Significance
Induction of RNAi through shRNA expression holds great potential for biomedical research. Ideal shRNAs exhibit high specificity, potency, and safety, but options for rationally designing such shRNAs remain limited. One ensuing concern is unintended perturbation of gene expression by shRNA sense or antisense strands (off-targeting). Here, we report a novel solution that specifically counteracts adverse activity of shRNA sense strands via their sequestration by tough decoy RNAs (TuDs). Using hepatitis C virus as a clinically relevant example, we demonstrate how TuD coexpression from an Adeno-associated viral (AAV) gene therapy vector concurrently decreases shRNA off-targeting and enhances on-target inhibition. The versatility of TuDs and AAVs makes our new strategy useful for improving specificity and safety of a wide array of RNAi experiments.
Keywords: RNA interference, short hairpin RNA, off-targeting, Adeno-associated viral vector, AAV
Abstract
Exogenous RNAi triggers such as shRNAs ideally exert their activities exclusively via the antisense strand that binds and silences designated target mRNAs. However, in principle, the sense strand also possesses silencing capacity that may contribute to adverse RNAi side effects including off-target gene regulation. Here, we address this concern with a novel strategy that reduces sense strand activity of vector-encoded shRNAs via codelivery of inhibitory tough decoy (TuD) RNAs. Using various shRNAs for proof of concept, we validate that coexpression of TuDs can sequester and inactivate shRNA sense strands in human cells selectively without affecting desired antisense activities from the same shRNAs. Moreover, we show how coexpressed TuDs can alleviate shRNA-mediated perturbation of global gene expression by specifically de-repressing off-target transcripts carrying seed matches to the shRNA sense strand. Our combination of shRNA and TuD in a single bicistronic gene transfer vector derived from Adeno-associated virus (AAV) enables a wide range of applications, including gene therapies. To this end, we engineered our constructs in a modular fashion and identified simple hairpin design rules permitting adaptation to preexisting or new shRNAs. Finally, we demonstrate the power of our vectors for combinatorial RNAi strategies by showing robust suppression of hepatitis C virus (HCV) with an AAV expressing a bifunctional TuD against an anti-HCV shRNA sense strand and an HCV-related cellular miRNA. The data and tools reported here represent an important step toward the next generation of RNAi triggers with increased specificity and thus ultimately safety in humans.
Despite the recent advances in gene-engineering technologies (1), RNAi remains one of our most versatile and powerful tools for studying and manipulating gene expression in eukaryotic organisms (2). Part of its attraction stems from the simplicity with which it is triggered, namely, by introducing small, interfering double-stranded RNAs that mimic processing products of endogenous microRNAs (miRNAs) and thus engage the cellular RNAi silencing machinery. One specific variant is shRNA that consists of a stem composed of an antisense (or guide) strand that is complementary to a target mRNA and a sense (or passenger) strand that ideally is inert and merely completes the double-stranded molecule. The two strands are linked by a loop that is eliminated by the cellular RNase III enzyme Dicer during shRNA maturation, before the resulting small RNA duplex is loaded into an RNA-induced silencing complex (RISC). After removal of the sense strand, the remaining single-stranded antisense arm of the shRNA directs the RISC to a fully complementary target mRNA, which then is cleaved. The ability to express shRNAs from RNA polymerase II or III promoters offers sundry choices for cell- or tissue-specific and ectopically regulated RNAi applications ranging from genome annotation to therapeutic silencing. Moreover, the compatibility of shRNA expression cassettes with the plethora of available nonviral or viral gene transfer vectors further extends the options for controlled and targeted RNAi silencing in cells, animals, and humans. Examples illustrating these assets are our previous reports of potent and persistent inhibition of hepatitis B virus (HBV) in HBV-transgenic mice, using liver-specific Adeno-associated viral vectors of serotype 8 (AAV8) for shRNA delivery (3, 4).
Despite these and many other promising data, a seminal concern that continues to hamper the academic or clinical use of RNAi is the specificity and hence the safety of gene silencing. This concern is fueled by findings that, in addition to binding their designated perfectly matched target, shRNA antisense strands also can recognize and degrade other mRNAs with imperfect complementarity. Akin to the natural miRNA pathway, this “off-targeting” down-regulates the affected gene by various mechanisms, including mRNA destabilization. Although the ensuing effects typically are milder than those of shRNA-induced mRNA cleavage, adverse off-targeting can substantially confound the results of RNAi experiments and even contribute to cellular toxicity. Off-targeting therefore also poses a challenge for genomewide high-throughput RNAi screens, which are particularly vulnerable to pervasive off-target effects and resulting false-positive results, as highlighted by numerous reports (e.g., refs. 5–10). In addition, a few studies consistently showed that not only the antisense but also the sense strand can be loaded into the RISC, thus exacerbating concerns about off-target silencing. We and others have found that both shRNA strands can be immunoprecipitated with Argonaute proteins (key components of the RISC) and that either strand can inhibit reporter constructs with artificial complementary binding sites (11–13). These observations are accompanied by accumulating data indicating that naturally occurring miRNAs also use both strands and that the miRNA sense (also called the “star”) strand can play essential roles in cellular physiology and pathology (14–16).
From a biological viewpoint, these findings are intriguing, because they continue to expand our understanding of the complexity of mammalian RNAi processes. Complementing early notions that strands with a lower 5′ thermodynamic stability are preferentially incorporated into RISC (17–19), recent data now suggest that many other parameters, such as the position of the loop, the sequence and structure of the stem, the relative abundance of different Argonaute proteins, or competitive Argonaute-2 binding of the 5′ nucleotide of either strand, influence strand selectivity (12, 14, 15, 20). Moreover, there is ongoing controversy over whether Dicer and associated proteins are required or dispensable for asymmetric RISC loading (21–23). No matter the exact mechanism, it is clear that off-targeting is unintended in the context of RNAi applications and must be alleviated. Alleviation is accomplished readily with siRNAs, synthetic counterparts of fully processed shRNAs that can be chemically and molecularly engineered toward preferred loading of the antisense strand (24). However, achieving the same alleviation with vector-encoded shRNAs is far more challenging, because they are expressed intracellularly and hence are not amenable to ectopic optimization. Nonetheless, several groups have elucidated shRNA design rules that promise a bias toward one shRNA strand, including asymmetric hairpin configurations with mismatches or bulges at the 5′ end of the antisense strand or bioinformatical preselection of shRNAs with minimal off-targeting potential within a given cellular genome (25–28). Furthermore, various laboratories, including ours, have devised shRNA variants that skip Dicer processing and are loaded directly into Argonaute-2, which then cleaves one of the two strands and predominantly leaves the other as a guide (29–34).
Still, the RNAi field urgently needs new strategies to counteract sense strand off-targeting, because rules for designing highly specific shRNAs remain elusive. In addition, all previously reported concepts require de novo creation of optimized shRNAs, but frequently it would be beneficial to improve the specificity of existing and prevalidated molecules. The use of such molecules is particularly indicated when the target region is confined, as is the case for viruses that typically present a limited number of conserved and thus targetable sequences or for allele-specific disorders in which a few nucleotides distinguish healthy from diseased cells. Therefore we developed a conceptually novel strategy which is based on the coexpression of an shRNA together with a competitive RNA decoy that specifically binds and inactivates the shRNA sense strand. Unlike all prior approaches, ours is fully compatible with preexisting shRNAs and requires no modifications or recrafting, other than cloning of the decoy following simple design rules that we established concurrently. Using an shRNA against a conserved region in hepatitis C virus (HCV) for proof of concept, we demonstrate the power of our decoy strategy to suppress shRNA sense strand activity effectively and to alleviate global off-targeting, resulting in a marked increase in RNAi specificity. The high versatility of the new approach reported here implies that it could be widely adapted and used in the future to improve the stringency of many types of RNAi experiments, including vector-mediated therapeutic silencing in humans.
Results
shRNA Sense Strand Activity Can Be Counteracted by Coexpression of Tough Decoys.
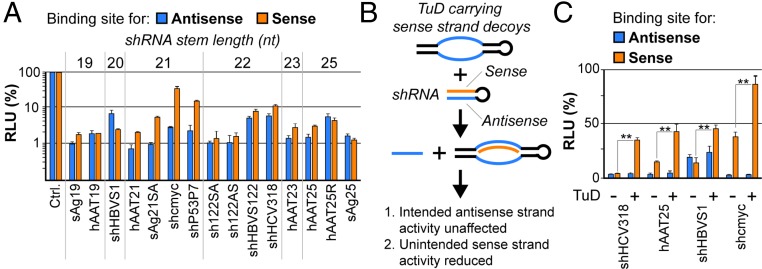
We initially sought to assess the frequency and extent of shRNA sense strand activity comprehensively by studying a set of 15 previously published or newly designed shRNAs (see Materials and Methods for details). To this end, we created dual luciferase reporters carrying binding sites for either the shRNA sense or antisense strand in the 3′ UTR of one of the two luciferase genes (Renilla luciferase) (Fig. S1). These reporters then were cotransfected into HEK293T cells together with each shRNA, and luciferase knockdowns were compared with the parental reporter construct lacking binding sites. Strikingly, regardless of sequence, length, or target, all tested shRNAs exhibited considerable sense strand activity, frequently even matching that of the antisense strand (Fig. 1A).
Fig. S1.
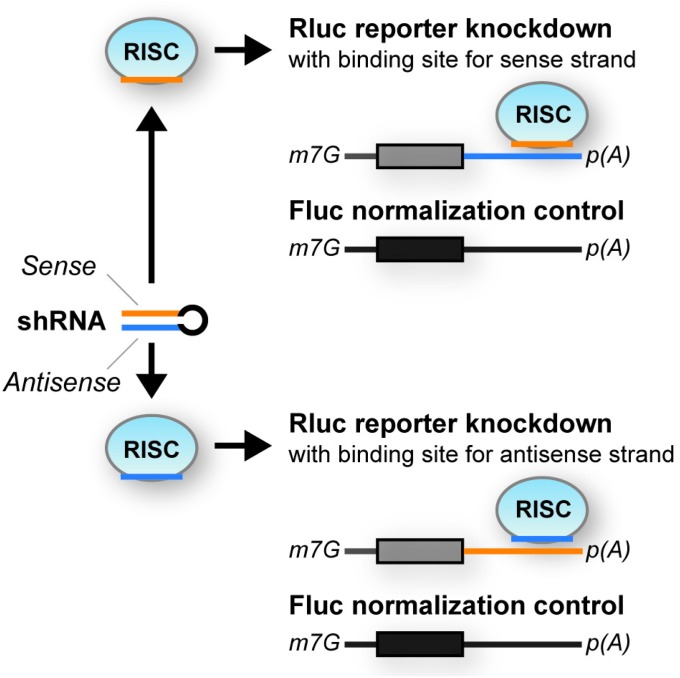
Schematic depiction of the dual luciferase assay used throughout this work. Plasmid psiCheck-2 coencodes Renilla and Firefly luciferases. Renilla was tagged with specific shRNA- or miR-122–binding sites because of its higher sensitivity, and Firefly was used for normalization. m7G, 7-methylguanylate cap (5′ end of mRNAs); R/Fluc, Renilla/Firefly luciferase; RISC, RNA-induced silencing complex; p(A), poly(A) tail.
Fig. 1.
shRNA sense strand activity and its counteraction by RNA decoys. (A) Results from cotransfection of HEK293T cells with a range of shRNAs and dual luciferase reporters carrying binding sites for either the shRNA antisense (blue) or sense (orange) strand in the 3′ UTR of Renilla luciferase. (Firefly luciferase was used for normalization; see also Fig. S1.) Plotted are relative light units (RLUs) compared with a control sample (ctrl); i.e., for each shRNA strand, the identical reporter was cotransfected with an irrelevant shRNA, and resulting RLUs always were set to 100%). (B) Schematic representation of sense strand counteraction by coexpression of TuD RNA decoys. (C) Validation of TuD functionality in HEK293T cells that were triple-transfected with a subset of shRNAs from A, dual luciferase reporters to detect antisense or sense strand activity, and a TuD specific for the sense strand of each shRNA. **P < 0.01; two-sided t-test. Error bars in A and C represent SDs of at least three independent experiments.
