Significance
Cellular energy production is a function of the abundance of the small circular DNA molecules in mitochondria. Mitochondrial DNA is replicated in both dividing and nondividing cells, and encoding ribonuclease H1 (RNase H1) is essential to this process. Here, we define its mechanistic role: the removal of the RNA primers used for mitochondrial DNA replication. In the absence of RNase H1, primers are fixed in both template strands of mitochondrial DNA. The retained primers are a major impediment to mitochondrial DNA polymerase γ, leading to the formation of persistent DNA gaps that are catastrophic for subsequent rounds of replication. Moreover, primer retention provides unambiguous identification of RNA-DNA transition sites in the control region of mitochondrial DNA, thereby defining two major origins of replication.
Keywords: DNA replication, RNase H, mitochondrial DNA, replication priming, origin of replication
Abstract
Encoding ribonuclease H1 (RNase H1) degrades RNA hybridized to DNA, and its function is essential for mitochondrial DNA maintenance in the developing mouse. Here we define the role of RNase H1 in mitochondrial DNA replication. Analysis of replicating mitochondrial DNA in embryonic fibroblasts lacking RNase H1 reveals retention of three primers in the major noncoding region (NCR) and one at the prominent lagging-strand initiation site termed Ori-L. Primer retention does not lead immediately to depletion, as the persistent RNA is fully incorporated in mitochondrial DNA. However, the retained primers present an obstacle to the mitochondrial DNA polymerase γ in subsequent rounds of replication and lead to the catastrophic generation of a double-strand break at the origin when the resulting gapped molecules are copied. Hence, the essential role of RNase H1 in mitochondrial DNA replication is the removal of primers at the origin of replication.
Human mitochondrial DNA (mtDNA) is most frequently organized as 16.5-kb circles of duplex DNA, whose two strands are called heavy (H) and light (L), based on their nucleotide composition. The human mitochondrial genome is gene dense, with only one substantial noncoding region (NCR) of approximately 1 kb (1). Based on electron microscopy (2), 5′ end mapping of DNA ends (3–5) and later 2D agarose gel electrophoresis (2D-AGE) studies (6, 7) it was inferred that replication of mammalian mtDNA is frequently unidirectional, commencing within the NCR.
In mammalian mitochondria, initiation of DNA synthesis occurs preferentially at or near two specific sites. On the leading strand, initiation has been assigned to a prominent free 5′ end of DNA, designated as Ori-H, located at nucleotide position 16,034 on the map of the mouse mitochondrial genome. Ori-H lies downstream of the light-strand promoter (LSP), and RNA species spanning from LSP to approximately Ori-H have been detected in mammalian mitochondria, albeit not covalently linked with DNA (8). LSP has been robustly mapped both in vivo (9) and in vitro (10) and is assumed to be used by mitochondrial RNA polymerase (POLRMT) to synthesize polycistronic transcripts of the light strand and to generate much shorter products terminating in the NCR, which serve as primers for leading (H-)strand DNA synthesis. POLRMT is also implicated in the synthesis of a primer at a short spacer DNA amid a cluster of five tRNA genes (termed Ori-L) that is a major site of initiation of lagging-strand DNA synthesis (11, 12). A role for encoding ribonuclease H1 (RNase H1) in generating, processing, or removing primers at Ori-H and Ori-L has been inferred but has not been experimentally tested.
Other data indicate that the initiation of mtDNA replication is more complex, because the NCR contains a second putative origin of replication, termed cluster II, or Ori-b (7, 13). Moreover, other sites of second-strand DNA synthesis than Ori-L have been inferred both by atomic force microscopy (14) and neutral 2D agarose gel electrophoresis (7, 13). Mitochondrial DNA replication can also result in the formation of fully duplex DNA intermediates that appear to be products of conventional coupled leading- and lagging-strand DNA synthesis (6), and replication of this type has been inferred to originate across a broad initiation zone extending well beyond the NCR (15, 16).
RNA has another role in mtDNA replication. Processed transcripts are incorporated into replication intermediates throughout the lagging strand, and these RNA-containing species are in a precursor-product relationship with mature, fully replicated mtDNA (7, 17). The mechanisms by which RNA/DNA hybrid-containing replication intermediates are generated and processed remain unknown, but a role for ribonucleases, in particular RNase H1, is again plausible (18, 19).
Enzymes with RNase H activity degrade RNA only when it is hybridized to DNA. One such enzyme, RNase H1, is essential for mtDNA maintenance in mice, as its ablation leads to embryonic lethality, owing to mtDNA depletion (18). In many organisms and plasmids, RNase H processes RNA of RNA/DNA to create the preferred substrates of DNA polymerases, subsequently removes these RNA primers after initiation of DNA synthesis (20), and degrades others to prevent DNA initiation (21).
Here, analysis of mtDNA molecules from mouse embryonic fibroblasts (MEFs) lacking the exons V–VII of the Rnaseh1 gene revealed prominent DNA ends at four discrete sites in the mouse mitochondrial genome, located in the vicinity of the major replication initiation sites. These findings indicate an essential role for RNase H1 in the removal of RNA primers. In the absence of RNase H1, persistent H-strand RNA primers in the NCR are not efficiently sealed by ligation. Initiation of a second round of replication on such nicked or gapped molecules creates intermediates with a double-strand break at the origin, whose replication cannot be completed, instead generating dead-end products.
Results and Discussion
Rnaseh1 Ablation in Mouse Cells Creates Prominent Linear Ends of mtDNA Mapping to the NCR.
