Significance
The link between species diversification and adaptation has long interested biologists working on multicellular eukaryotes, but remains poorly understood in prokaryotes, in which diversity is much greater. We tested the hypothesis that diversification is associated with environmental adaptation in Thaumarchaeota, an ancient and abundant microbial group and key player in the global nitrogen cycle. We provide evidence that the Thaumarchaeota underwent a major radiation event hundreds of millions of years ago that coincided with a major period of pH adaptation. Subsequently, these microbes have maintained high rates of diversification, potentially because of the high rate at which new terrestrial niches arise. This study provides a framework for comparing dynamics of evolutionary processes across the tree of life.
Keywords: archaea, ammonia oxidation, phylogeny, diversification, Thaumarchaeota
Abstract
The Thaumarchaeota is an abundant and ubiquitous phylum of archaea that plays a major role in the global nitrogen cycle. Previous analyses of the ammonia monooxygenase gene amoA suggest that pH is an important driver of niche specialization in these organisms. Although the ecological distribution and ecophysiology of extant Thaumarchaeota have been studied extensively, the evolutionary rise of these prokaryotes to ecological dominance in many habitats remains poorly understood. To characterize processes leading to their diversification, we investigated coevolutionary relationships between amoA, a conserved marker gene for Thaumarchaeota, and soil characteristics, by using deep sequencing and comprehensive environmental data in Bayesian comparative phylogenetics. These analyses reveal a large and rapid increase in diversification rates during early thaumarchaeotal evolution; this finding was verified by independent analyses of 16S rRNA. Our findings suggest that the entire Thaumarchaeota diversification regime was strikingly coupled to pH adaptation but less clearly correlated with several other tested environmental factors. Interestingly, the early radiation event coincided with a period of pH adaptation that enabled the terrestrial Thaumarchaeota ancestor to initially move from neutral to more acidic and alkaline conditions. In contrast to classic evolutionary models, whereby niches become rapidly filled after adaptive radiation, global diversification rates have remained stably high in Thaumarchaeota during the past 400–700 million years, suggesting an ongoing high rate of niche formation or switching for these microbes. Our study highlights the enduring importance of environmental adaptation during thaumarchaeotal evolution and, to our knowledge, is the first to link evolutionary diversification to environmental adaptation in a prokaryotic phylum.
Ammonia oxidation, the first and rate-limiting step in nitrification, is central to the global nitrogen cycle. Ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) perform this biochemical transformation, converting as much as 70% of the 100 Tg of nitrogen fertilizer applied annually (1) and generating significant nitrous oxide (2). Although only distantly related, AOA and AOB perform similar ecosystem functions by catalyzing the conversion of ammonia to nitrite (via hydroxylamine) by using ammonia monooxygenase, a multimeric enzyme comprising the subunits AmoA, AmoB, and AmoC (3). Although AOA were initially classified as Crenarchaeota, improvements in phylogenetic methods and genomic sampling of uncultured archaeal diversity led to their placement within a new archaeal phylum, the Thaumarchaeota (4, 5). These organisms are ubiquitous, with archaeal amoA genes frequently outnumbering those of AOB (6). Furthermore, AOA are the principal drivers of ammonia oxidation in many soils, particularly those with low pH (7, 8). Environmental pH is a major factor affecting the distribution of AOA in terrestrial ecosystems (3, 9), and most terrestrial Thaumarchaeota can be phylogenetically assigned to one of five pH-adapted lineages, two being acidophilic, two alkalinophilic, and one neutrophilic (9).
Despite great progress in understanding of thaumarchaeotal ecology and physiology (10), little is known about the evolutionary mechanisms that have generated their great diversity in nature. In multicellular eukaryotes, the fossil record can provide insight into evolutionary processes over geological timescales, but such approaches are not possible for many prokaryotes because of the lack of an informative fossil record. In this context, recently developed probabilistic methods geared toward reconstructing the dynamics of species diversification and trait evolution using molecular phylogenies (11–13) have great potential. A recent Bayesian method that explicitly aims to characterize and quantify heterogeneity in evolutionary rates (13–15) holds particular promise for modeling prokaryotic evolution, whereby features such as massive population size, frequent lateral gene transfer (LGT), and fast growth all facilitate rapid increases in diversity, suggesting that complex evolutionary regimes may be the norm. Here, we apply these recently developed methods to test a key hypothesis about the ecological drivers of thaumarchaeotal diversification through deep evolutionary time. Based on the documented importance of soil pH in structuring modern AOA communities (9), we tested the hypothesis that adaptation to pH is an important driver of diversification in the Thaumarchaeota by comparing associations between pH and a range of other environmental factors with diversification in Thaumarchaeota.
Results
Bayesian Phylogenetic Reconstruction.
