Significance
A small number of mutations to the viral hemagglutinin are sufficient to permit aerosol transmission, in a ferret model of human infection, of highly pathogenic avian H5N1 influenza A viruses. Here, we show how an antibody (H5.3) against hemagglutinin 5 (H5) recognizes both WT and variant H5 proteins. H5.3 retains germ-line characteristics, most remarkably a conformationally flexible combining site, consistent with an antibody that has not been through multiple cycles of affinity maturation. Many antibodies against H5 are lightly mutated and may arise from naive B cells, explaining the low antigenicity of H5N1 vaccines relative to seasonal influenza vaccines and supporting the idea that multiple exposures are necessary to develop a strong immune response to H5N1 strains.
Keywords: structural biology, immunity, antigen recognition, affinity maturation, influenza
Abstract
Antigenic drift of circulating seasonal influenza viruses necessitates an international vaccine effort to reduce the impact on human health. A critical feature of the seasonal vaccine is that it stimulates an already primed immune system to diversify memory B cells to recognize closely related, but antigenically distinct, influenza glycoproteins (hemagglutinins). Influenza pandemics arise when hemagglutinins to which no preexisting adaptive immunity exists acquire the capacity to infect humans. Hemagglutinin 5 is one subtype to which little preexisting immunity exists and is only a few acquired mutations away from the ability to transmit efficiently between ferrets, and possibly humans. Here, we describe the structure and molecular mechanism of neutralization by H5.3, a vaccine-elicited antibody that neutralizes hemagglutinin 5 viruses and variants with expanded host range. H5.3 binds in the receptor-binding site, forming contacts that recapitulate many of the sialic acid interactions, as well as multiple peripheral interactions, yet is not sensitive to mutations that alter sialic acid binding. H5.3 is highly specific for a subset of H5 strains, and this specificity arises from interactions to the periphery of the receptor-binding site. H5.3 is also extremely potent, despite retaining germ line-like conformational flexibility.
Influenza remains a major public health concern because seasonal influenza infects 600 million to 1.1 billion people annually, resulting in 3–5 million cases of severe disease, and 250,000–500,000 deaths (1). By comparison, the four influenza pandemics of the 20th century, caused by novel influenza strains infecting the immunologically naive human population, resulted in 50–100 million deaths (1–4). Influenza A immunity is principally mediated by the antibody response to the viral glycoprotein, hemagglutinin (HA) (5). HA is expressed as a preprotein, HA0, assembled as a trimer on the viral envelope, and cleaved by host proteases into HA1 and HA2. HA1 is a largely globular domain responsible for receptor binding, and HA2 is a rod-shaped helical bundle responsible for membrane fusion (Fig. 1A) (5). There are 18 genetically distinct subtypes of influenza A HA (H1–H18), of which only H1 and H3 currently circulate among humans (1, 6–9).
