Significance
Monoclonal antibody FLD194 isolated from a Vietnamese H5N1 survivor neutralizes all three clades of H5N1 viruses that have so far caused human infections. It is, therefore, a candidate for use in antiviral immunotherapy. Structural analysis of the HA-Fab complex shows the antibody-binding site is adjacent to, but does not involve, the sialic acid-binding site. The antibody neutralizes infectivity by restricting the access of receptors to HA using its Fc region in a way that may also be used by numerous other antibodies that bind at a distance from the receptor-binding site. The HA-Fab complex contains an HA subunit which has some of the features of HA in the conformation that is required for membrane fusion activity.
Keywords: influenza virus, neutralizing antibody, H5N1
Abstract
H5N1 avian influenza viruses remain a threat to public health mainly because they can cause severe infections in humans. These viruses are widespread in birds, and they vary in antigenicity forming three major clades and numerous antigenic variants. The most important features of the human monoclonal antibody FLD194 studied here are its broad specificity for all major clades of H5 influenza HAs, its high affinity, and its ability to block virus infection, in vitro and in vivo. As a consequence, this antibody may be suitable for anti-H5 therapy and as a component of stockpiles, together with other antiviral agents, for health authorities to use if an appropriate vaccine was not available. Our mutation and structural analyses indicate that the antibody recognizes a relatively conserved site near the membrane distal tip of HA, near to, but distinct from, the receptor-binding site. Our analyses also suggest that the mechanism of infectivity neutralization involves prevention of receptor recognition as a result of steric hindrance by the Fc part of the antibody. Structural analyses by EM indicate that three Fab fragments are bound to each HA trimer. The structure revealed by X-ray crystallography is of an HA monomer bound by one Fab. The monomer has some similarities to HA in the fusion pH conformation, and the monomer’s formation, which results from the presence of isopropanol in the crystallization solvent, contributes to considerations of the process of change in conformation required for membrane fusion.
The initial steps in influenza virus infection involve sialic acid receptor binding and membrane fusion, both of which are functions of the hemagglutinin (HA) virus membrane glycoprotein. Anti-HA antibodies that block these functions neutralize virus infectivity. Such antibodies are induced by infection and by vaccination, and the immune pressure that they impose on subsequently infecting viruses is responsible for the antigenic drift for which influenza viruses are notorious. Zoonotic infections, which can lead to new pandemics, occur periodically, and H5N1, H7N9, and H10N8 avian viruses are recent examples of this sort. The threat that zoonotic infections present is based, in part, on the lack of immunity in the human population to the novel HAs that they contain. In attempts to substitute for this deficiency, human immune sera have been used successfully to treat severe infections (1), and monoclonal antibodies have been prepared from mice and from humans for potential use in immunotherapy.
Analyses of antibodies produced by cloned immune cells derived from infected patients have revealed that antibodies are induced that are either subtype- or group-specific and others that cross-react with HAs of both groups (2). To date, cross-reactive antibodies have been shown to recognize both membrane-distal and membrane-proximal regions of HA (3). Subtype-specific antibodies, on the other hand, bind to the membrane-distal region, covering the receptor-binding site and, in some cases, inserting into it (4, 5).
In the studies reported here, a human monoclonal antibody is described that recognizes the HAs of viruses of all three clades of the H5 subtype that have caused human infection and is shown to be effective in protecting mice from lethal challenge. EM and X-ray crystallography studies of HA-Fab complexes indicate that the antibody binds to a site containing residue 122, located on the membrane-distal surface of the HA trimer. We describe the antibody-binding site in detail to show that binding occurs at a distance from the receptor-binding site. Infectivity neutralization and receptor-binding experiments, together with these observations, lead to the conclusion that the antibody neutralizes viruses by blocking receptor binding in a way that is dependent on the Fc region of the bound antibody. We compare the site with similar sites reported by others (6–9) for antibodies that have not as yet given crystalline HA-Fab complexes.
Under the conditions that we obtain crystals of the HA-Fab complex, the HA dissociates and reveals the structure of a monomeric HA. We consider the structure of the monomer in relation to the structure that HA has been shown to assume after exposure to the pH of membrane fusion.
FLD194 Neutralizes Infectivity of a Broad Range of H5N1 Viruses, in Vitro and in Vivo
Blood samples from a Vietnamese adult (donor CL115) who had recovered from infection with a clade 1 H5N1 virus, were collected 15 mo after infection (10). IgG-memory B cells were immortalized with EBV (11). Cultures secreting neutralizing antibodies were identified by a microneutralization assay using the prototype H5N1 clade 1 virus, A/Vietnam/1203/04, and the antibodies were cloned by limiting dilution. Two H5N1-neutralizing antibodies were identified, FLD84 and FLD194. We have studied the latter in more detail.