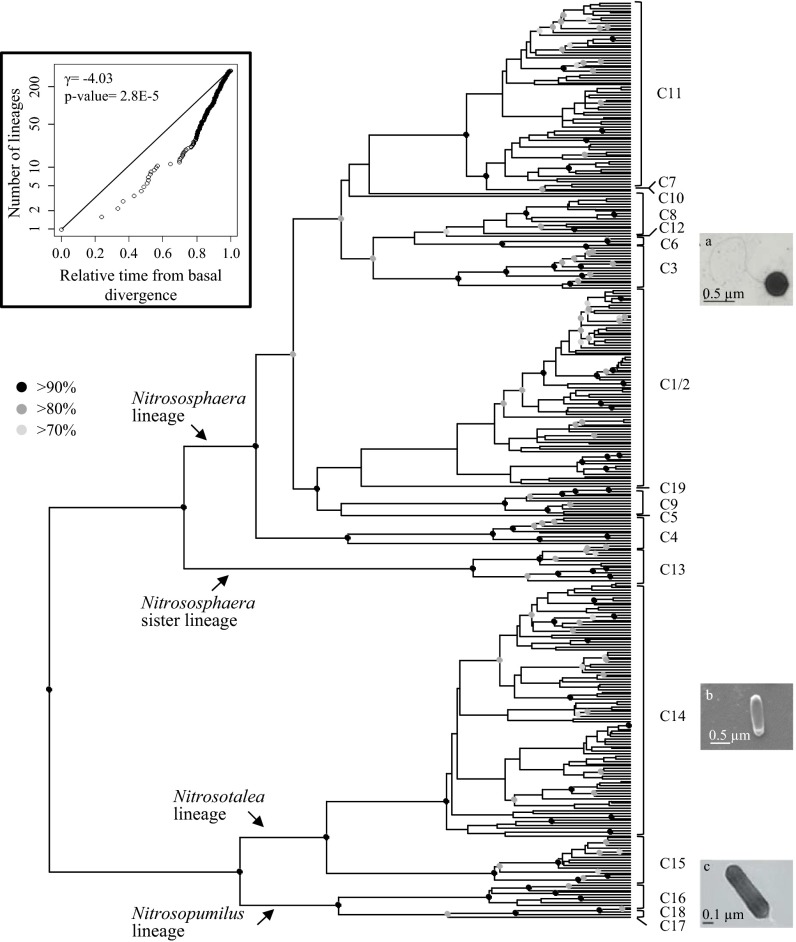
Bayesian phylogenetic analysis was performed on 370 amoA sequences, representing the full range of diversity within 47 UK soils (based on extensive 454 sequencing) spanning a large range of pH values (9, 16–18) (Fig. 1). Prior efforts were taken to account for the presence of recombination, leading us to remove putative recombinant sequences from the original, slightly larger amoA sequence alignment (n = 55 sequences removed; SI Appendix, Table S1). The resultant topology was highly congruent with previous phylogenetic analyses (refs. 9, 19; we use the classification proposed in ref. 9), with the exception that a small recognized hyperthermophilic Nitrosocaldus clade was absent from our data. Our new analysis incorporates a relaxed molecular clock model, which allows probabilistic inference of the tree root (20). Maximal support (posterior probability of 1) was recovered for a deep split of Nitrososphaera from a monophyletic group containing Nitrosotalea and Nitrosopumilus lineages (Fig. 1). Two notable differences from our previous study (9) were that (i) two clusters previously named C1 and C2 merged into a monophyletic neutroalkalinophilic cluster, hereafter named Nitrososphaera C1/2 (SI Appendix, Fig. S1); and (ii) the alkalinophilic Nitrososphaera C12 cluster was almost completely removed as a result of the detection of recombination. All other clusters were as previously described (9), and the pH preference of all clusters is highly congruent with previous findings (9) (SI Appendix, Fig. S2).
Fig. 1.
Bayesian phylogenetic tree of Thaumarchaeota based on the 370-sequence amoA alignment, incorporating a relaxed molecular clock model allowing statistical inference of the root. Posterior probabilities (PP) are indicated by a circle for each node (light gray, PP > 0.7; dark gray, PP > 0.8; black, PP > 0.9). Fig. 1 A–C reprinted with permission from refs. 16–18, respectively. (Inset, Upper Left) Log-transformed lineage-through-time plot with associated γ-statistic, along with a P value for the rejection of a pure-birth model of diversification.
We used the amoA tree in phylogenetic tests to reconstruct the evolutionary diversification of Thaumarchaeota. Because of a lack of fossil data, we did not temporally calibrate the analysis, and our tree is consequently scaled in relative time. In the absence of fossil data, LGT can provide useful information about the relative branching times of donor and recipient lineages. Petitjean et al. (21) inferred LGT of a DnaJ-ferredoxin fusion protein from the stem lineage leading to Viridiplantae (green algae and land plants) into the base of the Nitrososphaera/Nitrosopumilus clade. The divergence of Viridiplantae from the rest of Archaeplastida has been dated to approximately 950 Mya, with diversification within the group beginning approximately 200 million years later (21, 22). These dates were estimated by using fossil-calibrated relaxed molecular clock analyses and should be treated as approximate, with other analyses that used better models and more calibration points suggesting somewhat earlier dates (23). The inference of LGT from Viridiplantae to Thaumarchaeota also depends on the interpretation of a single gene tree, which can be notoriously difficult to resolve at these evolutionary timescales. Bearing these caveats in mind, these data suggest that the divergence of the Nitrososphaera from the Nitrosopumilus and Nitrosotalea lineages occurred ∼750–950 Mya (21) and 1.4 billion years ago at the earliest (23).
Evolutionary Analysis of Diversification.
Even without a firm species concept in prokaryotes (24), comparative phylogenetic methods provide insights into processes of lineage diversification and adaptation because they can test the null hypothesis that diversification occurs at a constant rate under a coalescent model (25, 26). For our thaumarchaeotal data, diversification tests based on the γ-statistic reject a rate-constant pure birth model (γ = −4.03; P = 8 × 10−5; Fig. 1, Inset) in favor of heterogeneous diversification through time. Use of a recently developed Bayesian method, Bayesian Analysis of Mixture Models (BAMM), which explicitly models variation in diversification rates across a tree (13–15), also provided strong support for heterogeneity in thaumarchaeotal diversification rates (Fig. 2 and SI Appendix, Fig. S3A).
