Significance
This work provides direct evidence of evolutionary rewiring of gene-regulatory circuitry accompanying divergence of two subclasses of echinoderm, the cidaroid and euechinoid sea urchins. These forms descend from a known common Paleozoic ancestor, and their embryos develop differently, offering an opportunity to probe the basic evolutionary process by which clade divergence occurs at the gene-regulatory network (GRN) level. We carried out a systematic analysis of the use of particular genes participating in embryonic skeletogenic cell specification, building on an established euechinoid developmental GRN. This study revealed that the well-known and elegantly configured regulatory circuitry that underlies skeletogenic specification in modern sea urchins is largely a novel evolutionary invention. The results dramatically display extensive regulatory changes in a specific developmental GRN, underlying an incidence of cladistic divergence at the subclass level.
Keywords: GRN evolution, network linkages, embryonic skeletogenesis, sea urchin embryogenesis
Abstract
Evolution of animal body plans occurs with changes in the encoded genomic programs that direct development, by alterations in the structure of encoded developmental gene-regulatory networks (GRNs). However, study of this most fundamental of evolutionary processes requires experimentally tractable, phylogenetically divergent organisms that differ morphologically while belonging to the same monophyletic clade, plus knowledge of the relevant GRNs operating in at least one of the species. These conditions are met in the divergent embryogenesis of the two extant, morphologically distinct, echinoid (sea urchin) subclasses, Euechinoidea and Cidaroidea, which diverged from a common late Paleozoic ancestor. Here we focus on striking differences in the mode of embryonic skeletogenesis in a euechinoid, the well-known model Strongylocentrotus purpuratus (Sp), vs. the cidaroid Eucidaris tribuloides (Et). At the level of descriptive embryology, skeletogenesis in Sp and Et has long been known to occur by distinct means. The complete GRN controlling this process is known for Sp. We carried out targeted functional analyses on Et skeletogenesis to identify the presence, or demonstrate the absence, of specific regulatory linkages and subcircuits key to the operation of the Sp skeletogenic GRN. Remarkably, most of the canonical design features of the Sp skeletogenic GRN that we examined are either missing or operate differently in Et. This work directly implies a dramatic reorganization of genomic regulatory circuitry concomitant with the divergence of the euechinoids, which began before the end-Permian extinction.
The mechanisms responsible for evolutionary divergence of animal body plans, as so extensively documented in the Phanerozoic fossil record, lie in alterations of the encoded genomic regulatory programs that direct development. This principle has long been evident a priori (1), and overwhelmingly, accumulating current evidence precludes any other general explanation (2). However, it still remains a challenge to adduce specific examples in which evolutionary rewiring of developmental gene-regulatory networks (GRNs) can be seen to account for observed differences in morphogenetic processes that distinguish descendants of a common ancestor. Knowledge of developmental GRNs remains insufficiently extensive, and it is not trivial to locate useful examples, which require comparison within a monophyletic clade at just sufficient distance so that the diverged morphology is clearly the output of homologous networks of developmental regulatory gene interactions.
In recent years, largely complete developmental GRN models have been solved that causally explain spatial specification in large domains of the embryo of the sea urchin Strongylocentrotus purpuratus (Sp), up to gastrulation (3–5). The explanatory power of these networks was demonstrated, in these pages, by a predictive computational analysis that showed that they contain sufficient information to regenerate the developmental course of events in silico, in automaton-like fashion (6). The present work stems from the almost irresistible opportunities that these same GRNs offer for approaching the basic evolutionary mechanisms of GRN divergence. Thus, here we focus on a sea urchin clade that descends from a common ancestor with Sp, but in which embryonic structures are generated differently from those to which the known GRNs pertain.
Sea urchins (class Echinoidea) are one of the five extant classes of echinoderms (the others are sea stars, brittle stars, sea cucumbers, and crinoids), and for more than a century, their embryos have served as major model systems for the study of early development; the initial high point was Boveri’s 1902–1908 demonstration that a complete set of chromosomes is required in every nucleus of the sea urchin embryo for embryonic development to work properly (7, 8). These and almost all subsequent experimental studies on sea urchin embryos, including all of the recent GRN analyses cited, have been carried out on species belonging to one of the two subclasses of sea urchins surviving in the post-Paleozoic world, the Euechinioidea. Relatively little is known of any aspect of developmental mechanism in their sister group, the subclass Cidaroidea. Although, as we briefly summarize below, the common Paleozoic ancestry of these echinoid subclasses is unequivocal, euechinoid and cidaroid sea urchins differ canonically in aspects of their body test plate organization and in other adult skeletal structures that develop in the juvenile immediately after morphogenesis (9). During embryogenesis, both euechinoid and cidaroid embryos produce geometrical systems of larval skeletal rods, displaying species-specific morphology. The skeleton provides the postembryonic echinoid larva with internal structural support and with mounting for the ciliated anterior larval arms that aid in motility and feeding. However, a striking distinction between cidaroid and euechinoid modes of embryonic skeletogenesis early on drew the attention of embryologists, in that the embryonic skeletons arise very differently. In euechinoids, four skeletogenic founder cells (large micromeres) segregate from all other fates near the very beginning of development, at fifth cleavage, and all descendants of these four vegetal pole cells exclusively execute skeletogenic specification and differentiation, according to a rigidly hierarchical, encoded network of regulatory gene interactions (4). In Sp embryos the cells of this lineage actively express skeletogenic genes during cleavage and blastulation (4). They divide exactly three times during this period, and then, well before gastrular invagination of the archenteron, they singly ingress into the blastocoel and divide one last time, and on the basis of internal ectodermal signal cues, they arrange themselves spatially within the blastocoel, form a syncytium, and progressively construct the skeleton during the remainder of embryogenesis (10–12). However, in cidaroid sea urchin embryos no precocious ingression of a skeletogenic micromere lineage occurs before gastrulation (13, 14). A variable number of micromeres, individual to individual, is formed at the vegetal pole early in cleavage. However, their ultimately skeletogenic descendants only emerge well after gastrulation is under way, together with a cloud of other mesodermal derivatives, by delamination from the tip of the midgastrular archenteron. As we see below, in cidaroid embryos specifically skeletogenic molecular functions are not transcriptionally executed in micromere descendants during cleavage. After emergence from the archenteron tip, the mesenchymal skeletogenic cells of cidaroid embryos migrate to the ectoderm and, late in embryonic development, proceed to construct the larval skeleton. We show here that the distinction in the mode of developmental origin of the larval skeleton in euechinoid vs. cidaroid embryogenesis is anything but a trivial heterochrony; rather, it is the morphological tip of an iceberg of fundamentally distinct GRN architecture.
The extant echinoderm classes were established in the Ordovician, if not earlier, and in major aspects of their body plans they have exemplified evolutionary stasis of definitive character suites for the ensuing 430 million years (my) (2, 15). For echinoids as a whole, these features include the globular test form and developmental rearrangements of the coeloms resulting in a stacked configuration in the juvenile (16, 17). Within these constraints, the fossil record displays a remarkable variety of early Paleozoic echinoid morphology. However, in the late Paleozoic, there arose an echinoid branch that is clearly ancestral to both the modern euechinoid and cidaroid subclasses, known as the archaeocidaroid lineage (18). A new high-resolution paleontological analysis (19) indicates that the last common archaeocidarid ancestor of both modern echinoid subclasses existed at the latest ∼268 my ago—i.e., at least 16 my before the Permian/Triassic extinction event, which terminated the Paleozoic and many of its canonical denizens. Since the Triassic, a curious and perhaps profound difference in evolutionary flexibility distinguishes euechinoid and cidaroid subclasses. The euechinoids have radiated prodigiously, diversifying into nearly 1,000 species of highly various morphology, whereas the cidaroids, comprised of only ∼100 species, have retained extremely conservative morphologies seemingly not far removed from their ancestral forms (18, 20). For example, during the Mesozoic the euechinoids evolved diverse clades displaying irregular morphology, such as sand dollars and heart urchins, whereas no such deviations from the ancestral symmetrical globular form have arisen in the cidaroid subclass. This fact generally biases the likelihood that novel features arising since divergence occurred in the euechinoid rather than the cidaroid lineage. Nonetheless, both subclasses display evolutionary innovations—i.e., subclass-specific, shared derived characters (apomorphies) with respect to the (fossilized) skeletal characters of their archaeocidarid ancestor, just as both display plesiomorphic morphological characters (9, 18, 19, 21).
