Significance
Actin is one of the most abundant and highly conserved eukaryotic proteins, but the basis for the exquisite sequence conservation in actin is not known. In contrast, bacterial actin-like proteins display almost no sequence conservation and form very different filaments. We have examined the filaments formed by an actin-like protein in the third kingdom of life, Archaea, and although they only have a single strand, the strand is very similar to each of the two strands in actin. This gives previously unidentified insights into the divergence of archaea and eukaryotes.
Keywords: helical polymers, variable twist, cytoskeletal filaments, crenactin
Abstract
The prokaryotic origins of the actin cytoskeleton have been firmly established, but it has become clear that the bacterial actins form a wide variety of different filaments, different both from each other and from eukaryotic F-actin. We have used electron cryomicroscopy (cryo-EM) to examine the filaments formed by the protein crenactin (a crenarchaeal actin) from Pyrobaculum calidifontis, an organism that grows optimally at 90 °C. Although this protein only has ∼20% sequence identity with eukaryotic actin, phylogenetic analyses have placed it much closer to eukaryotic actin than any of the bacterial homologs. It has been assumed that the crenactin filament is double-stranded, like F-actin, in part because it would be hard to imagine how a single-stranded filament would be stable at such high temperatures. We show that not only is the crenactin filament single-stranded, but that it is remarkably similar to each of the two strands in F-actin. A large insertion in the crenactin sequence would prevent the formation of an F-actin-like double-stranded filament. Further, analysis of two existing crystal structures reveals six different subunit–subunit interfaces that are filament-like, but each is different from the others in terms of significant rotations. This variability in the subunit–subunit interface, seen at atomic resolution in crystals, can explain the large variability in the crenactin filaments observed by cryo-EM and helps to explain the variability in twist that has been observed for eukaryotic actin filaments.
Actin is one of the most highly conserved as well as abundant eukaryotic proteins. From chickens to humans, an evolutionary separation of ∼350 million years, there are no amino acid changes in the skeletal muscle isoform of actin (1). There are at least six different mammalian isoforms that are quite similar to each other, and all seem to have diverged from a common ancestral actin gene (2). In contrast, we now know that bacteria have actin-like proteins that share a common fold (3–5) but have vanishingly little sequence similarity both among themselves and to eukaryotic actin (6).
Two recent crystal structures of a crenarchaeal actin, crenactin (7, 8), raise interesting questions about the structure of the crenactin filament and its evolutionary relationship to F-actin. In both crystals (with two different space groups) crenactin forms a single-stranded filament with eight subunits per ∼420-Å right-handed turn, with a rise and rotation per subunit, therefore, of ∼53 Å and 45°, respectively. In contrast, in F-actin there is a rise and rotation of ∼55 Å and ∼27° along each of the two long-pitch right-handed strands. It was stated (9) that outside of the crystal the crenactin filaments are double-stranded, based upon the suggestion (8) that a single-stranded filament would unlikely be stable and that power spectra from crenactin filaments showed a strong layer line at ∼1/(210 Å), half of the repeat in the crystals.