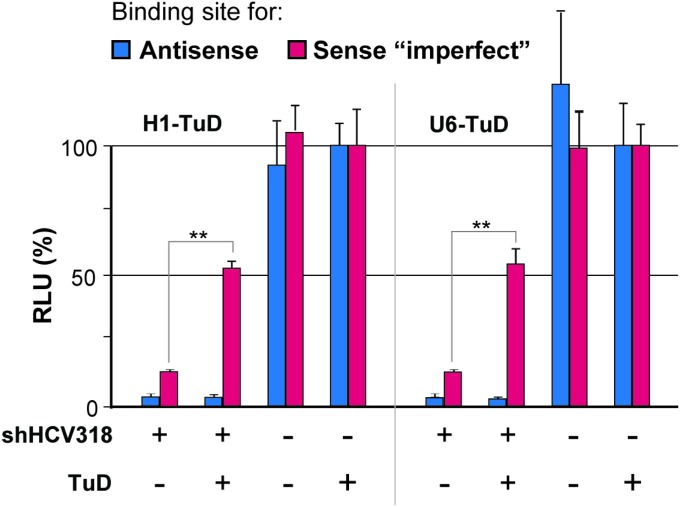
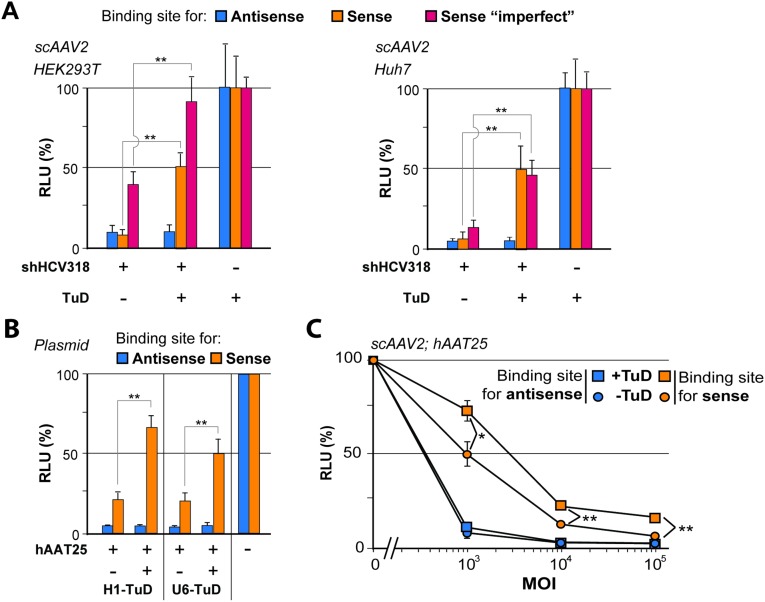
With the aim of counteracting these undesired silencing activities, we devised a novel approach that relied on selective sequestration and functional inactivation of the sense strand via codelivered decoy RNAs (Fig. 1B). Ideally, to provide maximal versatility and compatibility with preexisting shRNAs, these decoys should neither affect antisense strand activity nor require modifications of the original shRNA design. Specifically, we decided to explore “tough decoys” (TuDs) as competitive inhibitor RNAs because of their reported potency and small size (35, 36). Accordingly, we generated plasmids expressing H1 promoter-driven TuDs against the sense strands of a subset of the shRNAs in Fig. 1A and then determined sense and antisense activities using luciferase reporters carrying perfect binding sites for either strand. As hoped, coexpression of the TuDs caused a robust sense strand inhibition for all tested shRNAs (Fig. 1C, orange bars “+”). Importantly, antisense activities remained unaffected, indicating that TuD-mediated sense strand inhibition not only was potent but also was specific. Furthermore, we confirmed that TuD codelivery likewise permitted rescue of a sense strand target that mimicked an imperfect miRNA-like off-target and that there was no significant difference in efficiency when the TuD was driven by an H1 or U6 promoter under the tested conditions (Fig. S2).
Fig. S2.
Comparison of the H1 versus the U6 promoter for TuD expression. HEK293T cells were triple-transfected with the shHCV318-encoding plasmid (or with a plasmid with an irrelevant shRNA), dual luciferase reporters to detect shRNA antisense or sense strand activity, and a TuD specific for the sense strand of the shRNA. The shRNA always was driven by an H1 promoter. The TuD was under either an H1 or a U6 promoter, as indicated. **P < 0.01; two-sided t-test. Error bars represent SDs of at least three independent experiments.
Codelivery of shRNA and TuD Through Viral Vector-Mediated Transduction.
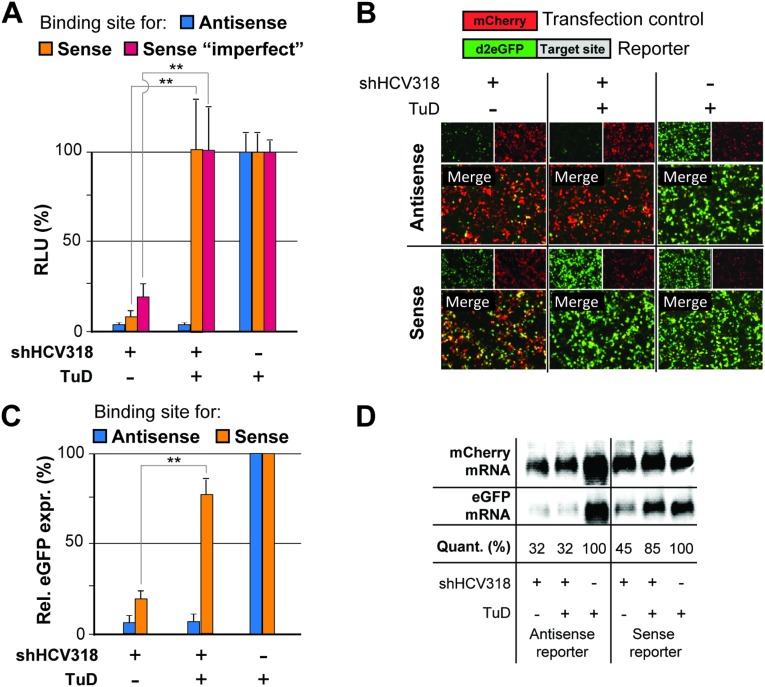
To improve the applicability of our strategy further, we next generated a cloning scaffold that allows easy generation of bicistronic vectors for simultaneous expression of shRNA and TuD from a single construct (Fig. 2). This scaffold was based on a recombinant Adeno-associated viral (AAV) vector construct, permitting packaging into AAV particles and hence providing the option for delivery into a large range of target cells in vitro or in vivo (37). The principal functionality of the bicistronic AAV-shRNA/TuD vector was validated by luciferase (Fig. S3A) and eGFP reporter assays (Fig. S3 B–D).
Fig. 2.
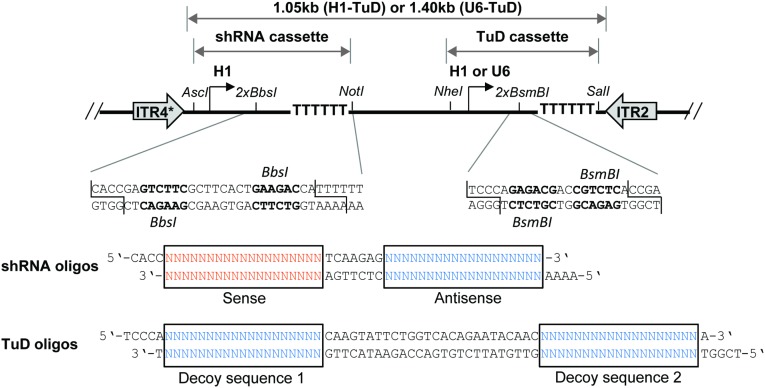
Structure and cloning of bicistronic AAV-shRNA/TuD vectors. (Upper) The general vector design with the two independent shRNA and TuD expression cassettes, together with selected unique restriction sites and stretches of six thymidines that serve as termination signal for RNA polymerase III promoters (H1 or U6). Both cassettes together are flanked by ITRs from AAV serotypes 2 or 4 [the asterisk indicates a truncation that mediates the self-complementary genotype (4)] for packaging into AAV particles. (Lower) A magnification of the two inverted BbsI or BsmBI sites (bold) that are used for cloning of shRNAs or TuDs, respectively, as annealed or extended oligonucleotides with the indicated overhangs.
Fig. S3.
Additional validation of potent sense strand inhibition from a single plasmid coexpressing shRNA and TuD. (A) Confirmation that both perfect and imperfect sense strand targets can be rescued by TuD coexpression as determined by cotransfection of HEK293T cells with the indicated shRNA/TuD plasmids or no-shRNA control and dual luciferase reporters containing the shown binding sites for the shRNA sense or antisense strand. (B–D) Validation that sense strand targets can be rescued by TuD coexpression using a fluorescent reporter system, as observed by fluorescence microscopy (B), quantitative FACS analysis (C), and mRNA Northern blot analysis (D) of HEK293T cells transfected with the indicated plasmids. In D, “Quant.” (quantification) indicates relative egfp band intensities normalized to mCherry intensities. **P < 0.01; two-sided t-test. Error bars in A and C represent SDs of at least three independent experiments.
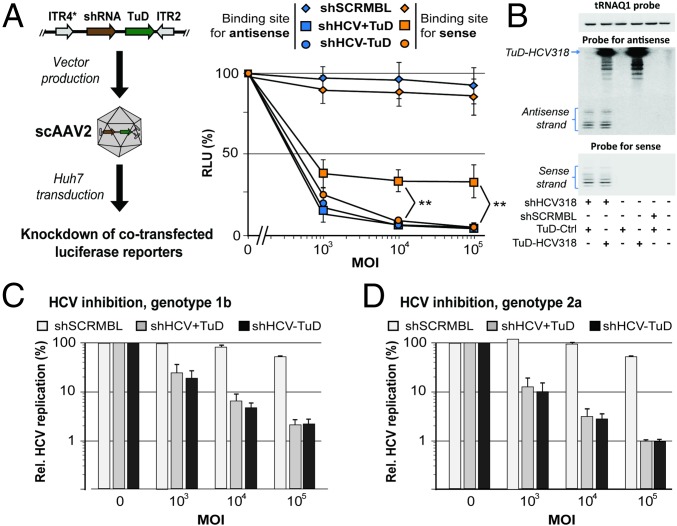
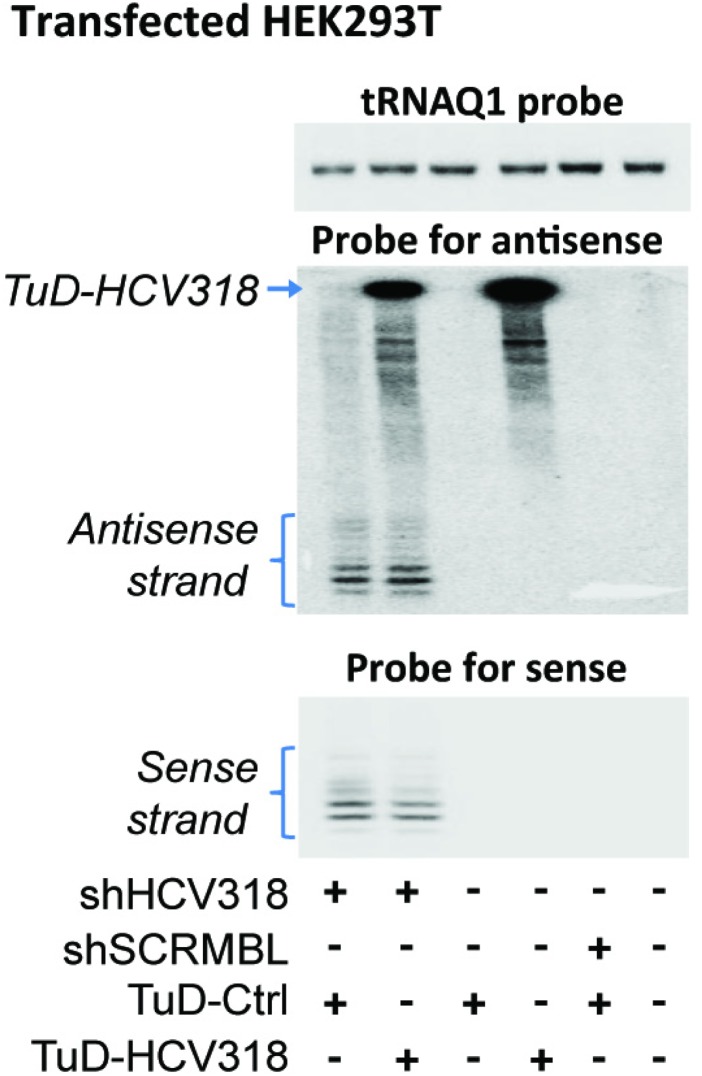
For a more thorough characterization of our approach, we focused on shHCV318 (Fig. 1A), which targets a conserved region in the 5′ UTR of the HCV genome and which is derived from a previously published shRNA (38). We selected this shRNA because (i) HCV is a clinically highly relevant target; (ii) it can be inhibited with RNAi (38–42); (iii) assays in R.B. and D.G. laboratories allowed a convenient read-out and made HCV a good model to validate our new approach; and (iv) shHCV318 had exhibited strong sense strand activity in our initial reporter assays (Fig. 1A). Bicistronic constructs carrying shHCV318 with or without sense strand TuD were packaged into self-complementary AAV vectors of serotype 2 [scAAV2, a potent AAV variant in cultured cells (43)] and then were used to transduce human Huh7 hepatocellular carcinoma cells. We observed strong sense strand-mediated knockdown of a cognate luciferase reporter that, remarkably, was counteracted efficiently by the TuD over a wide range of multiplicities of infection (MOIs; i.e., particle number per cell) without perturbing antisense strand activity (Fig. 3A). These findings were validated in another cell line and with a second luciferase reporter bearing an imperfect miRNA-like binding site for the shHCV318 sense strand (Fig. S4A). In addition, we independently confirmed the overall strategy with a second, unrelated shRNA (Fig. S4 B and C). Small RNA Northern blot analysis confirmed the expression of the sense strand TuD and moreover showed the presence of both mature sense and antisense strands of shHCV318 (Fig. 3B). An interesting observation was that the levels and lengths of both shRNA strands were unaltered compared with a TuD-negative control (lanes 1 and 2 in Fig. 3B and verified in a second cell line in Fig. S5). This finding implies that, in contrast to miRNAs, which typically are inhibited by TuDs through tailing and trimming (44), the type of anti-shRNA TuDs that we developed here inhibit shRNAs through stable sequestration rather than degradation.