The cross-reactivity of FLD194 was determined in neutralization assays using a panel of 13 human and avian pseudotype H5N1 viruses representative of clades 0, 1, 2.1.3, 2.2, 2.2.1, 2.3.1, 2.3.2, 2.3.2.1, 2.3.4, and 2.5. The IgG was found to be highly effective against all of the viruses tested. By comparison, two other anti-H5 antibodies, FLD20 [previously obtained from the same individual (10)] and FLD84, had lower potency and did not neutralize all of the strains (Table 1).
Table 1.
Neutralization breadth of a panel of monoclonal antibodies against pseudotype viruses containing a diverse set of H5 HAs
| Virus origin of HAs | Clade | IC90, μg/mL | ||
| FLD20 | FLD84 | FLD194 | ||
| A/Hong Kong/156/97 | 0 | 0.003 | 0.005 | 0.008 |
| A/Hong Kong/213/03 | 1 | 0.033 | 0.117 | 0.007 |
| A/Vietnam/1203/04 | 1 | 0.600 | 0.240 | 0.023 |
| A/Vietnam/1194/04 | 1 | 0.144 | 0.357 | 0.019 |
| A/Indonesia/05/05 | 2.1.3 | 0.149 | 0.280 | 0.033 |
| A/whooper Swan/Mongolia/244/05 | 2.2 | 0.345 | 0.293 | 0.008 |
| A/bar-headed goose/Qinghai/1A/05 | 2.2 | 0.010 | 0.015 | 0.036 |
| A/Egypt/3300-NAMRU3/08 | 2.2.1 | >20 | >20 | 0.020 |
| A/turkey/Turkey/1/05 | 2.2.1 | 0.355 | 0.117 | 0.019 |
| A/common magpie/HK/5052/07 | 2.3.2 | >20 | >20 | 0.046 |
| A/Hubei/01/10 | 2.3.2.1 | >20 | >20 | 0.013 |
| A/Anhui/1/05 | 2.3.4 | 0.607 | 0.195 | 0.131 |
| A/chicken/Korea/ES/03 | 2.5 | 0.010 | 0.018 | 0.042 |
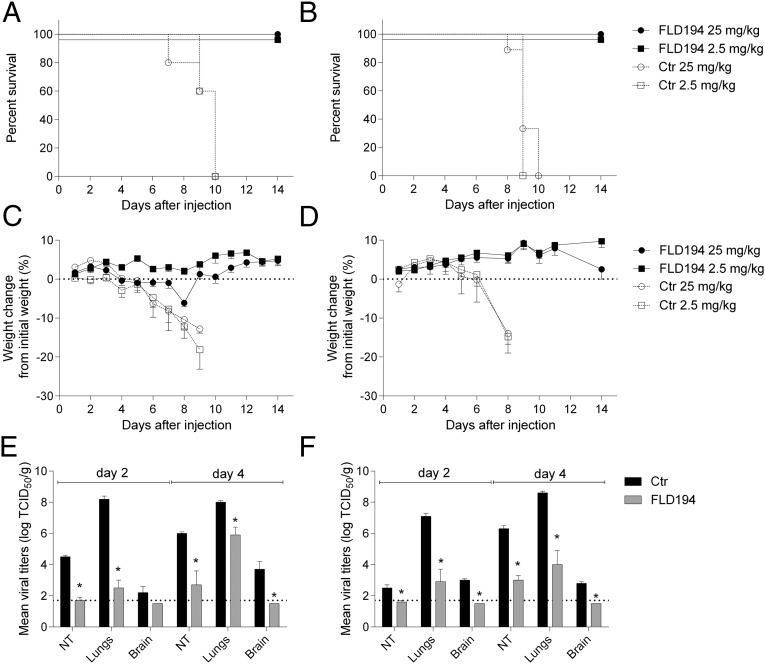
To determine whether the in vitro neutralizing activities of the human monoclonal antibodies would correlate with their prophylactic efficacy, BALB/c mice were inoculated i.p. with either 25 or 2.5 mg/kg of FLD194 or a control monoclonal antibody and were challenged 24 h later with 10 MLD50 (50% mouse lethal doses) of A/Vietnam/1203/04 (H5N1, clade 1) (Fig. 1 A and C) or A/Indonesia/5/05 (H5N1, clade 2) (Fig. 1 B and D). FLD194 at either concentration protected mice from lethal challenge with either H5N1 virus. To gain further insights into the kinetics of virus neutralization in vivo by FLD194, we measured the level of viral replication in nasal turbinates, the lungs, and the brains on days 2 and 4 post infection. The antibody significantly reduced virus titers in the nasal turbinates and lungs of mice infected with either H5N1 strain (Fig. 1 E and F) and restricted extrapulmonary dissemination of the virus. Prophylactic administration of FLD194 IgG (and FLD84) at 7.5 mg/kg also protected ferrets against lethal challenge from the clade 1 A/Vietnam/1203/04 virus (SI Appendix, Fig. 1).