We have been able to image and reconstruct at low resolution the filaments formed by crenactin. Surprisingly, these filaments contain only a single strand, as opposed to the two strands present in F-actin and in the filaments formed by a number of bacterial actin-like proteins (9–13). We show that the crenactin filament is consistent with the single strand seen in crystals (7, 8), and that this strand is quite similar to each of the two strands within F-actin. This supports arguments about the close phylogenetic proximity between eukaryotic actin and archaeal actin (14, 15) and highlights the substantial divergence that has taken place between the bacterial actin-like proteins and eukaryotic actin (6).
Results
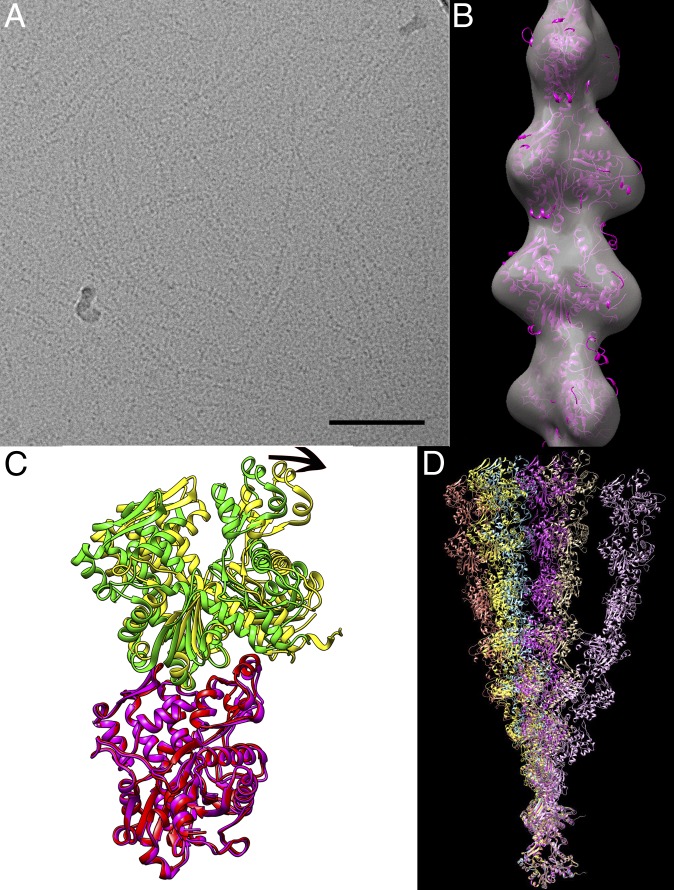
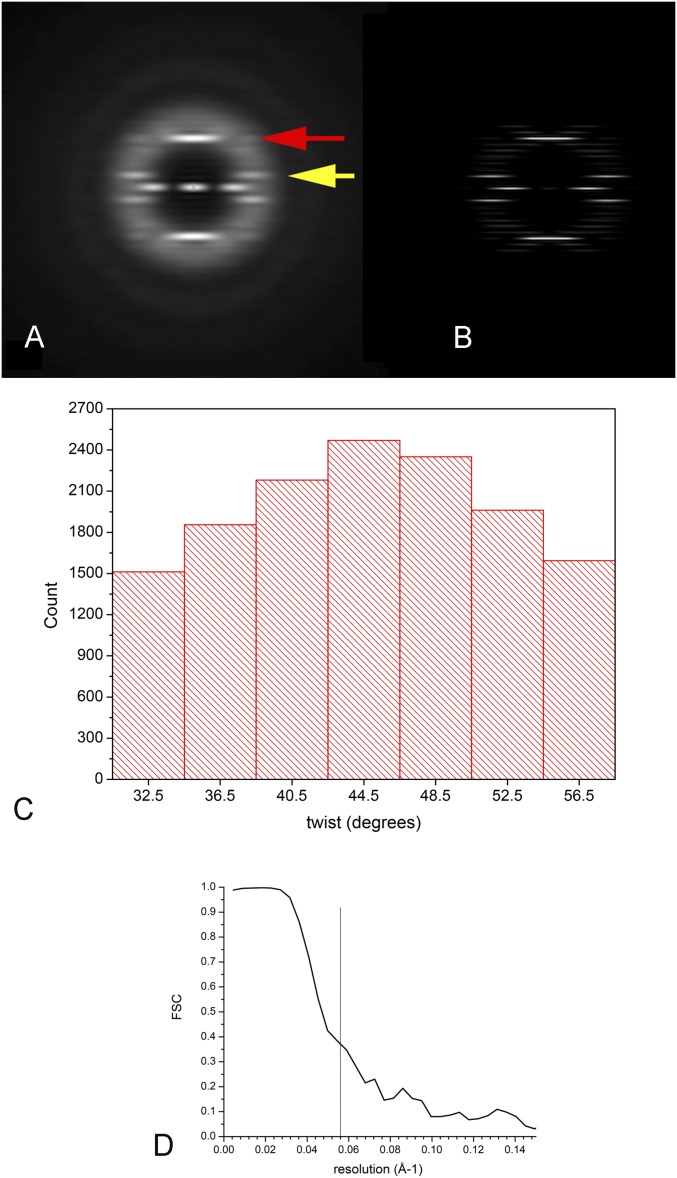
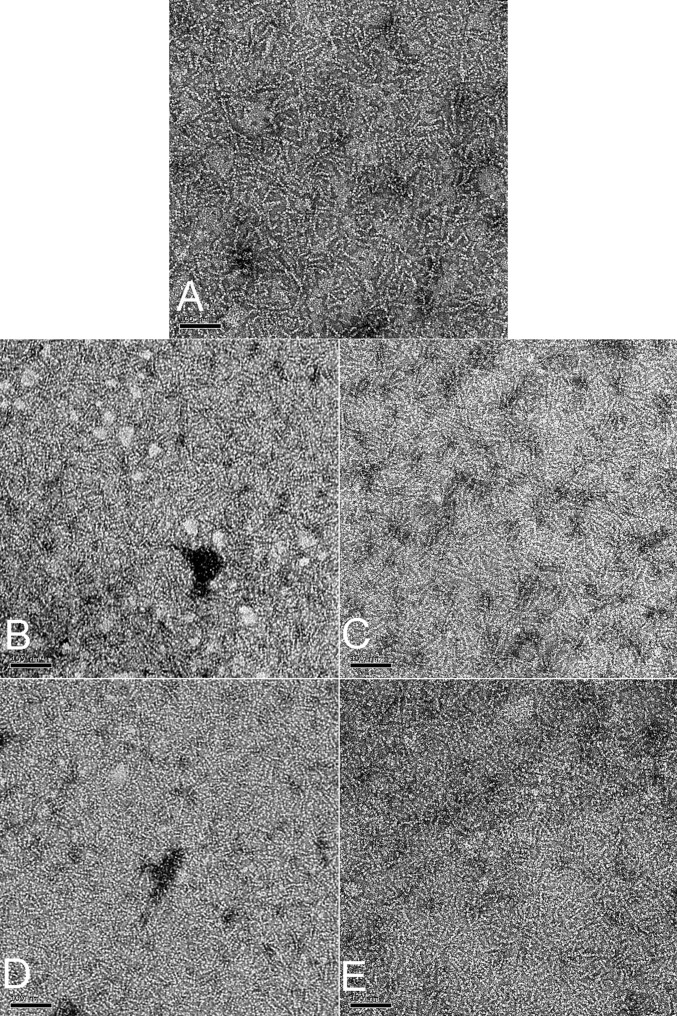
We have imaged the Pyrobaculum calidifontis crenactin filaments using electron cryomicroscopy (cryo-EM) (Fig. 1A). It can be seen that these filaments are quite flexible, which limits the resolution that may be achieved in 3D reconstructions. More importantly, analysis of segments reveals a remarkable variability in the pitch. Power spectra (Fig. S1) have been generated from multiple classes after sorting by twist (Fig. S1 and Movie S1), and these confirm that the classifications are really based upon differences in pitch, and are not an artifact. The power spectra can only be indexed as arising from a one-start helix with a mean pitch of ∼420 Å and with a mean twist of ∼8.1 subunits per turn. A reconstruction (Fig. 1B) at ∼18-Å resolution (Fig. S1D) from the crenactin filaments shows why the power spectra have a very strong n = 2 layer line and very weak n = 1 layer lines: The two major domains of the crenactin subunit are nearly equal in size and arranged symmetrically around the helical axis. All attempts to significantly extend the resolution beyond what is shown (such as by using shorter boxes) failed, likely owing to the fact that the local variability within these filaments is considerably greater than in F-actin. We also asked whether filament formation and morphology were sensitive to temperature. Samples polymerized or incubated at temperatures up to 90 °C seemed indistinguishable from room-temperature samples by negative staining (Fig. S2), because examining these samples by cryo-EM would likely be impossible owing to the higher temperatures preventing vitrification.
Fig. 1.
Crenactin filaments imaged by cryo-EM and crystallography. (A) A field of frozen-hydrated filaments over a hole in a lacey carbon grid. (Scale bar, 1,000 Å.) (B) The surface of a reconstruction is shown in transparent gray, and a crystal structure (magenta) of the crenactin subunit has been placed in the filament by rigid-body fitting. (C) The variability in filament-like subunit–subunit interfaces in crenactin crystal structures can be seen in this comparison between 4BQL chains D (magenta) and A (yellow) and chains C (red) and B (green) from the same crystal unit cell. Subunit C has been aligned to subunit D (0.8 Å rmsd), and the resulting transformation imposed on subunit B. The arrow at the top shows the considerable rotation between the two. (D) Six different filaments can be generated by imposing the six different interfaces in the two crenactin crystals. For example, the symmetry operation best relating chain A to chain B is found and then repeatedly applied to chain A to generate one filament, whereas a different filament will be generated by finding the symmetry operation relating chain B to the chain A above it. The filaments, from left to right (at the top) are 4CJ7_AB, 4BQL_BDs, 4BQL_AD, 4CJ7_ABs, 4BQL_ACs, and 4BQL_BC.
Fig. S1.