Fig. 3.
Efficient codelivery of shRNA and TuD by a single bicistronic scAAV vector. (A, Left) Experimental workflow. (Right) Results from the transduction of Huh7 cells with different MOIs of bicistronic scAAV2 vectors. Activities of the two shHCV318 strands were measured via cotransfected luciferase reporters carrying appropriate binding sites and were normalized to those obtained in the absence of the shRNA vector (MOI 0, set to 100%). shSCRMBL, scrambled negative control shRNA. **P < 0.01; two-sided t-test. (B) Small RNA Northern blot exemplifying expression of both shRNA strands and the TuD upon transduction of Huh7 cells (MOI 105). Human transfer RNA glutamine 1 (tRNAQ1) served as loading control. “TuD-Ctrl” was an empty TuD vector. The multiple weaker bands below the specific TuD signal are likely degradation products, visible because of the blot overexposure needed to permit codetection of the shRNA antisense strands. (C and D) Huh7.5 cells carrying subgenomic HCV replicons of genotype 1b (C) or 2a (D) were transduced with AAV vectors expressing shHCV318 shRNA with or without TuD at the shown MOIs. HCV replication was measured via replicon-encoded Firefly luciferase and normalized to control samples transduced at the same MOI with an irrelevant vector without shRNA (MOI 0 for each shRNA vector, set to 100%). Also shown are results with a scrambled control shRNA (shSCRMBL). Error bars represent SDs of at least three independent experiments.
Fig. S4.
Confirmation of functional sense strand inhibition with chimeric AAV-shRNA/TuD vectors. (A) HEK293T (Left) or Huh7 (Right) cells were transduced (MOI 105) with AAV vectors encoding the shRNA/TuD combinations shown and were cotransfected with luciferase reporters containing perfect or imperfect shRNA binding sites in their 3′ UTR. (B) Comparative analysis of H1 and U6 promoter-driven TuDs for an additional shRNA. HEK293T cells were double-transfected with plasmids encoding hAAT25 shRNA with or without sense strand TuD (or a plasmid without shRNA) and with dual luciferase reporters to detect shRNA antisense or sense strand activity. (C) Dose–response analysis in Huh7 cells transduced with different MOIs of scAAV2 vectors coexpressing hAAT25 shRNA and the indicated TuDs. The cells also were transfected with dual luciferase reporters containing binding sites for either shRNA strand in the 3′ UTR of Renilla luciferase. Note that this particular shRNA illustrates a case in which potent shRNA sense strand rescue required the use of the strong U6 promoter (instead of H1 used for shHCV318) for TuD expression in the AAV vector context. *P < 0.05; **P < 0.01; two-sided t-test. Error bars represent SDs of at least three independent experiments.
Fig. S5.
Northern blot to validate the data from Fig. 3B in another cell line (HEK293T). The cells were transfected with the indicated combinations of shRNA and TuD constructs, and both TuD and the two shRNA strands were detected using specific probes. Note that again there was no evidence for shRNA tailing and trimming, supporting our conclusion that the mechanism of TuD-induced shRNA inhibition is likely to be stable sequestration and thus is different from the previously reported TuD action on miRNAs (see main text).
Sense Strand Inactivation De-Represses Endogenous Off-Targets Without Perturbing On-Target Silencing.
Because reporter knockdown assays do not necessarily reflect how shRNAs silence their genuine targets, we next examined the effects of sense strand TuD expression on shHCV318-mediated inhibition of HCV replication. To this end, we transduced Huh7.5 cells carrying luciferase-encoding HCV replicons with scAAV2 vectors encoding shRNA and TuD at different MOIs. Notably, quantification of HCV replication via luciferase measurements showed a potent inhibition of HCV genotype 1b and 2a replication that was unaffected by coexpression of the sense strand TuD (Fig. 3 C and D). These data were congruent with our initial results from the reporter assays and thus confirmed that silencing of intended antisense targets is not afflicted by our sense strand-inactivation strategy.
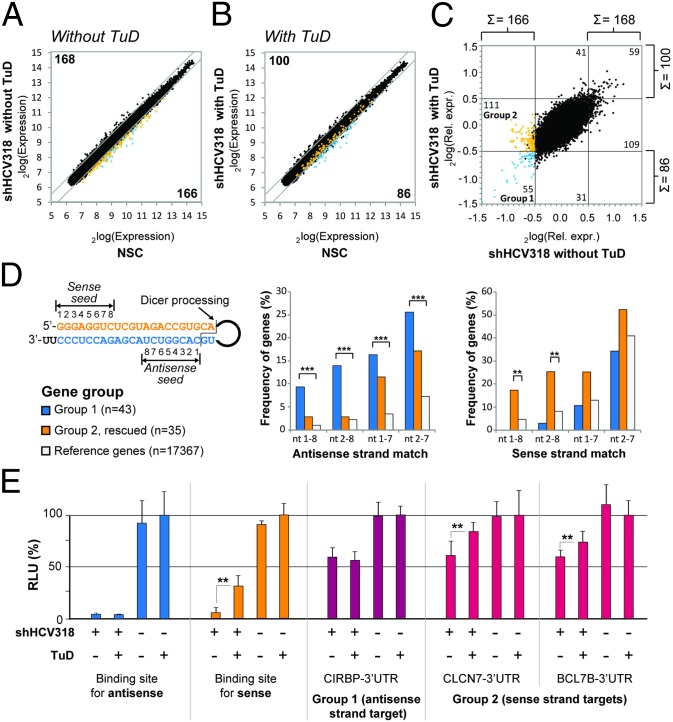
To study the off-targeting activity of shHCV318 in more detail and on a global level, we transduced Huh7 cells with scAAV2 (MOI 105) encoding either shHCV318 with or without sense strand TuD or a nonsilencing vector control for normalization, followed by mRNA expression profiling using microarrays that covered the whole human transcriptome. As shown in Fig. 4 A–C, the expression of shHCV318 without TuD resulted in 166 down- and 168 up-regulated genes (a total of 334 genes) compared with the nonsilencing vector control [P < 0.01; fold change >log2(0.5)]. Importantly, TuD coexpression significantly reduced the number of dysregulated genes to only 86 down- and 100 up-regulated candidates (a total of 186; Fig. 4 B and C). Together, these results suggested that shHCV318 had induced a perturbation of cellular mRNA expression which was partly alleviated by coexpression of the specific sense strand TuD.
Fig. 4.
TuD-mediated rescue of shRNA sense strand-mediated off-targeting. (A–C) Results from cDNA microarray analysis in scAAV2-transduced Huh7 cells (MOI 105) exemplifying that TuD codelivery reduced the number of genes that were dysregulated by shHCV318 expression. Numbers indicate genes that are significantly different from the nonsilencing control (NSC, lacking shRNA) in at least one of the shHCV318 samples [P < 0.01, two-sided t-test; fold-change cutoff at log2(0.5)]. Group 1 (blue) and group 2 (orange) genes are highlighted in each graph. (A) Absolute expression levels for shHCV318 without TuD, versus the NSC. (B) Absolute expression levels for shHCV318 plus specific TuD-HCV318, versus the NSC. (C) Relative expression levels for shHCV318 with specific TuD-HCV318, versus shHCV318 without TuD, each normalized to NSC. Effects of the TuD alone are shown in Fig. S6. (D) Frequency of shHCV318 sense or antisense strand seed sequence complementarities in two different gene groups from C. Depicted on the left is the shHCV318 shRNA with the location of the two seed sequences (nucleotides 1–8) in the sense or antisense strand. The cleavage site for Dicer processing which determines the seed position in the antisense strand is also shown. The two graphs illustrate the enrichment of full seed sequences (nucleotides 1–8) or portions thereof that correspond to the shRNA antisense or sense in the 3′ UTRs of two different gene groups. Frequencies (in percentages) were calculated as sequence counts per number of genes. Blue bars represent genes that were not affected by the TuD (group 1 in C). Orange bars represent sense strand off-targets that were rescued by TuD coexpression (group 2 in C). For both groups, only genes with annotated 3′ UTRs were analyzed. Moreover, in group 2, only genes that were significantly different between shHCV318 with or without TuD were taken into account (P < 0.01; one-sided t-test). Group 1 comprised 43 genes, and group 2 comprised 35 genes. Also shown are frequencies of antisense or sense strand matches in the entire gene population on the microarray (white bars). Significances were determined by Fisher’s exact test (cutoff: P = 0.01). **P < 0.01; ***P < 0.001. (E) Verification of off-target genes identified by microarray analysis (see C). Shown are three examples of genes from group 1 (CIRBP, antisense target) or group 2 (CLCN7 and BCL7B, sense targets) whose 3′ UTR was cloned behind a luciferase reporter. Knockdown and rescue of these constructs were analyzed in transfected Huh7 cells that were cotransduced with AAV vectors (MOI 105) expressing the shRNA/TuD combinations shown. Also included were controls carrying perfect binding sites for the shHCV318 antisense (blue bars) or sense (orange bars) strand. Note that the group 1 reporter behaved like the antisense reporter control and was repressed by shHCV318 equally well in the presence or absence of the TuD, whereas both group 2 reporters (shRNA sense strand targets) were significantly de-repressed by TuD coexpression, as predicted. **P < 0.01; two-sided t-test. Error bars represent SDs of at least three independent experiments.
Fig. S6.
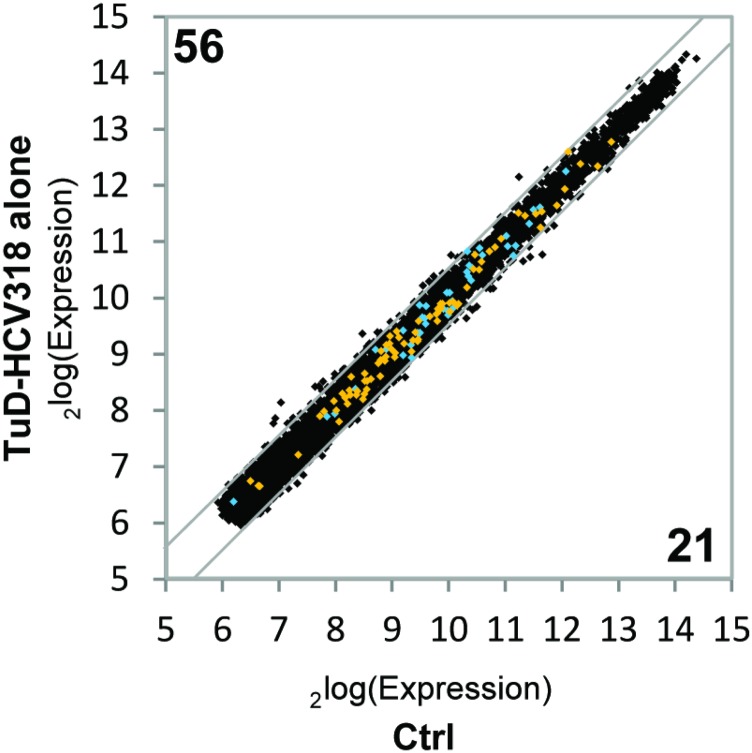
Expression of the TuD-HCV318 alone does not affect group 1 and group 2 genes. Absolute gene expression levels from cDNA microarray analysis in scAAV2-transduced Huh7 cells (MOI 105) expressing TuD-HCV318 were plotted versus Huh7 cells expressing neither shRNA nor TuD. Numbers indicate genes that are significantly different between samples. P < 0.01, two-sided t-test; fold-change cutoff at log2(0.5). The genes from group 1 (blue) and group 2 (orange) in Fig. 4 A–C are highlighted. Overall gene-expression changes are mild, and, importantly, none of the group 1 or group 2 genes is significantly dysregulated by TuD-HCV318 alone.
