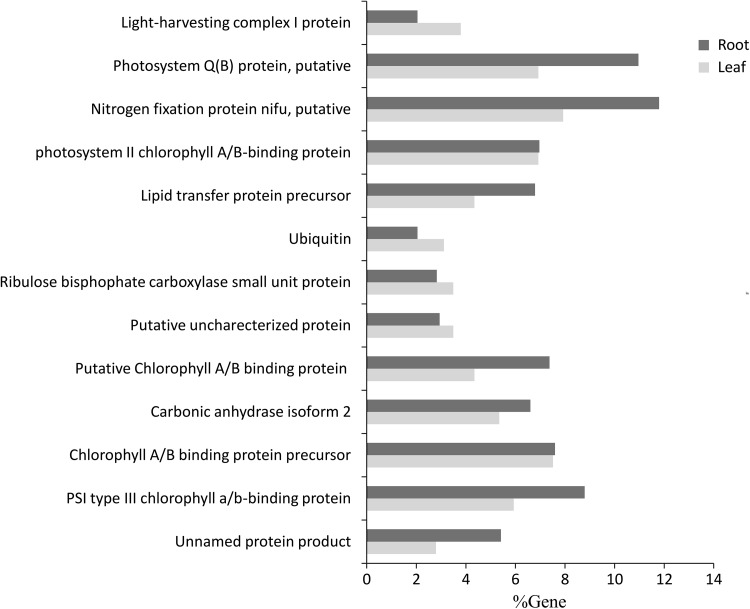
In general the photosynthesis related genes are expressed at a lower level in roots as compared to the expression in leaves. However, in this study the expression pattern is very different. Photosynthesis genes that are encoded separately in plastid and nuclear genome are responsible for photosynthesis core subunits, and light harvesting complex proteins and pigment biosynthesis; respectively (Gutierrez-Nava et al. 2004; Kino-oka et al. 2001). The genes related to chlorophyll a/b binding protein and photosystem-related proteins showed significant higher expression in roots and as compared to leaves (Fig. 4). It is well known that light and plastid development are both crucial for the expression of nuclear photosynthesis genes. Although in our analysis no biological process or biological pathways shows plastid related biosynthesis in root, we found that biological pathway related to “ribulose-1,5-bisphosphate carboxylase” (RuBisCO) was significantly high in leaves.
Fig. 4.
Graphical comparison of photosynthesis related genes in leaf and root
Defense genes dominantly expressed in roots as compared to leaves
Through long-term evolution and adaptation to extreme conditions, cotton has been found to be rich in resistance genes for abiotic stresses (Wendel and Cronn 2003). When cotton plants face water stress, due to the change in external environment, plants are able to activate a series of mechanisms to respond drought stress (Park et al. 2012; Ranjan et al. 2012a, b). Cells recognize external stresses and induce many signal proteins, including signaling receptor kinases, calcium-dependent protein kinases and G-proteins. In our results showed that the transcripts of these signal proteins accumulated in both tissues during drought stress. Genes for peroxidase, glutathione peroxidase, redox metabolism and superoxide dismutase, were predominantly expressed in root tissue (Table 5). These genes are involved in scavenging reactive oxygen species (ROS), and play a pivotal role as triggers of gene expression during abiotic stresses (Miller et al. 2010; Ranjan et al. 2012b).
Table 5.
Differentially expressed antioxidant genes in leaf and root tissues
| Contigs represent antioxidant genes | Contigs represent number of expressed contigs | Description | |
|---|---|---|---|
| Root | Leaf | ||
| Contig00283 | 305 | 0 | Peroxidase |
| Contig00097 | 74 | 60 | Peroxisomal enoyl-CoA hydratase |
| Contig00254 | 7 | 9 | Peroxiredoxin |
| Contig00468 | 9 | 20 | Cytoplasmic Cu/ZnSOD |
| Contig00520 | 5 | 16 | Glutamate synthase |
| Contig00525 | 11 | 1 | Metallothionein-like type 1 protein |
| Contig00718 | 1 | 21 | Cu/Zn superoxide dismutase |
| Contig00763 | 2 | 9 | Peroxiredoxin |
| Contig00867 | 8 | 2 | Glutathione S-transferase |
| Contig01025 | 10 | 5 | Glutathione S-transferase |
| Contig01228 | 8 | 0 | Peroxidase 17 |
| Contig01314 | 20 | 7 | Superoxide dismutase |
| Contig01348 | 22 | 0 | Superoxide dismutase |
| Contig01685 | 26 | 0 | Glutathione reductase |
| Contig01694 | 6 | 0 | Sulfite oxidase |
| Contig01756 | 5 | 1 | Thioredoxin H7 |
| Contig01852 | 8 | 3 | Catalase |
| Contig01916 | 2 | 4 | Peroxisomal targeting signal type 1 |
| Contig02183 | 3 | 3 | Ascorbate peroxidase |
| Contig02352 | 9 | 9 | Ascorbate peroxidase |
| Contig02464 | 1 | 2 | Glutathione S-transferase |
| Contig02486 | 2 | 0 | Glutathione S-transferase U1 |
| Contig02488 | 13 | 1 | Ascorbate peroxidase |
| Contig02585 | 1 | 1 | Lipoxygenase |
| Contig00114 | 95 | 0 | Superoxide dismutase |
| Contig00697 | 26 | 0 | Glutathione peroxidase |
| Contig03151 | 17 | 0 | Glutathione peroxidase |
| Contig02529 | 9 | 0 | Catalase |
Discussion