Fig. 1.
Antibody FLD194 protects BALB/c mice from challenge with H5N1 viruses. Mice received 2.5 or 25 mg/kg FLD194 or an irrelevant antibody as control, intraperitoneally 24 h before infection with 10 MLD50 of either A/Vietnam/1203/2004 (H5N1) (A and C) or A/Indonesia/5/2005 (H5N1) (B and D). Survival (A and B) and body weight (C and D) were recorded daily. The distribution of virus in different tissues following infection with 105 TCID50 (tissue culture infectious dose) of either A/Vietnam/1203/2004 (H5N1) or A/Indonesia/5/2005 (H5N1) is shown in E and F. Dotted lines indicate the limit of virus detection.
Structure of the FLD194 HA-Fab Complex
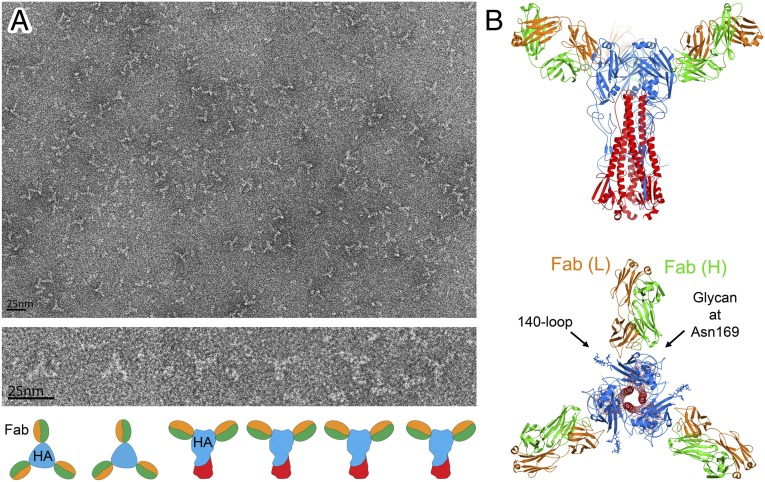
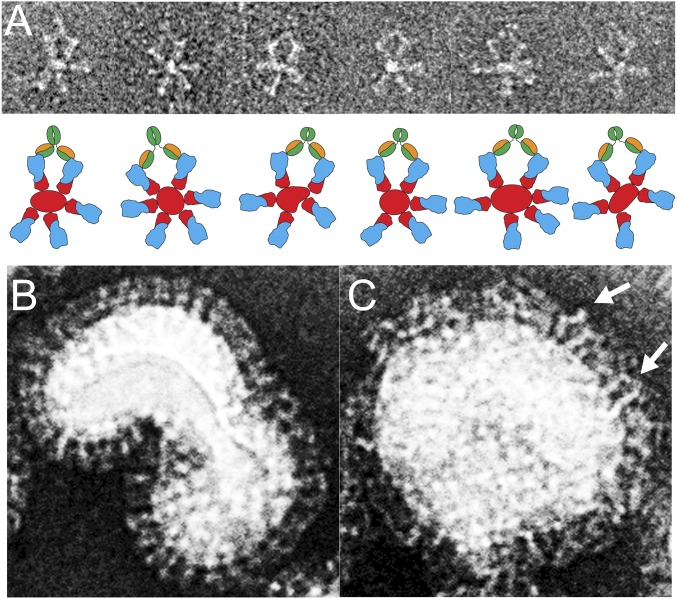
To characterize the interaction of the antibody with H5 HA, we measured binding of the Fab to insect cell-expressed H5 HA from the aerosol-transmissible mutant clade 1 virus (12, 13) by isothermal titration calorimetry. The binding is exothermic, with a KD of 36 nM and a stoichiometry close to 3 (SI Appendix, Fig. 2), suggesting that one Fab would bind to each subunit in an HA trimer. EM images of purified HA-Fab complexes also show a clear threefold symmetry indicating that three Fab fragments bind to each HA trimer.