(A) The averaged power spectrum computed from 5,820 segments, each 288 pixels long, after sorting segments by pitch. The layer line indicated by the yellow arrow is an n = 2 [at ∼1/(210 Å)] and the layer line indicated by the red arrow is an n = 0 [at ∼1/(53 Å)]. (B) The power spectrum from the projection of a reconstruction that is 800 pixels long. In the reconstruction, the n = 1 layer line between the equator and the strong n = 2 is so weak as to be invisible in this figure. (C) A histogram of the twist distribution from 192-pixel-long segments using a reference-based procedure with models having a fixed axial rise and a variable twist. The legitimacy of the sorting is shown in Movie S1, where the power spectra from these different bins behave as expected for a filament with a fixed rise and a variable twist. (D) The Fourier shell correlation (FSC) curve for the crenactin reconstruction. The vertical line is at 18-Å resolution.
Fig. S2.
Crenactin filaments at higher temperatures. (A) A control experiment shows crenactin filaments prepared at room temperature. Crenactin (7 µM) was polymerized for 2 h in 30 mM Hepes buffer (pH 7.5), 0.5 M KCl, 4 mM MgCl2, and 2 mM ATP. The sample was diluted to 3 μM with the same buffer, negatively stained with 2% (wt/vol) uranyl acetate, and examined in a Tecnai-12 electron microscope at 80 keV and with a nominal magnification of 30,000×. Images were recorded on a 1,000- × 1,000 CCD camera. (Scale bar, 1,000 Å.) (B–D) The samples were polymerized and diluted as in A but then incubated for 30–45 min at either 50 °C (B), 70 °C (C), or 90 °C (D) before staining with uranyl acetate. (E) The sample was polymerized and diluted as in A but at 90 °C and then stained with uranyl acetate heated to 90 °C.
Nevertheless, even at this limited resolution several points are clear. The filaments must be right-handed, as they are in the crystals, owing to the fact that the curvature of the subunit is great enough even at this resolution to establish the correct hand. Relating the subunit–subunit contacts in the filament to those within the crystals is interesting. There are actually six separate filament-like subunit–subunit interfaces in the two crystals, and they show considerable differences (Fig. 1 C and D). In 4CJ7.PDB (8) there is a crystallographic 41 screw axis with two chains (A and B) in the crystal asymmetric unit, together generating an approximate 81 screw. This 81 screw symmetry is a pseudosymmetry, because the interface between chains A and B within the asymmetric unit (1,093 Å2 of buried surface area) is not the same as the interface between chain B and chain A from the neighboring asymmetric unit (1,053 Å2 of buried surface area). Similarly, in 4BQL.PDB (7) there are four molecules of crenactin in the crystallographic asymmetric unit, and these generate four significantly different filament-like subunit–subunit interfaces (ranging from 1,096–1,291 Å2 of buried surface area).
To assess the difference among these six subunit–subunit interfaces, pairwise rmsds were calculated over 77 common interfacial residues. The rmsds (Table 1) range from 0.36 Å up to 1.75 Å and show that the difference between the interfaces is largest between subunit–subunit interfaces from different crystals. Of the four interfaces present in 4BQL, 4BQL_BC (interface between chain B and C) is the most dissimilar. 4BQL_BDs and 4BQL_AD are very similar with an interface rmsd of only 0.36 Å. Rotations and shifts between neighboring subunits in these six different crystal interfaces yield average values that are very close to the ones determined experimentally by cryo-EM (Table 2). The average rotation of two neighboring subunits in the crystals is 45.3°, whereas it is 44.4° for the filaments. The average rise of 52.4 Å in the crystals is close to the ∼52 Å seen by cryo-EM.
Table 1.
Pairwise interface rmsds between the different crenactin interfaces
| I_RMSD | 4BQL_ACs | 4BQL_AD | 4BQL_BC | 4BQL_BDs | 4CJ7_AB | 4CJ7_ABs |
| 4BQL_ACs | — | 0.50 | 0.87 | 0.68 | 1.44 | 1.23 |
| 4BQL_AD | 0.50 | — | 1.08 | 0.36 | 1.17 | 1.09 |
| 4BQL_BC | 0.87 | 1.08 | — | 1.19 | 1.75 | 1.50 |
| 4BQL_BDs | 0.68 | 0.36 | 1.19 | — | 1.07 | 0.98 |
| 4CJ7_AB | 1.44 | 1.17 | 1.75 | 1.07 | — | 1.24 |
| 4CJ7_ABs | 1.23 | 1.09 | 1.50 | 0.98 | 1.24 | — |
The interface rmsd (I_RMSD) was calculated over 77 residues located in the subunit–subunit interface.
Table 2.
Rotation and shift between neighboring subunits of different crenactin complexes
| Measurement | 4BQL_ACs | 4BQL_AD | 4BQL_BC | 4BQL_BDs | 4CJ7_AB | 4CJ7_ABs | Average |
| Rotation, ° | 43.2 | 45.9 | 45.2 | 47.8 | 47.5 | 42.4 | 45.3 |
| Rise, Å | 53.0 | 53.0 | 51.7 | 52.7 | 51.7 | 52.4 | 52.4 |
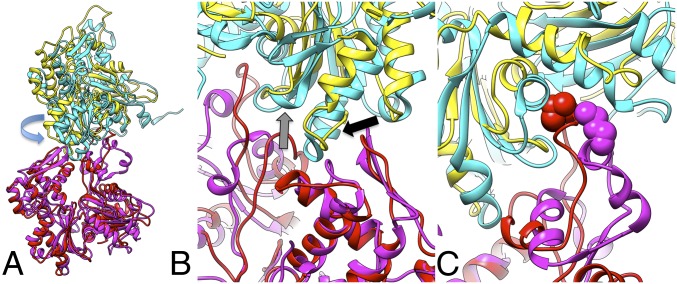
Given the limited resolution of the reconstruction, undoubtedly arising from the variability of the subunit–subunit interface within the filaments, it would be foolish to model this with a single subunit–subunit interface. Rather, we suggest that the observed filaments are consistent with the multiplicity of interfaces observed in the two crystals, and that what was thought to be “protofilaments” in the crenactin crystals are actually the crenactin filaments. The dimerization that occurs within both of the crenactin crystals results, we believe, from crystallographic symmetry constraints, because no sign of any such dimerization (i.e., a periodicity of ∼104 Å in addition to the ∼52-Å repeat) is seen by cryo-EM. A direct comparison between the atomic model for a single state of F-actin (16) and one interface in the crystals (chains A and D from 4BQL) shows the similarity in how subunits are stacked on top of each other in crenactin and in each of the two chains in F-actin (Fig. 2). Despite the ∼17° rotation (the difference between the ∼44° rotation per subunit in crenactin and the ∼27° rotation per subunit within one strand of F-actin) of a second crenactin subunit from the corresponding actin subunit (Fig. 2A) after the first subunits in each have been aligned, there is a very good correspondence between the elements in actin and in crenactin that make the interactions between these subunits (Fig. 2B). The loop 283–290 in actin corresponds to the loop 336–344 in crenactin, and these are both making an interaction between subdomain 3 (SD3) of the top subunit with subdomain 4 (SD4) of the bottom subunit (Fig. 2B, black arrow). The loop 166–172 in actin corresponds with the loop 192–199 in crenactin, and these are both making an interaction between SD3 of the top subunit with subdomain 2 (SD2) of the bottom subunit (Fig. 2B, gray arrow). Further, there is a conservation of Met44 in actin with Met52 in crenactin (7), and these are both making an interaction between SD2 of the bottom subunit with subdomain 1 (SD1) of the top subunit (Fig. 2C).