The loss of Rnaseh1 was previously shown to induce rapid mtDNA depletion (18). Because its substrate is RNA/DNA hybrid, this finding implies that processing or removal of one or more segments of RNA from replicating mtDNA molecules is essential for the replication process. To investigate this phenomenon at the molecular level and identify the precise steps at which RNase H1 activity is essential, we generated tamoxifen-inducible conditional knockout MEFs (Materials and Methods). After confirming that the gene was excised on addition of the drug, with loss of RNase H1 activity and mtDNA depletion (SI Appendix, Fig. S1), we analyzed the remaining mtDNA for structural abnormalities.
We initially focused on fragments of mouse mtDNA that included the NCR [nucleotides (nt) 15,450--16,300], as it contains origins of replication, as well as the replication terminus (2, 22). In MEFs retaining the Rnaseh1 gene, PstI/BamHI digestion yielded the expected fragment of 7.3 kb spanning nt 12,243–3,222 and a fainter shorter species of ∼4 kb whose 5′ end maps in the vicinity of Ori-H, based on its length and the probe applied (Fig. 1A, lanes 1 and 3). In Rnaseh1-ablated cells, a much more prominent band of ∼4 kb (with an end near Ori-H, or LSP) was evident (Fig. 1A, lane 4), and after treatment with RNase HI derived from Escherichia coli (Eco-RNase HI), two smaller fragments (Fig. 1A, lane 6) with 5′ ends at or close to Ori-H and Ori-b were seen (13). The straightforward explanation for the marked increase in abundance of fragments in the mtDNA of cells lacking Rnaseh1 with ends in the vicinity of two previously identified origins is retention of 5′ primer RNA at these locations. Furthermore, the decrease in signal of the full-length (7.3 kb) fragment suggests a substantial number of H-strands of mtDNA contain an RNA patch in the NCR contiguous with DNA at both ends, implying ligation of the 5′ end of the RNA to DNA.
Fig. 1.
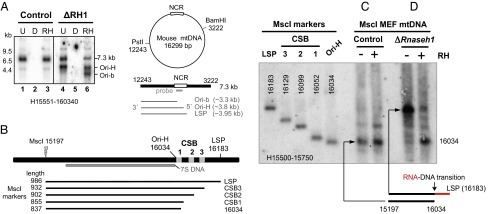
Loss of Rnaseh1 reveals two prominent origins of replication in the NCR and a primer starting at the light strand promoter. (A) MEF mtDNA, from cells treated for 9 d without (control) or with 4HT to excise the Rnaseh1 gene (ΔRH1), was digested with BamHI and PstI, treated with DNase (D) or Eco-RNase HI (RH) where indicated, denatured in 80% (vol/vol) formamide, 15 min at 85 °C, and separated by 1D-AGE (1% agarose Tris-acetate). λ-HindIII dsDNA ladder was run in parallel to provide size markers. Southern hybridization with a riboprobe to the 3′ end of the H-strand (H15551-160340) detected the full-length H-strand of 7.3 kb and one or two truncated species without or with Eco-RNase HI treatment (RH), respectively, whose lengths indicate their 5′ ends map to sites within the major NCR of mouse mtDNA (denoted LSP, Ori-H and Ori-b, see main text and B). (B) Map of the major NCR of murine mtDNA and the MscI digested markers and probe used for fine mapping of 5′ ends of DNA in the vicinity of Ori-H. MscI digested and denatured mtDNAs were fractionated by 1D-AGE and hybridized to riboprobe H15500-15750. Where indicated, samples were treated with (+) or without (−) Eco-RNase HI, before denaturation. DNA from mitochondria of MEFs treated without (C) or with 100 nM 4HT for 8 d (D). Interpretations of the RNase H sensitive species appear below C and D; red lines, RNA; black lines, DNA.
Loss of Rnaseh1 Results in Retention of an RNA Primer from LSP to OH.
To map the putative nascent strands of DNA more precisely and establish whether they retained an RNA primer, we applied high-resolution agarose gel fractionation to denatured DNA fragments. The approach was also designed to distinguish nascent strands of DNA mapping in the vicinity of Ori-H from the 7S DNAs of the mitochondrial D-loops (Materials and Methods and SI Appendix, Fig. S1 E and F). The 5′ end of the majority of nascent strands of mouse mtDNA longer than 7S DNA mapped to nucleotide position (np) 16,034 ± 20 (i.e., Ori-H). This map position was judged by migration of markers (Fig. 1B) with ends at LSP, the three conserved sequence blocks (CSBs), and np 16,034 (Fig. 1C and SI Appendix, Fig. S2 A and B). The major 5′ end of murine mtDNA detected by other methods also maps to np 16,034 (7, 8). A minor band, whose 5′ end mapped to CSB2, was also detectable in murine mtDNA samples, and it was slightly more distinct after Eco-RNase H treatment (Fig. 1C and SI Appendix, Fig. S2C, lanes 1 and 2).