Our experimental object was to pry open the genomic program innovations that underlie observed phenomenological distinctions in embryonic skeletogenesis between euechinoids and cidaroids. To approach this problem systematically, we carried out a large-scale investigation of developmental regulatory gene use in the embryonic endomesoderm of the cidaroid Eucidaris tribuloides (Et) (results from comparing development of endoderm and nonskeletogenic mesoderm in these embryos are reported separately). Et is the same species in which embryonic skeletogenic morphogenesis had been studied earlier (13, 14), and in which juvenile skeletogenesis was also investigated in our laboratory (9). We experimentally interrogated the Et skeletogenic specification system to determine the presence or absence of multiple distinct GRN circuit features that contribute decisively to embryonic skeletogenesis in Sp. Many relevant genes from the authenticated Sp skeletogenic specification GRN (4) were investigated, of which five essential participants are reported on in the following. These are the regulatory genes at the very top of the skeletogenic GRN hierarchy in Sp, the deployment of which our earlier work (22) predicted might have been the locus of the evolutionary changes that mobilized the skeletogenic network in the micromere lineage of the euechinoids. Experiments on another cidaroid species (23) have already cast doubt on the presence of one key component of this circuitry, the repressive paired box gene pmar1, which functions in a double negative transcriptional gate at the top of the skeletogenic GRN of Sp (4, 24). As described below, we show here that the pmar1 gene is indeed apparently not represented in the genome or in transcriptomes of Et. However, this turns out to be but one probably derivative feature of a very generally different regulatory architecture. The complete structure of the Et skeletogenic GRN is still a work in progress. The present study is more narrowly focused on evidence for evolutionary rewiring in this circuitry, which must have taken place following the separation of the surviving echinoid clades >260 my ago.
Results
Spatial Expression of Five Key Genes of the Euechinoid Skeletogenic GRN in Et Embryos.
Initial observations indicated a surprising lack of congruence between Sp and Et in the spatial domains of expression of four regulatory genes (i.e., genes encoding transcription factors) and of an essential signaling gene. These genes are of particular interest because of the important roles they play in the skeletogenic specification GRN of Sp. Even though their embryonic behavior is completely different from those of the skeletogenic micromere precursors of euechinoids, it had been shown by Wray and McClay (13) that the micromeres appearing early in Et cleavage do ultimately give rise to the postgastrular skeletogenic cells of this embryo. Thus, we could directly study expression of genes of the euechinoid GRN in known skeletogenic precursors of the cidaroid Et. We note here that the behavior of early ingressing skeletogenic micromeres of Sp is typical of many euechinoids, as supported by numerous observations on several different euechinoid species, both at morphological and molecular levels.
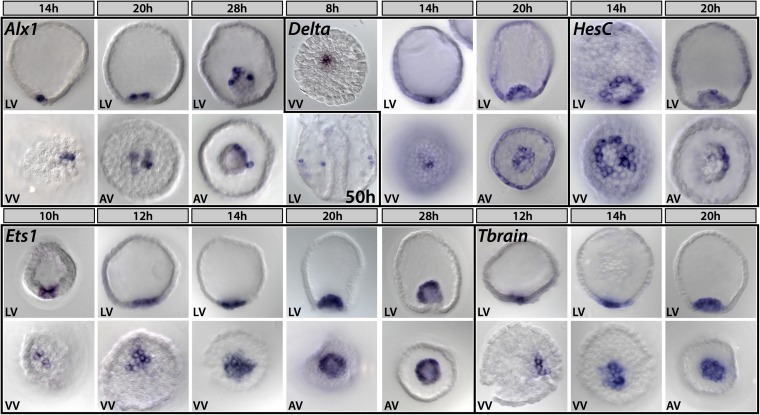
Detailed spatial expression of the genes reported on here had not previously been studied in Eucidaris embryos, and the whole-mount in situ hybridizations (WMISHs) of Fig. 1 provide an important baseline for consideration of their skeletogenic (or antiskeletogenic) functions. Each of the five genes was expressed differently in Et than would have been expected from the euechinoid examples.
Fig. 1.
Spatial expression of selected skeletogenesis genes in Et Alx1 expression is restricted to skeletogenic precursors throughout development; by 50 h, alx1-positive cells are seen migrating to the vegetal lateral clusters, where they will synthesize the larval skeleton. Delta is first expressed in micromere-descendants before hatching and is restricted to this lineage until late blastula stage (20 h), where it is expressed in scattered cells at the tip of the archenteron. Zygotic expression of hesC begins in a ring of cells that abut the micromere-descendants at the vegetal pole; by early gastrula, it is asymmetrically expressed in the archenteron [20 h, tip of archenteron/apical view (AV)]. Expression of ets1 begins in a few cells at the vegetal pole before hatching and expands to demarcate the whole mesodermal domain, eventually occupying the entire mesodermal bulb by early gastrula stage. Onset of zygotic tbrain expression occurs at the vegetal pole shortly after that of ets1, to which it exhibits very similar spatial expression. h, hours after fertilization; LV, lateral view; VV, vegetal view.
alx1.
The alx1 gene is a primary driver of skeletogenic specification and differentiation in sea urchin embryo and adult development, and it is a member of a family of homeodomain genes also used in vertebrate skeletogenesis (22, 25–27). In euechinoids alx1 is one of the initial set of positively acting transcriptional regulators that set up the skeletogenic regulatory state, and it is transcriptionally activated by a double-negative derepression subcircuit (24, 28), immediately upon segregation of the skeletogenic micromere founder cells early in cleavage (25). In Sp, this gene then participates in direct cross-regulation of the succeeding tiers of the skeletogenic specification GRN. However, it is immediately apparent that these features of alx1 regulation are not likely to exist in Et. Thus, in Et, alx1 is not even transcribed significantly until 4–6 h after the earliest micromere-specific genes are activated, (delta and ets1; Fig. 1 and Fig. S1), although alx1 expression is thereafter confined to micromeres and their skeletogenic descendants. Expression of the Et alx1 gene is, however, ultimately required for postgastrular skeleton formation to occur (Fig. 2), just as it is required for postembryonic skeletogenesis in both sea urchins and sea stars (22).
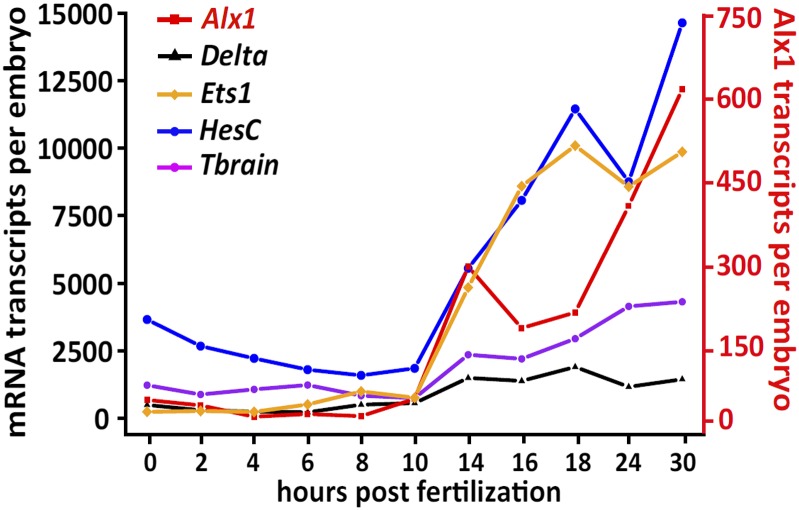
Fig. S1.
Time course of mRNA expression of Sp double-negative gate genes in this study. The left ordinate shows the estimated number of transcripts per embryo as determined by QPCR for delta, ets1, hesC, and tbrain. The right ordinate (shown in red) is estimated transcripts per embryo for alx1 only. The x axis shows the time points in development assayed, starting at fertilized egg (0 hpf) and ending when skeletogenic mesenchyme ingresses at early gastrula stage (30 hpf). A protocol for estimating the number of transcripts per embryo is described in SI Materials and Methods.
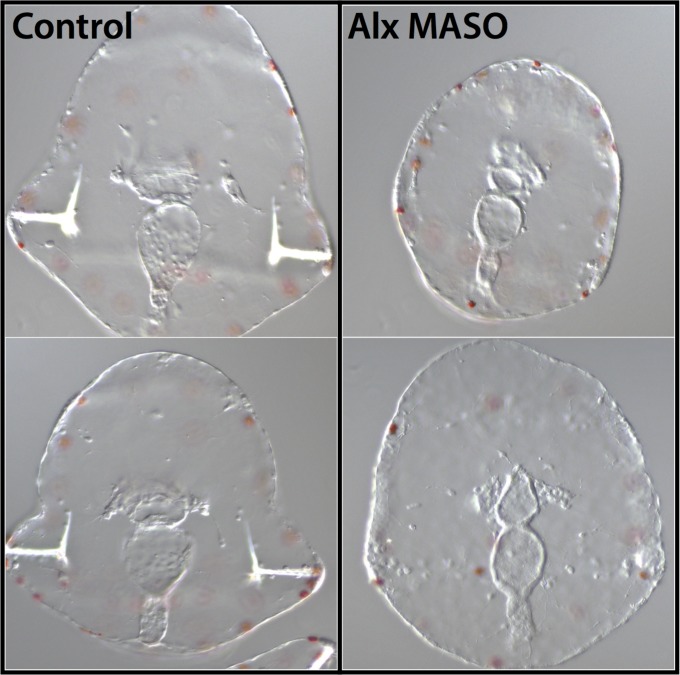
Fig. 2.
