Significance
Genetic factors play a major role in the development of human addiction. Identifying these genes and understanding their molecular mechanisms are necessary first steps in the development of targeted therapeutic intervention. Here, we have isolated the gene encoding Ras suppressor 1 (Rsu1) in an unbiased genetic screen for altered ethanol responses in the vinegar fly, Drosophila melanogaster. Our behavioral, genetic, and biochemical experiments show that Rsu1 links signaling from the integrin cell adhesion molecule to the small GTPase Rac1 in adult neurons to regulate actin dynamics and alcohol consumption preference. We also show that variants in human RSU1 associate with altered drinking and brain activation during a reward prediction task, thereby validating the predictive power of our approach.
Keywords: addiction, alcohol, genetics, actin
Abstract
Alcohol abuse is highly prevalent, but little is understood about the molecular causes. Here, we report that Ras suppressor 1 (Rsu1) affects ethanol consumption in flies and humans. Drosophila lacking Rsu1 show reduced sensitivity to ethanol-induced sedation. We show that Rsu1 is required in the adult nervous system for normal sensitivity and that it acts downstream of the integrin cell adhesion molecule and upstream of the Ras-related C3 botulinum toxin substrate 1 (Rac1) GTPase to regulate the actin cytoskeleton. In an ethanol preference assay, global loss of Rsu1 causes high naïve preference. In contrast, flies lacking Rsu1 only in the mushroom bodies of the brain show normal naïve preference but then fail to acquire ethanol preference like normal flies. Rsu1 is, thus, required in distinct neurons to modulate naïve and acquired ethanol preference. In humans, we find that polymorphisms in RSU1 are associated with brain activation in the ventral striatum during reward anticipation in adolescents and alcohol consumption in both adolescents and adults. Together, these data suggest a conserved role for integrin/Rsu1/Rac1/actin signaling in modulating reward-related phenotypes, including ethanol consumption, across phyla.
Alcohol consumption has a worldwide prevalence of 42% (1), and alcohol is the third most serious risk factor for health loss worldwide (2). The genetic contribution to the development of alcohol use disorders (AUDs) has been estimated at 40–60% based on family, adoption, and twin studies (3, 4). Although several studies in humans and model organisms have described genes and molecular pathways involved in alcohol responses (5, 6), our molecular understanding of how AUDs develop is still incomplete.
The vinegar fly, Drosophila melanogaster, is a genetically tractable organism used to model addiction-relevant, ethanol-induced behaviors (7, 8). When exposed to ethanol vapor, flies display biphasic behaviors similar to those elicited in humans. Low ethanol doses induce a state of disinhibition and increased locomotor activity, whereas higher doses lead to loss of postural control and sedation (9, 10). Flies also display addiction-like behaviors similar to mammals. In an ethanol consumption and preference assay (11), for example, flies gradually acquire alcohol preference and will overcome an aversive stimulus to consume alcohol (12).
In addition to the similarities that mammals and flies display in their behavioral responses to ethanol, numerous genes and signaling pathways affect alcohol-induced behaviors across organisms. In vitro and in vivo studies in Drosophila and mammals have revealed a link between alcohol and the actin cytoskeleton (13). When cultured primary mouse neurons are exposed to ethanol, there is a gradual decay in filamentous actin that correlates with decreased NMDA receptor current (14). Mice with a genetic KO of the actin-capping protein epidermal growth factor receptor kinase substrate 8 (EPS8), which displays reduced decay of both filamentous actin and NMDA receptor current in the presence of acute ethanol, show increased alcohol preference (14). Flies with mutations in the arouser gene, encoding an EPS8 homolog, also show an ethanol sensitivity phenotype (15).
A major regulator of actin cytoskeleton dynamics is the Rho family of small GTPases, including Rho, Rac, and Cdc42, and mutations in these genes affect alcohol-induced behaviors (13). Adult loss of Ras-related C3 botulinum toxin substrate 1 (Rac1) activity, for example, leads to enhanced sensitivity to alcohol-induced sedation, whereas loss of the Rac1 down-regulator RhoGAP18B causes reduced sensitivity (16). Although these studies have shown that Rho family GTPases play a role in alcohol responses, the upstream signaling pathways modulating their effects on actin cytoskeletal dynamics are not understood.
Here, we describe the identification and characterization of mutations in the icarus (ics) gene encoding Ras suppressor 1 (Rsu1), which exhibits reduced sensitivity to ethanol-induced sedation. Our experiments reveal that ics mediates normal behavioral responses to ethanol in the adult nervous system by regulating actin dynamics downstream of integrin and upstream of the Rac1 GTPase. Although WT flies gradually acquire ethanol consumption preference over several days, flies completely lacking Rsu1 show heightened naïve preference that does not increase further over the time of the assay. Conversely, flies lacking Rsu1 only in the mushroom bodies (MBs) show no naïve preference and also, fail to acquire preference over time, suggesting that distinct neural circuits mediate naïve and acquired ethanol preference. In humans, RSU1 was associated with frequency of lifetime drinking in an adolescent sample and alcohol dependence in an independent adult replication sample. In adolescents, RSU1 was also associated with altered functional MRI activation in the ventral striatum (VS) during reward anticipation. Our findings, thus, highlight Rsu1 and the integrin/Rsu1/Rac1 signaling pathway as important modulators of reward-related phenotypes, including ethanol consumption across phyla.
Results
ics Mutants Display Reduced Sensitivity to Ethanol-Induced Sedation.
To identify genes involved in ethanol-induced behaviors in Drosophila, we screened a collection of strains carrying random insertions of a transposable P element. We isolated one mutant that displayed reduced sensitivity to ethanol-induced sedation compared with controls (Fig. 1A and SI Appendix, Fig. S1A). DNA sequencing analysis revealed that the Gal4-containing P element in this line is inserted in the ics gene, and we, thus, labeled it icsG4. The ics gene had been previously identified because of its wing blister phenotype (17), and icsG4 mutant flies also exhibited wing blisters. The original mutant, icsBG, carrying a P element insertion at the 3′ end of ics exon 3 (SI Appendix, Fig. S1D) showed reduced sensitivity to ethanol-induced sedation similar to that of icsG4 (SI Appendix, Fig. S1A). Heterozygous ics flies showed no phenotype and were used as controls in some of the experiments below. To confirm that the transposon inserted in icsG4 was, indeed, responsible for the icsG4 ethanol phenotype, we mobilized the icsG4 P element by supplying the transposase enzyme. Precise excision (icsx23) of the P element reverted the mutant phenotype to the WT, whereas imprecise excision of the P element (icsx5; resulting in a deletion of 1,353 bp) (SI Appendix, Fig. S1D) showed the ics mutant phenotype (SI Appendix, Fig. S1B). Expression of the Rsu1 protein was absent in ics mutants (icsG4 and icsx5) and normal in the precise excision icsx23 (SI Appendix, Fig. S1C). The reduced ethanol sensitivity in ics mutants was not caused by altered pharmacokinetics, because ethanol absorption and metabolism were normal in icsG4, icsBG, and icsx5 flies compared with controls (SI Appendix, Fig. S1E). Flies carrying mutations in ics also showed normal locomotion (assessed by startle-induced phototaxis and negative geotaxis as well as spontaneous daily locomotion). These results suggest that ics mutations affect ethanol-induced behavior without generally disabling the flies.