Fig. 2.
Comparison of F-actin with a crenactin crystal structure. (A) Two subunits of F-actin from the same long-pitch strand are shown in red and yellow. Two subunits of crenactin (chains D and A, 4BQL) are shown in magenta and cyan, respectively. The bottom subunit of crenactin (chain D) has been aligned to the bottom subunit of actin. The arrow indicates the ∼18° rotation between the top subunit in crenactin (cyan) from the corresponding subunit in F-actin (yellow) after such an alignment. (B) A close-up of the interface seen from the back (where the view in A is the front). The black arrow points to the similarity of the loop in F-actin (residues 283–290) with the corresponding region in crenactin (336–344), which makes an insertion from SD3 of the top subunit into SD4 of the bottom subunit. The gray arrow points to the correspondence between the 166–172 loop in actin with the 192–199 loop in crenactin, and both make an insertion from SD3 of the top subunit into SD2 of the bottom subunit. (C) Met52 in crenactin (magenta spheres) makes a corresponding interaction from the top of SD2 of one subunit to the bottom of SD1 of another subunit as Met44 in actin (red spheres).
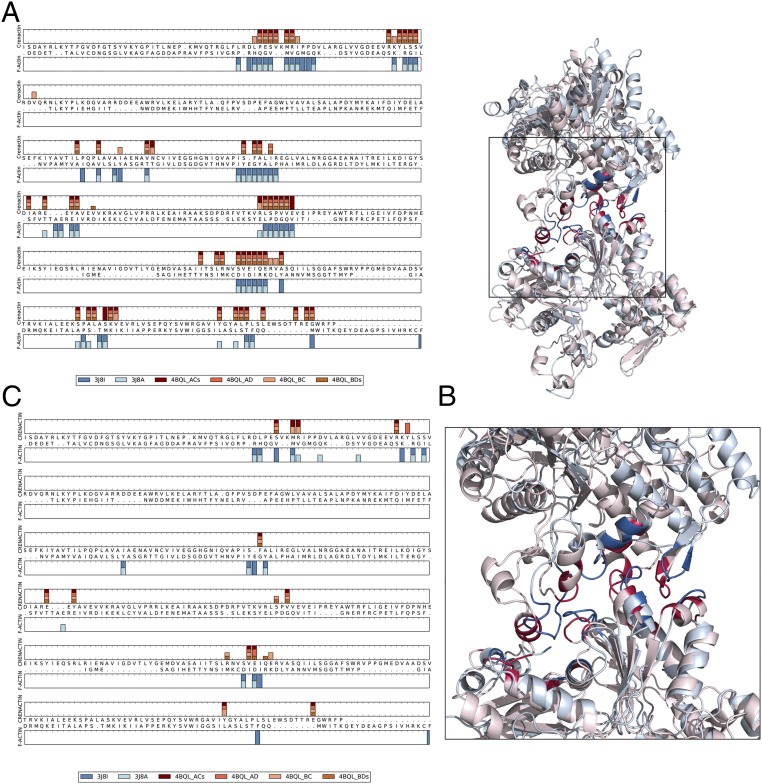
The remarkable similarity between the F-actin and crenactin interfaces becomes even more obvious when comparing their interfacial residues using a structural alignment (Fig. 3) of F-actin and one of the crenactin interfaces, 4BQL_BDs (the “s” at the end indicates that this interface arises from chains in two different asymmetric units). The interfacial residues of all four crenactin interfaces found in 4BQL and those in two recent F-actin models, 3J8I (16) and 3J8A (17), are highlighted. F-actin and crenactin share the same interaction sites, despite their low sequence identity. We focus on the interfaces present in 4BQL (7) because, according to the Protein Data Bank (PDB) validation reports (18), it is of higher quality than 4CJ7 (8). The side chain and Ramachandran outliers in 4CJ7, as well as clashes in the interfaces, might distort the conclusions.
Fig. 3.
Similarity of F-actin and crenactin interfaces. (A) Interfacial residues for the four different crenactin interfaces present in 4BQL and the two actin structures (blue) are mapped onto the structural alignment between actin and crenactin 4BQL_BDs (generated using the DaliLite webserver). Residues were defined as being part of the interface if they were within 8 Å (Cβ–Cβ distance) of a residue in the neighboring subunit. (B) The ribbon diagram for actin (3J8I) is shown in light blue, and crenactin (4BQL_BDs) is in light red. The interfacial residues identified in A are shown in dark blue for actin and dark red for crenactin. (C) Hot spots determined with computational alanine scanning for the four different crenactin interfaces present in 4BQL and the two actin structures are mapped onto the structural alignment between actin and crenactin. Hot spots are defined as residues having a change in binding energy ≥1 kcal/mol.
To identify residues with a dominant contribution to the free energy of subunit–subunit interactions, so-called binding hotspots, a computational alanine scanning (19–21) with the molecular modeling suite Rosetta (22, 23) was carried out for the interfaces of F-actin and crenactin. Fig. 3C shows the binding hot spots, defined as residues leading to a change in binding free energy ≥1 kcal/mol upon mutation to alanine (20, 21), for all investigated interfaces mapped onto the structural alignment between F-actin and crenactin 4BQL_BDs. The binding hotspots of crenactin and F-actin do not correlate as well as their interfacial residues, indicating that even though their binding interface is well conserved, different areas are responsible for contributing substantial binding energy. Table 3 shows the binding energies for the four interfaces in 4BQL and the interfaces in the two actin structures. The similarity among the different crenactin interfaces, as determined by their interface rmsd (Table 1), is reflected in the corresponding binding energies: 4BQL_AD and 4BQL_BDs show similar binding energies, and the binding energy of 4BQL_BC stands out the most. A comparison of the binding energies between F-actin and crenactin indicates that the binding between two crenactin subunits is substantially stronger than in the longitudinal interface of F-actin. This might explain why crenactin is able to form a single stranded filament whereas F-actin forms a double helix. In addition, the high binding energy might be responsible for the thermophilic characteristics of crenactin.
Table 3.