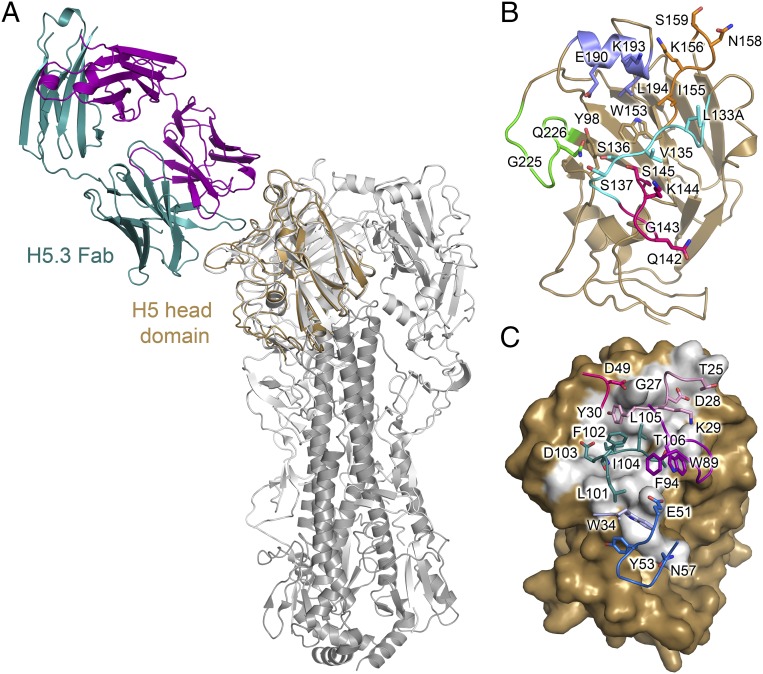
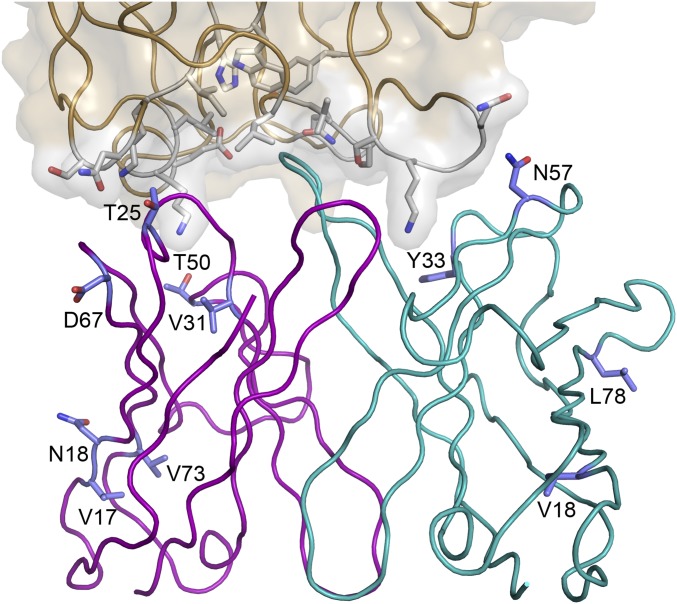
Fig. 1.
Human monoclonal antibody H5.3 recognizes the H5 receptor-binding site. (A) H5.3-wt_H5hd complex overlaid on the VN/1203 H5 trimer (PDB ID code 2FK0) showing H5hd in gold, the H5.3 light chain in purple, the H5.3 heavy chain in teal, and the H5 trimer (2FK0) in gray. (B) A cartoon diagram of H5hd showing HA residues contacted by H5.3 as sticks. The structural elements of the RBS are highlighted: the 130 loop is cyan, the 140 loop is pink, the 150 loop is orange, the 190 helix is blue, and the 220 loop is green. Trp153 forms the base and denotes the approximate center of the receptor-binding site. (C) A surface representation of H5hd in the same orientation as in B, with the solvent inaccessible interface shown in gray. H5.3 contact residues are labeled and shown as sticks and colored by CDR, with CDRH1 in light blue, CDRH2 in blue, CDRH3 in teal, CDRL1 in light pink, CDRL2 in dark pink, and CDRL3 in purple.
Despite the widespread presence of H5N1 influenza viruses in wild birds, the virus is not currently transmissible within the human population. Human-to-human transmission is inefficient and is partially restricted by the receptor specificity of the virus; human-type HAs preferentially recognize α2,6-linked sialic acid whereas avian-type HAs prefer α2,3-linked sialic acid (1, 10–12). However, there have been >600 human cases of H5N1 infection since 2004, resulting from the direct transmission of the virus from birds to humans, associated with an ∼60% mortality rate. There is the potential for a significant pandemic if H5 viruses develop the ability to spread efficiently between humans, which would necessitate specificity for α2,6-linked sialic acid (1–4, 13).
Receptor binding occurs in a shallow depression on the HA globular head domain, the edges of which are formed by four structural elements, the 190 helix and the 130, 150, and 220 loops (Fig. 1B), and the receptor binding site (RBS) base, which includes invariant hydrophobic residues Tyr98, Trp153, and Leu194 (5, 14, 15). Receptor specificity is critically influenced by position 226 on HA; Gln226-containing H3 strains are specific for α2,3 sialic acid linkages, and Leu226-containing H3 strains are specific for α2,6 sialic acid linkages (5, 16). In H5 strains, Leu226 enhances binding to α2,6-linked sialic acid receptors, but H5 viruses isolated from humans contain mutations at other sites that also promote use of α2,6-linked sialic acid receptors (11, 17, 18). Recent influenza pandemics have been caused by the acquisition of mutations that change the receptor preference to α2,6 sialic acid linkages, and recent studies with multiply passaged laboratory strains indicated that only a small number of mutations are necessary to introduce preference for α2,6 linkages into H5 strains (1, 6–9, 18–23). These viruses, termed respiratory droplet transmissible (rdt), typically have three mutations in or near the receptor binding site on HA (21, 22).