MASO perturbation of Alx1 disrupts skeletogenesis in Et larvae. Zygotes were injected with alx MASO, cultured for 5 d, and scored for the presence or absence of larval skeletal rods (11 of 16 lacked skeleton). Uninjected control groups were cultured and scored simultaneously (0 of 20 lacked skeleton).
delta.
The euechinoid delta gene is also an immediate transcriptional activation target of the Sp micromere double-negative gate subcircuit, and it continues to be expressed in this lineage until blastula stage, when its expression is extinguished there and instead appears in the surrounding nonskeletogenic mesoderm cells (24, 29, 30). In Et, delta expression occurs early in micromeres, 4–6 h before that of alx1, suggesting a primary function unconnected to later skeletogenic specification (Fig. 1). Expression of Et delta does not become nonskeletogenic until much later, when a complex pattern of ectodermal expression is installed (Fig. S2).
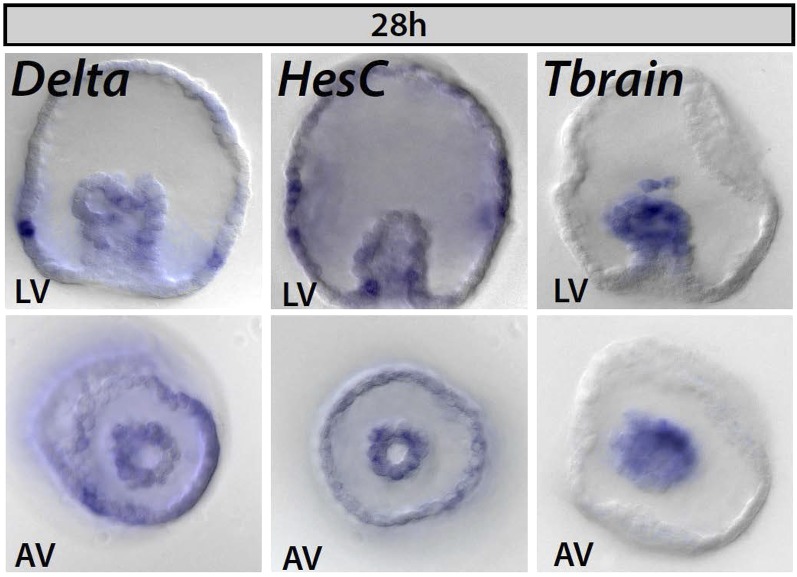
Fig. S2.
Spatial expression of delta, hesC, and tbrain at early gastrula stage in Et. Delta is observed in a few mesodermal cells at this stage and begins to be expressed in a scattered pattern in the ectoderm. HesC is expressed in the nonskeletogenic mesoderm and endoderm, is absent from the ecto-endodermal boundary, and is expressed in a diffuse, nonspecific pattern in the ectoderm. Tbrain expression is seen throughout the mesoderm and here can be seen in the ingressing spicule precursor cells at the tip of the archenteron.
hesC.
The most dramatically different functional implications revealed by Fig. 1 are to be seen in the expression in Et of the hesC gene. In the Sp GRN, HesC is the repressor controlling the initial skeletogenic regulatory state (i.e., including expression of alx1, delta, and ets1), and this state is controlled spatially by the transcriptional activity of the hesC gene. In the Sp GRN, the skeletogenic regulatory state is installed in micromeres by specific repression of the repressive hesC gene, executed by the micromere-specific repressor Pmar1, thus opening the double-negative gate subcircuit. HesC is transcriptionally expressed throughout the whole Sp embryo, except where this gene is turned off by pmar1 expression in the micromeres; thus, in Sp, hesC transcription and skeletogenic function are Boolean exclusives (4, 24, 31). However, in Et, hesC is expressed in micromeres at the same time as are ets1 and delta (by 10 h), in direct contrast to its double-negative gate function in Sp. Furthermore, the hesC gene is never vigorously expressed throughout the whole Et embryo as it is in Sp, and instead is strongly expressed (by blastula stage) only in the immediately surrounding nonskeletogenic mesoderm, as we see in more detail below.
ets1.
Zygotic transcription of ets1 is turned on as the double-negative gate is unlocked in early cleavage in the Sp GRN, and thereafter this gene provides powerful positive inputs to both regulatory and effector genes in skeletogenic specification, far into development (26, 28, 29). In Sp, there is also a prevalent store of maternal ets1 mRNA, but this is entirely missing in Et. As in Sp, the ets1 gene is activated as early in the micromeres as is the delta gene, but, strikingly, by 12–14 h (blastula stage), Et ets1 expression spreads to the nonskeletogenic mesoderm and is then extinguished in the micromere descendants altogether (Fig. 1). Thus, neither is this gene likely to function similarly in the cidaroid as in the euechinoid skeletogenic lineage.
tbr.
Finally, the tbr gene, which is required for and coopted to skeletogenic function in euechinoids (22, 24, 32, 33), is again expressed very differently in Et. Although the tbr gene is first activated in the micromeres, its expression quickly spreads to the entire nonskeletogenic mesodermal domain, where by double in situ hybridization, it can be seen to totally overlap that of ets1, and, in direct contrast to Sp, there is no evidence from its expression pattern that it ever plays a skeletogenic role.
Descriptive patterns of gene expression can never demonstrate the existence of given regulatory linkages, but they can certainly exclude their existence. Fig. 1 alone implies a very different cidaroid regulatory configuration than used in euechinoid skeletogenic specification.
Experimental Tests for Specific Linkages of the Euechinoid Skeletogenic GRN.
We now set about challenging Eucidaris regulatory linkages among the above and additional genes, with the specific intent of determining whether these linkages could be the same, or must be exclusive, of the linkages among these same genes in the Sp skeletogenic GRN.
Test for global confinement to skeletogenic lineage by HesC repression, of alx1, tbr, and ets1 transcription.
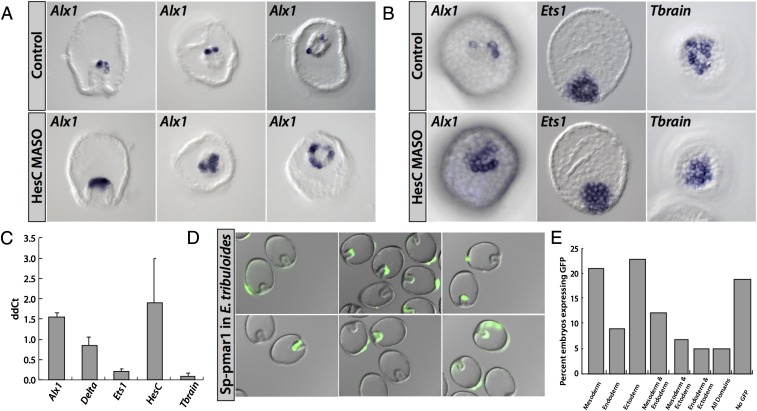
A dramatic demonstration of the function of the skeletogenic double-negative gate in Sp is afforded by either overexpression of the repressor Pmar1 or introduction into the egg of hesC morpholino antisense oligonucleotide (MASO), either of which results in global transformation of embryonic cells to skeletogenic fate, and in global expression of the double-negative target genes delta, alx1, and tbr (4, 24, 28, 31, 32). In Fig. 3 A, Left, and B, Left, we see the spatial effect of hesC MASO on alx1 expression in Et embryos. At blastula stage (Fig. 3B) expression of alx1 indeed expands but (reasonably enough) only to the extent of significant hesC expression, which, as evident from Fig. 1, is confined to the immediately surrounding nonskeletogenic mesoderm. At gastrula stage (Fig. 3A), alx1 expression expands to the immediately surrounding archenteron tip (mesoderm) cells. Thus, although hesC does repress alx1, it is not responsible for preventing alx1 expression throughout the embryo as in Sp, but only in the nonskeletogenic mesoderm. In Fig. 3B the effects of hesC MASO on spatial expression of tbr and ets1 are shown. Because Fig. 1 demonstrates the overlap of expression domains of hesc expression with those of ets1 and tbr genes, they are unlikely to be subject to HesC repression, and indeed, hesC MASO has no effect on their spatial expression, again in direct contrast with the case in Sp. In Fig. 3C these results are substantiated quantitatively in a quantitative PCR (QPCR) experiment, which shows that the only significant effects (i.e., >1.5× cycle number change, a log2 metric) are the modest increase in alx1 expression seen spatially above and in hesC transcript level itself; this gene apparently depresses its own transcription.
Fig. 3.