Analysis of mtDNA from MEFs lacking Rnaseh1 revealed a further fragment, barely detectable in control mtDNAs, whose 5′ end mapped at or close to LSP (Fig. 1D and SI Appendix, Fig. S2C). In vitro treatment with RNase H before denaturation greatly decreased the signal from this fragment while markedly increasing that of the fragment ending close to np 16,034. These findings are consistent with primer synthesis commencing at LSP and a major transition point to DNA synthesis at np ∼16,034 (Fig. 1 C and D and SI Appendix, Fig. S2C), with efficient primer removal in WT cells and tissue. Previous studies detected a prominent 5′ end of mouse mtDNA at np 16,034 (7), which the present data indicate to be a major RNA-DNA transition site for the initiation of mtDNA replication, Ori-H. Furthermore, this site coincides with the predominant 5′ end of mouse 7S DNA based on other analytical methods (8, 23). The persistence, in Rnaseh1-deficient cells, of a segment of RNA/DNA hybrid extending ∼150 nt upstream from Ori-H to LSP strengthens the view that mitochondrial RNA polymerase POLRMT is responsible for primer synthesis, as it is known to bind to LSP and initiate transcription from this site (10). The data are thus consistent with the proposition that both productive replication and D-loop formation involve primer initiation at LSP and transition to DNA synthesis at Ori-H in the mouse and that RNase H1 is required for removal of this primer.
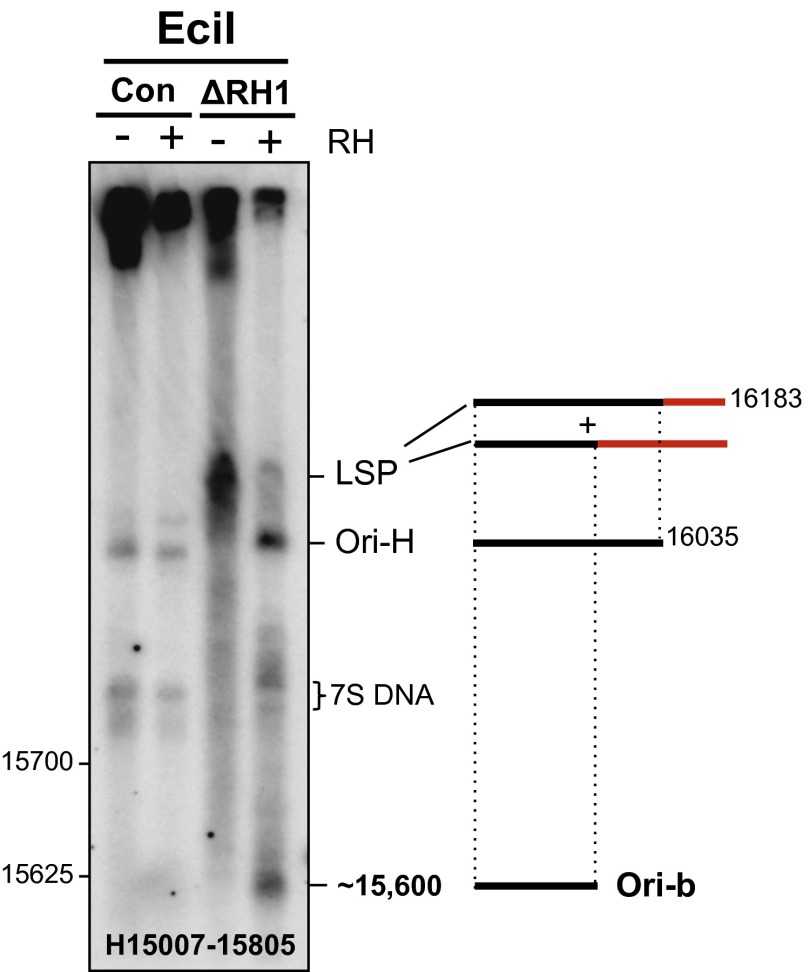
Ori-b Intermediates Accumulate in Rnaseh1−/− MEFs.
The 5′ end of the other prominent H-strand fragment found at elevated levels in Rnaseh1−/− cells (Fig. 1A) was located in the vicinity of the second proposed origin of replication inside the NCR, Ori-b (7, 13). Ori-b of mouse was previously mapped approximately by 2D-AGE, and the region was found to be associated with a prominent free 5′ end of DNA at nt 15,625 on the H-strand, based on ligation mediated (LM)-PCR analysis (7). To map accurately the fragment ends in the vicinity of Ori-b of Rnaseh1−/− cells high-resolution AGE was again used. In vitro Eco-RNase HI treatment of mtDNA of Rnaseh1−/− cells produced a fragment slightly shorter than a marker fragment terminating at np 15,625, indicating a retained RNA-DNA transition site at approximately np 15,600 (Fig. 2 and SI Appendix, Fig. S3 A and B), in addition to that at Ori-H. LM-PCR analysis of gel-extracted DNA fragments from Rnaseh1-ablated MEFs identified np 15,605 as the longest prominent free 5′ end of DNA, and thus we infer the major Ori-b RNA-DNA transition site to be located at this site (SI Appendix, Fig. S3C). The absence of any detectable H-strand DNA ends between Ori-H and Ori-b (SI Appendix, Fig. S4) suggests that RNA primers extend from LSP to either Ori-H or Ori-b (as illustrated to the right of the gel image in Fig. 2) and that these represent sites of transition to DNA synthesis.
Fig. 2.
Rnaseh1 ablation leads to the accumulation of free 5′ ends of DNA mapping to nucleotide position ∼15,600 of the heavy strand of mouse mtDNA. EciI digested and denatured [80% (vol/vol) formamide, 15 min at 85 °C] MEF mtDNAs were fractionated by 1D-AGE and blot hybridized to riboprobe H15007-15805. Where indicated, samples were treated with (+) or without (−) Eco-RNase HI before denaturation. ΔRH1, mtDNA from MEFs treated with 100 nM 4HT for 8 d; Con, control MEFs. Interpretations of the single-stranded products are illustrated beside the gel images; red lines, RNA; black lines, DNA. Markers are shown in SI Appendix, Fig. S3.