The most frequent potent neutralizing antibody response to HA arises from antibodies that target the receptor binding site and prevent virus attachment (5). Recent studies indicate that among RBS-directed antibodies, broad neutralization (across multiple isolates within a subtype or across subtypes) is achieved by insertion of a single complementary determining region (CDR) into the RBS to inhibit receptor binding (8). These broadly neutralizing antibodies (bnAbs) target conserved amino acids within the RBS and simultaneously avoid polymorphic sites on the ridges of the RBS. BnAbs may be relatively rare in human repertoires, and, as a consequence, current seasonal vaccine efforts focus on developing or boosting strain-specific responses to three or four currently circulating (“seasonal”) variants (2). Such strategies do not directly address the threat posed by noncirculating viruses with pandemic potential, such as H5 strains that circulate widely in wild bird populations and sporadically infect humans where they acquire mutations that enhance binding to human receptors (17). Instead, H5N1 vaccines against “prepandemic” strains have been developed commercially for future use in the case of a pandemic, illustrating that a prepandemic immunization program is feasible (24, 25).
The immune response against H5N1 vaccines in healthy adults is less robust than for most seasonal influenza strains, typically resulting in a response restricted to the strain used in the vaccine and to closely related variants (26–29). Notwithstanding this observation, we recently described a panel of human anti-H5 antibodies induced in response to vaccination of volunteers with an experimental H5N1 subunit vaccine (30) and here describe the structure and characterization of a human monoclonal antibody, H5.3, bound to A/Vietnam/1203/2004 (VN/1203) H5 and to two H5 rdt variants. H5.3 is an RBS-directed antibody that recapitulates many of the electrostatic interactions of the natural receptor, sialic acid, as well as forming additional interactions to the periphery of the RBS that provide specificity. H5.3 is potent and specific despite containing only 11 mutations from its unmutated common ancestor (UCA) and maintaining the structural flexibility typically associated with unmutated antibodies, as evidenced by significant rearrangement of CDRH3 and CDRL3. The structures determined here offer a chemical explanation for the evident trade-off between breadth and potency, and the germ-line characteristics highlight the role of lightly mutated antibodies in neutralization of new viral strains.
Results
Structure of H5.3:H5hd Complexes.
We determined the crystal structure of a human monoclonal antibody Fab, H5.3, in complex with VN/1203 H5 head domain (H5hd, PDB ID code 4XNM) to 2.5 Å (Fig. 1). We also determined the structures of H5.3 in complex with H5hd containing the rdt mutations from the Kawaoka (H5hd_rdt_Vn, PDB ID code 4XNQ) and Fouchier (H5hd_rdt_In, PDB ID code 4XRC) laboratory strains to 2.15 Å and 2.74 Å, respectively (18, 23). Refinement and data quality statistics are given in Table S1. In all cases, the asymmetric unit (ASU) contained two copies of the H5.3–H5hd complex (rmsd of 0.51, 0.52, and 0.63 Å between main chain atoms of the H5hd and antibody variable regions for the two copies of wt, rdt_Vn, and rdt_In, respectively). Both copies of the H5.3 Fab in the ASU were well-ordered in all crystals, except for residues 138–145 in the heavy chain constant region. The H5hd, which was expressed in bacteria, was well-resolved throughout the structure, except for one loop: residues 78–90 in chain C of the wt_H5hd, 79–82A in chain C of H5hd_rdt_In, and 77–82 in chain C of H5hd_rdt_Vn. With the exception of the poorly ordered loop, the rmsd between main chain atoms of H5hd and the trimeric ectodomain (PDB file 2FK0) (31) was 0.76 Å. An additional loop, described below, is disordered in the H5hd_rdt_Vn structure.