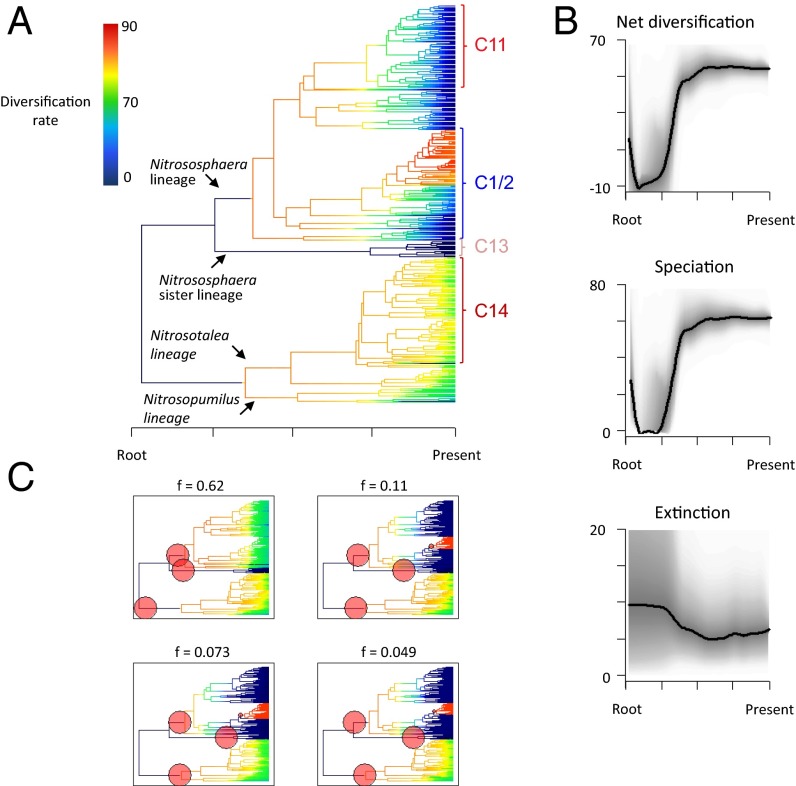
Fig. 2.
Evolutionary heterogeneity in diversification rates in terrestrial Thaumarchaeota inferred from the 370-sequence amoA tree within the BAMM framework. (A) Phylorate plot illustrating the heterogeneity in diversification rates along each branch of the phylogeny. The different color regimes depicted represent the mean of the marginal posterior density of diversification rates for distinct segments of the tree. (B) Plots of net diversification rate, speciation rate, and extinction rate (y axis) through time during thaumarchaeotal evolution. Shaded areas denote 90% Bayesian credibility intervals. (C) Plots of the four most probable diversification rate-shift configurations along with their individual contributions to the posterior distribution of all sampled BAMM models. All four plots include similar positive rate shifts (indicated by red filled circles).
According to the BAMM analysis, there was a ∼70-fold increase in diversification rate, the net product of speciation and extinction, during early thaumarchaeotal evolution (Fig. 2B), and major rate shifts were detected near the stem of Nitrososphaera and in the stem ancestral to Nitrosotalea and Nitrosopumilus (Fig. 2C). A third positive diversification rate shift was detected at the crown of the Nitrososphaera sister lineage, which was undergoing low rates of diversification at the time its sister group was undergoing higher diversification rates (Fig. 2A). BAMM models including the three previously described rate shifts constitute >0.95 of the posterior probability, and there was also evidence for a mixture of subsequent increases and decreases in diversification rate in more recent Thaumarchaeota lineages (Fig. 2C). Surprisingly, this analysis also suggests that the global diversification rate remained stably high, relative to its earliest level, during the latter half of thaumarchaeotal evolution (Fig. 2B), a period we estimate spans between 400 (21) and 700 (23) Mya. Although there was support for variation in diversification rate within the major pH-adapted clusters (Fig. 2A), the average rate of diversification within each of these clusters was statistically indistinguishable from the overall mean for the phylum, as evidenced by overlapping Bayesian credibility intervals for these estimates (SI Appendix, Fig. S4).
It is important to consider how these results might be affected by incomplete sampling of thaumarchaeotal diversity. Therefore, we repeated the BAMM analysis assuming that the overall diversity in our samples represented a range of different proportions of the true global diversity. The rapid initial increase in global diversification rate and associated rate shifts at the base of the amoA phylogeny were robust to these sampling assumptions (SI Appendix, Fig. S5). However, the stability in diversification rate across the latter half of thaumarchaeotal evolution was affected. Specifically, when <90% of sampling diversity was assumed to be recaptured, a slow, steady increase in diversification rate was observed during the same time period (SI Appendix, Fig. S5). However, this change was invariably less rapid and of a lesser magnitude than the initial increase (SI Appendix, Fig. S5). As an independent measure of confidence, we merged our 454 amoA sequence dataset with the complete diversity of nonredundant published amoA sequences. This led to an alignment of 613 unique sequences with better representation of Nitrosotalea and Nitrosopumilus, presumably recapturing known diversity from marine and estuarine environments that was not sampled in our soil study. These sequences were used in Bayesian phylogenetic analyses, and the resulting tree was used in a BAMM diversification analysis, which was highly congruent with results based on 454 data only (SI Appendix, Fig. S6).