The advent of pyrosequencing has enabled to obtain the transcriptional changes of entire genome of a particular plant or specific plant organs in response to various stresses. However, the whole genome sequences are still not available for G. herbaceum species of cotton, leading to a dependence on mRNA sequences by pyrosequencing or collections of ESTs assembled from random cDNA libraries. In this study, we used the 454 pyrosequencing platform for transcriptome sequencing of cotton (G. herbaceum) leaf and root tissues. It was observed that GujCot-21 and RAHS-14 showed substantial differences in root growth under osmotic stress (Fig. 1a, b) and leaf senescence under drought stress condition (Fig. 1c, f). The leaf and root both tissues shared some common gene expression during stress, with the purpose of enhancing protective systems. However, these two tissues appeared to respond differently to drought conditions. Leaves induced more genes than root but those genes were scattered in many processes, most significantly were productions of osmoprotectant and senescence associated genes. The genes expressed in roots mainly associated with antioxidant and photosynthesis related pathways (Table 5; Fig. 4). Chen et al. (2013) recently reported down regulated expression of chloroplast-/plastid-related genes in leaf tissue of drought tolerant cultivar of cotton as compare to drought sensitive cultivar, while Bowman et al. (2013), observed higher expression of genes in root associated with starch and sugar metabolism is similar to our results. Besides, the two tissues can affect each other via the signaling and transportation system. Through long-term selection and adaptation to extreme conditions, cotton has been found to be rich in resistance genes for a range of abiotic stresses, including drought, high temperature and salinity (Ranjan et al. 2012a; Zhou et al. 2014). Chen et al. (2013) results also suggested that high number of genes are expressed temporally and specifically in plant response to drought treatment.
Transcription regulation plays a central role in stress signal transduction pathways. In this study, we found 29 group of transcription factor genes differentially regulated at different levels in roots and leaves in response to drought stress. Among these, TF genes some were unique to leaf and root tissues. This finding reveals that a large group of TF genes are involved in the transcription regulation in response to drought stress in leaves and roots in a specific manner when cotton plant face drought stress. Our study also showed that a large proportion (289 in leaves and 278 in roots) of expressed unigenes have no functional annotation or have unknown function. It is also noteworthy that many of these unigenes showed remarkable expression changes in response to drought stress, especially with some having a unique expression in leaf and root tissues. Our results confirm differential expression of transcription factors in response to water-deficit stress, as has been observed in many plant species, including cotton (Park et al. 2012, Bowman et al. 2013). Therefore, it is possible to identify some drought responsive genes unique to cotton. Further analysis of the functions and expression controlling mechanism of these genes in cotton would not only supply the opportunity of isolation and identification of novel genes, but also enhance our further understanding of specific mechanism of drought tolerance in cotton.
Conclusions
To date, a limited number of studies on drought stress mediated gene expression in cotton have been reported. In this study we described an analysis of gene expression in cotton leaves and roots in response to drought stress and intended to carry out a comparative transcript profiling between the leaves and roots. We focused on a set of transcripts that exhibited unique expression in leaves and roots of cotton under drought stress. This is the first trial to comprehensively explore the genome-wide gene expression patterns in leaf and root of drought responsiveness in cotton. Our results show that cotton leaves are distinct from roots in terms of molecular mechanisms for responses to drought stress.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (XLSX 1935 kb). Additional information-1 Blast detail of leaf and root contigs
Supplementary material 2 (XLSX 637 kb). Additional information-2 Uniquely and differentially expressed genes in leaf and root tissue
Acknowledgments
We thank the Council of Scientific and Industrial Research (CSIR) India for supporting research grant.
Conflict of interest
No conflicts of interest.