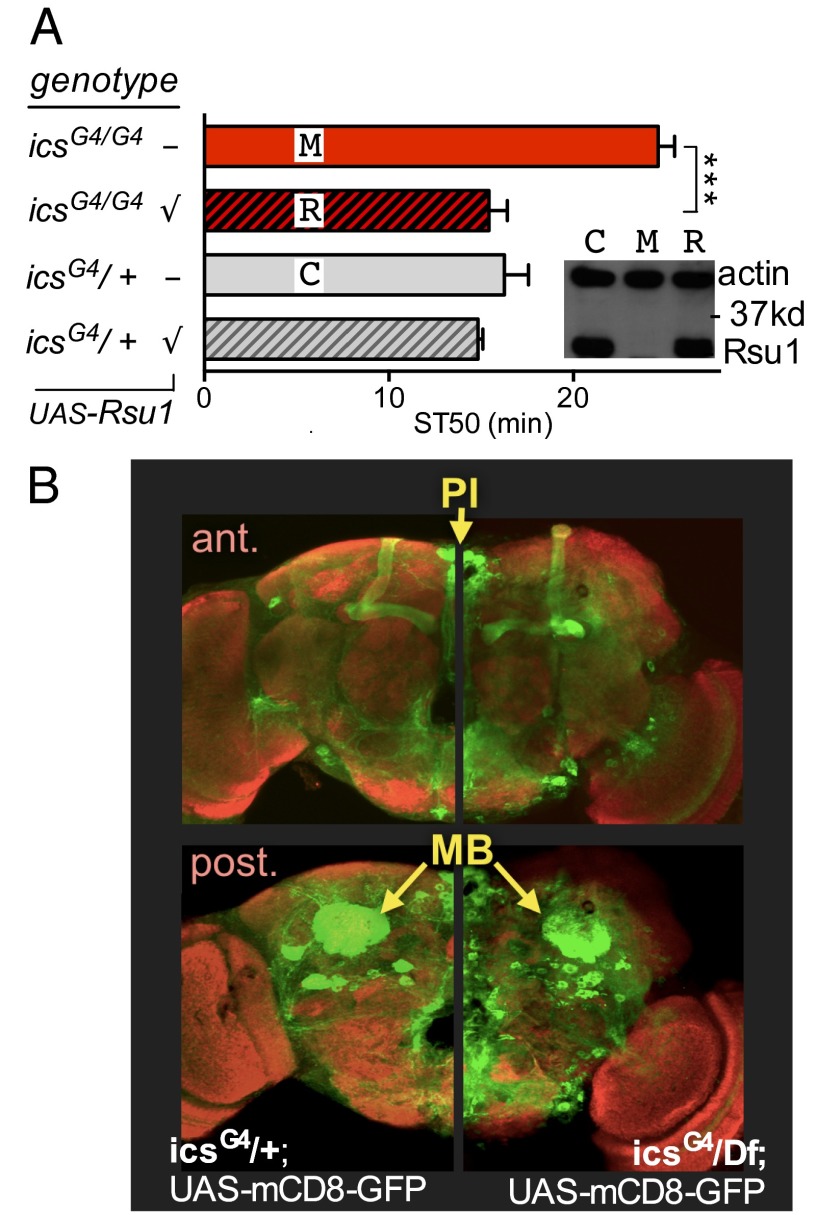
Fig. 1.
ics, Encoding Rsu1, is required for normal ethanol responses. Here, flies were exposed to a 130:20 ethanol:airflow rate, and bars represent means ± SEM. ST50 stands for the median sedation time; increased ST50 indicates reduced ethanol sensitivity. (A) Mutant icsG4 flies show reduced sensitivity to ethanol-induced sedation. This phenotype and (Inset) the loss of Rsu1 protein are rescued with expression of Rsu1 cDNA (UAS-Rsu1; transgene presence indicated by √; n = 8). C, control; M, mutant; R, rescue. ***P < 0.001. (B) Brain expression pattern of icsG4 revealed by a membrane-bound GFP reporter (UAS-mCD8-GFP; green). B shows (Upper) anterior (ant.) and (Lower) posterior (post.) confocal stacks of icsG4 (Left) heterozygous WT and (Right) homozygous mutant flies. Expression includes neurosecretory cells in the pars intercerebralis (PI) as well as the MBs. Neuropil is counterstained with anti-Brp nc82 antibody (red).
Rsu1 Is Required in the Adult Nervous System for Normal Ethanol Sensitivity.
To confirm that the reduced ethanol sensitivity of ics mutants was caused by loss of Rsu1 protein expression, we restored expression of Rsu1 by using the Gal4/UAS system (18) and introducing a UAS-Rsu1 cDNA transgene. We drove expression of Rsu1 in icsG4 mutant flies by taking advantage of the transcriptional activator Gal4 contained within the inserted P element, which disrupts Rsu1 expression while also expressing Gal4 under the control of the endogenous ics promoter and enhancers. Homozygous icsG4 flies carrying the Gal4-transactivated UAS-Rsu1 transgene showed WT ethanol sensitivity and restoration of WT Rsu1 protein expression levels (Fig. 1A). icsG4 drove expression of a UAS-GFP reporter in the brain, including in the MBs and neurosecretory cells of the pars intercerebralis; there were no obvious differences between icsG4 mutant and WT flies (Fig. 1B).
To investigate if icsG4-driven expression in the nervous system was necessary for normal ethanol responses, we suppressed the expression of the UAS-Rsu1 cDNA in neurons using a panneuronal inhibitor of Gal4, elav-Gal80 (19). Neuronal suppression of Rsu1 expression prevented rescue of the icsG4 phenotype by the UAS-Rsu1 transgene (Fig. 2A). To ask whether exclusive expression of Rsu1 in the nervous system was sufficient to rescue the ics mutant phenotype, we used the neuron-specific driver elav-Gal4 to drive expression of UAS-Rsu1 in the ics mutant background. As shown in Fig. 2B, reduced ethanol sensitivity of icsx5 was restored to WT levels when we expressed Rsu1 exclusively in neurons. Taken together, these data indicate that Rsu1 functions in the nervous system to regulate ethanol-induced behavior.
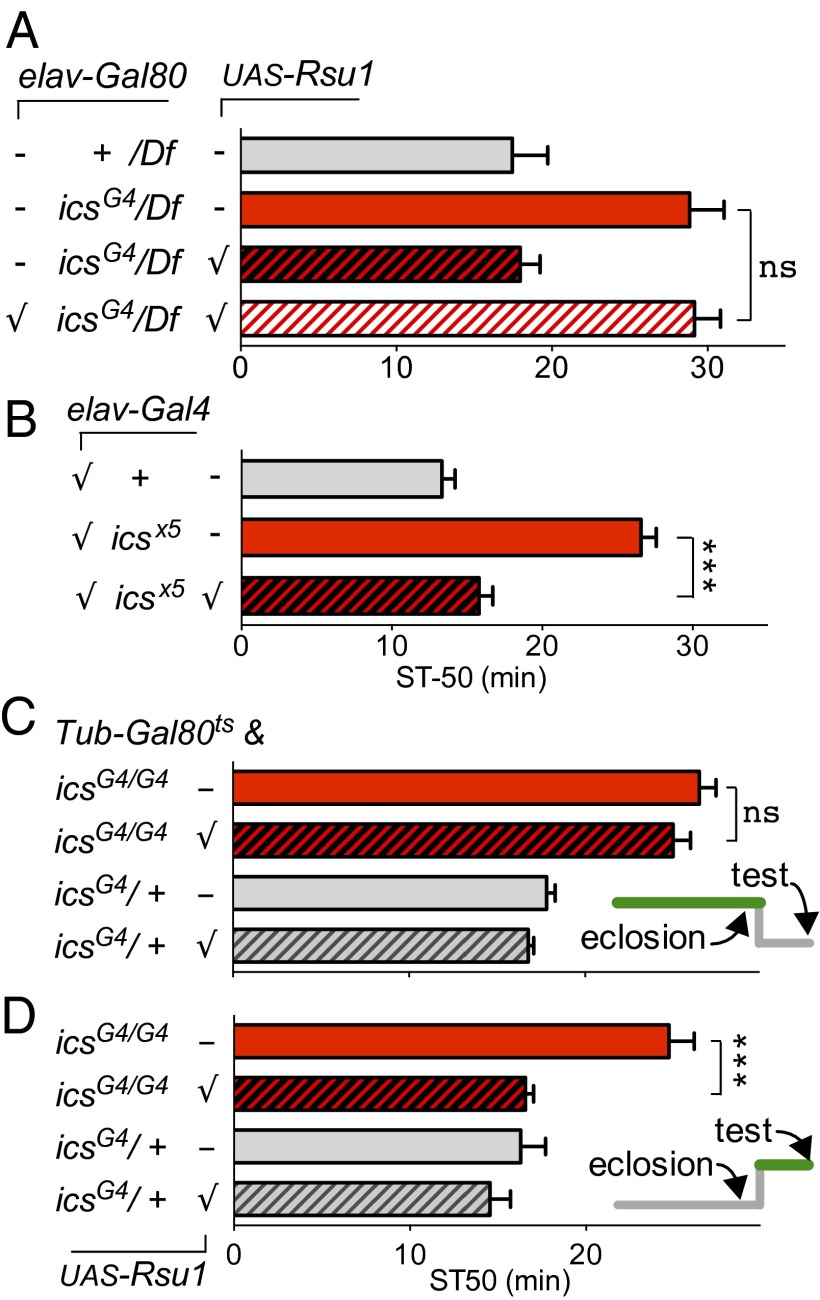
Fig. 2.
Rsu1 is required in the adult nervous system for normal ethanol responses. (A) Suppression of Gal4 and UAS-Rsu1 expression in the nervous system with elav-Gal80 abrogates the behavioral rescue (n = 6–7). Df, genetic deficiency Df(2L)BSC147 completely removing the ics gene locus; ns, not significant (P > 0.91). (B) Rsu1 expression exclusively in the nervous system through elavc155-Gal4 completely rescues the reduced ethanol sensitivity phenotype of icsx5 mutant flies. ***P < 0.001 (n = 7–9). (C and D) Adult expression (D) posteclosion but (C) not throughout development rescues the reduced ethanol sensitivity phenotype of icsG4 mutant flies. UAS-Rsu1 expression was suppressed using ubiquitously expressed Gal80ts, which inhibits Gal4 (and therefore, Rsu1 expression) at (Inset) 18 °C (gray) but not 29 °C (green). Flies were kept for 3 d at the test temperature before ethanol exposure. ns, Not significant (P > 0.29); ST50, median sedation time. ***P < 0.001 (n = 6–9).