To prove that the observed attenuating TuD effect indeed was caused by inhibition of the shRNA sense strand, we analyzed the enrichment of specific sequences in the 3′ UTRs of two different gene groups. The first were genes that always were down-regulated by shHCV318 regardless of whether the sense strand TuD was coexpressed (group 1 in Fig. 4 A–C, blue); accordingly, these genes should show an enrichment of antisense strand seed (nucleotides 1–8) matches in their 3′ UTRs. The second group of interesting genes was down-regulated by shHCV318 only in the absence of the TuD, suggesting that TuD coexpression had resulted in their de-repression (group 2 in Fig. 4 A–C, orange; only genes whose de-repression by the TuD was statistically significant were analyzed). We predicted that this group would represent endogenous off-targets of the shRNA sense strand that were rescued by the TuD, and therefore we expected to find an enrichment of sense strand seed matches in the 3′ UTRs of genes in group 2. For both groups, frequencies of full or partial antisense and sense seed matches in the 3′ UTRs were compared with the background frequencies of each respective sequence in the unbiased set of all 3′ UTRs from the genes contained on the microarrays. Notably, we indeed observed a highly significant enrichment of antisense strand seed matches in the 3′ UTRs of group 1 but not of group 2 (Fig. 4D, Left). The strongest enrichment, over ninefold, was noted for the full octamer seed match. Furthermore, confirming our predictions, sense strand seed matches were increased significantly in group 2 genes with a concomitant nonsignificant decrease in group 1 (Fig. 4D, Right). Again, the most pronounced enrichment (nearly fivefold) of complementary sense strand sequences in group 2 was found for complete matches of the seed sequence, i.e., nucleotides 1–8.
For further validation, we cloned one 3′ UTR of a group 1 gene that contained an antisense seed match and two 3′ UTRs of group 2 genes that carried sense seed matches into a dual luciferase reporter. Huh7 cells subsequently were transduced with the same scAAV2 vectors and MOI (105) as before and also were transfected with the three on-/off-target reporters. As expected, the group 1 antisense seed reporter was silenced to a similar degree by shHCV318 with and without TuD (Fig. 4E, CIRBP). Further, in line with the microarray data, both group 2 sense strand seed reporters were down-regulated with shHCV318 alone and were significantly rescued by sense strand TuD coexpression (Fig. 4E, CLCN7 and BCL7B).
As a whole, these data confirmed that the sense strand-specific TuD reduced endogenous off-target effects produced by the shRNA sense strand and thereby alleviated global dysregulation of cellular gene expression.
5′ but Not 3′ Strands of shRNAs Can Be Counteracted by TuDs.
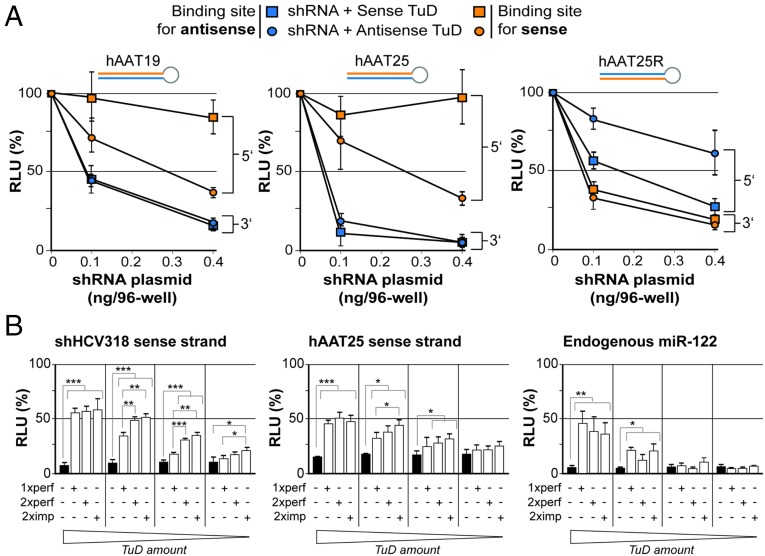
An interesting question at this point was whether TuD-mediated counteraction of shRNA sense strands was dependent on the exact shRNA configuration. We therefore tested TuD-mediated inhibition of a subset of shRNAs that had the same target site but differed in either stem length or strand order. Specifically, we analyzed human alpha-1 antitrypsin 19 (hAAT19) and hAAT25, which have 19- or 25-mer stems, respectively, and which both carry the antisense strand at the 3′ end of the hairpin (Fig. 5A). In contrast, hAAT25R has a 25-mer stem with the antisense strand located at the 5′ end of the hairpin. We then generated two TuDs directed against either strand of each shRNA and examined which combination of shRNA and TuD yielded highest TuD efficiencies. Curiously, the 5′ strand of all three shRNAs could be counteracted potently, but the 3′ strand could not, even at the lowest shRNA dose (Fig. 5A). From this result, we concluded that stem length is not critical but that the success of our strategy depends on a sense strand positioned at the 5′ end of the shRNA molecule. Notably, this particular configuration, 5′-sense-loop-antisense-3′, is, in fact, preferred by commercial shRNA design algorithms (e.g., www.invivogen.com/sirnawizard/design.php) and also was used in all our own previous work (including most of the shRNAs in Fig. 1A).
Fig. 5.
Determination of optimal shRNA and TuD design parameters. (A) Results from transfection of HEK293T cells with increasing doses of three shRNAs that differed in length and strand order but shared the same target. The cells were cotransfected with constant doses of specific TuDs and dual luciferase reporters as indicated. For each combination of TuD and reporter, luciferase reads were normalized to those measured in the absence of shRNA (set to 100%). (B) Functional analysis of three different TuD designs with one perfect, two perfect, or two imperfect binding site(s). (Left and Center) Results from HEK293T cells that were cotransfected with the indicated shRNAs and TuDs, together with a dual luciferase reporter to detect shRNA sense strand activity. (Right) Results from Huh7 cells that were cotransfected with a TuD against the 5′ arm of endogenous miR-122 and a corresponding dual luciferase reporter. In each experiment, four different TuD doses were evaluated: 100, 25, 6.25, and 1.56 ng per well (from left to right, as indicated below the graphs). Also shown in all panels are results obtained with an empty control (black bars), i.e., a TuD plasmid lacking specific binding sites for any of the three small RNAs. ***P < 0.001; **P < 0.01; *P < 0.05; two-sided t-test. Error bars in A and B represent SDs of at least three independent experiments.
Single Perfectly Matched Binding Sites Are Sufficient for Robust TuD Activity.
Having identified the ideal shRNA configuration, we also aimed at improving the design for TuD construction. The structure that was proposed originally and that we had adapted previously consists of two binding sites with central mismatches to the target RNA (36). Because the rules for placing these mismatches were rather obscure, we wanted to simplify this design and thus constructed new TuDs with two or one single perfectly matched binding site(s) for the sense strands of shHCV318 and hAAT25. Moreover, to determine whether the requirements are different for shRNA sense strands and the active arm of miRNAs, we also generated a set of TuDs against the 5′ strand of liver-specific miR-122. Each TuD then was tested at four different doses against a fixed amount of its cognate target and using a constant amount of shRNA (Fig. 5B). For all three targets, we generally observed a TuD dose dependency that was most pronounced for miR-122, congruent with published in vivo data for this particular miRNA (44). Compared with the standard TuD design with two imperfectly matched binding sites, the TuDs carrying two perfectly complementary binding sites were similarly efficient in inhibiting the 5′ strands of both shRNAs and the miRNA at all doses. Importantly, at the highest TuD dose, even the TuD carrying only one perfect binding site gave robust efficiencies that were identical to the conventional TuD design with two imperfect sites. We thus conclude that a largely simplified design—a single binding site with perfect complementarity—can provide high TuD activity that results in pronounced inhibition of the shRNA sense strand and miRNA.
Combinatorial Activity of shRNA and TuD Improves the Inhibition of HCV Replication.
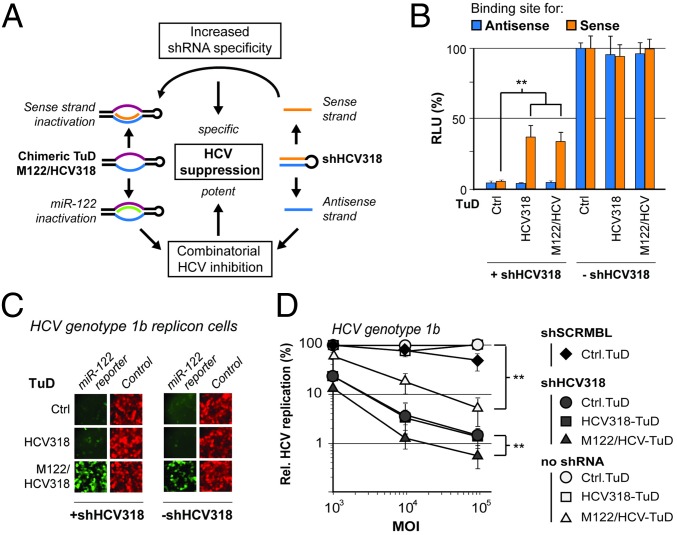
The aforementioned conclusion not only simplifies future TuD generation but also paves the way for the creation of chimeric TuDs with two different binding sites designed to counteract two independent target RNAs concurrently. To validate this concept, we again exploited the inhibition of HCV replication as a model. Based on a wealth of preclinical and clinical data showing that liver-specific miR-122 enhances HCV replication and that inhibition of this miRNA holds therapeutic value (45–49), we reasoned that TuD-mediated miR-122 sequestration could complement HCV suppression by RNAi efficiently. We thus generated a trifunctional scAAV2 vector carrying shHCV318 and a chimeric M122/HCV318-TuD that should counteract the shRNA sense strand and the miR-122 5′ arm simultaneously (Fig. 6A). Ideally, coexpression of shRNA and miR-122-TuD would lead to more potent HCV inhibition than the expression of either individually, and the sense strand TuD concomitantly would increase shRNA specificity.
Fig. 6.
Potent combinatorial HCV inhibition with an shRNA and a chimeric TuD. (A) Schematic depiction of the underlying rationale: Coexpression of shHCV318 and a chimeric TuD targeting miR-122 and the shHCV318 sense strand should result in combinatorial HCV inhibition and concomitantly increased shRNA specificity. (B) Results from the transduction of Huh7 cells with scAAV2 (MOI 105) expressing the shRNA/TuD combinations shown. Activities of shRNA sense and antisense activities were measured via cotransfected dual luciferase reporters carrying binding sites for the respective shRNA strand after the Renilla luciferase. (C) Results from the transduction of Huh7.5 cells carrying HCV 1b replicons with scAAV2 encoding the different shRNA/TuD combinations shown (MOI 105) as in B. The activity of endogenous miR-122 was measured by a cotransduced reporter vector (MOI 104) carrying a gfp gene with a perfect miR-122 binding site and untagged mCherry as the transduction control. (D) Results from the transduction of Huh7.5 cells carrying HCV 1b replicons with scAAV2 (MOI 105) expressing the shRNA/TuD combinations shown, as in B and C, and including a scrambled shRNA negative control (shSCRMBL). HCV replication was measured via replicon-encoded luciferase. **P < 0.01; two-sided t-test. Error bars represent SDs of four independent experiments (n = 4 each) that were averaged for the graph (resulting in a total n = 16). In B–D, the control was the empty TuD cloning scaffold lacking specific small RNA-binding sites.