To further test the robustness of our findings, we repeated the BAMM analysis by using a 16S rRNA dataset (508 aligned sequences) combining a comprehensive nonredundant representation of published sequence diversity with a 454 dataset we had recently generated (27). To provide the most direct comparison between 16S rRNA and amoA, we used a phylogenetic approach to ensure that the included 16S rRNA genes were sampled from lineages in which amoA genes are known to be present (SI Appendix, SI Materials and Methods). In striking support of the amoA findings, the 16S rRNA analysis also suggested an initial rapid increase in global diversification rate that was associated with similar major rate shifts toward higher diversification at the base of the thaumarchaeotal radiation (SI Appendix, Fig. S7). Again, the 16S rRNA analysis provides support for an initial radiation event that was followed by maintenance of relatively high diversification rates across the remainder of thaumarchaeotal evolution (SI Appendix, Fig. S7B). In slight contrast to amoA, the 16S rRNA analysis also supported a modest decrease in diversification rate approaching the present day (SI Appendix, Fig. S7B). However, if we assume that less global diversity was represented (<0.75; SI Appendix, Fig. S8), the diversification rate is inferred to be stable or slightly increasing, as observed for amoA. This scenario is likely true, in view of the reduced sequencing depth used in the 16S rRNA 454 approach (these primers also targeted non-AOA thaumarchaeotal groups and, to a lesser extent, some Euryarchaeota) (27), reducing the present AOA diversity obtained and thereby undersampling the true modern diversity.
Thus, with the use of independent gene markers, there is support for an early radiation within Thaumarchaeota, with stably high subsequent diversification rates over hundreds of millions of years of evolution.
Coupling of Environmental Adaptation to Diversification.
As pH has a major impact on the ecology of extant Thaumarchaeota (9), we postulated that this environmental variable contributed to the thaumarchaeotal diversification regime. We therefore reconstructed rates of pH adaptation across thaumarchaeotal evolution in the BAMM framework (13–15). Our reconstructions of adaptation were based on estimates of pH preference of extant Thaumarchaeota, considered as the pH of the soil from which the amoA sequences were isolated. This approach requires the assumptions that the organisms are most likely to be present in soils with pH close to their optimal pH for growth and that the fitness of sampled organisms is reflected in amoA sequence abundance; in other words, amoA sequences that represent free DNA or dormant cells should be highly underrepresented in comparison with well-adapted replicating organisms. Further, we assumed that the high level of sampling sufficiently reduced the significance of potential effects of spatial and temporal heterogeneity in soil pH.
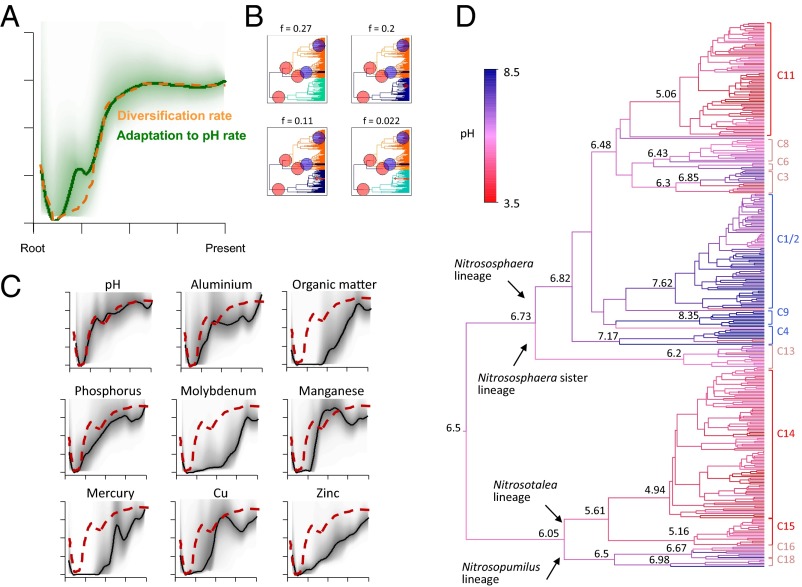
Remarkably, with the 370-sequence amoA dataset, our BAMM analyses indicated that the rate of change of pH adaptation was tightly coupled to thaumarchaeotal diversification, including the same three basal rate shifts described earlier (Fig. 3 A and B and SI Appendix, Fig. S3B). However, it is important to note that pH and a number of environmental factors may be interdependent, potentially confounding inferences surrounding the role of pH per se. For 181 of the 370 amoA sequences in our dataset, a broader range of quantitative environmental data were available that are potentially linked to pH (SI Appendix, Table S2). These sequences were spread across the amoA tree, enabling us to test the hypothesis that pH adaptation rates are more tightly coupled with diversification rates than other environmental variables by performing additional BAMM analyses on this reduced 181-sequence phylogeny. Importantly, the diversification and pH adaptation analyses were congruent across the 181- and 370-sequence trees, indicating that the removal of modern diversity from the larger tree did not affect the observed association between diversification and pH adaptation (Fig. 3C). Although all other tested environmental factors also showed an increase in the rate of putative adaptation across the period of thaumarchaeotal evolution, none tracked diversification rate as clearly as pH (Fig. 3C). For several factors, including organic matter, manganese, molybdenum, copper, and mercury, a rapid period of phenotypic change is inferred, but, unlike pH, this occurred long after the initial thaumarchaeotal radiation event (Fig. 3C). For aluminum, phosphorus, and zinc, rates of phenotypic adaptation initially increase at the same time as the onset of the initial radiation, but at much lower rates than pH, which closely matched the rate of diversification (Fig. 3C). Interestingly, although not as clearly correlated with diversification as pH, rates of phenotypic evolution linked to aluminum and phosphorus levels tracked the global diversification rate across thaumarchaeotal evolution (Fig. 3C).
Fig. 3.
Evolutionary heterogeneity in rates of environmental adaptation in terrestrial Thaumarchaeota inferred from the amoA 370-sequence tree within the BAMM framework. There is evidence that diversification (Fig. 2) is coupled to pH adaptation at two levels: (A) comparing the global rate of pH adaptation through time (green line) to the equivalent data for diversification rates (orange dotted line; taken from Fig. 2B) and (B) comparing the most probable phenotypic rate-shift configurations with equivalent diversification rate-shift data (Fig. 2C). (C) Equivalent analyses were performed for eight additional environmental factors on a smaller amoA tree of 181 sequences. (D) Ancestral pH preferences are shown for the 370-sequence amoA tree based on a ridge regression approach (28), with estimates of ancestral pH provided at key nodes.