Functional tests for presence in Et of known regulatory linkages of the Sp skeletogenic GRN. Test for global HesC repression of skeletogenic regulatory state. (A) The 28-h embryos injected with hesC MASO exhibit an expanded domain of the skeletogenic lineage marker alx1. (B) The 20-h embryos injected with hesC MASO showing no global expression of dominant-negative gate genes. Alx1 expands locally only, whereas ets1 and tbrain are unaffected. (C) Quantitative effects of hesC MASO on mRNA abundance at 18 h on expression of skeletogenic genes alx1, delta, ets1, hesc, and tbrain. Alx1 and hesC mRNAs are significantly up-regulated. The difference in cycle number (ddCt) with respect to an uninjected control group is shown on the ordinate. Error bars represent the SD of two independent experiments. (D) Spatially nonrestricted expression of Sp pmar1 expression construct on injection into Et eggs. (E) Quantitation of multiple expression domains observed in (D).
These experiments preclude the global control of skeletogenesis by hesC repression in Et, which is its prominent role in the Sp skeletogenic GRN. They also preclude any control in Et of either ets1 or tbr by hesC repression. We have already seen that neither of these genes is likely to have anything to do with skeletogenesis after cleavage in Et in any case.
Lack of evidence for existence of the pmar1 gene in Et.
A complete genomic sequence has been obtained for Et, although it is not annotated and has been assembled only to contigs of several kilobase median length. In addition, a mixed embryonic transcriptome has been sequenced and analyzed (data from Human Genome Sequence Center). Despite the unfinished genomics analyses, these genomics resources sufficed for identification of >95% of a large set of Sp protein-coding genes. However, we were unable to find any sequence whatsoever in either the Et genome or transcriptome databases indicating the existence of any genes resembling Sp pmar1. The Sp genome includes at least six clustered paralogues of this divergent paired box gene, and two of these genes, for which cis-regulatory evidence has also been obtained, are directly similar to the pmar1 transcripts that we functionally characterized earlier (31, 34). Because failure to identify pmar1 genes in the Et genome or embryo transcriptomes is not an entirely convincing result, we embarked on an additional, although indirect, approach and asked whether the regulatory state of Et micromeres (or indeed of any polar early cleavage Et cells) would support transcription of an Sp pmar1 gene. An accurately expressing, recombineered pmar1 BAC construct bearing a knocked-in GFP marker had previously been constructed and authenticated in gene-transfer experiments (34). It responds at known cis-regulatory sites to the two transcription factors that in Sp constitute the localized input responsible for endogenous pmar1 expression as soon as micromeres form (4). These are a Tcf input, which uses for its spatial activation function maternally localized β-catenin (35), and Otxα transcription factor, which is also transiently localized to the micromeres in Sp (36). However, when this pmar1 reporter construct was injected into Et eggs, no localized expression could be seen, and instead the construct expressed more or less equivalently in all domains of the embryo. This result is shown in Fig. 3 D and E. Additionally, we checked whether a localized Otx factor might be used for early control of the skeletogenic regulatory state in Et, even if this effect were not mediated by a pmar1 gene (or a recognizable pmar1 gene). A sequence encoding the maternal Otx factor was truncated to produce a dominant-negative form, which was shown to be functional by its effect on endoderm genes when the mRNA was injected into Et eggs. However, injection of this mRNA into Et eggs had no effect whatsoever on expression levels of any of the micromere genes, such as alx1 or ets1, as assessed by QPCR.
The minimum conclusion from these experiments is that the combinatorial localization system that in Sp provides the β-catenin/Tcf and Otxα transcriptional inputs causing micromere pmar1 expression does not exist in Et. In all probability, neither does pmar1, the lynchpin upstream gene of the double-negative gate, even exist in the Eucidaris genome. Together with the foregoing hesC MASO experiments, it can be concluded that the double-negative gate circuitry of the euechinoid micromere lineage does not control the skeletogenic regulatory state in Et. Absence of this circuit feature was also inferred for another cidaroid embryo (23).
What Does Specify the Ultimate Skeletogenic Fate of Micromeres in Et?
The alx1 gene is clearly not an initial regulatory mediator of skeletogenic specification in Et micromeres as it is in the Sp skeletogenic micromere GRN, because, as we show here, it is not even transcribed during the period of activation of the initial set of micromere-specific genes. However, alx1 is ultimately just as clearly a canonical driver of later skeletogenic differentiation in Et (Fig. 2), as it is also in euechinoids (25). Thus, we can use its expression as a faithful indicator of skeletogenic fate, unlike genes such as tbr and ets1, which, although expressed early in Et micromeres, apparently end up having little to do with skeletogenesis.
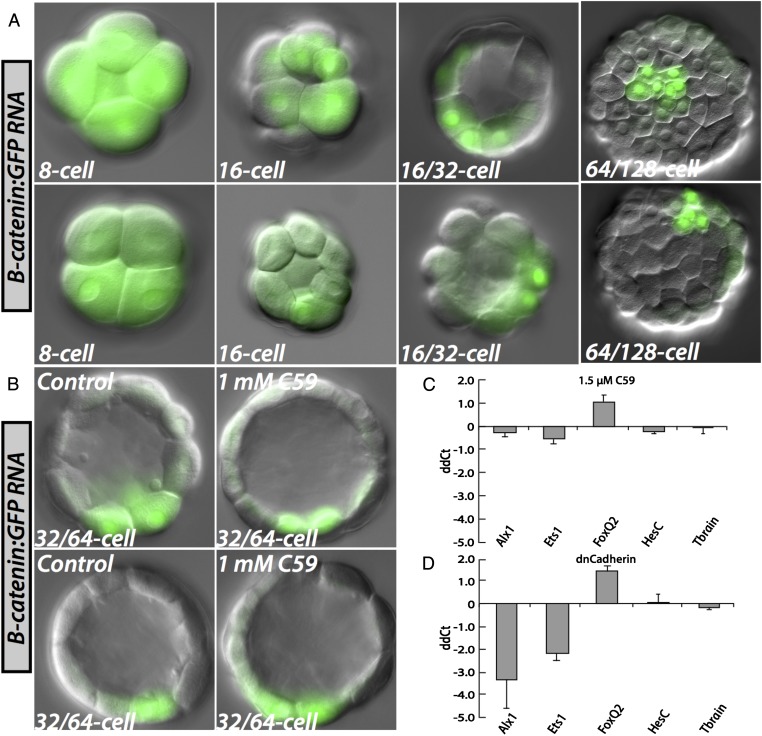
A not entirely unexpected clue as to the nature of the initial molecular input specifying skeletogenic fate devolves from the experiments in Fig. 4A, although they raise as well as answer a mechanistic question. As shown there, when mRNA encoding GFP-tagged β-catenin is injected into Et eggs, it is ubiquitously translated, but then over the next few cleavages, this protein is asymmetrically degraded (37), leaving it concentrated, dramatically and exclusively, in the micromeres. This negative cytoplasmic localization system is mediated by the β-catenin protein sequence per se, and the behavior of the tagged construct perfectly reflects the early highly localized retention of native β-catenin in euechinoid sea urchin micromeres, as observed immunocytologically (38) (we cannot of course be certain whether the kinetics of asymmetric clearance in Et are affected by the GFP tag). The responsible localization system does not depend on asymmetric Wnt signaling in the cleavage stage embryo; thus, the same localization of GFP-tagged β-catenin occurs in the presence of a potent antagonist of all canonical and noncanonical Wnt signaling, “C59” (Fig. 4B). C59 works by inhibiting Porcupine-dependent Wnt mobilization and secretion, and is both effective and specific in sea urchin embryos (39); detailed evidence for sea urchin embryos and references to its specificity and mode of action in other bilaterian systems are to be found in this reference. It follows from the results of experiments such as those reproduced in Fig. 4 A and B, that the β-catenin localization system of early Et embryos is a property of the oocyte/egg cytoplasmic localization system, which falls into the category of anisotropic deposition of molecules of gene-regulatory significance, a general feature of very early animal eggs (2). The main import of Fig. 4, however, is in the QPCR experiment of Fig. 4D. Here we see that there is virtually no expression of alx1 in micromeres (<8% of control values), if maternal β-catenin is sequestered by introduction of excess cadherin fragment, even though alx1 transcription is a late cleavage event. Furthermore, these effects depend not at all on Wnt signaling, even as late as 15 h. Thus, we are confronted with a missing link: β-catenin construct localization is complete in Et by the seventh cleavage (Fig. 4A), and a significant time gap of several hours separates this event from activation of the alx1 gene. The actual transcriptional mediator of alx1 activation that responds to the localized β-catenin/Tcf cue therefore remains unknown. We cannot yet experimentally either exclude or support the possibility that the initial transducer of the β-catenin/Tcf input is the cis-regulatory system of the Et ets1 gene, which is activated hours earlier than alx1 at approximately the right time. It may be significant that cis-regulatory analysis of alx1 expression in Sp showed it to be subject initially to obligatory ets1 activation (28). This dependence, however, remains to be demonstrated for Et alx1.