References
- Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem. 2006;281(49):37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- Agarwal PK, Agarwal P, Reddy MK, Sopory SK. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006;25(12):1263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- Arndt SK, Clifford SC, Wanek W, Jones HG, Popp M. Physiological and morphological adaptations of the fruit tree Ziziphus rotundifolia in response to progressive drought stress. Tree Physiol. 2001;21(11):705–715. doi: 10.1093/treephys/21.11.705. [DOI] [PubMed] [Google Scholar]
- Bowman MJ, Park W, Bauer PJ, Udall JA, Page JT et al (2013) RNA-seq transcriptome profiling of upland cotton (Gossypium hirsutum L.) root tissue under water-deficit stress. PLoS ONE 8(12):e82634 [DOI] [PMC free article] [PubMed]
- Caldeira CF, Bosio M, Parent B, Jeanguenin L, Chaumont F, Tardieu F (2014) A hydraulic model is compatible with rapid changes in leaf elongation rate under fluctuating evaporative demand and soil water status. Plant Physiol 164:1718–1730 [DOI] [PMC free article] [PubMed]
- Chen Y, Liu Z-H, Feng L, Zheng Y, Li D-D et al (2013) Genome-wide functional analysis of cotton (Gossypium hirsutum) in response to drought. PLoS ONE 8(11):e80879 [DOI] [PMC free article] [PubMed]
- Cohen D, Bogeat-Triboulot MB, Tisserant E, Balzergue S, Martin-Magniette ML, Lelandais G, Ningre N, Renou JP, Tamby JP, Le Thiec D, Hummel I. Comparative transcriptomics of drought responses in Populus: a meta-analysis of genome-wide expression profiling in mature leaves and root apices across two genotypes. BMC Genom. 2010;11:630. doi: 10.1186/1471-2164-11-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias AP, Brown J, Bonello P, Grotewold E. Metabolite profiling as a functional genomics tool. Methods Mol Biol. 2003;236:415–426. doi: 10.1385/1-59259-413-1:415. [DOI] [PubMed] [Google Scholar]
- Garcia-Alcalde F, Garcia-Lopez F, Dopazo J, Conesa A. Paintomics: a web based tool for the joint visualization of transcriptomics and metabolomics data. Bioinformatics. 2011;27(1):137–139. doi: 10.1093/bioinformatics/btq594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Nava Mde L, Gillmor CS, Jimenez LF, Guevara-Garcia A, Leon P. Chloroplast biogenesis genes act cell and noncell autonomously in early chloroplast development. Plant Physiol. 2004;135(1):471–482. doi: 10.1104/pp.103.036996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T. A genetic regulatory network in the development of trichomes and root hairs. Ann Rev Plant Biol. 2008;59:365–386. doi: 10.1146/annurev.arplant.59.032607.092949. [DOI] [PubMed] [Google Scholar]
- Jansen L, Hollunder J, Roberts I, Forestan C, Fonteyne P, Van Quickenborne C, Zhen RG, McKersie B, Parizot B, Beeckman T. Comparative transcriptomics as a tool for the identification of root branching genes in maize. Plant Biotechnol J. 2013;11(9):1092–1102. doi: 10.1111/pbi.12104. [DOI] [PubMed] [Google Scholar]
- Kino-oka M, Nagatome H, Taya M. Characterization and application of plant hairy roots endowed with photosynthetic functions. Adv Biochem Eng Biotechnol. 2001;72:183–218. doi: 10.1007/3-540-45302-4_7. [DOI] [PubMed] [Google Scholar]
- Libault M, Farmer A, Brechenmacher L, Drnevich J, Langley RJ, Bilgin DD, Radwan O, Neece DJ, Clough SJ, May GD, Stacey G. Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 2010;152(2):541–552. doi: 10.1104/pp.109.148379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. Root architecture and plant productivity. Plant Physiol. 1995;109(1):7–13. doi: 10.1104/pp.109.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010;33(4):453–467. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- Milner MJ, Mitani-Ueno N, Yamaji N, Yokosho K, Craft E, Fei Z, Ebbs S, Zambrano MC, Ma JF, Kochian LV (2014) Root and shoot transcriptome analysis of two ecotypes of Noccaea caerulescens uncovers the role of NcNramp1 in Cd hyperaccumulation. Plant J 78:398–410 [DOI] [PubMed]
- Naika M, Shameer K, Sowdhamini R. Comparative analyses of stress-responsive genes in Arabidopsis thaliana: insight from genomic data mining, functional enrichment, pathway analysis and phenomics. Mol BioSyst. 2013;9(7):1888–1908. doi: 10.1039/c3mb70072k. [DOI] [PubMed] [Google Scholar]
- Narsai R, Castleden I, Whelan J. Common and distinct organ and stress responsive transcriptomic patterns in Oryza sativa and Arabidopsis thaliana. BMC Plant Biol. 2010;10:262. doi: 10.1186/1471-2229-10-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003;34(2):137–148. doi: 10.1046/j.1365-313X.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- Parent B, Suard B, Serraj R, Tardieu F. Rice leaf growth and water potential are resilient to evaporative demand and soil water deficit once the effects of root system are neutralized. Plant Cell Environ. 2010;33(8):1256–1267. doi: 10.1111/j.1365-3040.2010.02145.x. [DOI] [PubMed] [Google Scholar]