Neurons expressing Gal4 in icsG4 mutant brains seemed no different from behaviorally normal icsG4/+ heterozygotes (Fig. 1B), suggesting that Rsu1 is not needed to properly set up ethanol response neuronal circuits. Given that ics mutant flies show a developmental wing blister phenotype (17), it was possible that we could have missed subtle developmental defects. We, therefore, wished to directly test the requirement for Rsu1 in adult flies using Gal80ts, which allows for temperature-dependent suppression of Gal4 driver activity (20). Using this system, the expression of Gal4 is suppressed at 18 °C but not at 29 °C. We first asked whether expression of UAS-Rsu1 cDNA during development only was able to restore normal ethanol-induced sedation to ics mutant adults. We reared flies (icsG4 UAS-Rsu1;Tub-Gal80ts) at 29 °C, allowed developmental expression of Rsu1, and then, suppressed expression during adulthood by shifting the flies to 18 °C for 3 d after eclosion. Expression of Rsu1 in this manner was unable to rescue the reduced ethanol sensitivity of icsG4 mutants (Fig. 2C). Conversely, when we raised flies at 18 °C, blocking Rsu1 expression during development but allowed Rsu1 expression in adulthood by shifting the flies to 29 °C for 3 d after eclosion, the phenotype of icsG4 mutants was completely rescued to WT levels (Fig. 2D). These data suggest that Rsu1 functions in the adult fly to regulate normal ethanol-induced behaviors and that Rsu1 is not required for the developmental wiring of neural circuits involved in regulating ethanol responses.
Rsu1 Functions Downstream of Integrin Signaling.
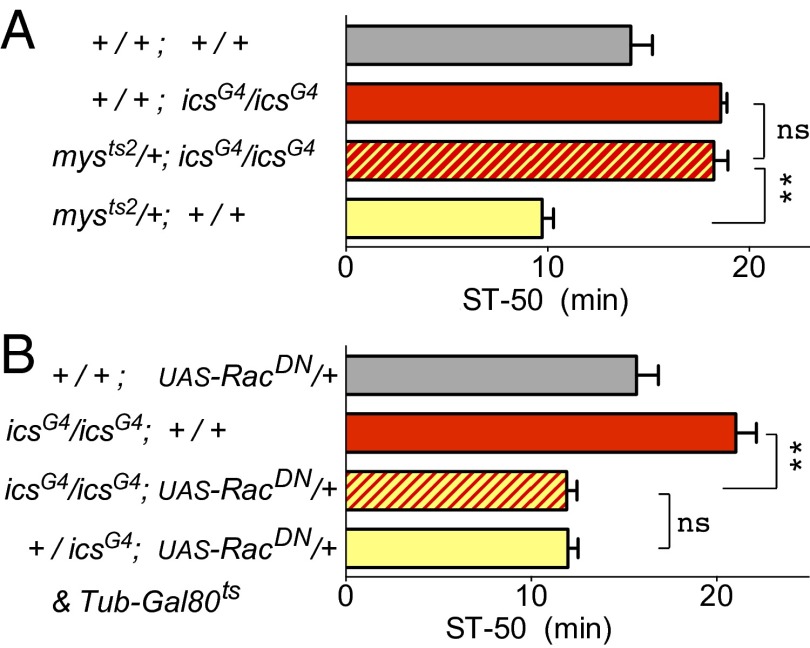
Developmental experiments show that Rsu1 acts in concert with the scaffolding protein PINCH (particularly interesting new cysteine-histidine-rich protein) to inhibit the JNK signaling pathway downstream of the integrin signaling receptor (17). We, therefore, investigated whether perturbation of the integrin signaling pathway in Drosophila would alter ethanol sensitivity. As previously reported (21), flies heterozygous for mutations in the β-integrin–encoding gene myospheroid (mysts2) showed increased sensitivity to the sedating effects of ethanol compared with the WT (Fig. 3A) (mysts2 homozygotes are not viable). When we introduced the icsG4 mutation into this genetic background, the mysts2 icsG4 double-mutant flies showed the same reduced ethanol sensitivity as icsG4 mutant flies (Fig. 3B), suggesting that Rsu1 controls ethanol-induced behavior downstream of the integrin receptor. We also observed genetic interactions between ics alleles and mutants in the genes encoding PINCH and integrin-linked kinase (ILK) (SI Appendix, Fig. S2), further supporting our hypothesis that Rsu1 affects ethanol-induced behaviors by regulating the integrin signaling pathway.
Fig. 3.
Rsu1 links β-integrin to Rac1 signaling. (A) icsG4 homozygous mutants combined with heterozygous β-integrin loss-of-function mutants (mysts2) are as resistant to ethanol-induced sedation as icsG4 mutants alone, indicating that Rsu1 functions downstream of β-integrin. Females were grown at 29 °C for maximum mysts2 effect. ns, Not significant (P > 0.69); ST-50, median sedation time. **P < 0.01 (n = 7–9). (B) icsG4 homozygous mutants combined with dominant negative Rac1 (UAS-Rac1DN) are as sensitive as Rac1DN mutants alone, suggesting that Rac1 functions downstream of Rsu1. Unexpressed UAS-RacDN/+ lacking a Gal4 driver served as a control. ns, Not significant (P > 0.92). **P < 0.001 (n = 8–10).
Rsu1 Acts Upstream of Rac1 and Affects Actin Dynamics.
Because Rsu1 acts in concert with PINCH to inhibit JNK activity during development (17), we tested for potential genetic interactions between mutations in ics and basket (encoding JNK). We were unable to find any such interaction or a sedation phenotype in basket mutants, which is consistent with two previous studies reporting the absence of an ethanol sedation phenotype in basket mutants (22, 23). Other than JNK, other downstream targets of integrin signaling include Rho family GTPases. Depletion of human Rsu1 in a human breast cancer cell line elevates the levels of activated Rac (Rac.GTP) (24), suggesting that Rsu1 reduces Rac1 activation. We, therefore, investigated whether Rsu1 functions through Rac1 to affect ethanol-induced responses. Expressing dominant negative (DN) Rac1 in ics-Gal4–expressing cells (icsG4/+;UAS-Rac1DN/+) resulted in increased sensitivity to ethanol-induced sedation (Fig. 3B). This increased sensitivity remained the same in the icsG4 homozygous mutant background, suggesting that Rac1 regulates ethanol responses downstream of Rsu1.
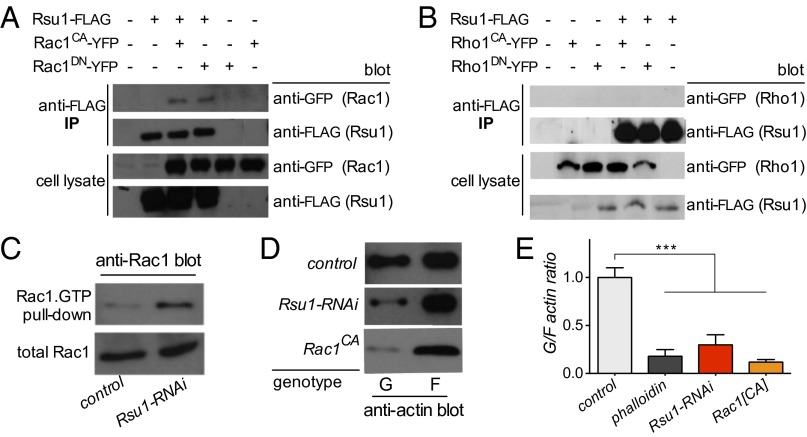
We next determined whether Rsu1 physically interacts with Rac1 by cotransfecting Drosophila Schneider (S2) cells with FLAG-tagged Rsu1 and various Rho GTPases tagged with YFP. Immunoprecipitation with an anti-FLAG antibody pulled down both GTP-locked Rac1G12V [constitutively active (CA)] and GDP-bound Rac1T17N (DN) (Fig. 4A). It did not, however, coimmunoprecipitate Rho1 (Fig. 4B) or Cdc42, suggesting that Rsu1 is a specific binding partner for Rac1 in the Rho family of small GTPases.
Fig. 4.
Rsu1 binds to Rac1 and affects actin dynamics in Drosophila S2 cells. (A and B) Rsu1 binds to (A) both the GTP-locked forms of Rac1 (Rac1CA) and GDP-locked forms of Rac1 (Rac1DN) but (B) not to Rho1 GTP- (Rho1CA) or GDP-locked (Rho1DN) forms. (C) Rac1.GTP pull-down experiments shows that RNAi-mediated knockdown of Rsu1 leads to increased Rac1.GTP loading. (D and E) Globular (G) to filamentous (F) actin assay measuring the ratio of actin in free globular to assembled filamentous form showing that RNAi-mediated knockdown of Rsu1 causes an approximately threefold decrease in G/F actin ratio, whereas overexpression of constitutive active Rac1CA causes an approximately ninefold decrease in G/F actin ratios compared with controls. The actin stabilizer phalloidin also decreases the G/F ratio and served as a positive control. IP, immunoprecipitation. ***P < 0.001 (n = 4–9).