Fig. 4.
Requirement for a Wnt-signal-independent β-catenin polar localization system. (A) Early cleavage Et embryos demonstrating progressive spatial restriction of an injected β-catenin:GFP mRNA to the vegetal pole. Before 16-cell stage, this mRNA is found in all cells of the embryo. At fourth cleavage, the mRNA comes to be restricted to micromere- and micromere-abutting nuclei at the vegetal pole. Several cleavages later, it is only found in a few cells at the vegetal pole, the only likely identity of which is the micromeres because they are disposed exactly as are the cells expressing micromere genes (Fig. 1). (B) Early cleavage embryos treated with C59, a reagent inhibiting porcupine-dependent Wnt signaling. C59 does not effect spatial restriction of β-catenin:GFP mRNA. (C) Quantitative effects, measured by QPCR, of treatment with 1.5 μM C59 on relevant genes in 15-h Et embryos. The difference in cycle number (ddCt) with respect to an uninjected control group is shown on the ordinate. Error bars represent the SD of four independent experiments. (D) QPCR analysis of effects at 16 h in Et embryos of injected dominant-negative Cadherin mRNA. Error bars represent the SD of two independent experiments.
The Basal Role of hesC in Mesoderm Specification.
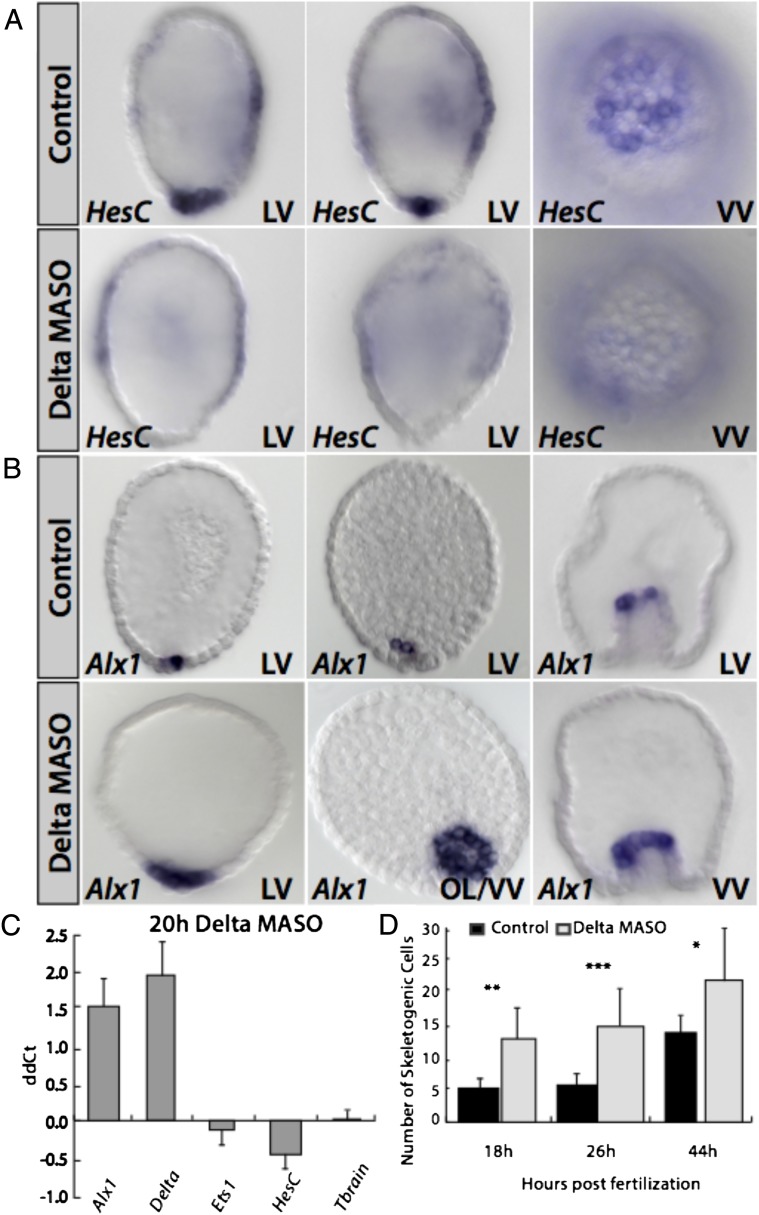
The relation between hesC and delta expression is a well-known constant of Notch signaling systems (40, 41). Although there are countless variations, in simple form, the Delta ligand promotes Notch receptor activation with the consequence that the immediate transcriptional effector, Su(H), activates Notch signal transduction target genes. Among these are very often genes encoding bHLH repressors of the same family as hesC. The expression of these repressors enforces the distinction between Delta signal-sending and Notch signal-receiving genes by transcriptional exclusion of delta transcription in the Notch signal-receiving cells. A beautiful illustration of this relationship can be seen in Fig. 5A. As we report elsewhere, Notch signaling is taking place in the Et embryo, but aside from the following negative relationship, it plays no role whatsoever in specification or differentiation of skeletogenic cells per se.
Fig. 5.
Spatial role of hesC in Et embryos. (A) Effects of delta MASO on hesc at the vegetal pole. In the presence of delta MASO, hesC expression is extinguished specifically at the vegetal pole, whereas its weak expression in certain regions of the ectoderm is unaffected. (B) Presence of delta MASO in Et causes an expansion of the skeletogenic marker alx1 to the surrounding nonskeletogenic mesoderm domain. (C) Quantitative effects of delta MASO measured by QPCR in 20-h embryos of Et. Delta and alx1 are significantly up-regulated in the presence of delta MASO. HesC is barely affected due to its background presence in the ectoderm. Ets1 and tbrain are unaffected. The difference in cycle number (ddCt) with respect to an uninjected control group is shown on the ordinate. Error bars represent the SD of three independent experiments. (D) Histogram shows the number of alx1-positive, i.e., skeletogenic, cells at three time points of Et development in embryos injected with delta MASO vs. uninjected controls. (18 and 26 h, n = 7; 44 h, n = 6). ***P < 0.0001; **P < 0.001; *P < 0.01 (all as determined by Student’s t test). LV, lateral view; OL/VV, oblique lateral/vegetal view; VV, vegetal view.
Fig. 5A shows that hesC expression, by this time in the surrounding nonskeletogenic mesodermal cells, is entirely dependent on Delta expression in the immediately adjacent skeletogenic micromeres. That is, as in the canonical case, Notch signal transduction of the micromere Delta signal results in hesc expression in these nonskeletogenic mesodermal cells. Because, as shown above, HesC is the repressor excluding alx1 expression from the nonskeletogenic mesoderm, if Delta expression is prevented, alx1 expression spreads to the nonskeletogenic mesoderm, now involving twice as many cells (Fig. 5 B–D). We perhaps see here the original role of micromere delta expression in sea urchin embryos, the spatial separation of skeletogenic from nonskeletogenic mesodermal specification. This result capsulizes the depth of the differences between the euechinoid and cidaroid specification systems; whereas Sp uses its double-negative gate circuitry to position skeletogenic function in the micromere lineage, Et uses Delta\Notch signaling for this purpose.
Discussion
Our main and specific objective was to assess at least the minimum evolutionary divergence that took place within a thoroughly known developmental GRN, during or soon after the last major cladistic split in the evolution of the echinoids. This divergence occurred in a late Paleozoic time interval that is constrained in real time by the fossil record. One uncertainty that could affect dynamic interpretation of the results is the possibility that the differences we observe between the test species of this work, Sp and Et, are actually in part the sum of changes that occurred only gradually—that is, during the Mesozoic (Triassic, Jurassic, and Cretaceous), subsequently to the split from which emerged the modern euechinoid and cidaroid subclasses. This would require, however, that the specific circuitry features we investigated vary among modern euechinoid orders that arose during the Mesozoic (18). However, although indeed incomplete, the evidence so far limits this possibility. Thus, a euechinoid belonging to an irregular euechinoid group (the Spatangoids), far removed from typical euechinoids such as Sp, also contains a pmar1 gene and also zygotically expresses the hesc gene all over the embryo except for the skeletogenic micromeres (42), exactly as in Sp. This global hesc expression, as we have seen, is in direct contrast to Et (we refer here only to the key shared linkages of interest, irrespective of the many and various other intraeuechinoid divergences that are also observed, but are irrelevant to skeletogenesis) (42). Therefore, key diagnostic features of the modern euechinoid (i.e., Sp) GRN are found in descendants of a euechinoid clade the last common ancestors of which with Sp arose anciently, perhaps at the beginning of the Jurassic (18). This result leaves untested only the most basal orders of euechinoids, but because those clades emerged directly from the subclass split per se, they limit the temporal argument pertaining to postdivergence events. Similarly, on the cidaroid side, as noted above, the orders composing this Subclass have displayed remarkably invariant and conservative morphology ever since their appearance (18). Consistent with this, as we have seen, Et indeed shares with a distant cidaroid the key property of lacking the double-negative skeletogenic specification gate (23). Therefore, with the caveat of the yet-unexamined most basal euechinoid orders, we can tentatively assume that we are here assaying genomic wiring features typical of almost the whole euechinoid subclass vs. those typical of the whole cidaroid subclass. These must be differences that indeed arose during the late Paleozoic at the divergence between these clades and/or in the earliest subsequent phases of euechinoid divergence—differences that have ever since been inherited by descendants of the crown group ancestors of each branch.