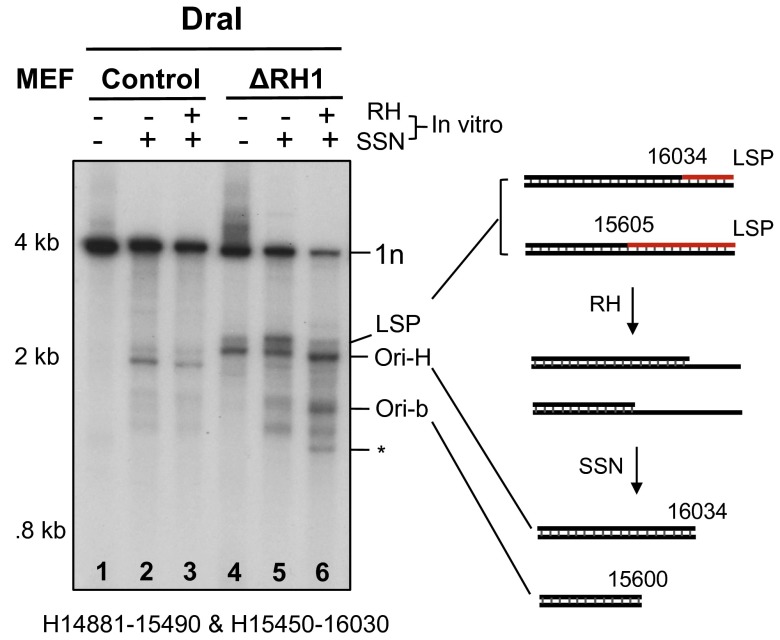
Increases in the levels of DNA fragments with ends at Ori-H and Ori-b were also evident when digestions with RNase H1 and single-strand specific nuclease (SSN) were successively applied to restriction fragments of mtDNA from Rnaseh1-ablated MEFs (Fig. 3, lane 6). Species detected in controls (Fig. 3, lanes 1–3) were considered unrelated to cellular RNase H1 activity where the loss of Rnaseh1 did not increase their abundance. SSN alone enhanced the double-stranded DNA fragments with an end near LSP (Fig. 3, lane 5 compared with lane 4). These results indicate not only the retention of primer RNA but also a persistent nick or gap opposite LSP in some molecules.
Fig. 3.
Persistent RNA patches after Rnaseh1 ablation revealed by a combined RNase H and single-stranded nuclease treatments. (A) MEF mtDNA from cells culture for 7 d without (control) or with (ΔRH1) 4HT was digested with DraI, and where indicated additionally with single stranded nuclease (SSN) and Eco-RNase HI (RH), before native 1D-AGE, 1% agarose TAE. Fragments were blot hybridized to riboprobes H14881-15490 and H15450-16030. Interpretations of the products are illustrated beside the gel images; red lines, RNA; black lines, DNA. A truncated fragment mapping to nt ∼15,200 (*) was not reproducible (SI Appendix, Fig. S5I).
RNA at Ori-L Is Fully Incorporated into Daughter Molecules.
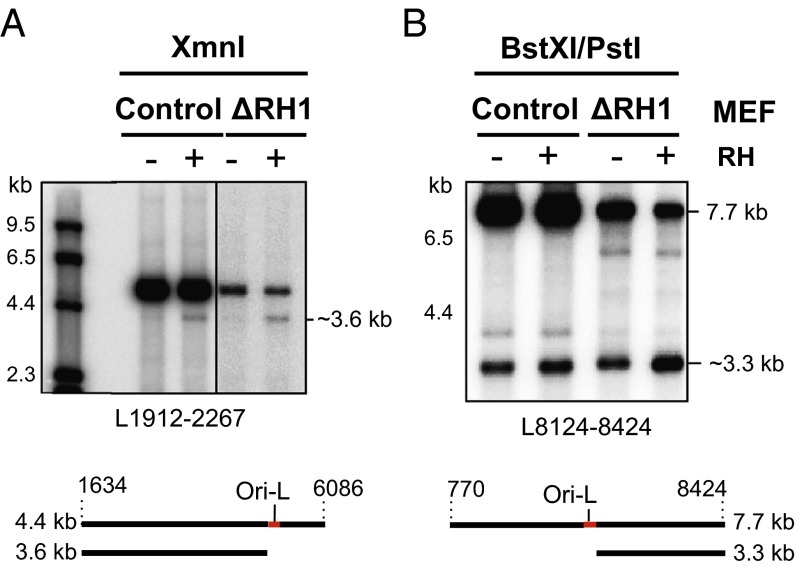
To determine whether RNase H1 also processed the primer at Ori-L, we carried out a similar analysis of the Ori-L region of mtDNA of Rnaseh1-ablated and control MEFs. In vitro Eco-RNase HI treatment truncated a small fraction of the XmnI fragments of MEF mtDNA spanning np 1,634–6,036, and this population was a greater proportion of the total after ablation of the Rnaseh1 gene (Fig. 4A), which is commensurate with increased retention of a primer in this region of murine mtDNA as a consequence of loss of Rnaseh1. The length of the product, detected by a probe specific for the L-strand (L1912-2267), was ∼3.6 kb, consistent with a primer at Ori-L (np 5,160–5,191). Likewise, digestion of mtDNA with BstXI and PstI and application of probe L8124-8424 revealed a 3.3-kb fragment spanning approximately Ori-L to np 8,424 (Fig. 4B). The increase in products flanking Ori-L after Eco-RNase HI treatment suggests the Ori-L primer was in many molecules covalently linked to the DNA, by ligation at its 5′ end, as well as contiguous with the DNA at its 3′ end.