Binding energy and interactions across different crenactin and actin interfaces
| Measurement | 4BQL_ACs | 4BQL_AD | 4BQL_BC | 4BQL_BDs | 3J8I | 3J8A |
| Rosetta | ||||||
| Binding energy (dG_separated), REU | −29.83 | −27.85 | −37.15 | −25.35 | −16.26 | −16.07 |
| PDBePISA | ||||||
| Solvation energy (ΔiG), kcal/mol | −13.70 | −12.60 | −16.80 | −15.1 | −16.60 | −15.10 |
| No. of H bonds | 16 | 17 | 14 | 13 | 5 | 8 |
| No. of salt bridges | 1 | 1 | 0 | 0 | 4 | 10 |
| Interface area, Å2 | 1,124.6 | 1,113.7 | 1,269.6 | 1,099.2 | 1,149.0 | 1,093.2 |
The binding energy (dG_separated) was calculated with Rosetta for the relaxed interface structures. The binding energy ΔiG, the numbers of hydrogen bonds and salt bridges across the different interfaces, and the interface area were determined using the PISA webserver. REU, Rosetta energy units.
The PDBePISA webserver (24) provided additional information about the crenactin and actin interfaces. Whereas the crenactin interfaces have 13–17 hydrogen bonds between the two subunits, the actin interfaces only have 5 and 8. In actin, the number of salt bridges is much higher, whereas in crenactin almost no salt bridges are present, suggesting that the interfaces in crenactin and actin are stabilized quite differently.
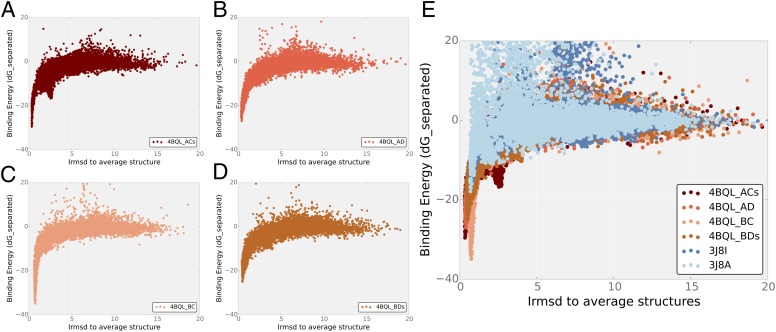
To computationally assess the flexibility of the crenactin filaments observed in our experiments, protein–protein dockings were carried out using the Rosetta docking protocol, RosettaDock (25, 26) (Fig. 4). The native conformation was identified for each docking run. Dockings for 4BQL_ACs and 4BQL_BC result in very narrow and deep energy funnels, suggesting that the binding is rather rigid and fixed. Dockings for 4BQL_BDs and 4BQL_AD result in much wider energy funnels with weaker energy, suggesting that these interfaces are more flexible than the other two. The lower binding energy might be tolerated to gain more flexibility. The results of the docking correlate well with the overall similarity between the interfaces according to interface rmsd and binding energy. The crenactin dockings can be compared (Fig. 4E) to dockings for F-actin using the two F-actin structures, 3J8I and 3J8A. Both actin docking experiments were able to identify the native conformation of the corresponding actin structure. It can clearly be seen that the binding of actin is significantly weaker than that of crenactin. Furthermore, it can be observed that the energy funnel is very narrow for actin, indicating that the binding is less flexible than it is in crenactin.
Fig. 4.
Docking of crenactin and actin interfaces. Plots of binding energy (dG_separated) vs. interface rmsd (Irmsd) for local docking of all four crenactin interfaces in 4BQL: (A) 4BQL_ACs, (B) 4BQL_AD, (C) 4bql_BC, and (D) 4BQL_BDs. (E) Superposition of the same calculations for two independent F-actin models (3J8A and 3J8I) onto the crenactin plots. For each crenactin docking, the same residues are used for rmsd calculation, as was done for the actin docking runs. The Irmsd of the crenactin structures is calculated to the average structure of all four crenactin interfaces present in 4BQL. Similarily, the Irmsd of the actin structures is calculated to the average structure of 3J8I and 3J8A.
Discussion
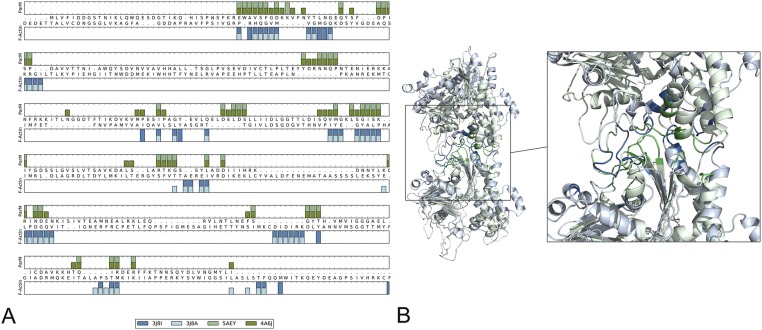
The remarkable diversity of filaments formed by bacterial actin-like proteins (such as the bacterial cell-shape–determining protein MreB, plasmid partitioning protein ParM, actin-like segregation protein AlfA, and magnetosome cytoskeleton protein MamK) has become clear (9–13, 27–31). This seems to reflect the large sequence divergence of the bacterial actin-like proteins (32), in striking contrast to the anomalous sequence conservation of eukaryotic actins. It has been shown, for example, that the filaments formed by the R1 plasmid ParM protein (10, 29–31) are extremely different from the filaments formed by the pSK41 ParM (13). Surprisingly, the pSK41 ParM subunit showed stronger structural similarity to the archaeal Thermoplasma acidophilum actin-like protein Ta0583, than to R1 ParM (13). Most recently, it has been shown that bacterial MreB forms double-stranded filaments where the two protofilaments are antiparallel (9). Although an initial paper (4) suggested that bacterial ParM filaments shared the same filament structure and subunit–subunit interfaces as F-actin, it was subsequently shown that the model for the ParM filament could not be correct because the filaments actually have the opposite helical hand to that in F-actin (10). Whereas in F-actin two subunits along the same long-pitch helical strand are related by a rotation of ∼27°, in ParM two subunits are related by a rotation of ∼−30°. So, it would be hard to imagine a quasi-equivalence of the interactions when there was a rotation of ∼57° between the two filaments. More importantly, however, it was noticed (4) that all of the elements in actin that are involved in polymerization have either diverged considerably (such as the four corners of the subunit) or are absent (the hydrophobic plug) in ParM (Fig. S3), which was surprising if it is assumed that they formed similar filaments. A paper reporting a reconstruction of a ParM filament (33) has suggested that there is a conservation of subunit–subunit contacts between ParM and actin, but this involves arguments such as that residues in a helix in ParM (Arg257 and Lys258) are equivalent to residues in a loop in actin (Glu270 and Gly268) that has the opposite polarity, raising questions about this equivalence. When ParM is aligned to actin and the longitudinal interfacial residues in both are mapped onto this alignment (Fig. S2) it can be seen how different the two are.