Changes in pH Specialization at the Base of the Thaumarchaeotal Radiation.
To further explore the hypothesis of a link between pH adaptation and diversification, we reconstructed ancestral soil pH adaptation across the 370-sequence amoA tree with the use of a recently developed method that models phenotypic evolution for every branch in a tree by using ridge regression, an extension of ordinary least-squares regression that accounts for phylogenetic structure (28). This analysis suggests that the thaumarchaeotal common ancestor was adapted to a pH of approximately 6.5 and that there were early shifts toward lower and higher pH preferences in the respective ancestors of the Nitrosotalea/Nitrosopumilus and Nitrososphaera lineages (Fig. 3D). Further, these results suggest that the specialized acidophilic and alkalinophilic clusters (9) arose independently from neutrophilic ancestors more recently in thaumarchaeotal evolution (Fig. 3D).
Discussion
It is clearly important to understand the evolutionary processes contributing to the long-term ecological success of microbes and their associated ecosystem functions. Experimental evolution and population genomics are providing new insights into how mutation, selection, and drift shape microbial evolution over short timescales, but these approaches have less to say about long-term evolutionary dynamics over geological timescales, or the microbial equivalents of the patterns that paleontologists trace in the fossil record. In this work, we integrated recently developed Bayesian comparative phylogenetic approaches with contemporary environmental data on soil composition to test a key hypothesis about thaumarchaeotal evolution. Modern AOA communities are structured accordingly to environmental pH (9), raising the hypothesis that pH was an important factor in the long-term evolution of terrestrial Thaumarchaeota, one of the most abundant of the 10 currently recognized archaeal phyla (29).
Our results support this hypothesis as, of nine environmental factors tested, pH adaptation was the only one strikingly coupled to the rate of diversification (Fig. 3). This is consistent with a scenario in which pH adaptation was a positive factor contributing to an early radiation during thaumarchaeotal evolution. However, our results cannot eliminate the possibility that other physiological innovations arose at a similar time that promoted diversification that were correlated to or unrelated to pH. Indeed, our comparative phylogenetic analyses point to periods of environmental adaptation involving several factors theoretically linked to pH, although these periods were invariably more detached from the major initial thaumarchaeotal radiation (Fig. 3). It is important to note here that our sampling was designed to represent a wide range of soil pH, presumably recapturing a broad range of physiological adaptation to pH. Conversely, for the other factors tested, the range of environmental variation in our samples might be suboptimal for the detection of adaptation. Therefore, further work that more fully explores the environmental drivers of thaumarchaeotal evolution would be a welcome addition to the present findings focused on pH.
Looking more deeply at the hypothesis that pH was an important factor promoting thaumarchaeotal diversification, our results suggest that the lineage ancestor was a neutrophile that initially split into two major lineages that showed preferences toward acid (Nitrosopumilus/Nitrosotalea) and alkaline (Nitrososphaera) conditions (Fig. 3D). Subsequently, pH adaptation was inferred to have occurred rapidly, which may have promoted niche exploitation and allopatric isolation and hence new lineage formation. Interestingly, as two other archaeal phyla likely arose at near-neutral pH (Euryarchaeota; pH ≥ 6) or in moderately acidic environments (Crenarchaeota; pH 4–6) (30), we can now infer that the common ancestor of archaea was probably a hyperthermophile (31, 32) with neutral pH preference.
As these analyses represent one of the first applications of Bayesian modeling of long-term evolutionary processes in prokaryotes, it is important to keep associated caveats in mind. In particular, our analyses may have been affected by biased or incomplete sampling of extant thaumarchaeotal diversity, although independent approaches suggested this was not a major concern. Sensitivity analyses (SI Appendix, Fig. S5) suggested that our main findings were robust to random incomplete sampling. In addition, our conclusions were consistent across diversification analyses performed on a full range of sequences available within public databases using two independent phylogenetic markers: archaeal amoA (SI Appendix, Fig. S6), which is unique to ammonia oxidizing archaea; and 16S rRNA (SI Appendix, Figs. S7 and S8). Nevertheless, certain environments (e.g., hyperthermophilic, halophilic, or heavy metal-contaminated) may contain additional uncharacterized AOA diversity. A second potential caveat concerns variation in extinction rates during thaumarchaeotal evolution. Extinction rates are difficult to model on phylogenetic trees, even for multicellular eukaryotes, for which extinction is a frequent and reasonably well-characterized process (33, 34). This is compounded by our poor understanding of extinction in the prokaryotic world, which may be less frequent because of large population sizes, propensity for LGT, fast evolutionary rates, ease of dispersal, and the ability to form dormant or resistant states (e.g., ref. 35). Further, we currently lack a quantitative understanding of the factors that might influence extinction rates and how they vary among prokaryotic groups or between prokaryotes and eukaryotes. Importantly, the Bayesian methods we used provide a natural framework for incorporating uncertainty about these rates, and indeed the large credibility intervals surrounding these parameter estimates (Fig. 2B) suggest that our inferences were robust to a large range of extinction rates. Nevertheless, we cannot exclude the possibility that high extinction rates during early thaumarchaeotal evolution impacted the shape of our amoA phylogeny and therefore the inference of diversification rates through time. Nonetheless, it seems less likely that these issues would have affected reconstructions of environmental adaptation, which also strongly depend on contextual data measured within extant lineages.