For a most stringent proof of concept, we focused on analyzing this construct in Huh7.5 HCV 1b replicon cells, because this genotype (together with HCV 1a) predominates in the United States and in Europe (50, 51). Accordingly, we transduced these cells, which naturally express high levels of miR-122 (45), with scAAV2 vectors encoding different combinations of shHCV318 and TuDs. To validate the function of the chimeric TuD, we first performed control experiments in which we cotransfected luciferase reporters sensitive to either the shRNA sense or antisense strand. In the presence of the M122/HCV318-TuD, we noted a strong and specific de-repression of the shHCV318 sense strand luciferase reporter comparable to that seen with the regular single TuD, proving potent inhibition of the shRNA sense strand by the chimeric TuD (Fig. 6B). Its functionality was confirmed further in parallel in cells that were cotransduced with the shRNA/TuD vectors and an scAAV2 expressing an eGFP reporter with a perfect miR-122 binding site in its 3′ UTR and with mCherry as a transduction control. The chimeric M122/HCV318-TuD efficiently counteracted miR-122 activity (Fig. 6C), verifying that it can concurrently block the shRNA sense and the miR-122 antisense strands. As expected, the single HCV318-TuD did not affect the miR-122–sensitive eGFP reporter, once again proving the specificity of the approach.
Finally, we measured the dose-dependent effects of our various constructs on HCV replication in transduced Huh7.5 HCV 1b replicon cells. Individual expression of either shHCV318 or M122/HCV318-TuD substantially reduced HCV 1b replication compared with corresponding negative controls (Fig. 6D). Remarkably, over the entire 100-fold dose range, the combination of shHCV318 and M122/HCV318-TuD (filled triangles in Fig. 6D) resulted in a significant further improvement of HCV 1b inhibition compared with each molecule alone. In fact, at the highest vector MOI, we reduced HCV 1b replication by more than two orders of magnitude. This reduction represents the strongest effect noted in this entire work (compare also Fig. 3C) and highlights the great potential for clinical applications of our novel trifunctional AAV vector comprising the chimeric TuD.
Discussion
In this study, we developed and validated a new strategy to reduce shRNA sense strand activity, with the goal of increasing RNAi specificity without compromising silencing by the shRNA antisense strand. Even after one and a half decades of RNAi research (52), this aim remains vital, because unwanted off-target effects continue to hamper the application of shRNAs and other ectopic RNAi silencers. Curiously, although the overall problem has long been recognized, the field has rarely considered the shRNA sense strand as a major source of off-targeting. In fact, only few studies to date have investigated loading the two different shRNA strands into an RISC and studying their individual potential for gene silencing. These studies include notable findings from the Davidson and Kay laboratories that shRNA sense strands can principally act as a surrogate guide that directs the RISC to cleave reporters with an artificial target (12, 53, 54). Our results here validate and extend these data, because we also observed pronounced sense strand activity against specific reporters for 15 different shRNAs that we tested. In addition, our cDNA profiling data together with our analysis of distinct 3′ UTRs confirm that unintentional shRNA sense strand activity also can widely perturb the expression of endogenous off-targets. Taken together, these findings underscore the necessity to take concerns about sense strand activity seriously and to develop potent counter measures.
Toward this goal, we chose to exploit TuDs based on their reported superior efficiency in blocking miRNA function, compared with other small RNA inhibitors (35, 36, 55). Contrary to all previous studies that used TuDs to inhibit antisense strands of endogenous RNAi triggers (miRNAs) and thus to relieve suppression of their cellular on-targets, we advanced this technology by repurposing TuDs to block the sense strand of exogenous RNAi triggers (shRNAs) and thereby de-repress their off-targets. Indeed, we found that TuD coexpression significantly and specifically rescued the knockdown of reporters that were sensitive to an shRNA sense strand. Notably, we demonstrated that this rescue not only applies to artificial targets but also can be used to correct off-targeting of endogenous genes, as exemplified by our finding that TuD expression reduced the number of dysregulated genes in shRNA-expressing cells by nearly 50% (186 versus 334) (Fig. 4 A–C). This finding substantiates our belief that specific inhibition of shRNA sense strand off-targeting, as now facilitated by our new concept, should be a primary goal during future in vitro and in vivo RNAi applications.
With this goal in mind, we considered it essential to make our strategy as accessible, versatile, and widely applicable as possible. Therefore we designed our plasmid and AAV vector constructs in a modular fashion, permitting rapid and simple cloning of custom TuDs as well as exchange of the entire shRNA or TuD expression cassette(s). It was encouraging to find that both shRNA and TuD can be coexpressed in cis from a single AAV vector, thus further simplifying forthcoming applications. Concurrently, given the natural diversity of AAV isolates with different specificities and the feasibility of creating designer AAV vectors with desired properties (43, 56), the modular design paves the way for the translation of our strategy into manifold other cell types and organs ex vivo or in vivo. Furthermore, users of our approach should benefit from the following lessons that we learned during TuD optimization and that will guide the future design of custom vectors.
First, we found that only the 5′ shRNA strand can be counteracted by TuDs; the 3′ strand is mostly inert. This finding suggests that the two shRNA strands are functionally distinct, as is consistent with recent data that 5′ and 3′ shRNA arms differ in their maturation and association with Argonaute proteins (12, 57). Interestingly, shRNAs also differ from endogenous RNAi triggers because both the 5′ and 3′ strands of miRNAs can be counteracted efficiently by TuDs (35, 36). Moreover, our titration data (Fig. 5B) suggest that shRNAs and miRNAs are likewise distinct with respect to their TuD dose dependency, because rescue of the miR-122 reporter required higher TuD amounts than the two shRNAs. However, another difference between shRNAs and miRNAs is that TuDs seem to sequester stably and thus inactivate shRNA strands (Fig. 3B and Fig. S5), whereas they induce tailing and trimming of mature miRNA strands, as reported in mouse livers (44). Therefore an intriguing aim for future work is to delineate further the molecular reasons for all these unique behaviors of shRNAs versus miRNAs that have become apparent through our original use of TuD technology; doing so will improve our understanding of RNAi mechanisms. Fortunately, as noted before, the shRNA configuration with the sense strand at the 5′ arm predominates among published shRNAs and also is widely recommended by academic and industrial groups. We thus are convinced that our strategy of using TuDs to inhibit the 5′ sense strand specifically will be applicable to the great majority of existing or future shRNAs.
Second, we were pleased to find that two perfect binding sites work with efficiency similar to that of two imperfect sites and that even a single binding site can yield robust inhibition of shRNA or miRNA 5′ strands (depending on TuD dose; see below). These two observations further simplify the generation of new TuDs, particularly as compared with the initial report on their use for miRNA inhibition, which used two imperfect binding sites (36). The possibility of encoding a single perfect site obviates uncertainties about the ideal position of the artificial mismatches and thus facilitates TuD design. Notably, two recent studies support this notion by showing that miRNAs also can be inhibited potently by a perfectly matched TuD (35) and that one or two miRNA binding sites work equally well (58). In turn, this finding emphasizes that our AAV vectors for custom TuD expression are also useful for specific inhibition of cellular miRNAs, as we already have exemplified in the present study with miR-122.
Third, we found at least two lines of evidence indicating that the overall intracellular TuD concentration is important. The first is that, for a fixed amount of shRNA (or miRNA), increasing TuD doses resulted in improved rescue of a cognate reporter plasmid (Fig. 5B). The second is that, for one of our shRNAs, potent sense strand inhibition required TuD expression from a U6 promoter (at least in an AAV context) (Fig. S4C), which is inherently stronger than H1, which was used in all other cases. The ensuing conclusion that TuD concentration can be limiting under certain circumstances again agrees with reported findings for miRNAs, in which constructs expressing up to four TuD repeats outperformed those with fewer (58). We thus generally recommend testing various combinations of promoters (H1 or U6) to drive shRNA and cognate TuD to strike an optimal balance between on-target knockdown and rescue of off-targeting. Again, the modular design of our AAV vectors will facilitate and accelerate this process. Moreover, it may be interesting to adapt a strategy from the Mikkelsen laboratory, which expressed clustered TuDs from a RNA polymerase II promoter (55, 58), to gain spatiotemporal control over RNAi specificity.
An example of a rational application of these design rules is provided by our demonstration of combinatorial HCV suppression through the concerted action of shHCV318 and the chimeric miR-122/shHCV318 TuD. Our observation of an additive inhibition of HCV genotype 1b, an HCV variant that has poor response to IFN-based therapy (50, 51), highlights the power and potential of multivalent AAV-shRNA/TuD vectors. Particularly noteworthy from a clinical standpoint is that the juxtaposition of two independent molecular mechanisms—shRNA knockdown and TuD-based miR-122 inactivation—should reduce HCV’s chances for mutational escape. This outcome is strongly implied by a plethora of consistent data from chronically HCV-infected chimpanzees (the only available physiologically relevant and immunocompetent HCV animal model) and human patients who were treated with locked nucleic acid inhibitors of miR-122 and who experienced substantial drops in HCV viremia with little development of viral resistance (46–48). This unique benefit of our novel vectors, combining specific shRNA knockdown with inhibition of cellular miRNAs, should stimulate their application in treating other diseases with a miRNA component, such as many cancers or HIV infection. The counteraction of the shHCV318 sense strand also is interesting from a biological viewpoint. Although HCV is a positive-sense RNA virus, it replicates through a negative-sense RNA strand, which therefore represents an intriguing therapeutic target (38, 59). However, previous failures to target the negative-sense HCV strand with siRNAs or shRNAs suggest that it is immune to RNAi (60, 61). Our own finding that HCV inhibition was not relieved by blocking our shRNA’s sense strand with the TuD indicates that HCV knockdown was mediated entirely by the shHCV318 antisense strand. We hence conclude that, despite its high sense strand activity (e.g., in reporter assays) (Fig. 1A), our shRNA also did not target the HCV negative-sense strand. This example exemplifies how our vectors also can be exploited as a new tool to dissect the molecular interactions between RNAi triggers and their targets, or, in this particular case, to study virus biology.
In conclusion, our work confirms increasing concerns that shRNA sense strands are a major source of RNAi off-targeting and concurrently proposes an original avenue toward enhanced specificity that is potent, easy to customize, compatible with a wealth of old and new shRNAs, and applicable for combinatorial shRNA/miRNA regulation. Ideally, in the future, this approach will be combined with further strategies to increase on-targeting or to prevent off-targeting, such as bioinformatical identification of “safe” sequences, rational design of shRNAs with inherent strand biases, AAV capsid engineering, or TuD concatamerization for better inhibition (25, 26, 43, 58). Moreover, our technology readily complements high-throughput strategies that also aim at identifying potent shRNAs with reduced off-targeting potential, such as a massively parallel sensor assay or screening of ultracomplex, high-coverage shRNA libraries (20, 62–64). Collectively, this approach should help to create a new generation of RNAi vectors with an optimal balance of efficiency, specificity, and safety and thus foster the current renaissance of the enthusiasm for a clinical translation of RNAi technologies.
Materials and Methods
Details of GFP reporter assays and mRNA northern blot analysis of egfp and mCherry expression are given in SI Materials and Methods.
Plasmid Construction.
Plasmids pBS-H1-TuD-empty-GFP and pBS-U6-TuD-empty-GFP are bicistronic constructs that contain an RSV promoter-driven GFP expression cassette together with an H1 or U6 promotor for expression of custom TuDs. They were constructed by insertion of annealed oligonucleotides TuD-empty F and R (see Table S1 for all oligonucleotides and primers) carrying two inverted BsmBI sites into BbsI-digested pBS-H1-GFP or pBS-U6-GFP, respectively (4, 65). pBS-H1-TuD-empty-GFP and pBS-U6-TuD-empty-GFP are ideal for cases in which a TuD should be expressed without shRNA. To create vectors coexpressing shRNA and TuD, the entire H1- or U6-TuD-empty cassette was PCR-amplified (primers H1 F and TuD R or U6 F and TuD R; Table S1) along with a stuffer sequence (Stuffer F and R; Table S1), and both fragments were joined by overlap extension PCR. The resulting stuffer-H1-TuD or stuffer U6-TuD products were cut with XbaI/SalI and inserted into NheI/SalI-digested pBS-H1-GFP, yielding pBS-H1-H1-TuD-empty or pBS-H1-U6-TuD-empty, respectively. In both cases, the first H1 promoter can be exploited for cloning and expression of custom shRNAs, as reported before (65). As a result of this vector design, the shRNA cassette remains excisable by unique AscI/NotI sites, and the TuD cassette can be isolated via unique NheI/SalI sites (Fig. 2).
Table S1.