Fig. 4.
Rnaseh1 ablation leads to the accumulation of fully incorporated primers at Ori-L. MEF mtDNA from cells culture for 10 (A) or 9 (B) d without (control) or with (ΔRH1) 4HT was restriction digested with XmnI (A) or BstXI and PstI (B), denatured, and fractionated by 1D-AGE (1% agarose Tris-acetate). After transfer the fragments were hybridized with riboprobes (A) L1912-2267 or (B) L8124-8424. RH, samples were treated with (+) or without (−) Eco-RNase HI. (A) All samples were fractionated on the same gel, a longer exposure of the ΔRH1 samples is shown to compensate for unequal loading. (A and B) Below the gel images are interpretations of the RNase H sensitive species (a black line of DNA with a red RNA patch).
Accumulation of Incorporated RNA in the Vicinity of LSP on the L-Strand of Rnaseh1−/− MEF mtDNA.
Next the entire mitochondrial genome was screened for RNA patches by analyzing both strands of three large overlapping fragments of mtDNA. Fragment ends consistent with primers ending at Ori-H, Ori-b, and Ori-L were present at high abundance in mtDNA samples from Rnaseh1-ablated MEFs (SI Appendix, Fig. S5, I and II). The only other prominent species associated with loss of RNase H1 was a truncated L-strand fragment with a 5′ end of DNA located in the NCR (Fig. 5A and SI Appendix, Fig. S5I). Fine mapping placed it at nucleotide position 16,220 ± 20 nucleotides, close to LSP (np 16,183; Fig. 5B and SI Appendix, Fig. S6). Eco-RNase HI treatment increased the signal from this species but did not appreciably alter its mobility, suggesting that the fragment was released from much longer molecules; i.e., the full-length fragment.
Fig. 5.
Loss of Rnaseh1 leads to retention of a RNA patch on the L-strand close to LSP. MEF mtDNA from cells cultured for 10 (A) or 8 (B) d with 4HT (ΔRH1) or from MEFs cultured without drug was digested with BamHI and PstI (A) or HincII (B), denatured for 15 min at 85 °C, in 80% (vol/vol) formamide, fractionated by 1D-AGE for 6 h at 11 V/cm, and blot hybridized to the indicated L-strand probe. RH, samples were treated with (+) or without (−) Eco-RNase HI. (A) 1% agarose Tris-acetate gel. (B) 2.3% (wt/vol) agarose sodium borate gel. Markers are based on PCR products of defined lengths (SI Appendix, Fig. S6).
Our analysis indicates persistent RNA patches on both strands in the vicinity of LSP in the absence of RNase H1 (Figs. 1D, 2, and 5). To probe the structure of this region in more detail, we digested mtDNA from Rnaseh1-ablated MEFs with BclI, which cuts at nt 16,179, very close to LSP (nt 16,183). In addition to the usual limit-digest products of 4.15 and 2.45 kb, there was a fragment of 6.6 kb, indicating nondigestion at nt 16,179 (Fig. 6A). Gel extraction, Eco-RNase HI digestion, and denaturing gel electrophoresis of the 6.6-kb fragment revealed two L-strand species of ∼2.5 and ∼4.1 kb (Fig. 6 B and C), consistent with the presence of a fully incorporated RNA patch close to LSP, in approximately half of the molecules.
Fig. 6.
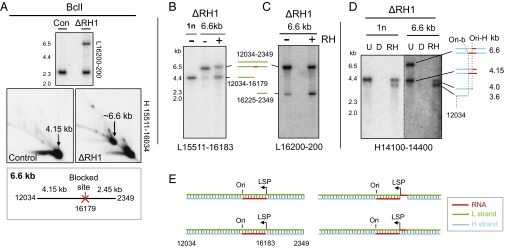
Loss of Rnaseh1 leads to the retention of RNA patches on the H- and L-strands close to LSP, some of which are contiguous with DNA at both ends. BclI-digested mtDNA from MEFs cultured for 9 d without (control) or with (ΔRH1) 4HT was fractionated by (A) 2D-AGE or (B–D) 1D-AGE and blot hybridized to the indicated probes. For 1D-AGE, samples were denatured before loading. (Inset below A) Blocked BclI site at np 16,179 predicts a fragment of 6.6 kb. The BclI product of ∼6.6. kb was gel extracted, denatured, and refractionated after treatment without or with Eco-RNase HI (RH) or (D)Nase (B–D); next to the gel images are interpretations of the single-stranded products and (E) the possible double-stranded combinations, with Ori-H and Ori-b conflated to Ori. (B) Gel-extracted 4.15-kb fragment (1n) provided an additional marker, and in D, the same 1n material enabled comparison of the relative amounts of Ori-H and Ori-b ends of the 1n and 6.6-kb fragments.
H-strand species of 4.1 kb were also released by denaturation of the 6.6-kb fragment (Fig. 6D), corresponding with those identified earlier as retaining primer RNA from LSP to Ori-H or Ori-b (Fig. 1D, 2, and 3). Eco-RNase HI modified the remainder of the H-strand material, enhancing the 4.1-kb products seen on denaturing gels (Fig. 6D). Thus, the 6.6-kb fragment systematically contains incorporated H-strand RNA across the restriction site, approximately half of which is covalently joined to DNA at both ends. The presence or absence of an RNA patch on the L-strand and the presence or absence of a gap associated with the H-strand RNA patch predicts four possible double-stranded species, according to whether these structures are found on the same or different molecules (Fig. 6E).