Our genetic data indicated that Rsu1 acts upstream of Rac1 to oppose the latter’s activity. We, therefore, hypothesized that, in the absence of Rsu1, there would be increased Rac1 activation. We found that knockdown of Rsu1 with RNAi in S2 cells (SI Appendix, Fig. S3) increased levels of Rac.GTP loading (Fig. 4C). In addition, both overexpression of Rac1CA and Rsu1 knockdown caused a decrease in the globular to filamentous actin ratio (Fig. 4 D and E). Taken together, these data indicate that Rsu1 binds to Rac1 and destabilizes actin filaments through Rac1 inhibition.
ics Mutants Show Increased Alcohol Preference in Drosophila.
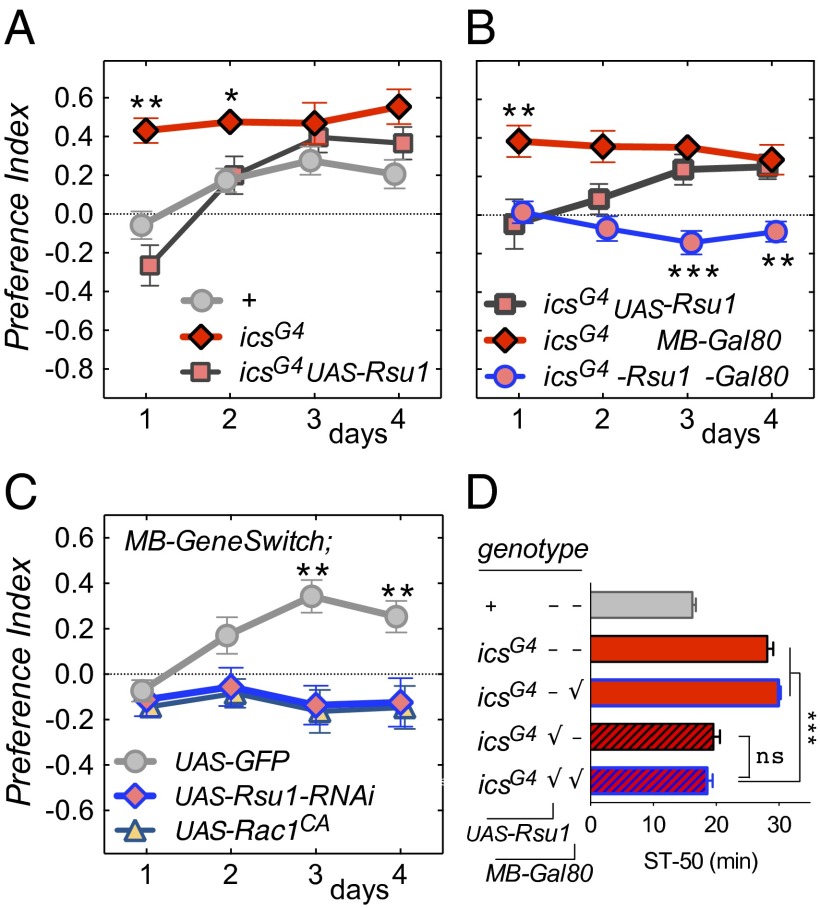
We next asked whether ics mutant flies exhibit an alcohol-drinking phenotype. Flies were tested in an ethanol consumption preference assay [capillary feeder (CAFÉ)] (11, 12). WT flies gradually acquire preference for ethanol over 3 d, showing that alcohol is reinforcing consummatory behavior (SI Appendix, Fig. S4), which likely involves reward pathways. Conversely, icsG4 mutant flies showed significant naïve preference for ethanol on day 1, which remained unchanged over 4 d of the assay (Fig. 5A). This enhanced preference in ics mutants was caused by an increase in ethanol consumption, whereas the total food consumption volume was no different from the WT (SI Appendix, Fig. S4). Introducing UAS-Rsu1 driven by icsG4 into the mutant flies restored this phenotype to WT (i.e., gradual acquisition of preference over the first few days of the assay) (Fig. 5A and SI Appendix, Fig. S4).
Fig. 5.
Alcohol consumption preference phenotypes in flies lacking Rsu1. (A) ics Mutant flies show increased naïve ethanol preference compared with control in the two-bottle choice CAFÉ assay. This phenotype is rescued by expression of UAS-Rsu1 in all icsG4-expressing cells. (B and D) ics Rescue flies lacking Rsu1 expression in the MBs only (icsG4 UAS-Rsu1 MB-Gal80) do not develop acquired ethanol preference but (B) have normal naïve preference on day 1 and (D) ethanol-induced sedation. (C) Adult MB-specific knockdown of Rsu1 or overexpression of Rac1CA causes loss of acquired ethanol preference. The transgenes were expressed in adults using a mifepristone-inducible MB-GeneSwitch driver. ns, Not significant; ST-50, median sedation time. *P < 0.05; **P < 0.01 (icsG4 vs. control); ***P < 0.001.
The MBs are a brain center in Drosophila involved in higher-order processing, such as associative olfactory learning (25) and ethanol-reinforced odor preference (26). We next asked whether Rsu1 was required in the MB for normal ethanol preference in the CAFÉ assay. Using an MB-Gal80 transgene, we inhibited MB expression of Rsu1 in icsG4;UAS-Rsu1 flies (SI Appendix, Fig. S5) (27). Like the WT, these flies showed no naïve ethanol preference, but unlike the WT, they did not acquire ethanol preference over the 4-d span of the experiment (Fig. 5B). To confirm that loss of Rsu1 from the MB caused this lack of acquired ethanol preference, we knocked down Rsu1 expression specifically in adult MB. Using a mifepristone-inducible MB-GeneSwitch driver (28), we found that adult expression of both UAS-Rsu1-RNAi and UAS-Rac1CA overexpression led to a complete loss of ethanol preference (Fig. 5C). Together, our data show that flies globally lacking Rsu1 display high naïve preference that does not change over time. Conversely, flies lacking Rsu1 only in the MB show neither naïve nor acquired preference. Both are in contrast to WT flies, which show no naïve preference but gradually acquire preference in the CAFÉ assay over a few days. Flies lacking Rsu1 in the MB only showed normal ethanol-induced sedation (Fig. 5D). These findings indicate that naïve responses to ethanol, such as naïve preference and sensitivity to sedation, are mediated by Rsu1 in neurons outside the MB, whereas within the MB, Rsu1 is essential for gradual acquisition of preference.
RSU1 Genotypes Are Associated with Reward Anticipation and Alcohol Consumption in Human Adolescents.
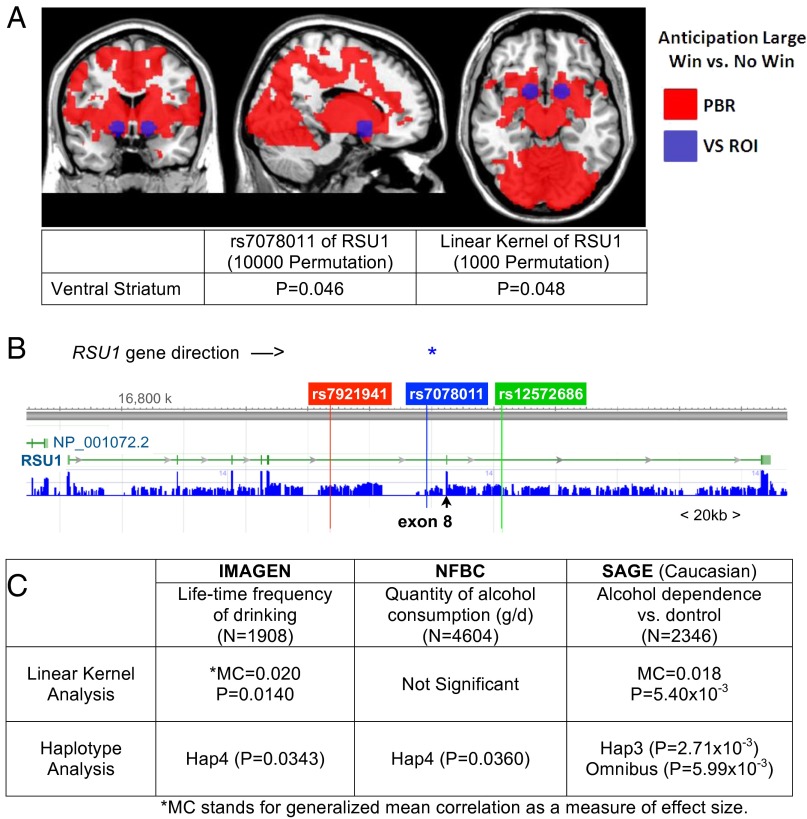
We next sought to translate our Drosophila findings to humans. Alcohol drinking activates the reward system, and alcohol preference and drinking behavior are associated with reward anticipation (29–35). Reward anticipation can be reliably measured during the monetary incentive delay (MID) task (36), where subjects must press a button on seeing an object on the screen. The form of the object determines whether subjects can accrue a large, small, or no monetary win if pressing the button in time. To test a possible association of SNPs in human RSU1 with reward anticipation, we measured brain activation with functional MRI blood oxygenation level dependent (BOLD) responses during the MID task. We first conducted neuroimaging analyses in 1,303 adolescents of the IMAGEN cohort who were assessed at age 14 y old. We observed extensive activation in the brain comparing the anticipation of a large win with no win, including in the VS, a region crucial for reward processing (37). In this region of interest, we detected an association of the minor T allele of rs7078011 in RSU1 with increased VS activation, which remained significant after controlling for the 70 SNPs present at the RSU1 locus in the IMAGEN dataset (P10,000permutation = 0.046) (Fig. 6A). However, we did not detect association of rs7078011 with frequency of lifetime drinking at 14 y old in the IMAGEN sample. Because rs7078011 is localized in the seventh intron of human RSU1 (Fig. 6B), we hypothesized that it may be a marker for an unidentified linked causal variant in the vicinity. Of 70 SNPs identified in RSU1, 22 SNPs were in linkage disequilibrium (LD) with rs7078011. These SNPs covered the eighth exon of the gene (Fig. 6B and SI Appendix, Fig. S6). To investigate if rare variants are present in the gene locus covered by these SNPs, we analyzed whole-genome sequencing data of the eighth exon in reference datasets (National Heart, Lung, and Blood Institute and 1000 Genomes Phase III). Here, we detected several rare variants (minor allele frequency < 1%) with a predicted disruption of protein function. These variants are either missense [i.e., rs144428707 (SNP), rs375646999 (SNP), rs375416941 (SNP), rs372364335 (SNP), and rs199904406 (SNP)] (SI Appendix, Table S1) or splicing related [i.e., rs373104238 (indel)]. However, our datasets did not have sufficient power to allow stable association analyses of these potentially causative polymorphisms.