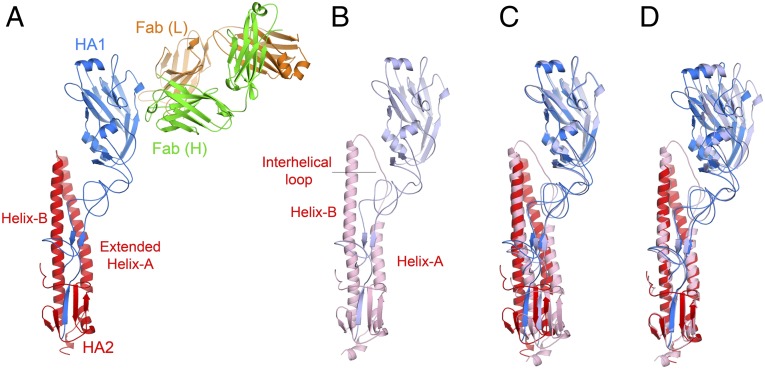
To define the epitope of FLD194 in atomic detail, we crystallized the HA-Fab complexes from the preparation used for EM and determined their structures by X-ray crystallography. Unlike the EM observations, the crystalline HA-Fab complex consists of a 1:1 complex of the Fab with monomeric HA (Fig. 2A) instead of the 3:3 trimeric complex. Because the H5 HA crystallizes at pH 7.5 as trimers (12), it is likely that the inclusion of isopropanol in the solvent on this occasion led to dissociation of the HA trimer and promoted crystallization. The known ability of isopropanol to enhance the formation of α-helices by proteins that are in a helix-coil equilibrium (14) is consistent with a role in the formation of the HA monomer. Gel-filtration analyses indicate that about 50% of the H5 aerosol-transmissible mutant HA dissociates into smaller species on incubation for 8 h in 20% isopropanol.
Fig. 2.
Crystal structure of the FLD194 HA-Fab complex. (A) Cartoon representation of the structure of the Fab-H5–transmissible mutant HA complex showing HA1 (blue) and HA2 (red), and the heavy (green) and light (orange) chains of the antibody. The HA in the complex is a monomer, and helix-A in HA2 is extended by refolding of the interhelical loop. (B) HA monomer taken from the native HA trimer for comparison with A. (C) An alignment of the HA structures in A and B obtained by superpositioning of the RBDs of HA1. (D) An alignment of the HA structures in A and B obtained by superpositioning of the helices B of HA2 shows that there is a slight reorientation of RBD in the HA-Fab complex structure comparing to that in the structure of a trimeric HA subunit.
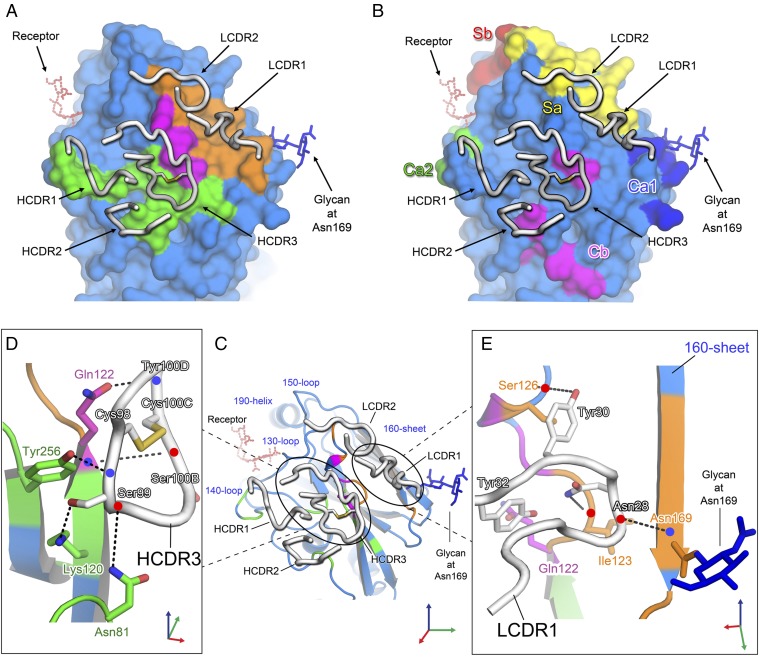
Structural alignment of the receptor-binding domain (RBD) of the HA-Fab complex with that of the trimeric structure of the H5 HA (12) gives an rmsd of 0.39 Å (Fig. 2C), indicating that the structure of the RBD is almost unchanged in the crystalline monomer. Consistent with this, it is possible to dock three FLD194 Fabs onto an HA trimer using the monomeric RBD of the HA-Fab complex as a guide (Fig. 3B). Viewed down the HA threefold axis, it is evident that the Fab binds to the membrane distal domain of HA between the 140-loop and the carbohydrate side-chain attached to residue 169 (Fig. 3B). The Fab heavy chain (green) covers the “140 side” of this region, and the Fab light chain (orange) contacts the “169 side.”
Fig. 3.
Solution structure of the FLD194 HA-Fab complex. (A) Negative stain EM of FLD194 Fab-H5 transmissible mutant HA complex. Most particles exhibit threefold symmetry (Top); selected particles are magnified (Middle) and depicted with schematic drawings (Bottom). (B) Models of a transmissible mutant HA trimer on which three FLD194 Fabs are docked, based on a combination of crystallographic (Fig. 2A) and EM (A) data. A view from the side and a view down the threefold axis of symmetry are shown.