Inferences Regarding the Role of RNase H1 in mtDNA Replication.
The detrimental effects of Rnaseh1 ablation enable us to reconstruct a number of key events in mtDNA replication. Primer RNA is synthesized, presumably by POLRMT, from LSP to Ori-H or Ori-b, where the transition to DNA synthesis occurs. Ordinarily RNase H1 rapidly removes these primers. The RNA primer synthesized at the major lagging-strand DNA initiation site Ori-L is also processed by RNase H1, as is a previously unreported patch of primer RNA on the L-strand mapping in the vicinity of LSP. Synthesis of both of these primers is also most likely attributable to POLRMT as the enzyme is capable of priming at Ori-L in vitro (11), and LSP has been previously suggested to act bidirectionally (8). Although initiation of second-strand DNA synthesis has been proposed previously to be the only such event required for replicating the L-strand of mammalian mtDNA (24), the discovery of a RNA patch on the L-strand at LSP suggests the final third is the product of a distinct priming event.
In the absence of RNase H1, no other enzyme is able to process or remove the primers in the NCR efficiently. However, RNase H1 cannot remove every last ribonucleotide of an RNA/DNA hybrid, and therefore other factors should also be required, such as those implicated in Okazaki fragment processing in the nucleus, that have also been found in mitochondria (25–27), and, possibly, MGME1 (28).
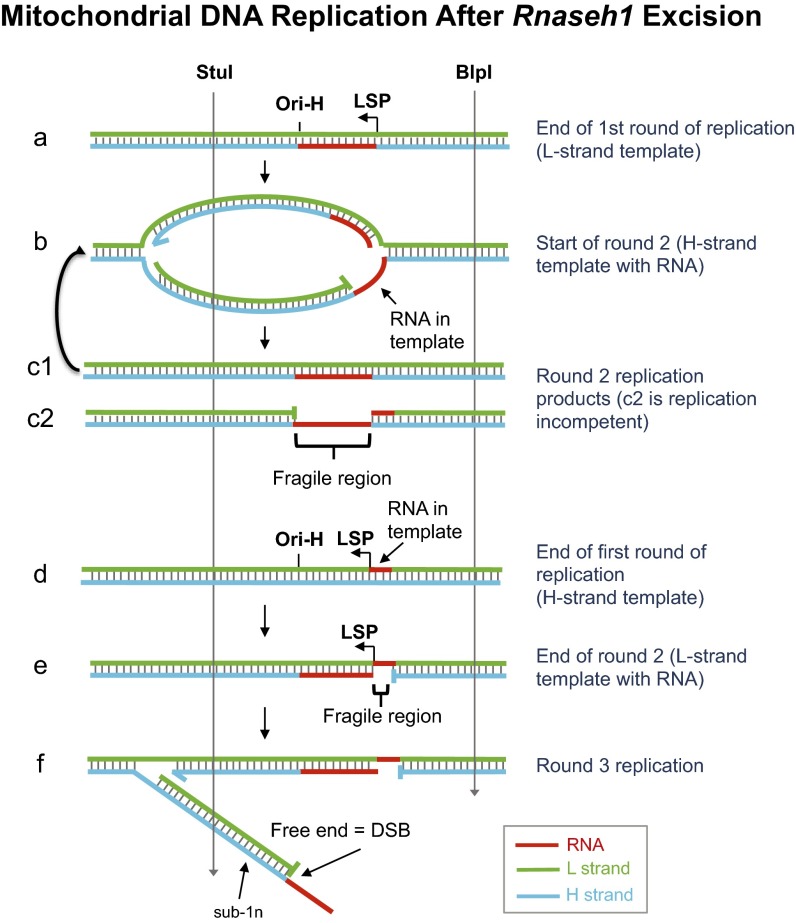
The various types of gapped and closed RNA-containing mtDNA molecules that accumulate in the absence of RNase H1 (Fig. 6E) are consistent with a relatively straightforward model, under which primer retention is not an insurmountable obstacle to the completion of mtDNA replication. The persistent RNAs are capable of being fully incorporated into mtDNA. However, although mitochondrial DNA polymerase [DNA polymerase γ (POLG)] copes well with a single ribonucleotide in a DNA template, more extensive RNA patches are a major impediment to its ability to synthesize DNA (29). Thus, retained RNA primers in mtDNA are expected to cause replication stalling and arrest and prevent the completion, and ultimately the initiation, of subsequent rounds of replication. In the case of the incorporated H-strand RNA primers in the NCR, POLG will be faced with a stretch of ∼150 or 550 ribonucleotides (Fig. 7, species b and c). The retained primer at Ori-L (Fig. 4), although shorter (11), is expected to cause similar problems. Failure to synthesize across these stretches of RNA will result in fragile molecules highly susceptible to breakage, especially in the case of initiation events at Ori-b. It is almost inconceivable that a daughter molecule with single-stranded RNA of this length in the NCR will be fit for further rounds of replication, and as such, they are likely to be rapidly degraded. The situation is no better in the case of molecules retaining an L-strand primer at LSP (Fig. 7, species d). Although the retained primer is much shorter than those on the H-strand, and hence the fragile region formed in the second round of replication much narrower (Fig. 7, species e), it is located close to the origin, and in the third round of replication, in the absence of RNase H1, the displaced H-strand will be free and will generate a double-strand break at the origin of replication (Fig. 7, species f). Such breaks would result in intermediates that are shorter than the unit length restriction fragment (sub-1n) after digestion with an enzyme(s) whose restriction sites flank the origin. Breakage in the single-stranded RNA region of species c2 or e would yield essentially the same product. Fragments truncated to approximately Ori-H and Ori-b, commensurate with species f, and broken forms of species c2 or e were detected when 1D- (Fig. 3, lane 4) or 2D-AGE (SI Appendix, Fig. S7) was applied to mtDNA samples from Rnaseh1-ablated MEFs digested with DraI or BlpI/StuI.