Fig. 6.
Genetic studies in humans. (A) Whole-brain analysis of reward anticipation large win vs. no win during the MID task shows positive BOLD response (PBR) during reward anticipation (family-wise error P < 0.05). The location of the VS (±15, 9, −9; 9-mm radius) is depicted in blue. The results of association analyses between VS and RSU1 gene are summarized. ROI, region of interest. (B) Exon/intron schematic of the RSU1 gene. The first SNP of haplotype block 6 (rs7921941; red), the last SNP of haplotype block 5 (rs12572686; green), and the main SNP (rs7078011; asterisk and blue) are highlighted. The eighth exon is indicated with an arrow. (C) Summary of genetic analyses of alcohol drinking in the human datasets IMAGEN, SAGE, and NFBC1966.
We, therefore, carried out a linear kernel-based association analysis (38) of the SNPs in strong linkage disequilibrium with rs7078011. Kernels combine the contribution of genetic variations, thus enabling detection of genetic effects that cannot be represented by a single SNP alone (39). Although kernel analyses do not indicate directionality of an association, they are particularly sensitive in reliably detecting associations with potentially causal rare variants. We found significant associations of the RSU1 kernel with both VS activation [generalized mean correlation (mc) = 0.020, P1,000permutations = 0.0480] (Fig. 6A and SI Appendix, Fig. S7A) and the frequency of lifetime drinking (mc = 0.020, P1,000permutations = 0.0140) in the IMAGEN sample at 14 y old (Fig. 6C and SI Appendix, Fig. S7B).
To investigate if RSU1 might be a risk factor for alcohol addiction, we analyzed 1,149 alcohol-dependent patients and 1,360 controls of Caucasian descent (SI Appendix, Table S2) from the Study of Addiction: Genetics and Environment (SAGE) (40). We found a significant association of the RSU1 kernel with alcohol dependence (mc = 0.018, P10,000permutations = 5.40 × 10−3) (Fig. 6C and SI Appendix, Fig. S7C). We also measured association of the RSU1 kernel with alcohol drinking in 4,604 adults age 31 y old of the population-based North Finish Birth Cohort 1966 (NFBC1966) (41). However, we found no significant association with the quantity of alcohol consumption (Fig. 6C and SI Appendix, Fig. S7D).
In addition, we carried out an analysis of haplotype block 5 involving rs7078011. It is noteworthy that the allele frequencies of NFBC are very different from those of IMAGEN (P = 2.03 × 10−48, χ2df = 21 = 286.19) and the SAGE (P = 1.35 × 10−52, χ2df = 20 = 303.80), whereas the latter two are very similar (P = 0.922, χ2df = 21 = 12.59) (SI Appendix, Table S3), indicating distinct genetic backgrounds of the samples. There was a nominally significant association of haplotype phase 4 (Hap4) of block 5 with increased frequency of drinking in the IMAGEN sample at age 14 y old (P = 0.0343) and a significant association of the RSU1 haplotypes with alcohol dependence in the SAGE dataset (omnibus test; P = 5.99 × 10−3 from 10,000 permutations). Although the association of the RSU1 haplotypes with alcohol dependence in the SAGE dataset was driven by Hap3 (P = 2.71 × 10−3), there was a trend for an association of Hap4 (P = 0.0856) (Fig. 6C and SI Appendix, Table S4). We also found a nominally significant association of Hap4 with quantity of alcohol consumption in the NFBC1966 dataset at age 31 y old (P = 0.0360) (Fig. 6C and SI Appendix, Table S4).
Last, we evaluated gray matter volume of the VS and white matter connectivity of brain structures related to the reward system and associative learning, which are both known to contribute to the development of addiction (42). There was no association with rs7078011 or the RSU1 kernel with VS volume (P1,000permutations = 0.449) or fractional anisotropy measures of diffusion tensor imaging in fiber tracts linking the hippocampus with the limbic system (fornix crescent: P1,000permutations = 0.554; fornix body: P1,000permutations = 0.711; VS: P1,000permutations = 0.176) (SI Appendix, Fig. S8). These anatomical findings suggest that the RSU1 variants alter behavior without changing human neuroanatomy, consistent with our findings in Drosophila, where no obvious developmental abnormality was observed in fly brains lacking Rsu1, thus underscoring the concordance of our Drosophila and human findings.
Discussion
Role of Rsu1 and Integrin Signaling in Ethanol Sensitivity.
The Rho family of small GTPases is known to regulate ethanol-induced behaviors (13), but the upstream pathways that signal to these GTPases in this context are largely unknown. In this report, we characterize the effects of ics/RSU1 on ethanol-related behaviors. We isolated mutations in the Drosophila ics gene because of their reduced sensitivity to ethanol-induced sedation. Drosophila Rsu1, like its human homolog RSU1, is a 32-kDa protein with a C-terminal domain that contains seven leucine-rich repeats and binds to the integrin effector PINCH to inhibit JNK signaling. In flies, absence of Rsu1 leads to abnormalities during wing development and dorsal closure (17). Indeed, the ics mutants that we isolated exhibit wing blisters similar to the ones caused by the loss of integrin, PINCH, and ILK, suggesting that Rsu1 acts in concert with these proteins in integrin-dependent cell adhesion (17). Our data indicate that, in the regulation of adult ethanol behaviors, Rsu1 acts downstream of integrin to antagonize integrin signaling, which was suggested by the fact that loss of Rsu1 leads to reduced ethanol sensitivity, whereas loss-of-function mutations of integrin, PINCH, and ILK result in the opposite phenotype: enhanced ethanol sensitivity. Thus, Rsu1 has modulatory roles on integrin signaling that are context-dependent. During wing development, Rsu1 mediates integrin signaling to antagonize JNK (17), whereas in the adult nervous system, Rsu1 antagonizes integrin signaling to suppress Rac1 activity.
Rsu1 Regulates Actin Dynamics.
We were unable to observe any genetic interaction between Rsu1 and JNK mutants. We, therefore, hypothesized that Rsu1 might act through the small Rho family GTPase Rac1 to regulate ethanol-induced behaviors, because depletion of Rsu1 enhanced Rac1 activation and cell migration (24). We found that Rsu1 acts upstream of Rac1 to antagonize Rac1 activity in both flies and cell culture. Rsu1 coimmunoprecipitated specifically with Rac1 (but not Rho1 or Cdc42) from Drosophila S2 cells but did not show a preference for either GTP- or GDP-bound Rac1. Because Rsu1 does not contain a potential Rac-inactivating, GTPase-activating (GAP) domain, we hypothesize that Rsu1 prevents Rac1 from interacting with its relevant activators and/or effectors, possibly by sequestering them or occluding binding sites within Rac1. We show that normal ethanol-induced behaviors, including sedation sensitivity and consumption preference, require proper Rsu1 and Rac1 function in the adult nervous system. Together, our findings suggest that integrin signals to Rac1 through Rsu1 to regulate actin dynamics, which is known to be required for proper synaptic function (43) as well as behavioral responses to drugs of abuse (13). It also establishes integrin/Rsu1 as an important functional input into the regulation of actin dynamics with behaviorally manifest consequences.