Our analyses stand in contrast to the limited number of previous studies that examined diversification in microorganisms. Analyses that used the γ-statistic (36) suggest that diversification rates have decreased during the evolution of many free-living bacteria and archaea (37). Another approach, the likelihood of branching times, suggested a rapid early diversification followed by a decrease in rate for the obligate endosymbiont Borrelia burgdorferi clade (38). Such results suggest that previously available niches have been filled by microorganisms, a situation recaptured in experimental evolution studies with bacteria (39, 40). This is akin to the classic adaptive radiation model described in multicellular eukaryotes, whereby an initial burst of speciation is followed by declining diversification through time as new niches become exhausted (41, 42). The analyses that used amoA sequences invariably suggested that diversification rates have remained stable or even steadily increased from the apex rates of the original radiation event (43) (Fig. 1, Inset, Fig. 2B, and SI Appendix, Figs. S5 and S6). Although 16S rRNA analysis did imply a minor decrease in diversification rate, this result was likely a product of undersampling (SI Appendix, Figs. S7 and S8). With realistic sampling assumptions for 16S rRNA, our results were entirely congruent with the pattern observed for amoA (SI Appendix, Fig. S6). Such long-term maintenance of high diversification rates does not support a classical model of niche filling (44) and may reflect a higher rate at which niches become available for prokaryotes. Given the long-term co-occurrence of high diversification and apparent environmental adaptation rates after early radiation (Fig. 3), our data lend support to this idea in terms of terrestrial niches associated with variable ecological characteristics. Another interesting possibility is that high ongoing diversification rates in Thaumarchaeota are associated with high levels of niche switching without extensive specialization, which has been suggested for certain eukaryotes (45–47), or high levels of energy source switching by assimilating different substrates such as inorganic or organic nitrogen compounds (48, 49). Our work therefore demonstrates how approaches that combine contemporary molecular and environmental data can be used to compare evolutionary processes between prokaryotes and eukaryotes, and raises important questions about the similarities and differences in the long-term drivers of diversification across the tree of life.
Materials and Methods
Phylogenetic Analyses.
Because very few thaumarchaeotal genomes have been published, we used the Roche 454 platform to sequence thaumarchaeotal amoA (9) and 16S rRNA diversity (27) from a range of UK soils. These data were used to generate three separate sequence alignments, two for amoA (370 and 613 sequences; both with 388 aligned nucleotide sites) and one for 16S rRNA (508 sequences; 568 aligned nucleotide sites). These alignments were built from the 454 data used alone (amoA 370-sequence alignment) or after merging with a wider representation of nonredundant sequences available from GenBank or the Silva database (50) (amoA 613-sequence alignment and 16S rRNA alignment; SI Appendix). Separate amoA alignments were necessary because downstream analyses required environmental data unavailable in the public datasets. Before phylogenetic analysis, redundant sequences were dereplicated at defined cutoffs by using Uclust (51) (SI Appendix), and the presence of recombination was checked with RDP4 (52) before recombinant sequences were removed if necessary. A recombination event was accepted in RDP4 when statistically significant (P < 0.01) in three of the four following methods: RDP, GENECONV, MaxChi, and Bootscan. Bayesian relaxed molecular clock phylogenetic analysis were performed in BEAST (Bayesian Evolutionary Analysis Sampling Trees) version 1.7 (53) after screening alignments for mutational saturation (54) and selecting the best substitution model in PartitionFinder (55). For amoA, the third codon position (CP) was excluded because of evidence of saturation, and CP1 and CP2 were separately modeled under GTR+G with equal base frequencies. The 16S rRNA was modeled under GTR+G+I with equal base frequencies. A Yule speciation prior (56) and uncorrelated lognormal relaxed clock model (20) was set, and two Markov chain Monte Carlo (MCMC) chains were run for between 100 and 500 million steps, sampling every 10,000 steps. Convergence was confirmed by using Tracer version 1.5 (tree.bio.ed.ac.uk/software/tracer/), and effective sample sizes exceeded 200 for all parameters after the first 10% of steps were removed. Maximum clade credibility trees from converged MCMC runs were generated by using TreeAnnotator version 1.7 (53).
The pH specialization of phylogenetic clusters was characterized as described previously (9) using generalized additive modeling with the mgcv package (57) in R (58).
Comparative Phylogenetics.
A lineage-through-time plot and γ-statistic analysis were performed with the 370-sequence amoA tree using LASER (59) in R (58). Four separate phylogenetic trees were then analyzed by using the BAMM program (13, 14). This included the three trees described earlier (i.e., 370- and 613-sequence amoA tree and 508-sequence 16S rRNA tree) and an additional amoA tree generated by pruning the 370-sequence tree. For the original 370-sequence amoA tree derived from 454 sequencing (9), the pH of soil samples was evenly distributed across the full range (pH 3.48–8.74), but this was not the case for samples on which the 613-sequence amoA and 508-sequence 16S rRNA trees were based. In addition, for 181 sequences embedded within the 370-sequence amoA tree, a wider range of environmental data were available from the same sampled soils (quantitative measures of organic matter and aluminum, phosphorus, molybdenum, manganese, mercury, copper, and zinc levels; SI Appendix, Table S2). Therefore, a new 181-sequence amoA tree was obtained by removing tips with missing environmental data, which were distributed across the whole tree. Separate BAMM analyses of diversification rates were performed for all four trees and phenotypic evolutionary rates for the 370- and 181-sequence amoA trees, using the full range of contemporary environmental data available. All phenotypic rate analyses were based on soil mean values defined for each tip of the tree based on dereplicated sequences. MCMC simulations in BAMM were run for 100 million and 1,000 million steps for diversification and phenotypic analyses, respectively, sampling parameters every 10,000 or 100,000 steps, respectively. A series of scripts was used to analyze the MCMC data and confirm convergence of the chains within the R package BAMMtools (60). pH preferences were reconstructed at each node of the 370-amoA tree using the RidgeRace algorithm (28), a method that explicitly allows heterogeneity in phenotypic rates across all branches in a tree.