Fig. 7.
Model of aberrant mitochondrial DNA replication in the absence of RNase H1. Primers on the H-strand from the LSP to the origin of replication Ori-H (or Ori-b, not illustrated) persist when RNase H1 is absent, and these are fully incorporated into mtDNA at the end of the replication cycle. In the next round of replication, the mitochondrial DNA polymerase γ (POLG) encounters a stretch of 150 (or 550) ribonucleotides of which it is expected to be able to synthesize only one or a few ribonucleotides, as it is a poor RNA-dependent DNA polymerase (29). RNase H1 ablation revealed a RNA patch on the L-strand near LSP that will prevent completion of H-strand synthesis once it is incorporated in the template strand. In the subsequent round of replication, the gap in the template H-strand effectively creates a double-strand break (DSB) at the origin (irrespective of RNA in the H-strand template).
Loss of Rnaseh1 Perturbs the Bootlace Mechanism of mtDNA Replication.
The processing of long RNAs (transcripts) hybridized to the lagging strand template during the protracted delay between first- and second-strand synthesis of mtDNA (7, 17) could potentially involve a role for RNase H1 (19). However, we found no evidence of an accumulation of RNA/DNA hybrid-containing replication intermediates in the absence of RNase H1. Nevertheless, the loss of RNase H1 did perturb the bootlace mechanism of replication, as incorporated transcripts were no longer evident in mtDNA samples from Rnaseh1−/− cells; instead, the mitochondrial replication intermediates were fully resistant to Eco-RNase HI, which contrasted markedly with mtDNAs from MEFs retaining the gene (SI Appendix, Fig. S8). Exactly how this perturbation arises remains to be established, but it likely contributes to the mtDNA depletion caused by the loss of RNase H1 (SI Appendix, Fig. S1D).
Ori-b Is Potentially the Primary Origin of Replication.
We previously mapped ends of LM-PCR products to Ori-b and detected an origin in the NCR distinct from Ori-H based on 2D-AGE (7, 13); more recently, massive parallel sequencing has lent support to Ori-b being an important cis-element for mammalian mtDNA (30). The marked increase in nascent strands of DNA mapping to Ori-b in Rnaseh1-ablated samples, evident after in vitro RNase HI treatments, might indicate that failure to process rapidly the H-strand RNA primers from LSP increases the probability of POLRMT continuing beyond Ori-H to Ori-b. This interpretation envisages Ori-b as a feature of some sort of failsafe mechanism to ensure (bootlace) replication initiates in the NCR. Alternatively, Ori-b could be the major origin of replication in normal circumstances: once RNase H1 has removed the primer to Ori-b, this would permit DNA synthesis across the gap from Ori-H to Ori-b. In this scheme, although Ori-b is the origin, it is highly transient and therefore is not nearly as evident as Ori-H (the replication terminus) at steady state in control cells and tissues.
Materials and Methods
Full methods are described in SI Appendix. MEFs were prepared as described previously (31). Embryo genotypes were Rnaseh1fl/fl Gt(ROSA)26Sortm1(Cre/Esr1)Nat homozygous (32). Conditional gene knockout in murine cells was via the action of cre-recombinase on LoxP sites flanking exons V–VII of the Rnaseh1 gene and confirmed using an in-gel activity assay for RNase H (33). Cell culture, mitochondrial DNA isolation, nucleic acid digestion, modification, and analysis were described previously (7, 13, 34, 35). Additionally, single-stranded mtDNAs were fractionated and analyzed after denaturation, in some cases using sodium borate agarose gels (36).
Supplementary Material
Acknowledgments
J.B.H. was an National Institutes of Health (NIH) CamGrad Scholar (2006–2010). This study was funded by the Medical Research Council, NIH Grant GM31819, the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, Academy of Finland, Juselius Foundation, and Tampere University Hospital Medical Research Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1503653112/-/DCSupplemental.