Table S1.
Data collection, phasing, and refinement statistics
| H5.3-H5hd | H5.3-H5hd-Vietnam | H5.3-H5hd-Indonesia | |
| Data collection | |||
| Space group | C2 | C2 | C2 |
| Cell dimensions | |||
| a, b, c, Å | 250.9, 51.4, 127.2 | 250.4, 51.2, 127.7 | 248.4, 51.1, 127.6 |
| α, β, γ, ° | 90.0, 99.8, 90.0 | 90.0, 100.2, 90.0 | 90.0, 99.9, 90.0 |
| Wavelength | 0.97856 | 1.00394 | 0. 97856 |
| Resolution, Å* | 2.5 (2.59–2.51) | 2.15 (2.19–2.15) | 2.74 (2.81–2.74) |
| Rsym*† | 10.9% (66%) | 4.8 (60.0) | 8.5 (59.8) |
| I/σI* | 14.2 (1.7) | 26.2 (2.4) | 14.5 (2.2) |
| Completeness, %* | 97.6 (94.3) | 96.9 (91.9) | 99.8 (99.8) |
| Redundancy* | 4.9 (3.7) | 4.0(3.9) | 5.1 (4.5) |
| Refinement | |||
| Resolution, Å* | 50–2.51 (2.57–2.51) | 50–2.0 (2.05–2.0) | 50–2.74 (2.81–2.74) |
| No. of reflections* | 53,874 (3,221) | 97,220 (3,778) | 42,101 (2,847) |
| Rwork/Rfreeठ| 21.4/24.6 | 19.7/21.1 | 19.7/25.1 |
| No. of atoms | |||
| Protein | 9,644 | 9,614 | 9,704 |
| Ligand/glycans | 38 | 38 | 38 |
| Water | 85 | 415 | 102 |
| B-factors, Å2 | |||
| Protein | 85.8 | 59.7 | 69.2 |
| Ligand/glycans | 110.5 | 87.0 | 107.4 |
| Water | 64.9 | 54.0 | 51.3 |
| rms deviations | |||
| Bond lengths, Å | 0.003 | 0.003 | 0.002 |
| Bond angles, ° | 0.7 | 0.8 | 0.8 |
| Ramachandran, % | |||
| Outliers | 0.08 | 0.08 | 0.16 |
| Allowed | 2.27 | 1.9 | 1.9 |
| Favored | 97.7 | 98.1 | 97.9 |
Parentheses indicate outer-shell statistics.
Rsym = ∑hkl∑i|Ihkl,i −〈Ihkl|/∑hk∑iIhkl,i. Ihkl,i is the scaled intensity of the ith measurement of reflection h, k, l.〈Ihkl〉is the average intensity for reflection I, and N is the number of observations of I.
Rcryst = ∑hkl|Fo − Fc|/∑hkl|Fo|, where Fo and Fc are the observed and calculated structure factors.
Rfree was calculated as for Rcryst, but using 5% of data, randomly chosen and excluded from refinement.
H5.3 forms an extensive interface with H5hd, using all six CDR loops and burying ∼808 Å2 on H5hd and ∼795 Å2 on H5.3 (Fig. 1 B and C). The interface is roughly centered on the H5 RBS. CDRH3 inserts into the highly conserved RBS, and CDRs L1, L2, L3, H1, and H2 contact variable residues on the periphery of the RBS, including the 130 loop, 150 loop, 220 loop, and 190 helix. CDRs L1 and L2 interact with the 190 helix on the rim of the RBS; and CDRs H1 and H2 interact with the 140 loop of H5hd (Fig. 1C). The interface (∼800 Å2 per partner) is slightly smaller than a typical protein–protein interface, yet H5.3 is, to our knowledge, the most potent human antibody described for H5N1 strains, with a viral neutralization IC50 of 0.02 µg/mL and a Kd of 5 nM (for recombinant H5.3 Fab binding to recombinant HA from strain VN/1203) (Fig. S1) (30, 32). CDRH3 is critical for this interaction, and mutations to CDRH3 abolish binding (Fig. S1). As described in the section Recognition of H5 RDT Variants, the HA rdt mutations are largely outside the contact interface.