The Fab contacts the HA surface using five of the six complementarity determining regions (CDRs) (Fig. 4A). The contact area spans multiple antigenic sites previously defined using mouse monoclonal antibody-selected antigenic variants of H1 and natural antigenic variants of H3 HAs (15, 16) (Fig. 4B). Heavy chain CDR1 (HCDR1) contacts the 140-loop, site A (Ca2), and HCDR2 contacts residues near residue 81, site E (Cb). HCDR3 contacts a region between residues Gln-122 and Ser-126, sites E (Cb) and B (Sa), respectively. Two light chain CDRs (LCDRs) make contacts near residue 128, at site B (Sa), and LCDR1 also contacts residue 122 in site E (Cb). Binding of the CDRs accounts for a buried surface area of 777 Å2 on HA.
Fig. 4.
Binding site of the FLD194 antibody. (A) Footprint of FLD194 on the H5-transmissible mutant HA surface. Areas contacted by heavy, light, and both chains of the FLD194 Fab fragment are colored green, orange, and purple, respectively. Heavy and light chain CDRs are shown and colored white. The receptor-binding site is indicated by a modeled receptor in red dashed bonds. (B) Surface representation of H5-transmissible mutant HA with previously identified antigenic sites of group 1 HA (16). (C) Cartoon representation of H5-transmissible mutant HA and bound FLD194 CDR loops; interactions made by HCDR3 and LCDR1 are shown in detail in D and E, with slightly reoriented views. Blue and red dots indicate backbone amide and carbonyl groups, respectively.
Based on the structure, the major part of the epitope recognized by FLD194 consists of residues 120–128 that form the center of the antibody-binding site (SI Appendix, Fig. 3). This part of the site has a surface area of 439 Å2, comprising 56% of the total buried surface area of HA. At the epitope’s C terminus, it adopts a short helical structure containing residue 125B, the second of two inserted residues that are specific features of group 1 HAs at this position. Three residues, Gln-122, Pro-125, and the Ser-125B are contacted by both heavy and light chains (colored purple in Fig. 4 A and C; also see SI Appendix, Fig. 3). Of the three contacted residues, binding to Gln-122 contributes the largest buried surface area of 89 Å2. This is consistent with the observations that the Gln-122Arg substitution in H5 HA severely restricts the neutralization activity of FLD194 and that the only viruses tested that were not neutralized had arginine or lysine at residue 122 (SI Appendix, Table 1 and Fig. 4) A bioinformatics analysis using H5 sequences from GISAID and NCBI databases (SI Appendix, Figs. 5 and 6) revealed that residue 122 is highly conserved, 99%, as glutamine in human isolates, including those obtained recently from Egypt and Cambodia, and less conserved, 91%, in avian H5 isolates.
FLD194 binding involves extensive interactions between HCDR3 and the 140-loop side of the epitope (Fig. 4C) and LCDR1 to the side containing residue 169. Together, they form a clamp around the 120–128 strand. The extended HCDR3 loop may be stabilized by a disulfide bond between Cys-98 and Cys-100C (Fig. 4 C and D) in a similar way to what has been observed in the HCDR3 of an anti-HIV envelope glycoprotein antibody where the disulfide was proposed to fix the CDR loop in a particular conformation for epitope recognition (17). The Cys-98 side of HCDR3 penetrates the cleft between the 140-loop and the 120–128 strand of HA, and this penetration allows Ser-99 of HCDR3 to form hydrogen bonds with residues located in the cleft, including Lys-120 in the 120–128 strand of HA and Tyr-256 and Asn-81 of adjacent structures (Fig. 4D). The Cys-100C side of HCDR3 covers the 120–128 strand, and both Ser-100B and Tyr-100D form hydrogen bonds with Gln-122. On the Asn-169 side of the site (Fig. 4E), Asn-28 and Tyr-30 of LCDR1 form hydrogen bonds with Ile-123 and Ser-126, respectively, and Tyr-32 covers Gln-122. These interactions are also stabilized by hydrogen bonds between Asn-28 and Asn-169.
Comparison with Similar Anti-H5 HA Human Monoclonal Antibodies
The binding sites of several other human monoclonal antibodies have been reported to be located at similar surface areas of HA to those recognized by FLD194. These antibodies have not been characterized by X-ray crystallography, but the epitopes recognized by them have been identified using different mapping techniques. Most of the antibodies bind to the 120–128 strand (SI Appendix, Fig. 7), and FLA5, FLD21 (6) (SI Appendix, Fig. 7B), AVFluIgG01, AVFluIgG03 (7) (SI Appendix, Fig. 7C), and C65C6 (8) (SI Appendix, Fig. 7SD) select escape mutants containing HA amino acid substitutions near Pro-125 (SI Appendix, Fig. 7D). The escape mutant selected by the antibody 100F4 (9) was also mapped to Glu-225, a residue that is contacted by the HCDR3 of FLD194.