References
- 1.Bayona-Bafaluy MP, et al. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003;31(18):5349–5355. doi: 10.1093/nar/gkg739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kasamatsu H, Vinograd J. Unidirectionality of replication in mouse mitochondrial DNA. Nat New Biol. 1973;241(108):103–105. doi: 10.1038/newbio241103a0. [DOI] [PubMed] [Google Scholar]
- 3.Chang DD, Hauswirth WW, Clayton DA. Replication priming and transcription initiate from precisely the same site in mouse mitochondrial DNA. EMBO J. 1985;4(6):1559–1567. doi: 10.1002/j.1460-2075.1985.tb03817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crews S, Ojala D, Posakony J, Nishiguchi J, Attardi G. Nucleotide sequence of a region of human mitochondrial DNA containing the precisely identified origin of replication. Nature. 1979;277(5693):192–198. doi: 10.1038/277192a0. [DOI] [PubMed] [Google Scholar]
- 5.Tapper DP, Clayton DA. Mechanism of replication of human mitochondrial DNA. Localization of the 5′ ends of nascent daughter strands. J Biol Chem. 1981;256(10):5109–5115. [PubMed] [Google Scholar]
- 6.Holt IJ, Lorimer HE, Jacobs HT. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100(5):515–524. doi: 10.1016/s0092-8674(00)80688-1. [DOI] [PubMed] [Google Scholar]
- 7.Yasukawa T, et al. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25(22):5358–5371. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang DD, Clayton DA. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci USA. 1985;82(2):351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ojala D, Crews S, Montoya J, Gelfand R, Attardi G. A small polyadenylated RNA (7 S RNA), containing a putative ribosome attachment site, maps near the origin of human mitochondrial DNA replication. J Mol Biol. 1981;150(2):303–314. doi: 10.1016/0022-2836(81)90454-x. [DOI] [PubMed] [Google Scholar]
- 10.Chang DD, Clayton DA. Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell. 1984;36(3):635–643. doi: 10.1016/0092-8674(84)90343-x. [DOI] [PubMed] [Google Scholar]
- 11.Fusté JM, et al. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol Cell. 2010;37(1):67–78. doi: 10.1016/j.molcel.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 12.Wong TW, Clayton DA. In vitro replication of human mitochondrial DNA: Accurate initiation at the origin of light-strand synthesis. Cell. 1985;42(3):951–958. doi: 10.1016/0092-8674(85)90291-0. [DOI] [PubMed] [Google Scholar]
- 13.Yasukawa T, Yang MY, Jacobs HT, Holt IJ. A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol Cell. 2005;18(6):651–662. doi: 10.1016/j.molcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Brown TA, Cecconi C, Tkachuk AN, Bustamante C, Clayton DA. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev. 2005;19(20):2466–2476. doi: 10.1101/gad.1352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowmaker M, et al. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J Biol Chem. 2003;278(51):50961–50969. doi: 10.1074/jbc.M308028200. [DOI] [PubMed] [Google Scholar]
- 16.Reyes A, Yang MY, Bowmaker M, Holt IJ. Bidirectional replication initiates at sites throughout the mitochondrial genome of birds. J Biol Chem. 2005;280(5):3242–3250. doi: 10.1074/jbc.M411916200. [DOI] [PubMed] [Google Scholar]
- 17.Reyes A, et al. Mitochondrial DNA replication proceeds via a ‘bootlace’ mechanism involving the incorporation of processed transcripts. Nucleic Acids Res. 2013;41(11):5837–5850. doi: 10.1093/nar/gkt196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerritelli SM, et al. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol Cell. 2003;11(3):807–815. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 19.Holt IJ. Mitochondrial DNA replication and repair: All a flap. Trends Biochem Sci. 2009;34(7):358–365. doi: 10.1016/j.tibs.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Kornberg A, Baker TA. DNA Replication. 2nd Ed W. H. Freeman & Co.; New York: 1992. [Google Scholar]
- 21.Kogoma T, Subia NL, von Meyenburg K. Function of ribonuclease H in initiation of DNA replication in Escherichia coli K-12. Mol Gen Genet. 1985;200(1):103–109. doi: 10.1007/BF00383320. [DOI] [PubMed] [Google Scholar]
- 22.Holt IJ, Reyes A. Human mitochondrial DNA replication. Cold Spring Harb Perspect Biol. 2012;4(12):a012971. doi: 10.1101/cshperspect.a012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walberg MW, Clayton DA. Sequence and properties of the human KB cell and mouse L cell D-loop regions of mitochondrial DNA. Nucleic Acids Res. 1981;9(20):5411–5421. doi: 10.1093/nar/9.20.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28(4):693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 25.Kalifa L, Beutner G, Phadnis N, Sheu SS, Sia EA. Evidence for a role of FEN1 in maintaining mitochondrial DNA integrity. DNA Repair (Amst) 2009;8(10):1242–1249. doi: 10.1016/j.dnarep.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duxin JP, et al. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol Cell Biol. 2009;29(15):4274–4282. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kazak L, et al. Alternative translation initiation augments the human mitochondrial proteome. Nucleic Acids Res. 2013;41(4):2354–2369. doi: 10.1093/nar/gks1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornblum C, et al. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat Genet. 2013;45(2):214–219. doi: 10.1038/ng.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasiviswanathan R, Copeland WC. Ribonucleotide discrimination and reverse transcription by the human mitochondrial DNA polymerase. J Biol Chem. 2011;286(36):31490–31500. doi: 10.1074/jbc.M111.252460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ju YS, et al. ICGC Breast Cancer Group ICGC Chronic Myeloid Disorders Group ICGC Prostate Cancer Group Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. eLife. 2014;3 doi: 10.7554/eLife.02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. In: Ausubel FM, editor. Current Protocols in Molecular Biology. 2005. , et al. (Wiley, New York), Unit 28.1. [DOI] [PubMed] [Google Scholar]
- 32.Badea TC, Wang Y, Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci. 2003;23(6):2314–2322. doi: 10.1523/JNEUROSCI.23-06-02314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carl PL, Bloom L, Crouch RJ. Isolation and mapping of a mutation in Escherichia coli with altered levels of ribonuclease H. J Bacteriol. 1980;144(1):28–35. doi: 10.1128/jb.144.1.28-35.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pohjoismaki JL, et al. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J Mol Biol. 2010;397(5):1144–1155. doi: 10.1016/j.jmb.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reyes A, et al. Actin and myosin contribute to mammalian mitochondrial DNA maintenance. Nucleic Acids Res. 2011;39(12):5098–5108. doi: 10.1093/nar/gkr052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brody JR, Kern SE. Sodium boric acid: A Tris-free, cooler conductive medium for DNA electrophoresis. BioTechniques. 2004;36(2):214–216. doi: 10.2144/04362BM02. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.