GRN Linkages of the Embryonic Sp Skeletogenic GRN Shown Here to be Specifically Absent from the Embryonic Et Skeletogenic Specification System.
We can now list specific regulatory features encoded in Sp cis-regulatory sequence that contribute decisively to the architecture of the Sp skeletogenic GRN (6), but that do not operate at all or operate differently in Et (reference citations below all refer to cis-regulatory studies or other decisive studies in Sp). This provides a minimum but hard estimate of regulatory differences between the embryonic skeletogenic specification circuitries that have arisen since the last common ancestor from which these two genomes descend.
The hesC cis-regulatory system.
First, in Sp, the hesC gene responds to a powerful global embryonic activator (24), a feature totally lacking in Et. In Et, hesC transcription is spatially controlled by Delta/Notch signaling from the micromeres and hence is expressed only in mesoderm immediately adjacent to the micromeres [Delta/Notch signaling does still provide an additional cis-regulatory input to hesc in Sp (34)]. Second, in Sp, the hesC gene is negatively controlled at the transcriptional level by Pmar1 repression (24, 34). In Et, no pmar1 gene or similarly functioning gene appears to exist.
The tbr cis-regulatory system.
First, in Sp, this gene is negatively controlled by HesC and positively controlled by a ubiquitous activator (32). Second, in Sp, tbr is expressed in skeletogenic cells. In Et, none of these three inputs operates on tbr transcription.
The ets1/2 cis-regulatory system.
First, in Sp, this gene is expressed maternally. Second, in later development, it is expressed in differentiating skeletogenic cells (where it plays a major role in activating skeletogenic effector genes). However, in Et, neither is true.
The delta cis-regulatory system.
In Sp, early embryonic spatial expression of delta is negatively controlled by HesC, and Ets1 serves as a positive driver (24, 29). However, in Et, HesC provides no spatial input into delta expression (although for unknown reasons, hesc MASO somewhat increases delta mRNA levels); in Et, Ets1 does not provide any positive input into delta expression.
The pmar1 gene.
This key gene of the Sp skeletogenic specification system is almost certainly absent altogether from the Et genome.
The initial combinatorial Otxα:Tcfβ-catenin cis-regulatory micromere input.
In Sp, this combinatorial input is used to trigger pmar1 transcription in micromeres (4, 34), whereas in Et this combination is not functional in skeletogenic micromere specification by direct test, and Otxα is not used at all in skeletogenic specification. Although this transcriptional regulator is encoded maternally in Et as in Sp, its function remains undemonstrated.
In sum, here there are nine specific cis-regulatory inputs into genes operating in both species that function in Sp and are absent in Et, plus a key gene missing in Et (or small subfamily of genes), plus a key localized combinatorial cis-regulatory transcriptional input used in Sp by the gene that is absent in Et. Assuming the euechinoid network is the evolutionary novelty (see below), each of these regulatory inputs represents the appearance of a new GRN linkage that had to be encoded in cis-regulatory DNA of genes in the euechinoid lineage, a linkage that is lacking in the cis-regulatory sequences of the same genes in the cidaroid lineage. Perforce a minimum estimate, we see here something of the scale of genomic regulatory change required for architectural network evolution, even in a small, confined GRN dedicated to specification of one cell lineage. Canonically, this type of evolutionary process is far removed from the single cis-regulatory module divergences easily accessed in studies of intra- and interspecific adaptive variation (2).
Plesiomorphy and Polarity in the Echinoid Regulatory Linkages.
All of the changes enumerated above are gains of function with respect to the regulatory configuration of the Et system, with most of them involving multiple different inputs per cis-regulatory module. Although it is conventional to note that all such changes could also represent loss-of-function changes in the cidaroid lineage—meaning that the euechinoid regulatory system could equally be plesiomorphic—the evidence is no longer balanced; it is much more likely [just as intuitively assumed by past observers (13, 14)] that the euechinoid skeletogenic GRN is the derived, novel character shared among descendants of the common euechinoid ancestor. A crucial argument that now comes into view is that the gains of function are sequentially and logically nested. That is, a given change requires particular sets of sequential changes, which impose polarity on the process. For example, acquisition of cis-regulatory response to a global regulator in the hesc gene introduces the possibility of release of control of genes such as alx1 from a strictly mesodermal activator to control by a general global activator, and of the delta gene from its strictly Notch-dependent control also to that of a global activator. However, such relaxations of domain-specific positive regulatory constraint in turn make it necessary to control micromere expression by negative rather than positive means, as executed by the euechinoid double-negative gate. This is not to propose a specific pathway, but to point out that, whatever the pathway, we are dealing here with an internally sequential logic train, rather than a series of independent changes that indeed individually might be considered equally likely to be gain as loss of function. A second argument concerns the cooption of the tbr gene to skeletogenic function. This work shows that cooption to be a euechinoid novelty, because in Et tbr is not skeletogenic in function, and because we know from comparative studies that the plesiomorphic role of tbr in echinoderms is not skeletogenic either (33). Therefore, this cooption is a derived euechinoid character, and in Sp, tbr also is driven by a ubiquitous activator (32) so that its expression is made skeletogenesis-specific only by the double-negative gate. Third, and similarly, control of delta gene expression is executed by the Notch response system in Et, whereas addition of a global positive control input in the delta gene in Sp (24) is therefore also a euechinoid derivation. We conclude that all of the linkages of the skeletogenic control GRN that are found in Sp but are absent from Et are probably shared derived characters of the euechinoids.
However, if this is the case, there must also remain plesiomorphic aspects of the skeletogenic program that would have been identified in this work as shared features present in both Et and Sp. Indeed this logical expectation is fulfilled. The most prominent plesiomorphic GRN character is of course the dominant role of alx1 as a driver of skeletogenic differentiation. The role of alx1 is plesiomorphic for echinoderm skeletogenesis in general (22, 25). A second major plesiomorphy in circuit wiring is indicated by the retention in both systems of negative spatial control of alx1 by HesC repression. Similarly, a third plesiomorphic linkage is retention of negative cis-regulatory control of delta expression by HesC. This linkage, exactly like the HesC repression of alx1, is used in Sp for global control of expression, and in Et for control of skeletogenic vs. nonskeletogenic mesodermal expression.
Evolutionary Assembly of the Euechinoid Skeletogenic GRN.
Solution of the Et skeletogenic GRN will facilitate a rational reconstruction of the evolutionary path by which the euechinoid skeletogenic micromere specification GRN might likely have assembled from its starting configuration. Only some general propositions can be offered at this juncture. It is clear from this work that multiple genomic regulatory changes had to be installed in the euechinoid lineage, whatever the exact pathway, and it is obvious that these cannot have entered the system all at once, nor would piecemeal alterations have had functional utility. However, in this conundrum originates the most powerful argument for the polarity of the evolutionary train of events. The presumably plesiomorphic cidaroid skeletogenesis system has a fundamental, key feature that would have allowed the accumulation of the novel GRN linkages without at the same time destroying its needed function of programming embryo/larval skeletogenesis. This feature is that development of the cidaroid micromere cell lineage is in functional terms essentially a dual process. In Et, cleavage-stage micromere functions per se and skeletogenic functions per se are separate. The cleavage-stage micromeres do not execute skeletogenic specification, and instead their role is to emit Delta signals, which are used negatively in late cleavage to protect the nonskeletogenic mesoderm from skeletogenic differentiation fate. Skeletogenic specification occurs only subsequently (in micromere descendants), when and after alx1 is belatedly turned on. Skeletogenic differentiation takes place even later, mainly at the tip of the archenteron and subsequently in the blastocoel. Thus, the precocious skeletogenic functions controlled by the novel euechinoid skeletogenic GRN could have assembled over evolutionary time at the embryological address of the micromere lineage, during or soon after the period the cladistic cidaroid/euechinoid split was taking place, without interrupting any of the developmentally later skeletogenic functions on which the embryo of the euechinoid stem lineage would still have depended. In other words, in the plesiomorphic state the micromere lineage executed signaling but not skeletogenic functions during cleavage and blastulation, but during euechinoid divergence novel skeletogenic circuitry executed in the micromere lineage during early development could have been superimposed, without necessarily interfering with gastrular skeletogenesis until the latter became redundant.