- Park W, Scheffler BE, Bauer PJ, Campbell BT. Genome-wide identification of differentially expressed genes under water deficit stress in upland cotton (Gossypium hirsutum L.) BMC Plant Biol. 2012;12:90. doi: 10.1186/1471-2229-12-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, et al. Repeated polyploidization of gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492:423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- Payton P, Kottapalli KR, Kebede H, Mahan JR, Wright RJ, Allen RD. Examining the drought stress transcriptome in cotton leaf and root tissue. Biotechnol Lett. 2011;33(4):821–828. doi: 10.1007/s10529-010-0499-y. [DOI] [PubMed] [Google Scholar]
- Ranjan A, Nigam D, Asif MH, Singh R, Ranjan S, Mantri S, Pandey N, Trivedi I, Rai KM, Jena SN, Koul B, Tuli R, Pathre UV, Sawant SV. Genome wide expression profiling of two accession of G. herbaceum L. in response to drought. BMC Genom. 2012;13:94. doi: 10.1186/1471-2164-13-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A, Pandey N, Lakhwani D, Dubey NK, Pathre UV, Sawant SV. Comparative transcriptomic analysis of roots of contrasting gossypium herbaceum genotypes revealing adaptation to drought. BMC Genom. 2012;13:680. doi: 10.1186/1471-2164-13-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Blanco MJ, Alvarez S, Navarro A, Banon S. Changes in leaf water relations, gas exchange, growth and flowering quality in potted geranium plants irrigated with different water regimes. J Plant Physiol. 2009;166(5):467–476. doi: 10.1016/j.jplph.2008.06.015. [DOI] [PubMed] [Google Scholar]
- Sengupta D, Reddy AR. Water deficit as a regulatory switch for legume root responses. Plant Signal Behav. 2011;6(6):914–917. doi: 10.4161/psb.6.6.15340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58(2):221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- Smith S, De Smet I. Root system architecture: insights from arabidopsis and cereal crops. Philos Trans R Soc Lond B Biol Sci. 2012;367(1595):1441–1452. doi: 10.1098/rstb.2011.0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi I, Ranjan A, Sharma YK, Sawant S. The histone H1 variant accumulates in response to water stress in the drought tolerant genotype of Gossypium herbaceum L. Protein J. 2012;31(6):477–486. doi: 10.1007/s10930-012-9425-6. [DOI] [PubMed] [Google Scholar]
- Tuteja N, Sopory SK. Chemical signaling under abiotic stress environment in plants. Plant Signal Behav. 2008;3(8):525–536. doi: 10.4161/psb.3.8.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Blasing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, Fernie AR, Stitt M. Pageman: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinform. 2006;7:535. doi: 10.1186/1471-2105-7-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valliyodan B, Nguyen HT. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr Opin Plant Biol. 2006;9(2):189–195. doi: 10.1016/j.pbi.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Wendel JF, Cronn RC. Polyploidy and the evolutionary history of cotton. Adv Agron. 2003;78:139–186. doi: 10.1016/S0065-2113(02)78004-8. [DOI] [Google Scholar]
- Xiao X, Tang C, Fang Y, Yang M, Zhou B, Qi J, Zhang Y. Structure and expression profile of the sucrose synthase gene family in the rubber tree: indicative of roles in stress response and sucrose utilization in the laticifers. FEBS J. 2014;281(1):291–305. doi: 10.1111/febs.12595. [DOI] [PubMed] [Google Scholar]
- You J, Zhang H, Liu N, Gao L, Kong L, Yang Z. Transcriptomic responses to aluminum stress in soybean roots. Genome. 2011;54(11):923–933. doi: 10.1139/g11-060. [DOI] [PubMed] [Google Scholar]
- Yue G, Zhuang Y, Li Z, Sun L, Zhang J. Differential gene expression analysis of maize leaf at heading stage in response to water-deficit stress. Biosci Rep. 2008;28(3):125–134. doi: 10.1042/BSR20070023. [DOI] [PubMed] [Google Scholar]
- Zhou M, Sun G, Sun Z, Tang Y, Wu Y (2014) Cotton proteomics for deciphering the mechanism of environment stress response and fiber development. J Proteomics 105:74–84 [DOI] [PubMed]
- Zhu YN, Shi DQ, Ruan MB, Zhang LL, Meng ZH, Liu J, Yang WC. Transcriptome analysis reveals crosstalk of responsive genes to multiple abiotic stresses in cotton (Gossypium hirsutum L.) PLoS ONE. 2013;8(11):e80218. doi: 10.1371/journal.pone.0080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 (XLSX 1935 kb). Additional information-1 Blast detail of leaf and root contigs
Supplementary material 2 (XLSX 637 kb). Additional information-2 Uniquely and differentially expressed genes in leaf and root tissue