Involvement of Rsu1 in Higher Behaviors.
Our further characterization revealed different behavioral roles for Rsu1 in anatomically distinct neuronal circuits. For normal naïve responses to ethanol, Rsu1 functions in the nervous system outside of the MB. Absence of Rsu1 from these non-MB neurons resulted in reduced sensitivity to ethanol-induced sedation as well as naïve preference for ethanol in a choice assay. In contrast, loss of Rsu1 in MB led to normal naïve ethanol sedation sensitivity and consumption preference but caused a failure to acquire ethanol preference, suggesting that activated Rac1 in the MB prevents this behavioral plasticity. Indeed, when we overexpressed Rac1CA in adult MB, the flies failed to acquire ethanol preference. Conversely, flies lacking Rsu1 throughout the brain showed high naïve ethanol preference, suggesting that activation of Rac1 outside the MB promotes naïve preference. Thus, Rsu1 has opposite effects on ethanol preference depending on the affected circuits. Such opposite effects are reminiscent of mouse findings, where suppression of Rac1 in the nucleus accumbens promoted conditioned place preference (CPP) for cocaine (44), whereas global Kalirin7 KO (a Rac1 activator) led to reduced cocaine CPP (45). Our data expand on these findings by showing that (i) similar to mammals, gene function in distinct circuits can differentially affect drug preference in Drosophila, (ii) in addition to Kalirin7-mediated activation, integrin/Rsu1-regulated suppression is an important input into Rac1 regulation, and (iii) we extend the mouse Rac1 findings from effects on cocaine-mediated reinforcement (in CPP) to voluntary drug/alcohol consumption in both Drosophila and humans.
Previous studies have shown a remarkable conservation of genetic determinants of alcohol and substance use behavior across both species (5). We investigated whether RSU1 was involved in human reward processing and alcohol-drinking behaviors, including addiction, by analyzing several datasets, including the IMAGEN adolescent imaging genetics cohort (46), the SAGE alcohol dependence dataset (40), and the NFBC1966 (41). Like most other large genetic datasets, these samples have been analyzed in various different projects. Repeat analysis raises the question of a potentially greater false-positive rate, because correction for multiple testing is usually confined to the number of tests within one project. Although an increased risk for false-positives is a real possibility, we have mitigated it by (i) testing a very specific hypothesis, which has been experimentally supported in the Drosophila studies presented, and (ii) validating our results across different independent datasets.
Because we were interested in investigating the genetic basis of mechanisms that convey increased risk for alcohol-drinking behavior, we first analyzed the population-based IMAGEN sample of 14-y-old adolescents who did not meet criteria for AUDs. In this sample, a generic reward stimulus as presented in the MID task might be more salient and a more reliable activator of the reward system than alcohol-specific cues. Using this approach, we first carried out single-SNP analyses to identify a marker for the strongest genetic signal for VS activation during reward anticipation in the RSU1 gene. This analysis resulted in the detection of the association of VS activation during reward anticipation but not frequency of lifetime drinking with SNP rs7078011 localized in intron 7 of RSU1.
We hypothesized that rs7078011 might be a marker of potentially causative rare genetic variants. Indeed, analyzing whole-genome sequencing data, we detected several rare variants in the genomic locus delineated by 22 SNPs in strong linkage disequilibrium with rs7078011 that probably impair protein function of Rsu1. Although our datasets were underpowered to carry out a genetic association analysis of the rare variants detected, we were able to carry out a kernel-based association analysis with these 22 SNPs. Using the kernel method, we confirmed the association of RSU1 with VS activation during reward anticipation in the IMAGEN dataset, and we also found an association of the RSU1 kernel with frequency of lifetime drinking in the same sample. The fact that the association of rs7078011 with the investigated phenotypes was less stable than the association of the kernel is in keeping with the possibility of rare variants underlying the observed associations. When rare causal variants are present, their linkage disequilibrium with noncausal SNPs with higher frequencies might vary from sample to sample. This variability can be caused by recurrent rare mutations or admixture of populations with different genetic backgrounds. It is, thus, possible that the same rare variant can be linked with different alleles in different samples. Such differential linkage could lead to false-negative findings if only the same SNP was analyzed. Alternatively, different rare variants within the observed gene locus might associate with different phenotypes under study. Using a kernel analysis allowed us to overcome these problems.
Our kernel analyses in additional independent datasets revealed an association of RSU1 with adult alcohol dependence but not with adult drinking behavior in a general population sample. Because early substance use in adolescents is a risk factor for adult alcohol dependence (47), these results might indicate that the effect of Rsu1 on reward processing influences a risk drinking trajectory at very early stages of exposure to alcohol. However, one limitation of our study is that it is not possible to unambiguously rule out an association of adult alcohol drinking in the population with RSU1. The markedly different LD structure of RSU1 in the NFBC1966 cohort might have masked an association of the kernel. The observed nominal association of the RSU1 haplotype 4 with the amount of drinking might, indeed, indicate a weak signal in this locus.
The haplotypes included in the kernel are distributed around exon 8, which encodes one of seven leucine-rich repeats found in the Rsu1 protein that are crucial for its interaction with PINCH1 (48). In human glioma cells, an alternative splicing site has been described, which gives rise to an exon 8-deleted splice variant of RSU1 translating into a less stable protein with reduced function (49). It is possible that the rare variants detected might result in an impaired interaction of Rsu1 protein with PINCH and/or decreased protein stability. Such decreases might disrupt Rsu1 function in a way analogous to the knockdown of Rsu1 in Drosophila, causing the alcohol preference phenotype. However, additional investigations are required to analyze the effect of these variants on Rsu1 function and test their association with alcohol drinking in large metaanalyses.
Together, our data show that Rsu1 regulates reward-related behaviors, such as ethanol consumption, in flies and humans. We found no structural abnormalities associated with Rsu1 variants in either flies or humans but show that Rsu1 is required after development in adult flies for normal ethanol-induced behaviors. Our data from both species are, therefore, highly concordant. We hypothesize that the physiological process underlying these phenotypes is synaptic plasticity. In the integrin/Rsu1/Rac1 signaling cascade, both integrin (50) and Rac1 (13) are known to affect synaptic structure and plasticity. Our findings, thus, underscore the use of model organisms. For one, they are useful in elucidating the molecular mechanisms of genes mediating addiction-like behaviors. Also, they show remarkable predictive power with unbiased forward genetic screens in generating testable hypotheses that can be translated to human phenotypes.
Materials and Methods
Detailed methods can be found in SI Appendix, SI Materials and Methods.
Drosophila Experiments.
Flies were kept and assayed as described (51) with the CAFÉ modified from ref. 12. Standard cell culture and biochemistry approaches were used.
Human Data.
This project was approved by the ethics committee at King's College London as well as the local ethics committees at each recruitment site. Informed consent was obtained from each participant and at least one parent. A detailed description of recruitment and assessment procedures, as well as in/exclusion criteria, has previously been published (46). The IMAGEN cohort and assays are described in ref. 46, and the frequency of drinking phenotype was defined using an adapted version of the 2007 ESPAD (European School Survey Project on Alcohol and Other Drugs) questionnaire (www.espad.org), which assesses the number of times alcoholic drinks were consumed until 14 y of age. Individuals were ranked into seven categories from 0 (nondrinker) to 6 (over 50 times; mean = 2.0, SD = 1.8, median = 2.0 in males; mean = 2.1, SD = 1.7, median = 2.0 in females). The NFBC is described in ref. 41, and the quantity of drinking phenotype was determined using an adapted version of the 2007 ESPAD questionnaire (www.espad.org), which assesses “the quantity of alcohol consumed on a TYPICAL DAY when you are drinking.” In this cohort, the mean alcohol intake was 9.1 g alcohol per day (mean = 13.8, SD = 19.7, median = 7.8 in males; mean = 4.8, SD = 7.9, median = 2.2 in females). In our analysis, these original values were analyzed in a quantitative way without any presumed threshold for case–control split. The SAGE dataset is described in ref. 40, which integrates different case–control studies for alcohol dependence. Particularly, 2,509 Caucasian cohorts (case = 688, control = 404 in males; case = 461, control = 956 in females) were chosen in our study because of similar genetic background to the IMAGEN dataset. The descriptive statistics of all three datasets are summarized in SI Appendix, Table S2. We used kernel-generalized variance (38) to quantify the dependency between the functional BOLD and behavioral responses with RSU1 in three cohorts.