Fig. S3.
Interfacial residues of ParM and F-actin. (A) Interfacial residues for two ParM models (5AEY and 4A6J, green) and two actin models (3J8A and 3J8I, blue) are mapped onto the structural alignment between actin (3J8I) and ParM (5AEY) (generated using the DaliLite webserver). Residues were defined as being part of the interface if they were within 8 Å (Cβ–Cβ distance) of a residue in the neighboring subunit. (B) The ribbon diagram for actin (3J8I) is shown in light blue and ParM (5AEY) is in light green. The interfacial residues identified in A are shown in dark blue for actin and dark green for ParM.
Phylogenetic analysis has shown that whereas the crenarchaeal actins only share ∼20% sequence identity with eukaryotic actin they are much more closely related to actin than are the bacterial homologs (14). This has led to the suggestion that eukaryotic actin evolved from an ancestral archaeal actin (34). The recent report (15) that a lokiarchaeal actin has greater similarity to eukaryotic actin than it does to the crenactins provides strong support for this suggestion. Our finding that the crenarchaeal filament is single-stranded is surprising and unexpected (8, 9), and the fact that the one strand in the crenarchael actin filament is very similar to each of the two strands present in eukaryotic F-actin gives additional credibility to the suggestion that actin is much more closely related to crenactin than it is to the bacterial actin-like proteins (14). Our results cannot answer the question of whether actin evolved from crenactin, or whether both share a last common ancestor. For example, the hydrophobic plug in actin (residues 262–274) appears as an insertion in a single long helix present in bacterial ParM (4). However, in crenactin this insertion (residues 293–325) is much longer and involves 33 residues rather than the 13 in actin. This large loop in crenactin would provide steric hindrance to any second strand and explains why the subunit structure of crenactin is incompatible with a two-stranded F-actin-like filament. Given that a crystal structure of another archaeal actin (35) is very similar to ParM and has no hydrophobic plug whatsoever, and that actins from the newly discovered Lokiarchaeum have a hydrophobic plug similar to that found in eukaryotic actin (15), it is more parsimonious to assume that the longer hydrophobic plug arose as an insertion in crenactin, rather than the shorter plug being a deletion in actin. The Lokiarchaea provide strong support for the hypothesis that eukaryotic actin and the archaeal actins have a common ancestry (15), and that the proliferation of actin and actin-like proteins (e.g., actin-related proteins) already took place in an archaeal ancestor of eukaryotes. This would exclude horizontal gene transfer as an explanation for the commonalities seen. An interesting question that remains to be answered is whether the intense selective forces that have existed on almost every residue in eukaryotic actin arose with the presumably double-stranded actin filaments formed by the lokiarchaea, or whether they did not appear until eukaryotes existed.
The multiplicity of filament-like subunit–subunit interfaces seen in the two crenactin crystals (7, 8) is both interesting and unusual. Thus far, none of the more than 80 actin crystal structures has captured the subunit–subunit interface present in F-actin. Although it was believed that one actin crystal structure did contain the interface between SD4 of one subunit and SD3 of a subunit above it (36), comparison with new atomic models (16, 17) shows that the two interfaces are actually quite different, and one could not be simply converted to the other. The fact that the actual crenactin filament can be approximated by an 81 screw may be the most important factor allowing this filament to be crystalized, whereas F-actin cannot be approximated by a 21 screw despite early attempts to argue that a 21 screw axis in an actin crystal was the filament axis (37–40).
It was initially suggested that the variable twist in F-actin filaments was a rather unique property of actin (41), but subsequent studies have revealed that it is a quite general property of many polymers (42), including the bacterial actin-like filaments (10, 30). In F-actin the magnitude of this variability in twist is on the order of 6° per subunit, so it is interesting that the six different subunit–subunit interfaces in the two crenactin crystals (Fig. 1D) show rotations of similar magnitude. The multiplicity of these interfaces may simply reflect the fact that the main driving force in protein oligomerization and polymerization is burying surface area (43, 44), which allows for a good deal of promiscuity.
Materials and Methods
Recombinant crenactin from P. calidifontis was purified as described (7). Crenactin (10 µM) was polymerized in 30 mM Hepes-HCl (pH 7.4), 0.7 M KCl, 4 mM MgCl2, and 5 mM ATP for 1–2 h. It was diluted to 3–5 µM with the same buffer but with 0.25 mM KCl. The crenactin filaments were applied (2.5 μL) to lacey carbon grids and vitrified in a Vitrobot Mark IV (FEI, Inc.) at 95–100% relative humidity. The grids were imaged at 300 keV with 75,000× magnification in a Titan Krios (FEI, Inc.) using a Falcon 2 direct electron detector, resulting in a 1.05-Å-per-pixel sampling. The EPU software was used to control the microscope, and dose fractionation was used, with each full image containing seven frames. A defocus range of 1.4–5.0 μm was used, and 253 images were acquired. From these, 22,615 segments (each 384 pixels long) were cut out, using a shift of 75 pixels between adjacent segments (80% overlap). The integrated images (all seven frames) were used for cytoplasmic tubulo-filament determination with CTFFIND3 (45), as well as for filament boxing using e2helixboxer within EMAN2 (46). Reconstructions were generated using the first three fractions, containing a dose of ∼30 electrons per square angstrom. Owing to the limited resolution possible, images were decimated to 2.2 Å per pixel, and boxes were padded to 192 × 192 pixels for iterative helical real-space reconstruction (47). An averaged power spectrum from a single twist class (Fig. S1A) shows two very strong layer lines: an n = 0 meridional (red arrow), at ∼1/(53 Å) that arises from the rise per subunit, and an n = 2 layer line (yellow arrow) at ∼1/(220 Å). The power spectra can only be indexed as arising from a one-start helix with a mean pitch of ∼420 Å and with a mean twist of ∼8.1 subunits per turn. Multiple approaches to sorting and classification were tried, with none leading to any significant improvement in the reconstructions. A reference-based approach to sorting by the twist of the one-start helix (having a mean pitch of ∼420 Å) was used, where models were generated having a large range of pitch, but with a constant rise per subunit of 53 Å. This sorting was validated by looking at the power spectra from the classified segments (Movie S1). If segments all had a fairly fixed pitch but were being misclassified owing to a poor signal-to-noise ratio, then the resulting power spectra would all show a second-order (n = 2) near-equatorial layer line at ∼1/(210 Å) that was rather fixed. In fact, the power spectra show the variability expected, demonstrating that the classification worked properly. The final reconstruction was made from 1,749 segments, after excluding those with different pitch and with significant out-of-plane tilt. Reconstructions generated with more segments (involving a broader pitch distribution or more out-of-plane tilt) were not significantly better or worse.