DNA oligonucleotides used in this study
| Oligonucleotides for general cloning | |
| TuD empty F | caccGACGGCGCTAGGATCATCCCAGAGACGACCGTCTCACCGATGATCCTAGCGCCGTC |
| TuD empty R | aaaaGACGGCGCTAGGATCATCGGTGAGACGGTCGTCTCTGGGATGATCCTAGCGCCGTC |
| H1 F | GGTCCTGCTGGAGTTCGTGAGCTAGCCATATTTGCATGTCGCTATGTGTTC |
| U6 F | GGTCCTGCTGGAGTTCGTGAGCTAGCCGAGTCCAACACCCGTGGGAATC |
| TuD R | TTAGGTCGACCGGCCGCAAAAAAGACGGCGCTAG |
| Stuffer F | TTAGTACTAGTCACCGGGGTGGTGCCCATCCTG |
| Stuffer R | TCACGAACTCCAGCAGGACCATG |
| shRNA oligonucleotides, antisense sequence underlined | |
| hAAT19 F | caccGAAGCGTTTAGGCATGTTTTCAAGAGAAACATGCCTAAACGCTTC |
| hAAT19 R | aaaaGAAGCGTTTAGGCATGTTTCTCTTGAAAACATGCCTAAACGCTTC |
| hAAT25 F | caccGAAGCGTTTAGGCATGTTTAACATCTCAAGAGGATGTTAAACATGCCTAAACGCTTC |
| hAAT25 R | aaaaGAAGCGTTTAGGCATGTTTAACATCCTCTTGAGATGTTAAACATGCCTAAACGCTTC |
| sAg21SA F | caccGTTTACTAGTGCCATTTGTTCTCAAGAGGAACAAATGGCACTAGTAAAC |
| sAg21SA R | aaaaGTTTACTAGTGCCATTTGTTCCTCTTGAGAACAAATGGCACTAGTAAAC |
| shHCV318 F | caccGGGAGGTCTCGTAGACCGTGCATCAAGAGTGCACGGTCTACGAGACCTCCC |
| shHCV318 R | aaaaGGGAGGTCTCGTAGACCGTGCACTCTTGATGCACGGTCTACGAGACCTCCC |
| shHBVS1 F | caccGGTATGTTGCCCGTTTGTCTTCAAGAGAGACAAACGGGCAACATACC |
| shHBVS1 R | aaaaGGTATGTTGCCCGTTTGTCTCTCTTGAAGACAAACGGGCAACATACC |
| shHBVS122 F | caccGGTATGTTGCCCGTTTGTCTTCTCAAGAGGAAGACAAACGGGCAACATACC |
| shHBVS122 R | aaaaGGTATGTTGCCCGTTTGTCTTCCTCTTGAGAAGACAAACGGGCAACATACC |
| shcmyc F | caccGTCCTGAGACAGATCAGCAACATCAAGAGTGTTGCTGATCTGTCTCAGGAC |
| shcmyc R | aaaaGTCCTGAGACAGATCAGCAACACTCTTGATGTTGCTGATCTGTCTCAGGAC |
| shSCRMBL F | caccGAGCCGGTCAGCTGCGAGTGTATCAAGAGTACACTCGCAGCTGACCGGCTC |
| shSCRMBL R | aaaaGAGCCGGTCAGCTGCGAGTGTACTCTTGATACACTCGCAGCTGACCGGCTC |
| Oligonucleotides for 3′UTR target site reporters (S sequences are targeted by the antisense strand, and AS sequences are targeted by the sense strand) | |
| sAg-S perf F | tcgaCTCAGTTTACTAGTGCCATTTGTTC |
| sAg-S perf R | ggccGAACAAATGGCACTAGTAAACTGAG |
| sAg-AS perf F | tcgaGAACAAATGGCACTAGTAAACTGAG |
| sAg-AS perf R | ggccCTCAGTTTACTAGTGCCATTTGTTC |
| hAAT-S perf F | tcgaGAAGCGTTTAGGCATGTTTAACATC |
| hAAT-S perf R | ggccGATGTTAAACATGCCTAAACGCTTC |
| hAAT-AS perf F | tcgaGATGTTAAACATGCCTAAACGCTTC |
| hAAT-AS perf R | ggccGAAGCGTTTAGGCATGTTTAACATC |
| HCV318-S perf F | tcgaGGGAGGTCTCGTAGACCGTGCA |
| HCV318-S perf R | ggccTGCACGGTCTACGAGACCTCCC |
| HCV318-AS perf F | tcgaTGCACGGTCTACGAGACCTCCC |
| HCV318-AS perf R | ggccGGGAGGTCTCGTAGACCGTGCA |
| HCV318-AS imp F | tcgaTGCACGGTCTATGAGACCTCCC |
| HCV318-AS imp R | ggccGGGAGGTCTCATAGACCGTGCA |
| HBVS1-S perf F | tcgaGGTATGTTGCCCGTTTGTCT |
| HBVS1-S perf R | ggccAGACAAACGGGCAACATACC |
| HBVS1-AS perf F | tcgaAGACAAACGGGCAACATACC |
| HBVS1-AS perf R | ggccGGTATGTTGCCCGTTTGTCT |
| P53P7-S perf F | tcgaGGAGCTGAATGAGGCCTTAGA |
| P53P7-S perf R | ggccTCTAAGGCCTCATTCAGCTCC |
| P53P7-AS perf F | tcgaTCTAAGGCCTCATTCAGCTCC |
| P53P7-AS perf R | ggccGGAGCTGAATGAGGCCTTAGA |
| sh122-AS perf F | tcgaTGGAGTGTGACAATGGTGTTTGT |
| sh122-AS perf R | ggccACAAACACCATTGTCACACTCCA |
| shcmyc-S perf F | tcga GTCCTGAGACAGATCAGCAACA |
| shcmyc-S perf R | ggcc TGTTGCTGATCTGTCTCAGGAC |
| shcmyc-AS perf F | tcga TGTTGCTGATCTGTCTCAGGAC |
| shcmyc-AS perf R | ggcc GTCCTGAGACAGATCAGCAACA |
| CIRBP 3UTR F | AATAGTCGACAAACCCTTCCTGCTCAAGATCG |
| CIRBP 3UTR R | AATTAGCGGCCGCGTGCACGTCCACTGTCCCTGC |
| CLCN7 3′UTR F | GATAGTCGACGGCCCAGCCCTGCCCATAATGG |
| CLCN7 3′UTR R | GATTAGCGGCCGCGCCCTGTGAGGGCTAAGCAGGG |
| BCL7B 3′UTR F | GATAGTCGACCACCATCCCGGCCCTCCGCC |
| BCL7B 3′UTR R | GATTAGCGGCCGCTATGTCTTCACTCACTGTTGCCC |
| Oligonucleotides for TuD cloning via directly annealed DNA duplexes, decoy sites underlined | |
| TuD-HCV318 2ximp F | tcccaAGCACGGTCTATGAGACCTCCCCAAGTATTCTGGTCACAGAATACAACTGCACGGTCTATGAGACCTCCCa |
| TuD-HCV318 2ximp R | tcggtGGGAGGTCTCATAGACCGTGCAGTTGTATTCTGTGACCAGAATACTTGGGGAGGTCTCATAGACCGTGCTt |
| TuD-HCV318 2xperf F | tcccaTGCACGGTCTACGAGACCTCCCCAAGTATTCTGGTCACAGAATACAACTGCACGGTCTACGAGACCTCCCa |
| TuD-HCV318 2xperf R | tcggtGGGAGGTCTCGTAGACCGTGCAGTTGTATTCTGTGACCAGAATACTTGGGGAGGTCTCGTAGACCGTGCAt |
| TuD-hAAT 2ximp F | tcccaTTAAACATGCATAAACGCTTCCAAGTATTCTGGTCACAGAATACAACTTAAACATGCCGAAACGCTTCa |
| TuD-hAAT 2ximp R | tcggtGAAGCGTTTCGGCATGTTTAAGTTGTATTCTGTGACCAGAATACTTGGAAGCGTTTATGCATGTTTAAt |
| TuD-HBVS1 2ximp F | tcccaAGACAAACGTGCAACATACCCAAGTATTCTGGTCACAGAATACAACAGACAAACGAGCAACATACCa |
| TuD-HBVS1 2ximp R | tcggtGGTATGTTGCTCGTTTGTCTGTTGTATTCTGTGACCAGAATACTTGGGTATGTTGCACGTTTGTCTt |
| TuD-M122 2ximp F | tcccaCAAACACCATGCCAACACTCCACAAGTATTCTGGTCACAGAATACAACCAAACACCATGCCAACACTCCAa |
| TuD-M122 2ximp R | tcggtTGGAGTGTTGGCATGGTGTTTGGTTGTATTCTGTGACCAGAATACTTGTGGAGTGTTGGCATGGTGTTTGt |
| TuD-M122 2xperf F | tcccaCAAACACCATTGTCACACTCCACAAGTATTCTGGTCACAGAATACAACCAAACACCATTGTCACACTCCAa |
| TuD-M122 2xperf R | tcggtTGGAGTGTGACAATGGTGTTTGGTTGTATTCTGTGACCAGAATACTTGTGGAGTGTGACAATGGTGTTTGt |
| Oligonucleotides used as probes in small RNA Northern blots | |
| U6snRNA | TGTGCTGCCGAAGCGAGCAC |
| shHCV318 antisense | TCGAGGGAGGTCTCGTAGACCGTGCA |
| shHCV318 sense | TCGATGCACGGTCTACGAGACCTCCC |
| Oligonucleotides for TuD cloning via overlap extension, decoy sites underlined | |
| TuD-HCV318 1ximp F | AATAGGTCTCGTCCCAAGCACGATAAATGAGACCTCCCCAAGTATTCTGGTCACAGAATA |
| TuD-HCV318 1ximp R | AATAGGTCTCGTCGGTGGGAGGTCTCATAGACCGTGCAGTTGTATTCTGTGACCAGAATA |
| TuD-cmyc 1xperf F | AATACGTCTCGTCCCATGTTGCTGATCTGTCTCAGGACCAAGTATTCTGGTCACAGAATA |
| TuD-cmyc noBS R | AATACGTCTCGTCGGTTAGCTTATCGGACTGATGTTGAGTTGTATTCTGTGACCAGAATA |
| TuD-hAAT 1xperf F | AATACGTCTCGTCCCATTAAACATGCCTAAACGCTTCCAAGTATTCTGGTCACAGAATA |
| TuD-hAAT 1xperf R | AATACGTCTCGTCGGTGAAGCGTTTAGGCATGTTTAACGTTGTATTCTGTGACCAGAATA |
| TuD-hAATS 1xperf F | AATACGTCTCGTCCCAAGCGTTTAGGCATGTTTAACATCAAGTATTCTGGTCACAGAATA |
| TuD-HCV318 1xperf R | AATACGTCTCGTCGGTGGGAGGTCTCGTAGACCGTGCAGTTGTATTCTGTGACCAGAATA |
| TuD-M122 1xperf F | AATAGGTCTCGTCCCACAAACACCATTGTCACACTCCACAAGTATTCTGGTCACAGAATA |
All sequences are shown in 5′→3′ orientation. Where applicable, restriction sites are indicated in bold letters and overhangs in lowercase. Note that the first stem of the TuD is already included in our plasmid backbone and hence is not part of the oligonucleotides shown above. The advantage of this design is that it facilitates cloning of customized TuDs by permitting the use of shorter oligonucleotides.
To generate specific TuD-expressing vectors, the TuD plasmids described above were digested with BsmBI, and oligonucleotide duplexes comprising the decoy sequences were inserted by one of two strategies: either two long oligonucleotides were annealed and cloned directly or two short oligonucleotides with overlapping 3′ ends were elongated by PCR, digested with BsmBI, and cloned. The second method has the advantage of being cheaper and more versatile, because it allows easy shuffling of different decoy sites with the same set of oligonucleotides; however, it also is more time-consuming and laborious. (Table S2 indicates which method was used for the different TuDs). Plasmids coexpressing shRNAs were generated either by insertion of annealed oligonucleotides (Table S1 and ref. 4) into BbsI-digested pBS-H1-H1-TuD-empty or by transfer of an already existing shRNA cassette that was excised from either pBS-H1-GFP or pBS-U6-GFP by AscI/NotI digestion and inserted into the same sites of pBS-H1-H1-TuD-empty. In all constructs described above, the entire recombinant sequence was flanked by two AAV inverted terminal repeats (ITRs) for packaging as self-complementary AAV vector particles (Fig. 2).
Table S2.