Supplementary Material
Acknowledgments
We thank Antonio Lopez, Pranav Penninti, Geetha Kalahasti, Sean Khatami, Summer Acevedo, and the rest of the laboratory of A.R. for their help and advice with experiments. We thank Mike Groteweil, Gregg Roman, and the Bloomington Stock Center for fly stocks. We thank the late Paula Rantakallio [launch of North Finish Birth Cohort 1966 (NFBC1966)], Outi Tornwall, and Minttu Jussila (DNA Biobanking). We also acknowledge the contribution of the late Leena Peltonen. The DNA extractions, sample quality controls, biobank upkeep, and aliquotting were performed at the National Public Health Institute, Biomedicum Helsinki and supported financially by the Academy of Finland and Biocentrum Helsinki. This work was supported by NIH T32 Fellowship DA007290 (to S.A.O.) and Grants F31AA021340 (to S.A.O.), K08DK091316 (to A.R.R.), R01GM084103 (to J.L.K.), and R01AA019526 (to A.R.), and A.R. was supported by an Effie Marie Cain Scholarship in Biomedical Research from the University of Texas Southwestern Medical Center. This work was also supported by the European Union-funded FP6 Integrated Project IMAGEN LSHM-CT-2007-037286 (reinforcement-related behavior in normal brain function and psychopathology), FP7 Projects IMAGEMEND 602450 and Multidisciplinary Approaches to Translational Research In Conduct Syndromes 603016, Innovative Medicine Initiative Project European Autism Interventions - A Multicentre Study for Developing New Medications 115300-2, and Medical Research Council Programme Grant 93558 (developmental pathways into adolescent substance abuse). Additional support was provided by the Swedish Funding Agency Forskningsrådet för miljö, areella näringar och samhällsbyggande, the National Institute for Health Research Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, German Bundesministerium für Bildung und Forschung Grant eMED SysAlc 01ZX1311A (Forschungsnetz Addiction: Early Recognition and Intervention Across the Lifespan), and NIH Big Data to Knowledge U54 Grant Enhancing Neuro-Imaging Genetics Through Meta-Analysis Center for Worldwide Medicine, Imaging and Genomics. NFBC1966 received financial support from Academy of Finland Project Grants 104781, 120315, 129269, 1114194, 24300796, Center of Excellence in Complex Disease Genetics, and SALVE; University Hospital Oulu, Biocenter, University of Oulu Grant 75617; National Heart, Lung, and Blood Institute Grant 5R01HL087679-02 through SNP Typing for Association with Multiple Phenotypes from Existing Epidemiologic Data Program 1RL1MH083268-01; NIH/National Institute of Mental Health Grant R01MH63706:02; ENGAGE Project and Grant HEALTH-F4-2007-201413; European Union FP7 Euro Health Aging Grant 277849; and Medical Research Council, United Kingdom Grants G0500539, G0600705, G1002319, and Prevention of the Metabolic Syndrome by New Lifestyle Intervention Methods/SALVE.
Footnotes
Conflict of interest statement: T.B. served in an advisory or consultancy role for Hexal Pharma, Lilly, Medice, Novartis, Otsuka, Oxford Outcomes, PCM Scientific, Shire, and Viforpharma. T.B. received conference attendance support and conference support or speaker’s fees from Lilly, Medice, Novartis, and Shire. T.B. is/has been involved in clinical trials conducted by Shire and Viforpharma. This work is unrelated to those grants and relationships. J.G. has received research funding from AstraZeneca, Eli Lilly & Co., Janssen-Cilag, and Bristol-Myers Squibb and speaker’s fees from AstraZeneca, Janssen-Cilag, and Bristol-Myers Squibb.
A complete list of The IMAGEN Consortium can be found in the SI Appendix.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417222112/-/DCSupplemental.
Contributor Information
Collaborators: Lisa Albrecht, Charite, Mercedes Arroyo, Cambridge University, Eric Artiges, INSERM, Semiha Aydin, Physikalisch-Technische Bundesanstalt, Christine Bach, Central Institute of Mental Health, Tobias Banaschewski, Central Institute of Mental Health, Alexis Barbot, Commissariat à l'Energie Atomique, Gareth Barker, King’s College, Arun Bokde, Trinity College Dublin, Zuleima Bricaud, INSERM, Uli Bromberg, University of Hamburg, Ruediger Bruehl, Physikalisch-Technische Bundesanstalt, Christian Büchel, University of Hamburg, Anna Cattrell, King’s College, Patricia Conrod, King’s College, Katharina Czech, Charite, Jeffrey Dalley, Cambridge University, Sylvane Desrivieres, King’s College, Tahmine Fadai, University of Hamburg, Herta Flor, Central Institute of Mental Health, Vincent Frouin, Commissariat à l'Energie Atomique, Jürgen Gallinat, Charite, Hugh Garavan, Trinity College Dublin, Fanny Gollier Briand, INSERM, Penny Gowland, University of Nottingham, Bert Heinrichs, Deutsches Referenzzentrum für Ethik, Andreas Heinz, Charite, Thomas Hübner, Technische Universität Dresden, Albrecht Ihlenfeld, Physikalisch-Technische Bundesanstalt, Alex Ing, King's College, Bernd Ittermann, Physikalisch-Technische Bundesanstalt, Tianye Jia, King’s College, Jennifer Jones, Trinity College Dublin, Eleanor Kennedy, King's College, Dirk Lanzerath, Deutsches Referenzzentrum für Ethik, Mark Lathrop, McGill University, Claire Lawrence, University of Nottingham, Hervé Lemaitre, INSERM, Katharina Lüdemann, Charite, Steven Lubbe, King’s College, Christine Macare, King's College, Karl Mann, Central Institute of Mental Health, Adam Mar, Cambridge University, Jean-Luc Martinot, INSERM, Eva Mennigen, Technische Universität Dresden, Fabiana Mesquita de Carvahlo, King’s College, Kathrin Müller, Technische Universität Dresden, Frauke Nees, Central Institute of Mental Health, Charlotte Nymberg, King’s College, Marie-Laure Paillere, INSERM, Tomas Paus, University of Toronto, Zdenka Pausova, University of Toronto, Jean-Baptiste Poline, Commissariat à l'Energie Atomique, Luise Poustka, Central Institute of Mental Health, Erin Quinlan, King's College, Jan Reuter, Charite, Stephan Ripke, Technische Universität Dresden, Trevor Robbins, Cambridge University, Gabriel Robert, King's College, Sarah Rodehacke, Technische Universität Dresden, Barbara Ruggeri, King’s College, Dirk Schmidt, Technische Universität Dresden, Sophia Schneider, University of Hamburg, Florian Schubert, Physikalisch-Technische Bundesanstalt, Michael Smolka, Technische Universität Dresden, Wolfgang Sommer, Central Institute of Mental Health, Rainer Spanagel, Central Institute of Mental Health, Claudia Speiser. GABO:milliarium mbH & Co. KG, Tade Spranger, Deutsches Referenzzentrum für Ethik, Alicia Stedman, University of Nottingham, Dai Stephens, University of Sussex, Nicole Strache, Charite, Andreas Ströhle, Charite, Maren Struve, Central Institute of Mental Health, Naresh Subramaniam, Cambridge University, Amir Tahmasebi, University of Toronto, David Theobald, Cambridge University, Nora Vetter, TU Dresden, Helene Vulser, INSERM, Bernadeta Walaszek, Physikalisch-Technische Bundesanstalt, Robert Whelan, Trinity College Dublin, Steve Williams, King’s College, Bing Xu, King's College, Juliana Yacubian, University of Hamburg, Veronika Ziesch, and Technische Universität Dresden
References
- 1.World Health Organization . Global Status Report on Alcohol. World Health Organization; Geneva: 2004. pp. 65–66. [Google Scholar]
- 2.Lim SS, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990-2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2224–2260. doi: 10.1016/S0140-6736(12)61766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gelernter J, Kranzler HR. Genetics of alcohol dependence. Hum Genet. 2009;126(1):91–99. doi: 10.1007/s00439-009-0701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dick DM, et al. Endophenotypes successfully lead to gene identification: Results from the collaborative study on the genetics of alcoholism. Behav Genet. 2006;36(1):112–126. doi: 10.1007/s10519-005-9001-3. [DOI] [PubMed] [Google Scholar]
- 5.Schumann G, et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci USA. 2011;108(17):7119–7124. doi: 10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joslyn G, et al. Glypican gene GPC5 participates in the behavioral response to ethanol: Evidence from humans, mice, and fruit flies. G3 (Bethesda) 2011;1(7):627–635. doi: 10.1534/g3.111.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodan AR, Rothenfluh A. The genetics of behavioral alcohol responses in Drosophila. Int Rev Neurobiol. 2010;91:25–51. doi: 10.1016/S0074-7742(10)91002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaun KR, Devineni AV, Heberlein U. Drosophila melanogaster as a model to study drug addiction. Hum Genet. 2012;131(6):959–975. doi: 10.1007/s00439-012-1146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H-G, Kim Y-C, Dunning JS, Han K-A. Recurring ethanol exposure induces disinhibited courtship in Drosophila. PLoS ONE. 2008;3(1):e1391. doi: 10.1371/journal.pone.0001391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolf FW, Rodan AR, Tsai LT-Y, Heberlein U. High-resolution analysis of ethanol-induced locomotor stimulation in Drosophila. J Neurosci. 2002;22(24):11035–11044. doi: 10.1523/JNEUROSCI.22-24-11035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ja WW, et al. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci USA. 2007;104(20):8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devineni AV, Heberlein U. Preferential ethanol consumption in Drosophila models features of addiction. Curr Biol. 2009;19(24):2126–2132. doi: 10.1016/j.cub.2009.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothenfluh A, Cowan CW. Emerging roles of actin cytoskeleton regulating enzymes in drug addiction: Actin or reactin’? Curr Opin Neurobiol. 2013;23(4):507–512. doi: 10.1016/j.conb.2013.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Offenhäuser N, et al. Increased ethanol resistance and consumption in Eps8 knockout mice correlates with altered actin dynamics. Cell. 2006;127(1):213–226. doi: 10.1016/j.cell.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Eddison M, et al. arouser reveals a role for synapse number in the regulation of ethanol sensitivity. Neuron. 2011;70(5):979–990. doi: 10.1016/j.neuron.2011.03.030. [DOI] [PubMed] [Google Scholar]