FLD194 Antibody Blocks Receptor Binding and Neutralizes Virus by Its Fc Region
A common mechanism of neutralization by strain-specific anti-influenza antibodies involves inhibition of receptor binding (18). The crystal structure of the complex formed by the FLD194 Fab with H5 HA shows that the receptor-binding site is not contacted by the antibody (Fig. 4). Nevertheless, the antibody is highly effective in neutralizing H5 viruses (IC90∼100 ng/mL) by comparison with HA2-reactive antibodies, such as FI6, which has very little receptor blocking activity (IC90, >1,000 ng/mL) (2). To investigate the mechanism of infectivity neutralization by FLD194 IgG, we tested its ability to block receptor binding by H5N1 virus using biolayer interferometry (SI Appendix, Fig. 8). The results show that at receptor analog loading levels that allow the virus to saturate the biolayer, a concentration of ∼150 nM FLD194 IgG is needed to block avian receptor binding by 100 pM wild-type H5 virus (SI Appendix, Fig. 8). Wild-type H5 virus binding to human receptor is weak, and comparable data could not be obtained. However, similar concentrations of FLD194 IgG are required to block binding of 100 pM of a mutant H5 virus that has measurable affinity for both avian and human receptors (19) (SI Appendix, Fig. 8). FLD194, therefore, inhibits both avian and human receptor binding. Given that there are ∼300–500 HA trimers per virus (i.e., 90–150 nM binding sites for 100 pM virus), the receptor-binding inhibition data suggest complete receptor-binding inhibition requires stoichiometric binding to each HA subunit. Under conditions where inhibition of receptor binding by FLD194 is incomplete, it is possible that neutralization at lower antibody occupancy could occur at the membrane fusion stage of virus entry as a consequence of HA cross-linking by antibody, as observed in Fig. 5.
Fig. 5.
Electron micrographs of FLD194 IgG bound to HA rosettes and virus. (A) Image of A/Vietnam/1194/2003 H5 HA rosettes in complex with FLD194 IgG (Upper); cartoon representations of the micrographs are shown (Lower). (B) A/Vietnam/1194 (RG14) H5N1 virus with a few IgG bound showing an ordered layer of HA on the virus surface. (C) A/Vietnam/1194 (RG14) H5N1 virus bound with FLD194 IgG. The bound antibody molecules form an extra layer above the HA molecules on the virus surface; the white arrows indicate examples of an IgG cross-linking two HAs.
When similar receptor-binding inhibition experiments were done using the FLD194 Fab fragment instead of FLD194 IgG, inhibitory activity was greatly reduced (SI Appendix, Fig. 8 B and D) and receptor binding was not fully blocked by up to 1500 nM Fab, which represents a reduction in receptor-binding inhibition by at least a factor of 100. The importance of the Fc region for the neutralizing activity of FLD194 is also indicated by the fact that FLD194 Fab does not neutralize virus even up to 0.1 mg/mL, representing a reduction of neutralization activity of at least 1,000-fold.
To understand how the Fc region of FLD194 blocks receptor binding and facilitates virus neutralization, we examined how FLD194 IgG binds to influenza virus and to HA-rosettes by EM (Fig. 5). The micrograph of H5 HA-rosette-IgG complexes indicates that FLD194 binds between two neighboring HAs and cross-links them (Fig. 5A). The orientation of the Fab fragment of IgG bound to HA is similar to that observed for Fab in the crystal structure of the HA-Fab complex. When FLD194 IgG binds to the virus, it forms a layer about 80 Å thick above the HA-decorated virus surface (Fig. 5 B and C). Because the Fab binds to HA almost horizontally, with a binding angle of about 100°, the thickness of the IgG layer can be attributed primarily to the projecting Fc regions of the bound antibodies. Together with our receptor blocking and infectivity neutralization data, it can also be concluded that this layer restricts the access of the virus to cell-surface sialic acid receptors, sterically preventing virus attachment. Bound Fab fragments would be much less effective inhibitors of attachment. These conclusions are similar to those made before for an infectivity neutralizing mouse monoclonal antibody, which binds outside the receptor-binding site. Removal of the Fc in that case also resulted in a 3-log reduction in neutralizing activity, and a similar conclusion was drawn that neutralization by the antibodies is a result of the Fc regions shielding virus HA from cellular receptors (20). In some infections and under lower effective concentrations of antibody, it is possible that virus–antibody complexes could enhance infection through cellular uptake following interaction with Fc receptors (21).This possibility should be explored before use of this or other antibodies as antiviral treatments is recommended.
Structure of HA in the FLD194 Fab-H5 HA Complex
Each subunit of the native HA trimer contains two polypeptides, HA1 and HA2. HA2 contains two prominent α-helices, helix-A and helix-B, with the longer helix-B forming a coiled-coil through interactions with helix-B in HA2 of the other subunits in the trimer (Fig. 2B). In an HA trimer, the C terminus of helix-A is linked to the N terminus of helix-B by an extended interhelical loop, HA2 residues 58–75 (Fig. 2B). In the HA structure found in the FLD194 Fab-H5 HA complex, both HA1 and HA2 are components of a monomer, interactions made between HA1 and the extended interhelical loop are lost, and helix-A is extended by five turns to form a continuous α-helix from residue 38–75 (Fig. 2A).