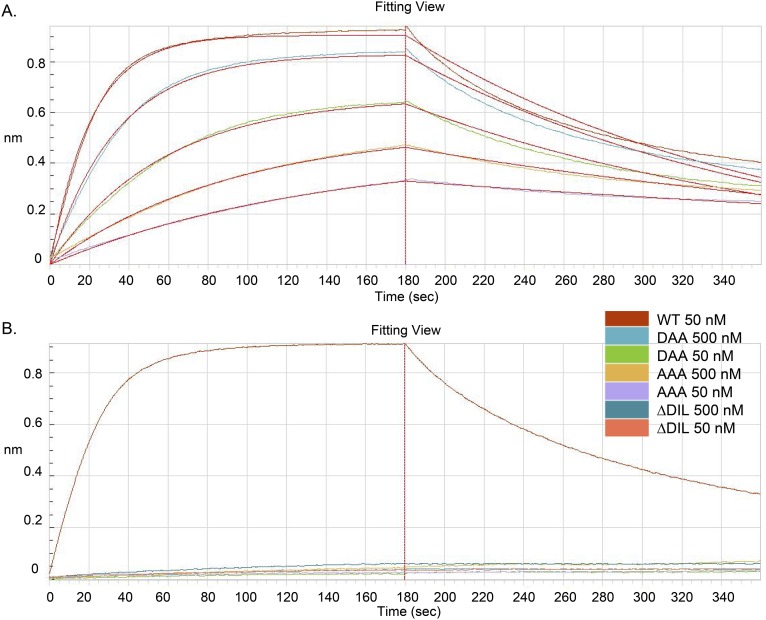
Fig. S1.
H5.3-H5hd affinities. Measured on an Octet interferometry instrument. (A) These data were fit with a 1:1 binding mode, yielding apparent Kd = 8 nM. The Fab concentrations tested in this experiment were 100 nM, 50 nM, 25 nM, 12.5 nm, 7.5 nM, and 3.6 nM. (B) The WT Fab concentration tested in this experiment was 50 nM. Mutant Fabs were tested at 500 nM and 50 nM. The affinity of all mutant Fabs was bellow the limit of detection. Mutants were made in the tip of CDRH3 (D103, I104, L105). The WT residues D103/I104/L105 (DIL) insert into the receptor binding site and are essential for binding.
Comparison with the Receptor and Other RBS-Directed Antibodies.
Because the H5.3 CDRH3 inserts into the RBS, we compared the H5.3–H5hd complexes with the structure of the avian receptor analog (α2,3-SLN; 3′-sialyl-N-acetyllactosamine) bound to A/Vietnam/1194/2004 H5 (PDB ID code 4BGY) (18) and to influenza HA-antibody complexes that similarly project CDRH3 into the RBS (PDB ID codes 3SM5, 4HKX, 4M5Z, 4O5I, 2VIR, and 1KEN) (33–38) (Fig. 2). H5.3 hydrogen bonding (H-bonding) interactions replace sialic acid hydrogen bonding in the H5.3–H5hd complex (Fig. 2 A and B), much as has been seen in other receptor mimetic antibodies, with the difference that H5.3 uses, exclusively, main chain H-binding residues.
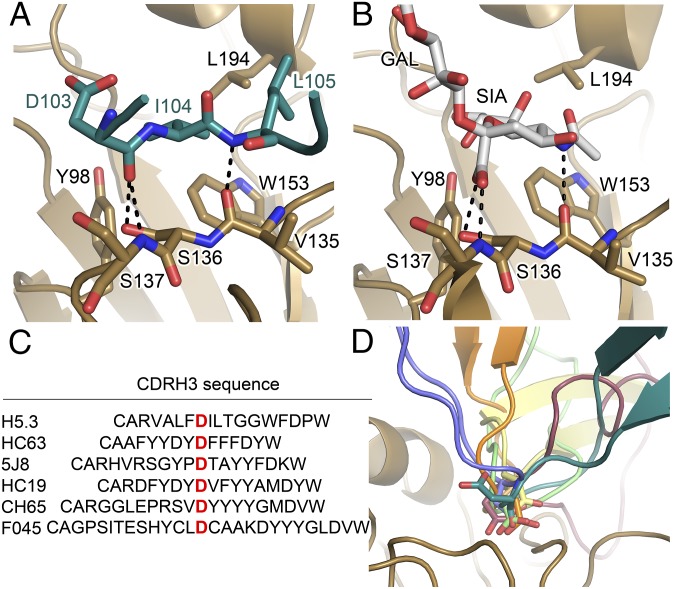
Fig. 2.