TuD constructs used in this study
| TuD name | Decoy sites | Cloned via | Used in figure |
| TuD-HCV318 2ximp1 | 2× imperfect for shHCV318 sense strand | Extension | 1C, 3 A–D, 4 A–E, 6 B–D, S2, S3 A–D, S4A, S5, S6 |
| TuD-HCV318 2ximp2 | 2× imperfect for shHCV318 sense strand | Annealing | 5B |
| TuD-HCV318 2xperf | 2× perfect for shHCV318 sense strand | Annealing | 5B |
| TuD-hAAT 1xperf/HCV318 1xperf | 1× perfect for hAAT sense strand & 1× perfect for shHCV318 sense strand | Extension | 5 A and B |
| TuD-hAAT 2ximp | 2× imperfect for hAAT sense strand | Annealing | 1C, 5B, S4 B and C |
| TuD-hAAT 2xperf | 2× perfect for hAAT sense strand | Extension | 5B |
| TuD-hAATS 1xperf/HCV318 1xperf | 1× perfect for hAAT antisense strand & 1× perfect for shHCV318 sense strand | Extension | 5 A and B |
| TuD-HBVS1-2ximp | 2× imperfect for shHBVS1 sense strand | Annealing | 1C |
| TuD-cmyc-1xperf | 1× perfect for shcmyc sense strand | Extension | 1C |
| TuD-M122 2ximp | 2× imperfect for miR-122-5p | Annealing | 5B |
| TuD-M122 2xperf | 2× perfect for miR-122-5p | Annealing | 5B |
| TuD-M122 1xperf/HCV318 1ximp | 1× perfect for miR-122-5p and 1× imperfect for shHCV318 sense strand | Extension | 5B, 6 B–D |
miR-122-5p denotes the 5′ arm of miR-122, i.e., the strand that is involved in HCV regulation.
Anti-hAAT, anti-sAg (HBV surface antigen), and anti-p53 shRNAs were available in the laboratory from previous studies (4, 66) (see the same references for all shRNA target sequences). An additional shRNA against HBV, shHBVS1, was cloned as described elsewhere (67), and shHCV318 (targeting the HCV genome) was adapted from a previously reported shRNA (38). A scrambled negative control shRNA (shSCRMBL) was generated based on shHCV318 using the “Scramble siRNA/shRNA” function in siRNA Wizard 3.1 (www.invivogen.com/sirnawizard/design.php). Dual luciferase reporter constructs carrying single shRNA target sites behind one of the two luciferases (Renilla) were generated by insertion of annealed oligonucleotides carrying the respective sequence (Table S1) into XhoI/NotI-digested psiCheck-2 (Promega). This particular configuration (see also Fig. S1) was chosen because the Renilla luciferase reporter yields higher photon counts than the coencoded Firefly luciferase (which was used for normalization), thus providing a broader and more sensitive window for detection and quantification of shRNA effects. Reporter constructs for shHCV318 off-target measurements were generated by PCR amplification of the 3′ UTR of selected genes or of parts thereof using primers CIRBP 3′ UTR F and R, CLCN7 3′ UTR F and R, or BCL7B 3′ UTR F and R (Table S1), followed by SalI/NotI digestion and insertion into XhoI/NotI-digested psiCheck-2. GFP reporter constructs were generated by cloning annealed oligonucleotides HCV318-S perf F and R, or HCV318-AS perf F and R (Table S1) into the eGFP 3′ UTR in a plasmid coexpressing eGFP and mCherry from a bidirectional CMV promoter.
Tissue Culture and Transfection.
HEK293T cells were cultured in DMEM supplemented with 10% (vol/vol) FCS, 1% penicillin/streptomycin (P/S), and 1% l-glutamine (l-Glu) (all reagents from PAA). Huh7 cells were cultured in DMEM supplemented with 10% (vol/vol) FCS, 1% P/S, 1% l-Glu, and 1% nonessential amino acids (NEAA) (PAA Laboratories). All cells were cultured under standard growth conditions [5% (vol/vol) CO2, 37 °C] and passaged upon confluency. For transfections, HEK293T cells were grown to 70–90% confluency and transfected using polyethylenimine (PEI) as described elsewhere (29). Huh7 cells were grown to 90–95% confluency and transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s guidelines.
Luciferase Reporter Assays.
Cells were grown in transparent 96-well tissue-culture plates (Greiner). Typically, unless stated otherwise, 2.5–5 ng (HEK293T) or 50 ng (Huh7) psiCheck-2 plasmid and 50–100 ng shRNA or TuD vector were used (total DNA amount per well was 200 ng). When shRNA and TuD were delivered from separate plasmids, constructs were transfected in a 1:1 molar ratio unless stated otherwise. Two days posttransfection, the medium was removed from each well, and the cells were lysed in 25 μL of the lysis buffer provided with the Dual-Luciferase assay kit (Promega) and were incubated for 15 min at room temperature. Five microliters of the lysate then was transferred into a white Lumitrac 200 plate (Greiner). Renilla and Firefly luciferase activities were measured after consecutive injection of 25 μL reconstituted luciferase assay buffer and Stop&Glow solution (supplemented with Rluc substrate) according to the Dual Luciferase kit using a Glomax 96 microplate luminometer (Promega).
Small RNA Northern Blot.
Huh7 cells were seeded at 2 × 105 cells per well in a six-well plate and were transduced on the following day with an MOI of 105 with the different shRNA/TuD-encoding scAAV2 vectors. Two days postinfection, the cells were lysed, and total RNA was prepared using Qiazol (Qiagen) according to the manufacturer’s instructions. Ten micrograms of total RNA was precipitated, washed once with 70% (vol/vol) EtOH, and dissolved in 5 μL RNA loading buffer [98% (vol/vol) formamide, 10 mM EDTA, 0.025% bromophenol blue, 0.025% xylene cyanol], briefly denatured at 70 °C, and resolved in a 15% (vol/vol) acrylamide/8 M urea gel. The gel was electroblotted onto a Nylon membrane (Hybond N+; Amersham), which then was UV-crosslinked (energy program, HL-2000 Hybrilinker, UVP) and hybridized in ExpressHyb hybridization buffer (Clontech) with radioactively labeled probes. Radioactive end-labeling of oligonucleotide probes was performed using T4 polynucleotide kinase (New England Biolabs) under standard reaction conditions. The oligonucleotides used as probes were anti-human transfer RNA glutamine 1 (tRNAQ1) as housekeeper, shHCV318 antisense, and shHCV318 sense (Table S1). The TuD was detected using the shHCV318 sense probe.
scAAV Purification by Iodixanol Density Gradient Ultracentrifugation.
HEK293T cells were triple-transfected with pVAE2AE4-5 (Adenovirus helper plasmid) (68), an AAV helper plasmid, p5E18, encoding AAV serotype 2 capsid proteins for in vitro transduction assays (69), and the AAV vector plasmid (as indicated) using the PEI transfection method. Three days posttransfection, cells were harvested using a cell scraper and were lysed by repeated freeze–thaw cycles. scAAV vector particles were purified via iodixanol (Optiprep; Sigma) density gradient ultracentrifugation for 2 h at 50,000 rpm at 4 °C in a Beckman 70.1Ti rotor and a Beckman L-90K ultracentrifuge as described previously (29). scAAV vector genome titers were determined by quantitative PCR against a plasmid standard curve (29, 70).
HCV Inhibition Assay.
Huh7.5-lucubineo-JFH1 (HCV genotype 1b) or -ET (genotype 2a) replicon cell lines (71) were cultured in DMEM supplemented with 10% (vol/vol) FCS, 1% P/S, 1% l-Glu, 1% NEAA, and 1 mg/mL or 500 μg/mL G418, respectively. On the day of scAAV transduction, 1 × 106 cells were seeded per 96-well plate and were inoculated directly thereafter with an scAAV vector MOI of 103, 104, or 105. Two days posttransduction, the growth medium was removed, and cells were lysed in 30 μL Fluc lysis buffer [0.1% Triton X-100, 25 mM glycylglycine (pH 7.8), 15 mM MgSO4, 4 mM EGTA (pH 7.8), 10% (vol/vol) glycerol, 1 mM DTT] per well and were frozen immediately at −20 °C. After thawing, 20 μL of the lysate was transferred to a Lumitrac 200 plate (Greiner) and was incubated for 15 min at room temperature in the dark with 50 μL Fluc assay buffer [25 mM glycylglycine, 15 mM KPO4 (pH 7.8), 15 mM MgSO4, 4 mM EGTA, 7 μM d-luciferin, 2 mM ATP, 1 mM DTT). Firefly luciferase activity was measured using a Glomax 96 microplate luminometer (Promega).
Expression Profiling by Microarray.
Huh7 cells were seeded at 2 × 105 cells per well in a six-well plate. The next day, the cells were transduced at an MOI of 105 with different shRNA/TuD scAAV2 vectors (Fig. 4 A–C). Two days posttransduction, the cells were lysed, and total RNA was prepared using Qiazol (Qiagen) according to the manufacturer’s instructions. RNA quality was assessed using a Bioanalyzer and a RNA Nano Chip (both from Agilent). Only RNAs with an RNA integrity number of at least 8.7 were used for further studies. The samples then were analyzed with a HumanHT-12 v4 Expression BeadChip (Illumina). Reverse transcription, hybridization, and data normalization to reference transcripts were conducted by the Genomics Core Facility of the German Cancer Research Center in Heidelberg. During data analysis, all transcripts whose expression was less than threefold higher than their SD were excluded (background subtraction). Dysregulated transcripts were identified by comparison of each shRNA sample with the cognate nonsilencing control (P < 0.01, unpaired, two-sided t-test, degrees of freedom = 14). Genes that were significantly rescued, i.e., less down-regulated in the presence of the TuD, were identified by comparing the two shRNA samples against each other (P < 0.01, unpaired, two-sided t-test). The respective genes were grouped as indicated in Fig. 4, and the 3′ UTR sequences of each group were extracted from the Ensembl database using the Biomart sequence retrieval tool (www.biomart.org). Because not all genes on the chip had extractable 3′ UTR sequences, the number of genes used for seed match analysis was lower than the total number of significant hits. Sequence matches were defined as reverse complements of the sequences indicated in Fig. 4D. Sequence enrichments in each gene group were calculated as follows: number of genes containing at least one sequences match in their 3′ UTRs divided by the total number of genes in the respective gene group. All genes with extractable 3′ UTR sequences present on the chip were used as background set. Statistical significance of the counting results was determined by Fisher’s exact test (in group 1: two-sided test for sense strand seed matches and one-sided test for antisense strand seed matches; in group 2: one-sided test for sense strand seed matches and two-sided test for antisense strand seed matches).
SI Materials and Methods
GFP Reporter Assays.
HEK293T cells were grown in 24-well plates (Greiner) and were cotransfected with 500 ng shRNA vector and 100 ng pSSV9-DualFluor target vector (containing binding sites for the shRNA sense or antisense strand in its 3′ UTR) per well. Two days posttransfection, the cells were analyzed by either fluorescence microscopy or FACS. For fluorescence microscopy, cells were imaged directly with an Olympus IX61 microscope (10× objective; exposure times: 50 ms for mCherry and 100 ms for eGFP; 100% light intensity). Pictures were colored and merged using ImageJ. For dual-color FACS, cells were trypsinized and washed once with 1× PBS and then were kept in 1× PBS on ice. The FACS analysis was performed using a FACScalibur device (BD Bioscience). Instrument settings were detector gain: FSC, E0 (lin); SSC, 454 (lin); GFP (FL-2), 425 (log); mCherry (FL-3), 535 (log); compensation: FL-3 = 99% FL-2. For each sample, at least 10,000 cells were analyzed. Live cells were selected via forward scatter/side scatter. The mCherry cutoff was set according to a nontransfected HEK293T control, and mean values of eGFP and mCherry signals were quantified for the mCherry-positive cell populations. Each eGFP intensity was normalized to its corresponding mCherry signal.
mRNA Northern Blot Analysis of egfp and mCherry Expression.
HEK293T cells were seeded at 5 × 105 cells per well in a six-well plate and were transfected the next day with 2 μg shRNA/TuD-expressing plasmid and 200 ng pSSV9-DualFluor target plasmid. Two days posttransfection, the cells were lysed, and total RNA was prepared using Qiazol (Qiagen) according to the manufacturer’s instructions. Two micrograms of total RNA were mixed with one third loading buffer (Ambion), denatured briefly at 65 °C, and then resolved in a 0.8% agarose/6.7% (vol/vol) formaldehyde gel in 1× 3-(N-morpholino)propanesulfonic acid buffer (Northernmax; Ambion). After transfer onto a Nylon membrane (Hybond N+; Amersham) by passive diffusion overnight and UV-crosslinking (time-program, CL-1000 UV crosslinker, UVP), the membrane was hybridized in PerfectHyb buffer (Sigma) as described previously (65). For probe generation, the eyfp cDNA (nearly identical to egfp and thus suitable as probe template) was excised from pIRESNeo-FLAG/HA-EYFP (72) by BamHI digestion; the mCherry cDNA was isolated from pSSV9-DualFluor by HindIII/SalI digestion. Following gel purification, 50 ng of template was used for probe generation via random priming, using the Prime-it Rmt Random priming kit (Stratagene) according to the manufacturer’s instructions. Signals specific for egfp and mCherry mRNAs were detected sequentially on the same membrane. Therefore, egfp mRNA was detected first, followed by stripping the egfp probes off the membrane by boiling in 0.1% SDS for 10 s and subsequent hybridization and detection of mCherry mRNA. Bands were quantified using the measure function in ImageJ after background subtraction. egfp signals first were normalized to respective mCherry signals, and then each sample was normalized to its cognate nonsilencing control.