Supplementary Material
Acknowledgments
The authors thank Dr. Anthony Travis (University of Aberdeen) for help with Bio-Linux. This work was funded by Natural Environment Research Council Fellowship NE/J019151/1 (to C.G.-R.) and European Research Grant ERC2010-AdG-268701 (to T.M.E. and T.A.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The alignment files and phylogenetic trees in this paper have been deposited in the Dryad Digital Repository, datadryad.org (10.5061/dryad.0nv00).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419329112/-/DCSupplemental.
References
- 1.Gruber N, Galloway JN. An Earth-system perspective of the global nitrogen cycle. Nature. 2008;451(7176):293–296. doi: 10.1038/nature06592. [DOI] [PubMed] [Google Scholar]
- 2.Oswald R, et al. HONO emissions from soil bacteria as a major source of atmospheric reactive nitrogen. Science. 2013;341(6151):1233–1235. doi: 10.1126/science.1242266. [DOI] [PubMed] [Google Scholar]
- 3.Nicol GW, Leininger S, Schleper C, Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol. 2008;10(11):2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- 4.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: Proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6(3):245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 5.Spang A, et al. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18(8):331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Leininger S, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442(7104):806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 7.Gubry-Rangin C, Nicol GW, Prosser JI. Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol. 2010;74(3):566–574. doi: 10.1111/j.1574-6941.2010.00971.x. [DOI] [PubMed] [Google Scholar]
- 8.Stopnišek N, et al. Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment. Appl Environ Microbiol. 2010;76(22):7626–7634. doi: 10.1128/AEM.00595-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gubry-Rangin C, et al. Niche specialization of terrestrial archaeal ammonia oxidizers. Proc Natl Acad Sci USA. 2011;108(52):21206–21211. doi: 10.1073/pnas.1109000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prosser JI, Nicol GW. Archaeal and bacterial ammonia-oxidisers in soil: The quest for niche specialisation and differentiation. Trends Microbiol. 2012;20(11):523–531. doi: 10.1016/j.tim.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 11.FitzJohn RG. Diversitree: Comparative phylogenetic analyses of diversification in R. Methods Ecol Evol. 2012;3(6):1084–1092. [Google Scholar]
- 12.Revell LJ. phytools: An R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3(2):217–223. [Google Scholar]
- 13.Rabosky DL. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS ONE. 2014;9(2):e89543. doi: 10.1371/journal.pone.0089543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rabosky DL, et al. Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nat Commun. 2013;4:1958. doi: 10.1038/ncomms2958. [DOI] [PubMed] [Google Scholar]
- 15.Rabosky DL, Donnellan SC, Grundler M, Lovette IJ. Analysis and visualization of complex macroevolutionary dynamics: An example from Australian scincid lizards. Syst Biol. 2014;63(4):610–627. doi: 10.1093/sysbio/syu025. [DOI] [PubMed] [Google Scholar]
- 16.Spang A, et al. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: Insights into metabolic versatility and environmental adaptations. Environ Microbiol. 2012;14(12):3122–3145. doi: 10.1111/j.1462-2920.2012.02893.x. [DOI] [PubMed] [Google Scholar]
- 17.Lehtovirta-Morley LE, et al. Characterisation of terrestrial acidophilic archaeal ammonia oxidisers and their inhibition and stimulation by organic compounds. FEMS Microbiol Ecol. 2014;89(3):542–552. doi: 10.1111/1574-6941.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437(7058):543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 19.Pester M, et al. amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol. 2012;14(2):525–539. doi: 10.1111/j.1462-2920.2011.02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4(5):e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petitjean C, Moreira D, López-García P, Brochier-Armanet C. Horizontal gene transfer of a chloroplast DnaJ-Fer protein to Thaumarchaeota and the evolutionary history of the DnaK chaperone system in Archaea. BMC Evol Biol. 2012;12:226. doi: 10.1186/1471-2148-12-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Douzery EJP, Snell EA, Bapteste E, Delsuc F, Philippe H. The timing of eukaryotic evolution: Does a relaxed molecular clock reconcile proteins and fossils? Proc Natl Acad Sci USA. 2004;101(43):15386–15391. doi: 10.1073/pnas.0403984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parfrey LW, Lahr DJG, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci USA. 2011;108(33):13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doolittle WF, Zhaxybayeva O. What is a prokaryote? In: Rosenberg E, et al., editors. The Prokaryotes – Prokaryotic Biology and Symbiotic Associations. Springer; Heidelberg: 2013. pp. 22–37. [Google Scholar]
- 25.Kingman JFC. On the genealogy of large populations. J Appl Probab. 1982;19A:27–43. [Google Scholar]
- 26.Zhaxybayeva O, Gogarten JP. Cladogenesis, coalescence and the evolution of the three domains of life. Trends Genet. 2004;20(4):182–187. doi: 10.1016/j.tig.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Vico-Oton E, Quince C, Nicol GW, Prosser JI, Gubry-Rangin C. Phylogenetic congruence and ecological coherence in terrestrial Thaumarchaeota. ISME J 2015 doi: 10.1038/ismej.2015.101. , 10.1038/ISMEJ.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kratsch C, McHardy AC. RidgeRace: Ridge regression for continuous ancestral character estimation on phylogenetic trees. Bioinformatics. 2014;30(17):i527–i533. doi: 10.1093/bioinformatics/btu477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rinke C, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499(7459):431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 30.Blank CE. Phylogenomic dating—the relative antiquity of archaeal metabolic and physiological traits. Astrobiology. 2009;9(2):193–219. doi: 10.1089/ast.2008.0248. [DOI] [PubMed] [Google Scholar]