H5.3 binds to the HA receptor binding site. (A and B) The CDRH3 of H5.3 (teal) is inserted into the HA (gold) receptor binding site and binds in the same location as the α2,3 sialoglycan receptor analog (PDB ID code 4BGY). The backbone of H5.3 CDRH3 forms the same pattern of hydrogen bonds with H5 as does sialic acid. Hydrogen bonds are shown as dashed black lines. (C) Sequence alignment of HA receptor-binding site targeting antibodies, highlighting the Asp at the tip of CDRH3 that often mimics the carboxylate of sialic acid. F045-092 is abbreviated as F045. (D) The Asp at the tip of the H5.3 CDRH3 (teal) is oriented away from the RBS, unlike the Asp at the tips of CDRH3 in HC63 (blue, 1KEN), 5J8 (orange, 4M5Z), HC19 (gray, 2VIR), CH65 (3SM5, green), F045-092 (4O5I, burgundy), which are inserted into the receptor binding site, mimicking the carboxylate group of sialic acid.
In H5.3, the backbone amide of Leu105 accepts a hydrogen bond from the carbonyl oxygen of HA1 Val135. The carbonyl oxygen of H5.3 Asp103 donates hydrogen bonds to the side chain hydroxyl of Ser136 and the backbone amide nitrogen of Ser137 in HA1 (Fig. 2A). In the receptor analog, the carbonyl oxygen of Val135 accepts a hydrogen bond from the amide of the acetamide group of sialic acid, essentially the same as the interaction between H5 Val135 and H5.3 Leu105. The carboxylate group of sialic acid accepts hydrogen bonds from the side chain hydroxyl of Ser136 and the backbone amide of Ser137, similar to the interaction between these residues and the H5.3 Asp103 main chain carbonyl oxygen (Fig. 2). In addition to this direct readout of the receptor hydrogen-bonding network, Ile104 from H5.3 contacts the hydrophobic RBS floor formed by HA1 Tyr98, Trp153, and Leu194 (Fig. 2A).
H5.3 recapitulates the hydrogen-bonding scheme of the α2,3-SLN receptor using only main chain antibody features. In this regard, H5.3 differs substantially from other examples of CDRH3-based RBS-directed antibodies, which use an Asp side chain to form the contacts made by the carboxylic acid of the receptor. Notably, H5.3 possesses an Asp (Asp103) on the tip of CDRH3, but this functional group is pointed away from the RBS and does not form similar contacts to the carboxylic acid in sialic acid (Fig. 2D).
Recognition of H5 RDT Variants.
H5.3 has been shown to bind H5 rdt variants presented in the VN/1203 background as efficiently as wt VN/1203 (30). To understand how an RBS-directed antibody is able to accommodate both human and avian receptor binding sites, we determined the structures of H5.3 in complex with two H5 rdt variants. Each H5N1 rdt virus contains three mutations in or near the RBS: Asn158Asp, Asn224Lys, Gln226Leu (H5hd_rdt_Vn) in the VN/1203 H5 (21) and Thr160Ala, Gln226Leu, Gly228Ser (H5hd_rdt_In) in the A/Indonesia/05/2005 H5 (22). We introduced both sets of rdt mutations into the VN/1203 background and determined the structures of each H5 rdt variant in complex with H5.3 Fab to 2.15 Å and 2.74 Å, for H5.3-H5hd_rdt_Vn and H5.3-H5hd_rdt_In, respectively. Overall, both rdt complex structures align well with the H5.3–H5hd complex structure, and H5.3 binds the variants in the same orientation as it binds H5hd (Fig. 3).