Interface Analysis.
Rotation and rise between two neighboring subunits of the different complexes were determined with the UCSF Chimera package (48). The binding energies of the interfaces were calculated using the Rosetta InterfaceAnalyzer as used in (49). The PDBePISA webserver (50) was used to determine the solvation energy, the number of hydrogen bonds, the number of salt bridges, and the interface area.
Structural Alignment and Interface Residue Mapping.
Structural alignments between F-actin and crenactin subunits were generated using the DaliLite webserver (51), available at www.ebi.ac.uk/Tools/structure/dalilite. Interfacial residues between two subunits were determined using the Rosetta InterfaceAnalyzer as used in ref. 49. Residues were considered to be part of the interface if they were within 8 Å of a residue in the neighboring subunit. The interfacial residues of both subunits present in an interface were subsequently mapped onto the structurally aligned monomers.
Computational Alanine Scanning.
Computational alanine scanning (20, 21) was carried out for each relaxed interface using the Robetta webserver available at robetta.bakerlab.org/alascansubmit.jsp. Binding hotspots were classified as residues showing an increase in binding free energy upon mutation to alanine by more than 1 kcal/mol as defined in refs. 20 and 21.
Computational Docking Experiments.
Global docking experiments for each minimized complex were carried out using the Rosetta Docking Protocol, called RosettaDock (25, 26). For each complex, 10,000 structures were generated and ranked according to their binding energy as calculated with the Rosetta InterfaceAnalyzer.
SI Materials and Methods
Structure Preparation for Use in Rosetta.
To prepare the crystal structures for use in Rosetta (22, 23), each structure was subjected to an all-atom refinement with all-heavy-atom constraints in the Rosetta force field using the Rosetta Relax protocol. The relax protocol minimizes Rosetta energy by searching the local conformational space and thereby removing errors due to irregularities (e.g., atom overlaps or amino acid romaters with unfavorable Rosetta energy) while keeping all heavy atoms as close as possible to their starting positions in the crystal structure. The relaxation was carried out 10 times and the structure with the lowest energy was selected for further analysis.
relax.mpi.linuxgccrelease -ex1 -ex2aro -use_input_sc -nstruct 10 \\
-in:file:s ${pdb} -relax:constrain_relax_to_start_coords \\
-relax:ramp_constraints false -flip_HNQ -no_optH false
Computational Alanine Scanning.
Computational alanine scanning (20, 21) was carried out for each relaxed interface using the Robetta webserver available at robetta.bakerlab.org/alascansubmit.jsp. The protocol calculates the binding free energy changes brought about by alanine mutation at each interface position separately. An automatic scan of all interface residues was carried out; interface residues were defined as residues that have at least one atom within a sphere of 4 Å radius of an atom belonging to the other partner of the chain or a residue that becomes significantly buried upon complex formation (20). Binding hotspots were classified as residues showing an increase in binding free energy upon mutation to alanine by more than 1 kcal/mol as defined in refs. 20 and 21.
Docking Experiments.
RosettaDock (25, 26) starts from a local perturbation of the input structure (∼3 Å, ∼8 °) and carries out a low-resolution (e.g., the side chains are replaced by a single unified pseudoatom) Monte Carlo search with adaptive rotational and translational steps. After this low-resolution stage, the lowest-energy structure generated during that stage is selected for high-resolution refinement (25). To keep the docking local, the orientations of both docking partners were not altered. For each complex, 10,000 structures were generated.
docking_protocol.mpi.linuxgccrelease -in:file:s ${pdb} \\
-in:file:native ${pdb} -nstruct 10000 \\
-randomize1 false -randomize2 false -spin false -ex1 -ex2aro \\
-docking:sc_min true \\
-evaluation:Ca_Irms -evaluation:Irms \\
-dock_pert 3 8
The docking results were ranked according to the binding energy. The binding energy (dG_separated) was calculated with the Rosetta InterfaceAnalyzer as used in ref. 49 by separating and repacking both subunits present in a complex.
InterfaceAnalyzer.mpi.linuxgccrelease -in:file:s ${pdb} \\
-nstruct 50 -pack_input true -pack_separated true \\
-out:file:score_only \\
-pose_metrics::interface_cutoff 10 \\
-out:file:scorefile ${pdb}_10.sc -ex1 -ex2aro
The interface rmsd (IRMS) was calculated over common interface residues. Interface residues were defined as being part of the interface if they were within 10 Å of a residue in the other monomer. In total, 77 residues were used for calculating the rmsd for the different crenactin interfaces and 68 residues for the actin interfaces.
Supplementary Material
Acknowledgments
We thank Kelly Dryden (University of Virginia) for assistance with the microscopy. This work was supported by NIH Grants GM081303, S10-RR025067, and S10-OD018149 (to E.H.E.) and Grant 621-2010-5551 from the Swedish Research Council (to K.V. and A.-C.L.). The cryo-EM work was conducted at the Molecular Electron Microscopy Core facility at the University of Virginia, which is supported by the School of Medicine and was built with NIH Grant G20-RR31199.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 9150.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509069112/-/DCSupplemental.