- 31.Di Giulio M. The universal ancestor and the ancestor of bacteria were hyperthermophiles. J Mol Evol. 2003;57(6):721–730. doi: 10.1007/s00239-003-2522-6. [DOI] [PubMed] [Google Scholar]
- 32.Groussin M, Gouy M. Adaptation to environmental temperature is a major determinant of molecular evolutionary rates in archaea. Mol Biol Evol. 2011;28(9):2661–2674. doi: 10.1093/molbev/msr098. [DOI] [PubMed] [Google Scholar]
- 33.Rabosky DL. Extinction rates should not be estimated from molecular phylogenies. Evolution. 2010;64(6):1816–1824. doi: 10.1111/j.1558-5646.2009.00926.x. [DOI] [PubMed] [Google Scholar]
- 34.Quental TB, Marshall CR. Diversity dynamics: Molecular phylogenies need the fossil record. Trends Ecol Evol. 2010;25(8):434–441. doi: 10.1016/j.tree.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Dykhuizen DE. Santa Rosalia revisited: Why are there so many species of bacteria? Antonie van Leeuwenhoek. 1998;73(1):25–33. doi: 10.1023/a:1000665216662. [DOI] [PubMed] [Google Scholar]
- 36.Pybus OG, Harvey PH. Testing macro-evolutionary models using incomplete molecular phylogenies. Proc Biol Sci. 2000;267(1459):2267–2272. doi: 10.1098/rspb.2000.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin AP, Costello EK, Meyer AF, Nemergut DR, Schmidt SK. The rate and pattern of cladogenesis in microbes. Evolution. 2004;58(5):946–955. doi: 10.1111/j.0014-3820.2004.tb00429.x. [DOI] [PubMed] [Google Scholar]
- 38.Morlon H, Kemps BD, Plotkin JB, Brisson D. Explosive radiation of a bacterial species group. Evolution. 2012;66(8):2577–2586. doi: 10.1111/j.1558-5646.2012.01598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138(6):1315–1341. [Google Scholar]
- 40.Brockhurst MA, Morgan AD, Rainey PB, Buckling A. Population mixing accelerates coevolution. Ecol Lett. 2003;6:975–979. [Google Scholar]
- 41.Rabosky DL, Lovette IJ. Explosive evolutionary radiations: Decreasing speciation or increasing extinction through time? Evolution. 2008;62(8):1866–1875. doi: 10.1111/j.1558-5646.2008.00409.x. [DOI] [PubMed] [Google Scholar]
- 42.McGuire JA, et al. Molecular phylogenetics and the diversification of hummingbirds. CurrBiol. 2014;24(8):910–916. doi: 10.1016/j.cub.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 43.Cao H, Auguet J-C, Gu J-D. Global ecological pattern of ammonia-oxidizing archaea. PLoS ONE. 2013;8(2):e52853. doi: 10.1371/journal.pone.0052853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price TD, et al. Niche filling slows the diversification of Himalayan songbirds. Nature. 2014;509(7499):222–225. doi: 10.1038/nature13272. [DOI] [PubMed] [Google Scholar]
- 45.Hardy NB, Otto SP. Specialization and generalization in the diversification of phytophagous insects: tests of the musical chairs and oscillation hypotheses. Proc Biol Sci. 2014;281(1795):20132960. doi: 10.1098/rspb.2013.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bush GL. Sympatric host race formation and speciation in frugivorous flies of the genus Rhagoletis (Diptera, Tephritidae) Evolution. 1969;23(6):237–251. doi: 10.1111/j.1558-5646.1969.tb03508.x. [DOI] [PubMed] [Google Scholar]
- 47.Zietara MS, Lumme J. Speciation by host switch and adaptive radiation in a fish parasite genus Gyrodactylus (Monogenea, Gyrodactylidae) Evolution. 2002;56(12):2445–2458. doi: 10.1111/j.0014-3820.2002.tb00170.x. [DOI] [PubMed] [Google Scholar]
- 48.Qin W, et al. Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci USA. 2014;111(34):12504–12509. doi: 10.1073/pnas.1324115111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mussmann M, et al. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc Natl Acad Sci USA. 2011;108(40):16771–16776. doi: 10.1073/pnas.1106427108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yilmaz P, et al. The SILVA and “all-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014;42(database issue):D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 52.Martin DP, et al. RDP3: A flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26(19):2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia X, Xie Z, Salemi M, Chen L, Wang Y. An index of substitution saturation and its application. Mol Phylogenet Evol. 2003;26(1):1–7. doi: 10.1016/s1055-7903(02)00326-3. [DOI] [PubMed] [Google Scholar]
- 55.Lanfear R, Calcott B, Ho SY, Guindon S. Partitionfinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29(6):1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 56.Gernhard T. The conditioned reconstructed process. J Theor Biol. 2008;253(4):769–778. doi: 10.1016/j.jtbi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 57.Wood SN. Fast stable direct fitting and smoothness selection for generalized additive models. J R Stat Soc Series B Stat Methodol. 2008;70(3):495–518. [Google Scholar]
- 58.R Development Core Team 2007 R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna), www.R-Project.org.
- 59.Rabosky DL. LASER: A maximum likelihood toolkit for detecting temporal shifts in diversification rates from molecular phylogenies. Evol Bioinform Online. 2006;2:273–276. [PMC free article] [PubMed] [Google Scholar]
- 60.Rabosky DL, et al. BAMMtools: An R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods EcolEvol. 2014;5(7):701–707. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.