Before the HA structure was determined by X-ray crystallography (22), HA2 residues 38–75 were predicted to have the propensity to form an α-helix (23), and this prediction was confirmed using synthetic peptides (24). Subsequently, comparison of the structures of HA in the neutral pH and the fusion pH conformations showed directly that helix-A, the interhelical-loop, and the N-terminal residues of helix-B (residues 76–105) form continuous α-helices in a trimeric coiled-coil at fusion pH (25). The extended helix-A observed here in the monomeric HA structure is, therefore, a component of the postfusion HA structure (Fig. 2A; also compare Fig. 2 A and B). This structure’s formation occurs as part of the process of trimer dissociation; the extension of helix-A itself may be responsible for trimer dissociation, or it may occur on dissociation and, once formed, it may prevent trimer reformation. By contrast, retention of the coiled-coil structure formed at the N terminus of helix-B in both neutral pH- and fusion pH-HA structures, has been taken to suggest that complete dissociation of the trimer and trimer reformation are not components of the structural transition required for membrane fusion (25). The observations here, however, may give support to the alternative possibility.
In relation to the structural changes required for membrane fusion, two other features of the monomer in the FLD194 HA-Fab complex can be considered. Firstly, the observation that the structure of the monomeric membrane distal domain, HA1, is highly similar to that in a subunit of the trimer (Fig. 2C) indicates that complete trimer dissociation can occur without denaturation of this domain. A similar observation was made for the structure of the monomeric membrane distal domain following its dissociation at fusion pH (26). Secondly, refolding of the interhelical loop results in loss of all of the interactions between the loop and HA1. As a consequence HA1 is less tightly packed against HA2 as well as being dissociated into monomers (Fig. 2D). The “fusion peptide,” by contrast, is retained in its neutral pH position, implying that neither dissociation nor partial HA1 detachment directly results in the fusion peptide’s extrusion.
Conclusion
We have characterized the structure, virus binding, and infectivity-neutralizing specificity of a human monoclonal antibody derived from an H5N1-infected survivor. The antibody is characterized by high infectivity-neutralizing potency and a broad spectrum neutralizing activity against human and avian H5N1 viruses in vivo and in vitro. Our structural analysis of the Fab complex with H5 HA reveals that the Fab binds outside the receptor-binding site to a relatively conserved epitope. EM analyses of IgG-HA and IgG-virus complexes, together with receptor inhibition and virus-neutralization assays, suggest that FLD194 neutralizes viruses by blocking receptor binding, shielding HAs from cellular receptors by the Fc parts of the antibody. The location of the epitope and the suggested role of antibodies bound to it are consistent with conclusions that antigenic variation can result from amino acid substitutions distributed over the whole membrane distal surface of HA and distant from the receptor-binding site. The relative sizes of the membrane distal domain and an antibody are consistent with a major role in infectivity neutralization of receptor-binding inhibition by the Fc regions of antibodies that bind in these locations. In the course of crystallization of the HA-Fab complex, we obtained a monomeric form of HA. A similar structure was reported before for H1 HA, which, by contrast, contained a reoriented HA1 and a disordered HA2 interhelical loop (27). In the monomeric HA described here, the interhelical loop refolds into an α-helix that extends helix-A in HA2, in a manner reminiscent of the structure that HA2 adopts at fusion pH.
Materials and Methods
Human monoclonal antibodies were isolated from cloned B cells, and their neutralization activities and in vivo efficacies were assessed as previously described. LysC-digested Fab fragments were incubated with transmissible H5 HA, and the complexes formed were purified from unbound Fab by gel-filtration. Purified complexes were examined by negative stain EM. Crystals of the Fab-H5 HA complex were obtained by sitting drop vapor diffusion and analyzed by cryocrystallography. The Fab-H5 HA structure was determined by molecular replacement. The atomic structure coordinates and structure factors have been deposited in the Protein Data Bank as PDB ID code 5A3I. Receptor-blocking activity was measured by biolayer interferometry using an Octet RED instrument (ForteBio) by immobilizing polymeric receptor analogs on streptavidin biosensors to which influenza virus binding was monitored in the absence or presence of varying concentrations of IgG or Fab. Additional detailed information is described in SI Appendix, Materials and Methods.