References
- 1.Hennessey ES, Drummond DR, Sparrow JC. Molecular genetics of actin function. Biochem J. 1993;291(Pt 3):657–671. doi: 10.1042/bj2910657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miwa T, et al. Structure, chromosome location, and expression of the human smooth muscle (enteric type) gamma-actin gene: Evolution of six human actin genes. Mol Cell Biol. 1991;11(6):3296–3306. doi: 10.1128/mcb.11.6.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van den Ent F, Amos LA, Löwe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413(6851):39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- 4.van den Ent F, Møller-Jensen J, Amos LA, Gerdes K, Löwe J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 2002;21(24):6935–6943. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petek NA, Mullins RD. Bacterial actin-like proteins: Purification and characterization of self-assembly properties. Methods Enzymol. 2014;540:19–34. doi: 10.1016/B978-0-12-397924-7.00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derman AI, et al. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: Regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol Microbiol. 2009;73(4):534–552. doi: 10.1111/j.1365-2958.2009.06771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindås AC, Chruszcz M, Bernander R, Valegård K. Structure of crenactin, an archaeal actin homologue active at 90°C. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 2):492–500. doi: 10.1107/S1399004714000935. [DOI] [PubMed] [Google Scholar]
- 8.Izoré T, Duman R, Kureisaite-Ciziene D, Löwe J. Crenactin from Pyrobaculum calidifontis is closely related to actin in structure and forms steep helical filaments. FEBS Lett. 2014;588(5):776–782. doi: 10.1016/j.febslet.2014.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van den Ent F, Izoré T, Bharat TA, Johnson CM, Löwe J. Bacterial actin MreB forms antiparallel double filaments. eLife. 2014;3:e02634. doi: 10.7554/eLife.02634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orlova A, et al. The structure of bacterial ParM filaments. Nat Struct Mol Biol. 2007;14(10):921–926. doi: 10.1038/nsmb1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozyamak E, Kollman J, Agard DA, Komeili A. The bacterial actin MamK: In vitro assembly behavior and filament architecture. J Biol Chem. 2013;288(6):4265–4277. doi: 10.1074/jbc.M112.417030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Popp D, et al. Polymeric structures and dynamic properties of the bacterial actin AlfA. J Mol Biol. 2010;397(4):1031–1041. doi: 10.1016/j.jmb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 13.Popp D, et al. Structure and filament dynamics of the pSK41 actin-like ParM protein: Implications for plasmid DNA segregation. J Biol Chem. 2010;285(13):10130–10140. doi: 10.1074/jbc.M109.071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ettema TJ, Lindås AC, Bernander R. An actin-based cytoskeleton in archaea. Mol Microbiol. 2011;80(4):1052–1061. doi: 10.1111/j.1365-2958.2011.07635.x. [DOI] [PubMed] [Google Scholar]
- 15.Spang A, et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521(7551):173–179. doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galkin VE, Orlova A, Vos MR, Schröder GF, Egelman EH. Near-atomic resolution for one state of f-actin. Structure. 2015;23(1):173–182. doi: 10.1016/j.str.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von der Ecken J, et al. Structure of the F-actin-tropomyosin complex. Nature. 2015;519(7541):114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gore S, Velankar S, Kleywegt GJ. Implementing an X-ray validation pipeline for the Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):478–483. doi: 10.1107/S0907444911050359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massova I, Kollman PA. Computational alanine scanning to probe protein-protein interactions: A novel approach to evaluate binding free energies. J Am Chem Soc. 1999;121(36):8133–8143. [Google Scholar]
- 20.Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein-protein interfaces. Sci STKE. 2004;2004(219):pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 21.Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc Natl Acad Sci USA. 2002;99(22):14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leaver-Fay A, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaufmann KW, Lemmon GH, Deluca SL, Sheehan JH, Meiler J. Practically useful: What the Rosetta protein modeling suite can do for you. Biochemistry. 2010;49(14):2987–2998. doi: 10.1021/bi902153g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 25.Chaudhury S, et al. Benchmarking and analysis of protein docking performance in Rosetta v3.2. PLoS ONE. 2011;6(8):e22477. doi: 10.1371/journal.pone.0022477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray JJ, et al. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331(1):281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 27.Polka JK, Kollman JM, Agard DA, Mullins RD. The structure and assembly dynamics of plasmid actin AlfA imply a novel mechanism of DNA segregation. J Bacteriol. 2009;191(20):6219–6230. doi: 10.1128/JB.00676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popp D, et al. Filament structure, organization, and dynamics in MreB sheets. J Biol Chem. 2010;285(21):15858–15865. doi: 10.1074/jbc.M109.095901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popp D, et al. Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability. EMBO J. 2008;27(3):570–579. doi: 10.1038/sj.emboj.7601978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galkin VE, Orlova A, Rivera C, Mullins RD, Egelman EH. Structural polymorphism of the ParM filament and dynamic instability. Structure. 2009;17(9):1253–1264. doi: 10.1016/j.str.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bharat TA, Murshudov GN, Sachse C, Löwe J. Structures of actin-like ParM filaments show architecture of plasmid-segregating spindles. Nature. 2015 doi: 10.1038/nature14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker E, et al. DNA segregation by the bacterial actin AlfA during Bacillus subtilis growth and development. EMBO J. 2006;25(24):5919–5931. doi: 10.1038/sj.emboj.7601443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gayathri P, Fujii T, Namba K, Löwe J. Structure of the ParM filament at 8.5Å resolution. J Struct Biol. 2013;184(1):33–42. doi: 10.1016/j.jsb.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernander R, Lind AE, Ettema TJ. An archaeal origin for the actin cytoskeleton: Implications for eukaryogenesis. Commun Integr Biol. 2011;4(6):664–667. doi: 10.4161/cib.16974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roeben A, et al. Crystal structure of an archaeal actin homolog. J Mol Biol. 2006;358(1):145–156. doi: 10.1016/j.jmb.2006.01.096. [DOI] [PubMed] [Google Scholar]
- 36.Kudryashov DS, et al. The crystal structure of a cross-linked actin dimer suggests a detailed molecular interface in F-actin. Proc Natl Acad Sci USA. 2005;102(37):13105–13110. doi: 10.1073/pnas.0506429102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schutt CE, Rozycki MD, Chik JK, Lindberg U. Structural studies on the ribbon-to-helix transition in profilin: Actin crystals. Biophys J. 1995;68(4) Suppl:12S–17S, discussion 17S–18S. [PMC free article] [PubMed] [Google Scholar]
- 38.Schutt CE, Rozycki MD, Myslik JC, Lindberg U. A discourse on modeling F-actin. J Struct Biol. 1995;115(2):186–198. doi: 10.1006/jsbi.1995.1043. [DOI] [PubMed] [Google Scholar]
- 39.Schutt CE, Lindberg U. Actin as the generator of tension during muscle contraction. Proc Natl Acad Sci USA. 1992;89(1):319–323. doi: 10.1073/pnas.89.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schutt CE, Lindberg U, Myslik J, Strauss N. Molecular packing in profilin: Actin crystals and its implications. J Mol Biol. 1989;209(4):735–746. doi: 10.1016/0022-2836(89)90603-7. [DOI] [PubMed] [Google Scholar]
- 41.Egelman EH, Francis N, DeRosier DJ. F-actin is a helix with a random variable twist. Nature. 1982;298(5870):131–135. doi: 10.1038/298131a0. [DOI] [PubMed] [Google Scholar]
- 42.Lu A, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156(6):1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perica T, Chothia C, Teichmann SA. Evolution of oligomeric state through geometric coupling of protein interfaces. Proc Natl Acad Sci USA. 2012;109(21):8127–8132. doi: 10.1073/pnas.1120028109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller S, Lesk AM, Janin J, Chothia C. The accessible surface area and stability of oligomeric proteins. Nature. 1987;328(6133):834–836. doi: 10.1038/328834a0. [DOI] [PubMed] [Google Scholar]
- 45.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142(3):334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 46.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85(4):225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 48.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 49.Lewis SM, Kuhlman BA. Anchored design of protein-protein interfaces. PLoS ONE. 2011;6(6):e20872. doi: 10.1371/journal.pone.0020872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krissinel E, Henrick K. Protein interfaces, surfaces and assemblies’ service PISA at the European Bioinformatics Institute. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 51.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16(6):566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.