Materials and Methods
Detailed materials and methods are available in SI Materials and Methods. Briefly, Et were acquired from Sea Life, Inc. Procedures for handling eggs and embryos of this species were developed in the course of this work and are detailed in SI Materials and Methods. WMISH was conducted essentially after Ransick (43), with modifications. Microinjection experiments in Et were done essentially as described elsewhere for euechinoids (44).
SI Materials and Methods
Animals and Embryo Cultures.
Et sea urchins were obtained from SeaLife and were maintained in room temperature (r/t) aquaria. Animals were spawned with 0.5 M KCl. Cultures were grown in Millipore-filtered sea water (MFSW) in an incubator set to 22 °C unless otherwise indicated.
Real-Time Quantitative PCR.
For each MASO-treated time point, injected and uninjected embryos were counted (∼70 embryos per timepoint were used), gently centrifuged, and lysed with Buffer RLT (Qiagen). Just before column chromatography, an equal amount of exogenous GFP RNA was added to both treatment and uninjected control samples to normalize for RNA preparation, which was carried out according to the manufacturer’s protocol (RNeasy; Qiagen). All samples were processed in concert. cDNA was synthesized for the entire sample (iScript; Bio-Rad). Approximately one embryo per reaction was assayed in triplicate by QPCR (SYBR Green; Life Technologies). Primer sequences to amplify QPCR products in this study are presented in Table S1.
Table S1.
Sequences of primer sets for QPCR detection
| Gene | QPCR forward primer | QPCR reverse primer |
| Alx1 | ATCCGGGTATGAAATGCCCA | TTCTGCAGATGCGGAGCATA |
| Delta | AAATGTAACGTGCCGTGTGAGCCA | TACAGCTCACATTGGTCGCACCT |
| Ets1 | TGAGTCATCACCGAACTCGAACCA | GGTGTCCGTCAAACGTGTCAAA |
| FoxQ2 | TACGCCTATCCTTCCACCATC | GTGAAGGCAGCGACGAATATG |
| HesC | ACGTCGAGCAAGAATCAACG | CACTCGACTGGGTCTGTAATTCCT |
| Tbrain | ATTCTCCAAGGTAGTGGGCTGCAT | GATGCGAGGTTGGTACTTGTGCAT |
WMISH.
Digoxigenin (DIG)- or fluorescein (FLU)-labeled RNA probes were prepared by cloning purified PCR product (0.7–1.2 kb in length) into PGEM-T vector (Promega). All plasmids were sequenced to confirm insert and orientation. Primer sequences used to amplify PCR product used for this study are presented in Table S2. Antisense RNA probe was synthesized with T7 or SP6 RNA polymerase (Roche) and purified by column chromatography (RNeasy; Qiagen). For fixation, embryos were fixed on ice in paraformaldehyde (PFA)-maleic acid buffer (MAB) fixation buffer [4% (wt/vol) PFA, 32.5% (vol/vol) MFSW, 32.5 mM maleic acid (pH 7),162.5 mM NaCl], left overnight (o/n) at 4 °C, and brought into hybridization buffer (HyB: 50% formamide, 5× Denhardt’s, 5× SSC, 1 mg/mL yeast tRNA, 100 mM NaCl, 0.1% Tween-20, and 50 μg/mL Heparin) by the following series: 10%, 25%, 50%, 75%, and 100%. Fixed embryos were washed twice in and also stored in HyB at −20 °C. For WMISH, a modifed version of a standard protocol (43) was used. Briefly, fixed embryos were incubated in HyB at 63 °C for 1 h. DIG- or FLU-labeled probes were added to a final concentration of 0.5–1 ng/μL and incubated o/n at 63 °C. Posthybridization washes were the following: HyB for 15 min, 50:50 HyB/2× SSC for 15 min, 2X SSC for 20 min, 0.2× SSC for 20 min, 0.1× SSC for 30–60 min. Embryos were washed 3× in Tris-buffered saline with Tween-20 (TBST) and blocked at r/t in blocking buffer 1 [80% TBST, 10% sheep serum, 1 mg/mL bovine serum albumin (BSA)] for 30 min, and subsequently blocking buffer 2 (89% TBST, 10% sheep serum, 0.1 mg/mL BSA) for 30 min. Anti-DIG or -FLU fab fragments (Roche) was added to a final concentration of 0.25 μg/mL, incubated for 1 h at r/t, and removed by washing 6× in TBST. Probes were detected by washing 2× in alkaline phosphatase (AP) Buffer and 1× in AP Buffer with 10% dimethylformamide and nitro blue tetrazolium (NBT)/5-chloro-4-bromo-3-indolyl phosphate (BCIP). Staining was halted with TBST/EDTA. Embryos were stored in 70% glycerol until imaging.
Table S2.
Sequences of WMISH primer sets used in this study
| Gene | WMISH forward primer | WMISH reverse primer |
| Alx1 | TGAAATGCCCATAGCTCCACGA | ATGCCCATGACTGAACTGTGCT |
| Delta | ACGGTGATACTAATCCTTCACTGG | AGACAGGTGTACCCGTCAGC |
| Ets1 | AATGAGGTTGGACGAGTGCTGTCA | GTCCGTCAAACGTGTCAAAGGGT |
| HesC | ACGCAAACGTCGAGCAAGAATC | GCCACATTTGTTTGGCAGCTGTTG |
| Tbrain | TGTTCCCTCAACTGGTCTTCAAGC | CATAGCGCCCTCTTGTGATAGGAT |
Microinjection of MASOs, Constructs, and RNA.
Unfertilized eggs of Et were prepared essentially as described (44). MASOs were synthesized by Gene Tools (Philomath), and their sequences are provided in Table S3. All MASO injection solutions were 1 mM, and each fertilized egg received ∼10 pL of injection solution. Embryos for WMISH or QPCR were collected and processed as described above. Dominant-negative Cadherin RNA—which blocks β-catenin nuclearization at the vegetal pole, as described (38)—was injected at a concentration of 1,000 ng/μL. For visualization of early, asymmetric nuclearization of β-catenin, RNA encoding a fused β-catenin:GFP product was synthesized by using SP6 mMessage Machine RNA polymerase (Life Technologies) and injected at a concentration of 3 μg/μL. For microinjection of the 2.59-kb sp-pmar1 minimal reporter construct from ref. 34, ∼1,500 molecules of reporter construct were injected per embryo, and injected embryos were scored at 26 h postfertilization (hpf).
Table S3.
Sequences of MASO antisense oligonucleotides in this study
| Gene | MASO sequence | Interferes with |
| Alx1 | AGTATTTCATCGTCTCCACCTTTTC | Splicing |
| Delta | ATAACATATAGCACGCCGAGAAGGC | Translation |
| HesC | AATCACAAGGTAAGACGAGGATGGT | Translation |
Treatment with C59 Inhibitor.
The concentration of the porcupine inhibitor C59 (C7641-2s; Cellagen Technology) at which to expose embryos of Et was established by dose–response. Shortly after fertilization, embryos were assigned to four treatment groups of the inhibitor: 0.3, 0.9, 3, and 9 μM. Phenotypes for these groups were assessed under a dissecting microscope. Based on these observations, experiments were carried out at 1.5 μM. Embryos were added to the C59-containing MFSW shortly after they were fertilized, cultured in the medium at 22 °C until the desired time, and processed for QPCR analysis as described above.
Acknowledgments
We thank Dr. Isabelle Peter for excellent discussions and for her creative suggestion of injecting the minimal Sp-pmar1 reporter construct; Ryan Range for providing plasmid containing β-catenin:GFP insert; and Miao Cui for providing C59 inhibitor and educating us in its use. We extend a special thanks to Andy Cameron and Parul Kudtakar at the Beckman Institute’s Center for Computational Regulatory Genomics for providing support with transcriptomics in Eucidaris. This work was supported by a National Science Foundation CREATIV Grant 1240626.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509845112/-/DCSupplemental.