All experiments involving highly pathogenic avian influenza viruses were conducted in a Biosafety Level 3 (BSL3) containment facility that was approved for use by the US Department of Agriculture and Centers for Disease Control and Prevention. Animal experiments were approved by the National Institutes of Health Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank the staff at the Diamond Light Source Synchrotron for assistance and beamline access under Diamond Light Source Proposal 7707. We thank Cameron Simmons (Oxford University Clinical Research Unit and the Hospital for Tropical Diseases) for collection of clinical samples and Silvia Maniero [Istituto Zooprofilattico Sperimentale delle Venezie (IZSVe)], Prof. Hussein A. Hussein (Cairo University), and Dr. Abdo Arafa (National Laboratory for Veterinary Quality Control on Poultry Production) for excellent technical assistance. This work was funded by the Medical Research Council through Programmes U117584222, U117512723, and U117570592. IZSVe activities were financially supported by the NoFlu project, Fondazione Cariplo Vaccine Program Grant 2009-3594. This research was supported, in part, by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Conflict of interest statement: A.L. is the scientific founder of Humabs BioMed SA. A.L. holds shares in Humabs BioMed SA. D.C. is currently Chief Scientific Officer of Humabs Biomed.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5A3I).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510816112/-/DCSupplemental.
References
- 1.Zhou B, Zhong N, Guan Y. Treatment with convalescent plasma for influenza A (H5N1) infection. N Engl J Med. 2007;357(14):1450–1451. doi: 10.1056/NEJMc070359. [DOI] [PubMed] [Google Scholar]
- 2.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333(6044):850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 3.Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu Rev Immunol. 2013;31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- 4.Whittle JR, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci USA. 2011;108(34):14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee PS, et al. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun. 2014;5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khurana S, et al. Antigenic fingerprinting of H5N1 avian influenza using convalescent sera and monoclonal antibodies reveals potential vaccine and diagnostic targets. PLoS Med. 2009;6(4):e1000049. doi: 10.1371/journal.pmed.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun L, et al. Generation, characterization and epitope mapping of two neutralizing and protective human recombinant antibodies against influenza A H5N1 viruses. PLoS ONE. 2009;4(5):e5476. doi: 10.1371/journal.pone.0005476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu H, et al. A human antibody recognizing a conserved epitope of H5 hemagglutinin broadly neutralizes highly pathogenic avian influenza H5N1 viruses. J Virol. 2012;86(6):2978–2989. doi: 10.1128/JVI.06665-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian M, et al. Unraveling of a neutralization mechanism by two human antibodies against conserved epitopes in the globular head of H5 hemagglutinin. J Virol. 2013;87(6):3571–3577. doi: 10.1128/JVI.01292-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simmons CP, et al. Prophylactic and therapeutic efficacy of human monoclonal antibodies against H5N1 influenza. PLoS Med. 2007;4(5):e178. doi: 10.1371/journal.pmed.0040178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Traggiai E, et al. An efficient method to make human monoclonal antibodies from memory B cells: Potent neutralization of SARS coronavirus. Nat Med. 2004;10(8):871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong X, et al. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature. 2013;497(7449):392–396. doi: 10.1038/nature12144. [DOI] [PubMed] [Google Scholar]
- 13.Imai M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486(7403):420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirota N, Mizuno K, Goto Y. Cooperative alpha-helix formation of beta-lactoglobulin and melittin induced by hexafluoroisopropanol. Protein Sci. 1997;6(2):416–421. doi: 10.1002/pro.5560060218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31(2 Pt 1):417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 16.Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289(5796):373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 17.Doria-Rose NA, et al. NISC Comparative Sequencing Program Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509(7498):55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knossow M, Skehel JJ. Variation and infectivity neutralization in influenza. Immunology. 2006;119(1):1–7. doi: 10.1111/j.1365-2567.2006.02421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong X, et al. Enhanced human receptor binding by H5 haemagglutinins. Virology. 2014;456-457(0):179–187. doi: 10.1016/j.virol.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleury D, et al. A complex of influenza hemagglutinin with a neutralizing antibody that binds outside the virus receptor binding site. Nat Struct Biol. 1999;6(6):530–534. doi: 10.1038/9299. [DOI] [PubMed] [Google Scholar]
- 21.Tamura M, Webster RG, Ennis FA. Antibodies to HA and NA augment uptake of influenza A viruses into cells via Fc receptor entry. Virology. 1991;182(1):211–219. doi: 10.1016/0042-6822(91)90664-w. [DOI] [PubMed] [Google Scholar]
- 22.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 23.Ward CW, Dopheide TA. Influenza virus haemagglutinin. Structural predictions suggest that the fibrillar appearance is due to the presence of a coiled-coil. Aust J Biol Sci. 1980;33(4):441–447. doi: 10.1071/bi9800441. [DOI] [PubMed] [Google Scholar]
- 24.Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73(4):823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 25.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371(6492):37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 26.Bizebard T, et al. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature. 1995;376(6535):92–94. doi: 10.1038/376092a0. [DOI] [PubMed] [Google Scholar]
- 27.Cho KJ, et al. Insight into structural diversity of influenza virus haemagglutinin. J Gen Virol. 2013;94(Pt 8):1712–1722. doi: 10.1099/vir.0.051136-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.