- 16.Rothenfluh A, et al. Distinct behavioral responses to ethanol are regulated by alternate RhoGAP18B isoforms. Cell. 2006;127(1):199–211. doi: 10.1016/j.cell.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 17.Kadrmas JL, et al. The integrin effector PINCH regulates JNK activity and epithelial migration in concert with Ras suppressor 1. J Cell Biol. 2004;167(6):1019–1024. doi: 10.1083/jcb.200408090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 19.Yang C-H, et al. Control of the postmating behavioral switch in Drosophila females by internal sensory neurons. Neuron. 2009;61(4):519–526. doi: 10.1016/j.neuron.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302(5651):1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 21.Bhandari P, Kendler KS, Bettinger JC, Davies AG, Grotewiel M. An assay for evoked locomotor behavior in Drosophila reveals a role for integrins in ethanol sensitivity and rapid ethanol tolerance. Alcohol Clin Exp Res. 2009;33(10):1794–1805. doi: 10.1111/j.1530-0277.2009.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corl AB, et al. Happyhour, a Ste20 family kinase, implicates EGFR signaling in ethanol-induced behaviors. Cell. 2009;137(5):949–960. doi: 10.1016/j.cell.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 23.Kapfhamer D, et al. JNK pathway activation is controlled by Tao/TAOK3 to modulate ethanol sensitivity. PLoS ONE. 2012;7(12):e50594. doi: 10.1371/journal.pone.0050594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dougherty GW, Jose C, Gimona M, Cutler ML. The Rsu-1-PINCH1-ILK complex is regulated by Ras activation in tumor cells. Eur J Cell Biol. 2008;87(8-9):721–734. doi: 10.1016/j.ejcb.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pitman JL, et al. There are many ways to train a fly. Fly (Austin) 2009;3(1):3–9. doi: 10.4161/fly.3.1.7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaun KR, Azanchi R, Maung Z, Hirsh J, Heberlein U. A Drosophila model for alcohol reward. Nat Neurosci. 2011;14(5):612–619. doi: 10.1038/nn.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S. Sequential use of mushroom body neuron subsets during drosophila odor memory processing. Neuron. 2007;53(1):103–115. doi: 10.1016/j.neuron.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao Z, Roman G, Zong L, Davis RL. Pharmacogenetic rescue in time and space of the rutabaga memory impairment by using Gene-Switch. Proc Natl Acad Sci USA. 2004;101(1):198–203. doi: 10.1073/pnas.0306128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stacey D, et al. IMAGEN Consortium RASGRF2 regulates alcohol-induced reinforcement by influencing mesolimbic dopamine neuron activity and dopamine release. Proc Natl Acad Sci USA. 2012;109(51):21128–21133. doi: 10.1073/pnas.1211844110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nees F, et al. IMAGEN Consortium Determinants of early alcohol use in healthy adolescents: The differential contribution of neuroimaging and psychological factors. Neuropsychopharmacology. 2012;37(4):986–995. doi: 10.1038/npp.2011.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrews MM, et al. Individuals family history positive for alcoholism show functional magnetic resonance imaging differences in reward sensitivity that are related to impulsivity factors. Biol Psychiatry. 2011;69(7):675–683. doi: 10.1016/j.biopsych.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beck A, et al. Ventral striatal activation during reward anticipation correlates with impulsivity in alcoholics. Biol Psychiatry. 2009;66(8):734–742. doi: 10.1016/j.biopsych.2009.04.035. [DOI] [PubMed] [Google Scholar]
- 33.Villafuerte S, et al. Impulsiveness and insula activation during reward anticipation are associated with genetic variants in GABRA2 in a family sample enriched for alcoholism. Mol Psychiatry. 2012;17(5):511–519. doi: 10.1038/mp.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wrase J, et al. Dysfunction of reward processing correlates with alcohol craving in detoxified alcoholics. Neuroimage. 2007;35(2):787–794. doi: 10.1016/j.neuroimage.2006.11.043. [DOI] [PubMed] [Google Scholar]
- 35.Yau W-YW, et al. Nucleus accumbens response to incentive stimuli anticipation in children of alcoholics: Relationships with precursive behavioral risk and lifetime alcohol use. J Neurosci. 2012;32(7):2544–2551. doi: 10.1523/JNEUROSCI.1390-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knutson B, Westdorp A, Kaiser E, Hommer D. FMRI visualization of brain activity during a monetary incentive delay task. Neuroimage. 2000;12(1):20–27. doi: 10.1006/nimg.2000.0593. [DOI] [PubMed] [Google Scholar]
- 37.Der-Avakian A, Markou A. The neurobiology of anhedonia and other reward-related deficits. Trends Neurosci. 2012;35(1):68–77. doi: 10.1016/j.tins.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bach FR, Jordan MI. Kernel independent component analysis. J Mach Learn Res. 2002;3(1):1–48. [Google Scholar]
- 39.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bierut LJ, et al. Gene, Environment Association Studies Consortium A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA. 2010;107(11):5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones PB, Rantakallio P, Hartikainen AL, Isohanni M, Sipila P. Schizophrenia as a long-term outcome of pregnancy, delivery, and perinatal complications: A 28-year follow-up of the 1966 north Finland general population birth cohort. Am J Psychiatry. 1998;155(3):355–364. doi: 10.1176/ajp.155.3.355. [DOI] [PubMed] [Google Scholar]
- 42.Torregrossa MM, Corlett PR, Taylor JR. Aberrant learning and memory in addiction. Neurobiol Learn Mem. 2011;96(4):609–623. doi: 10.1016/j.nlm.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cingolani LA, Goda Y. Actin in action: The interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev Neurosci. 2008;9(5):344–356. doi: 10.1038/nrn2373. [DOI] [PubMed] [Google Scholar]
- 44.Dietz DM, et al. Rac1 is essential in cocaine-induced structural plasticity of nucleus accumbens neurons. Nat Neurosci. 2012;15(6):891–896. doi: 10.1038/nn.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiraly DD, et al. Behavioral and morphological responses to cocaine require kalirin7. Biol Psychiatry. 2010;68(3):249–255. doi: 10.1016/j.biopsych.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schumann G, et al. IMAGEN consortium The IMAGEN study: Reinforcement-related behaviour in normal brain function and psychopathology. Mol Psychiatry. 2010;15(12):1128–1139. doi: 10.1038/mp.2010.4. [DOI] [PubMed] [Google Scholar]
- 47.Gil AG, Wagner EF, Tubman JG. Associations between early-adolescent substance use and subsequent young-adult substance use disorders and psychiatric disorders among a multiethnic male sample in South Florida. Am J Public Health. 2004;94(9):1603–1609. doi: 10.2105/ajph.94.9.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dougherty GW, Chopp T, Qi SM, Cutler ML. The Ras suppressor Rsu-1 binds to the LIM 5 domain of the adaptor protein PINCH1 and participates in adhesion-related functions. Exp Cell Res. 2005;306(1):168–179. doi: 10.1016/j.yexcr.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 49.Chunduru S, et al. Identification of an alternatively spliced RNA for the Ras suppressor RSU-1 in human gliomas. J Neurooncol. 2002;60(3):201–211. doi: 10.1023/a:1021130620178. [DOI] [PubMed] [Google Scholar]
- 50.Kramár EA, Lin B, Rex CS, Gall CM, Lynch G. Integrin-driven actin polymerization consolidates long-term potentiation. Proc Natl Acad Sci USA. 2006;103(14):5579–5584. doi: 10.1073/pnas.0601354103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peru y Colón de Portugal RL, et al. Adult neuronal Arf6 controls ethanol-induced behavior with Arfaptin downstream of Rac1 and RhoGAP18B. J Neurosci. 2012;32(49):17706–17713. doi: 10.1523/JNEUROSCI.1944-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.