References
- 1.Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 1971;46(2):111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- 2.Peter IS, Davidson EH. Genomic Control Process, Development and Evolution. Academic, Elsevier; Oxford: 2015. [Google Scholar]
- 3.Peter IS, Davidson EH. A gene regulatory network controlling the embryonic specification of endoderm. Nature. 2011;474(7353):635–639. doi: 10.1038/nature10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci USA. 2008;105(16):5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peter IS, Davidson EH. Modularity and design principles in the sea urchin embryo gene regulatory network. FEBS Lett. 2009;583(24):3948–3958. doi: 10.1016/j.febslet.2009.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peter IS, Faure E, Davidson EH. Predictive computation of genomic logic processing functions in embryonic development. Proc Natl Acad Sci USA. 2012;109(41):16434–16442. doi: 10.1073/pnas.1207852109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boveri T. Zellenstudien VI. Die Entwicklung dispermer Seeigeleier. Ein Beitrag zur Befruchtungslehre und zur Theorie des Kerns. Gustav Fischer; Jena: 1907. [Google Scholar]
- 8.Laubichler MD, Davidson EH. Boveri’s long experiment: Sea urchin merogones and the establishment of the role of nuclear chromosomes in development. Dev Biol. 2008;314(1):1–11. doi: 10.1016/j.ydbio.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao F, et al. Juvenile skeletogenesis in anciently diverged sea urchin clades. Dev Biol. 2015;400(1):148–158. doi: 10.1016/j.ydbio.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 10.Nishita M, et al. Interaction between Wnt and TGF-beta signalling pathways during formation of Spemann’s organizer. Nature. 2000;403(6771):781–785. doi: 10.1038/35001602. [DOI] [PubMed] [Google Scholar]
- 11.Armstrong N, McClay DR. Skeletal pattern is specified autonomously by the primary mesenchyme cells in sea urchin embryos. Dev Biol. 1994;162(2):329–338. doi: 10.1006/dbio.1994.1090. [DOI] [PubMed] [Google Scholar]
- 12.McIntyre DC, Lyons DC, Martik M, McClay DR. Branching out: Origins of the sea urchin larval skeleton in development and evolution. Genesis. 2014;52(3):173–185. doi: 10.1002/dvg.22756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wray GA, McClay DR. The origin of spicule-forming cells in a ‘primitive’ sea urchin (Eucidaris tribuloides) which appears to lack primary mesenchyme cells. Development. 1988;103(2):305–315. doi: 10.1242/dev.103.2.305. [DOI] [PubMed] [Google Scholar]
- 14.Schroeder T. Development of a primitive sea urchin, Eucidaris tribuloides. Biol Bull. 1981;161:141–151. [Google Scholar]
- 15.Bottjer DJ, Davidson EH, Peterson KJ, Cameron RA. Paleogenomics of echinoderms. Science. 2006;314(5801):956–960. doi: 10.1126/science.1132310. [DOI] [PubMed] [Google Scholar]
- 16.Mooi R, David B. Evolution within a bizarre phylum: Homologies of the first echinoderms. Am Zool. 1998;38:965–974. [Google Scholar]
- 17.Peterson KJ, Arenas-Mena C, Davidson EH. The A/P axis in echinoderm ontogeny and evolution: Evidence from fossils and molecules. Evol Dev. 2000;2(2):93–101. doi: 10.1046/j.1525-142x.2000.00042.x. [DOI] [PubMed] [Google Scholar]
- 18.Kroh A, Smith AB. The phylogeny and classification of post-Paleozoic echinoids. J Syst Palaeontology. 2010;8:147–212. [Google Scholar]
- 19.Thompson JR, et al. Reorganization of sea urchin gene regulatory networks at least 268 million years ago as revealed by oldest fossil cidaroid echinoid. Sci Rep. 2015 doi: 10.1038/srep15541. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopkins MJ, Smith AB. Dynamic evolutionary change in post-Paleozoic echinoids and the importance of scale when interpreting changes in rates of evolution. Proc Natl Acad Sci USA. 2015;112(12):3758–3763. doi: 10.1073/pnas.1418153112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith AB, Hollingworth NTJ. Tooth structure and phylogeny of the Upper Permian echinoid Miocidaris keyserlingi. Proceedings of the Yorkshire Geological Society. 1990;48:47–60. [Google Scholar]
- 22.Gao F, Davidson EH. Transfer of a large gene regulatory apparatus to a new developmental address in echinoid evolution. Proc Natl Acad Sci USA. 2008;105(16):6091–6096. doi: 10.1073/pnas.0801201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamazaki A, Kidachi Y, Yamaguchi M, Minokawa T. Larval mesenchyme cell specification in the primitive echinoid occurs independently of the double-negative gate. Development. 2014;141(13):2669–2679. doi: 10.1242/dev.104331. [DOI] [PubMed] [Google Scholar]
- 24.Revilla-i-Domingo R, Oliveri P, Davidson EH. A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc Natl Acad Sci USA. 2007;104(30):12383–12388. doi: 10.1073/pnas.0705324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ettensohn CA, Illies MR, Oliveri P, De Jong DL. Alx1, a member of the Cart1/Alx3/Alx4 subfamily of Paired-class homeodomain proteins, is an essential component of the gene network controlling skeletogenic fate specification in the sea urchin embryo. Development. 2003;130(13):2917–2928. doi: 10.1242/dev.00511. [DOI] [PubMed] [Google Scholar]
- 26.Rafiq K, Cheers MS, Ettensohn CA. The genomic regulatory control of skeletal morphogenesis in the sea urchin. Development. 2012;139(3):579–590. doi: 10.1242/dev.073049. [DOI] [PubMed] [Google Scholar]
- 27.Lyons DC, Martik ML, Saunders LR, McClay DR. Specification to biomineralization: Following a single cell type as it constructs a skeleton. Integr Comp Biol. 2014;54(4):723–733. doi: 10.1093/icb/icu087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Damle S, Davidson EH. Precise cis-regulatory control of spatial and temporal expression of the alx-1 gene in the skeletogenic lineage of s. purpuratus. Dev Biol. 2011;357(2):505–517. doi: 10.1016/j.ydbio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Revilla-i-Domingo R, Minokawa T, Davidson EH. R11: A cis-regulatory node of the sea urchin embryo gene network that controls early expression of SpDelta in micromeres. Dev Biol. 2004;274(2):438–451. doi: 10.1016/j.ydbio.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 30.Smith J, Kraemer E, Liu H, Theodoris C, Davidson E. A spatially dynamic cohort of regulatory genes in the endomesodermal gene network of the sea urchin embryo. Dev Biol. 2008;313(2):863–875. doi: 10.1016/j.ydbio.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliveri P, Davidson EH, McClay DR. Activation of pmar1 controls specification of micromeres in the sea urchin embryo. Dev Biol. 2003;258(1):32–43. doi: 10.1016/s0012-1606(03)00108-8. [DOI] [PubMed] [Google Scholar]
- 32.Wahl ME, Hahn J, Gora K, Davidson EH, Oliveri P. The cis-regulatory system of the tbrain gene: Alternative use of multiple modules to promote skeletogenic expression in the sea urchin embryo. Dev Biol. 2009;335(2):428–441. doi: 10.1016/j.ydbio.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minemura K, Yamaguchi M, Minokawa T. Evolutionary modification of T-brain (tbr) expression patterns in sand dollar. Gene Expr Patterns. 2009;9(7):468–474. doi: 10.1016/j.gep.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 34.Smith J, Davidson EH. Regulative recovery in the sea urchin embryo and the stabilizing role of fail-safe gene network wiring. Proc Natl Acad Sci USA. 2009;106(43):18291–18296. doi: 10.1073/pnas.0910007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weitzel HE, et al. Differential stability of beta-catenin along the animal-vegetal axis of the sea urchin embryo mediated by dishevelled. Development. 2004;131(12):2947–2956. doi: 10.1242/dev.01152. [DOI] [PubMed] [Google Scholar]
- 36.Chuang CK, Wikramanayake AH, Mao CA, Li X, Klein WH. Transient appearance of Strongylocentrotus purpuratus Otx in micromere nuclei: Cytoplasmic retention of SpOtx possibly mediated through an alpha-actinin interaction. Dev Genet. 1996;19(3):231–237. doi: 10.1002/(SICI)1520-6408(1996)19:3<231::AID-DVG6>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 37.Stamos JL, Weis WI. The β-catenin destruction complex. Cold Spring Harb Perspect Biol. 2013;5(1):a007898. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126(2):345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- 39.Cui M, Siriwon N, Li E, Davidson EH, Peter IS. Specific functions of the Wnt signaling system in gene regulatory networks throughout the early sea urchin embryo. Proc Natl Acad Sci USA. 2014;111(47):E5029–E5038. doi: 10.1073/pnas.1419141111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9(21):2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 41.Borggrefe T, Oswald F. The Notch signaling pathway: Transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66(10):1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamazaki A, Minokawa T. Expession patterns of mesenchyme specification genes in two distantly related echinoids, Glyptocidaris crenularis and Echinocardium cordatum. Gene Expr Patterns. 2015;17(2):87–97. doi: 10.1016/j.gep.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 43.Ransick A. Detection of mRNA by in situ hybridization and RT-PCR. Methods Cell Biol. 2004;74:601–620. doi: 10.1016/s0091-679x(04)74024-8. [DOI] [PubMed] [Google Scholar]
- 44.McMahon AP, et al. Introduction of cloned DNA into sea urchin egg cytoplasm: Replication and persistence during embryogenesis. Dev Biol. 1985;108(2):420–430. doi: 10.1016/0012-1606(85)90045-4. [DOI] [PubMed] [Google Scholar]