Acknowledgments
We thank Elena Senís for help with RNA preparations during the revision of this manuscript; Ellen Wiedtke for help with AAV vector production; Andrea Bauer from the Genomics Core Facility of the German Cancer Research Center for help with expression profiling; Christian Bender for help with determining sequence frequencies in the background gene set during the analysis of the microarray data; and various members of the D.G. laboratory and Kathleen Börner for critical reading of the manuscript and for helpful comments. This work was supported by the Cluster of Excellence CellNetworks (German Research Foundation, EXC81), including a PhD scholarship (S.M.), and by the Frontier Innovation Grant of Heidelberg University. S.M. and S.G. received support from the Hartmut Hoffmann-Berling International Graduate School of Molecular and Cellular Biology at Heidelberg University. R.B. was supported by the Deutsche Forschungsgemeinschaft (FOR1202, TP1).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510476112/-/DCSupplemental.
References
- 1.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457(7228):426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giering JC, Grimm D, Storm TA, Kay MA. Expression of shRNA from a tissue-specific pol II promoter is an effective and safe RNAi therapeutic. Mol Ther. 2008;16(9):1630–1636. doi: 10.1038/mt.2008.144. [DOI] [PubMed] [Google Scholar]
- 4.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 5.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9(1):57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 6.Jackson AL, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21(6):635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 7.Jackson AL, et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12(7):1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vickers TA, et al. Off-target and a portion of target-specific siRNA mediated mRNA degradation is Ago2 ‘Slicer’ independent and can be mediated by Ago1. Nucleic Acids Res. 2009;37(20):6927–6941. doi: 10.1093/nar/gkp735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol. 2012;14(3):318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schultz N, et al. Off-target effects dominate a large-scale RNAi screen for modulators of the TGF-β pathway and reveal microRNA regulation of TGFBR2. Silence. 2011;2:3. doi: 10.1186/1758-907X-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schürmann N, Trabuco LG, Bender C, Russell RB, Grimm D. Molecular dissection of human Argonaute proteins by DNA shuffling. Nat Struct Mol Biol. 2013;20(7):818–826. doi: 10.1038/nsmb.2607. [DOI] [PubMed] [Google Scholar]
- 12.Gu S, et al. Thermodynamic stability of small hairpin RNAs highly influences the loading process of different mammalian Argonautes. Proc Natl Acad Sci USA. 2011;108(22):9208–9213. doi: 10.1073/pnas.1018023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwak PB, Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat Struct Mol Biol. 2012;19(2):145–151. doi: 10.1038/nsmb.2232. [DOI] [PubMed] [Google Scholar]
- 14.Winter J, Diederichs S. Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol. 2011;8(6):1149–1157. doi: 10.4161/rna.8.6.17665. [DOI] [PubMed] [Google Scholar]
- 15.Winter J, Diederichs S. Argonaute-3 activates the let-7a passenger strand microRNA. RNA Biol. 2013;10(10):1631–1643. doi: 10.4161/rna.26424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okamura K, et al. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15(4):354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115(2):209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 18.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 19.Tomari Y, Zamore PD. Perspective: Machines for RNAi. Genes Dev. 2005;19(5):517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 20.Fellmann C, et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol Cell. 2011;41(6):733–746. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betancur JG, Tomari Y. Dicer is dispensable for asymmetric RISC loading in mammals. RNA. 2012;18(1):24–30. doi: 10.1261/rna.029785.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noland CL, Doudna JA. Multiple sensors ensure guide strand selection in human RNAi pathways. RNA. 2013;19(5):639–648. doi: 10.1261/rna.037424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noland CL, Ma E, Doudna JA. siRNA repositioning for guide strand selection by human Dicer complexes. Mol Cell. 2011;43(1):110–121. doi: 10.1016/j.molcel.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimm D. Asymmetry in siRNA design. Gene Ther. 2009;16(7):827–829. doi: 10.1038/gt.2009.45. [DOI] [PubMed] [Google Scholar]
- 25.Gu S, et al. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151(4):900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boudreau RL, Spengler RM, Davidson BL. Rational design of therapeutic siRNAs: Minimizing off-targeting potential to improve the safety of RNAi therapy for Huntington’s disease. Mol Ther. 2011;19(12):2169–2177. doi: 10.1038/mt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding H, Liao G, Wang H, Zhou Y. Asymmetrically designed siRNAs and shRNAs enhance the strand specificity and efficacy in RNAi. J RNAi Gene Silencing. 2007;4(1):269–280. [PMC free article] [PubMed] [Google Scholar]
- 28.McIntyre GJ, Yu YH, Lomas M, Fanning GC. The effects of stem length and core placement on shRNA activity. BMC Mol Biol. 2011;12:34. doi: 10.1186/1471-2199-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Börner K, et al. Robust RNAi enhancement via human Argonaute-2 overexpression from plasmids, viral vectors and cell lines. Nucleic Acids Res. 2013;41(21):e199. doi: 10.1093/nar/gkt836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu YP, Schopman NC, Berkhout B. Dicer-independent processing of short hairpin RNAs. Nucleic Acids Res. 2013;41(6):3723–3733. doi: 10.1093/nar/gkt036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang JS, et al. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci USA. 2010;107(34):15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang JS, Maurin T, Lai EC. Functional parameters of Dicer-independent microRNA biogenesis. RNA. 2012;18(5):945–957. doi: 10.1261/rna.032938.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cifuentes D, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328(5986):1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131(6):1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 35.Haraguchi T, et al. A potent 2′-O-methylated RNA-based microRNA inhibitor with unique secondary structures. Nucleic Acids Res. 2012;40(8):e58. doi: 10.1093/nar/gkr1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haraguchi T, Ozaki Y, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009;37(6):e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimm D, Kay MA. From virus evolution to vector revolution: Use of naturally occurring serotypes of adeno-associated virus (AAV) as novel vectors for human gene therapy. Curr Gene Ther. 2003;3(4):281–304. doi: 10.2174/1566523034578285. [DOI] [PubMed] [Google Scholar]
- 38.Krönke J, et al. Alternative approaches for efficient inhibition of hepatitis C virus RNA replication by small interfering RNAs. J Virol. 2004;78(7):3436–3446. doi: 10.1128/JVI.78.7.3436-3446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee CH, Kim JH, Lee SW. Prospects for nucleic acid-based therapeutics against hepatitis C virus. World J Gastroenterol. 2013;19(47):8949–8962. doi: 10.3748/wjg.v19.i47.8949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma H, et al. 2014. Formulated minimal-length synthetic small hairpin RNAs are potent inhibitors of hepatitis C virus in mice with humanized livers. Gastroenterology 146(1):63–66.
- 41.Pei Z, et al. Adenovirus vectors lacking virus-associated RNA expression enhance shRNA activity to suppress hepatitis C virus replication. Sci Rep. 2013;3:3575. doi: 10.1038/srep03575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yokota T, et al. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep. 2003;4(6):602–608. doi: 10.1038/sj.embor.embor840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grimm D, et al. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol. 2008;82(12):5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie J, et al. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat Methods. 2012;9(4):403–409. doi: 10.1038/nmeth.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309(5740):1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 46.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Janssen HL, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368(18):1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 48.Ottosen S, et al. In vitro antiviral activity and preclinical and clinical resistance profile of miravirsen, a novel anti-hepatitis C virus therapeutic targeting the human factor miR-122. Antimicrob Agents Chemother. 2015;59(1):599–608. doi: 10.1128/AAC.04220-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122--a key factor and therapeutic target in liver disease. J Hepatol. 2015;62(2):448–457. doi: 10.1016/j.jhep.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 50.Bühler S, Bartenschlager R. New targets for antiviral therapy of chronic hepatitis C. Liver Int. 2012;32(Suppl 1):9–16. doi: 10.1111/j.1478-3231.2011.02701.x. [DOI] [PubMed] [Google Scholar]
- 51.Welch NM, Jensen DM. Pegylated interferon based therapy with second-wave direct-acting antivirals in genotype 1 chronic hepatitis C. Liver Int. 2015;35(Suppl 1):11–17. doi: 10.1111/liv.12715. [DOI] [PubMed] [Google Scholar]
- 52.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 53.Boudreau RL, Martins I, Davidson BL. Artificial microRNAs as siRNA shuttles: Improved safety as compared to shRNAs in vitro and in vivo. Mol Ther. 2009;17(1):169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boudreau RL, Monteys AM, Davidson BL. Minimizing variables among hairpin-based RNAi vectors reveals the potency of shRNAs. RNA. 2008;14(9):1834–1844. doi: 10.1261/rna.1062908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bak RO, Hollensen AK, Primo MN, Sørensen CD, Mikkelsen JG. Potent microRNA suppression by RNA Pol II-transcribed ‘Tough Decoy’ inhibitors. RNA. 2013;19(2):280–293. doi: 10.1261/rna.034850.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kienle E, et al. Engineering and evolution of synthetic adeno-associated virus (AAV) gene therapy vectors via DNA family shuffling. J Vis Exp. 2012;Apr 2(62):3819. doi: 10.3791/3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dallas A, et al. Right- and left-loop short shRNAs have distinct and unusual mechanisms of gene silencing. Nucleic Acids Res. 2012;40(18):9255–9271. doi: 10.1093/nar/gks662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hollensen AK, Bak RO, Haslund D, Mikkelsen JG. Suppression of microRNAs by dual-targeting and clustered Tough Decoy inhibitors. RNA Biol. 2013;10(3):406–414. doi: 10.4161/rna.23543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson JA, Richardson CD. Hepatitis C virus replicons escape RNA interference induced by a short interfering RNA directed against the NS5b coding region. J Virol. 2005;79(11):7050–7058. doi: 10.1128/JVI.79.11.7050-7058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lisowski L, Elazar M, Chu K, Glenn JS, Kay MA. The anti-genomic (negative) strand of Hepatitis C Virus is not targetable by shRNA. Nucleic Acids Res. 2013;41(6):3688–3698. doi: 10.1093/nar/gkt068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith RM, Smolic R, Volarevic M, Wu GY. Positional effects and strand preference of RNA interference against hepatitis C virus target sequences. J Viral Hepat. 2007;14(3):194–212. doi: 10.1111/j.1365-2893.2006.00794.x. [DOI] [PubMed] [Google Scholar]
- 62.Bassik MC, et al. Rapid creation and quantitative monitoring of high coverage shRNA libraries. Nat Methods. 2009;6(6):443–445. doi: 10.1038/nmeth.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bassik MC, et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152(4):909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kampmann M, Bassik MC, Weissman JS. Integrated platform for genome-wide screening and construction of high-density genetic interaction maps in mammalian cells. Proc Natl Acad Sci USA. 2013;110(25):E2317–E2326. doi: 10.1073/pnas.1307002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grimm D, et al. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J Clin Invest. 2010;120(9):3106–3119. doi: 10.1172/JCI43565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beer S, et al. Low-level shRNA cytotoxicity can contribute to MYC-induced hepatocellular carcinoma in adult mice. Mol Ther. 2010;18(1):161–170. doi: 10.1038/mt.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen CC, et al. Long-term inhibition of hepatitis B virus in transgenic mice by double-stranded adeno-associated virus 8-delivered short hairpin RNA. Gene Ther. 2007;14(1):11–19. doi: 10.1038/sj.gt.3302846. [DOI] [PubMed] [Google Scholar]
- 68.Matsushita T, et al. Adeno-associated virus vectors can be efficiently produced without helper virus. Gene Ther. 1998;5(7):938–945. doi: 10.1038/sj.gt.3300680. [DOI] [PubMed] [Google Scholar]
- 69.Gao GP, et al. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci USA. 2002;99(18):11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grimm D. Production methods for gene transfer vectors based on adeno-associated virus serotypes. Methods. 2002;28(2):146–157. doi: 10.1016/s1046-2023(02)00219-0. [DOI] [PubMed] [Google Scholar]
- 71.Lohmann V. HCV replicons: Overview and basic protocols. Methods Mol Biol. 2009;510:145–163. doi: 10.1007/978-1-59745-394-3_11. [DOI] [PubMed] [Google Scholar]
- 72.Meister G, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15(2):185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]