Fig. 3.
Comparison of H5.3 interfaces in H5hd-rdt complexes. (A) Overlay of wt_H5hd (gold), H5hd_rdt_Vn (green), and H5hd_rdt_In (light pink). The 220 loop is highlighted in red in the wt_H5hd and H5hd_rdt_In structures; the 220 loop is not visible in the H5hd_rdt_Vn structure. The surface rendering of H5 is rotated 20° relative to A and highlights the solvent inaccessible interface of H5.3 on H5hd_rdt_Vn (B) and H5hd_rdt_In (C). The rdt residues in the H5.3 interface are highlighted in red. The rdt residue not in the interface is highlighted in blue (T160A in H5hd_rdt_In is not visible in this view of the structure). The CDR loops of H5.3 are shown as loops, with the residues involved in the interface highlighted as sticks, with the heavy chain in teal and the light chain in purple.
The most significant difference among these structures is that the 220 loop (residues 219–226 in one copy and 218–226 in the other copy in the crystallographic ASU) is disordered in H5hd_rdt_Vn (Fig. 3A). The 220 loop forms one rim of the receptor-binding site and contains two of the three rdt residues in H5hd_rdt_Vn (Asn224Lys and Gln226Leu). The loop is part of the H5.3-H5hd interface, as judged by solvent accessibility, although it makes only a single contact (a van der Waals contact in the H5.3-H5hd structure and a hydrogen bond in the H5.3-H5hd_rdt_In structure). The disorder is likely dependent on both the presence of the H5hd_rdt_Vn mutations and the loss of intersubunit contacts to the adjacent HA1 protomer (Fig. S2). Despite forming a rim of the RBS, and dictating receptor use, the 220 loop is not important for H5.3 binding.
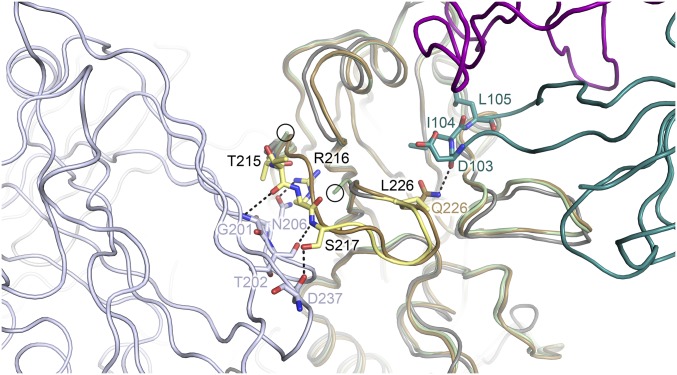
Fig. S2.
The loss of trimer contacts and the Gln226→Leu mutation contribute to destabilize the 220 loop in the H5.3-H5hd_rdt_Vn structure. In gray, with the 220 loop highlighted in yellow, is the trimeric VN/1203 rdt H5 ectodomain (PDB ID code 4BH2). Intersubunit H-bonds are shown as dotted lines between the 220 loop (yellow) and a neighboring protomer (blue-gray). These interactions are lost in monomeric head domain constructs. Additionally, the loss of the hydrogen bond between Gln226 and Asp103 likely further destabilizes the 220 loop. In the figure, the H5.3-H5hd_rdt_Vn structure (H5hd_rdt_Vn in light green, H5.3 heavy chain in teal, H5.3 light chain in purple) is aligned to both wt_H5hd (gold) and a protomer of the VN/1203 rdt trimeric H5 ectodomain (PDB ID code 4BH2) protomer (gray). Additional H5 ectodomain protomers are shown as blue-gray (front, left) and light gray (back, left). The last structured residues in the 220 loop from the H5.3-H5hd_rdt_Vn structure are circled.
Binding Determinants Outside the Receptor-Binding Site.
In addition to contacts between the H5.3 CDRH3 and H5, H5.3 forms important interactions with the 190 helix and 140 loop, elements that form the extreme edges of the RBS (Fig. 4). The 140 loop is recognized by residues from CDRs H1 and H2, and residues from CDRs L1 and L2 recognize the 190 helix. The 140 loop and 190 helix are sites of sequence divergence between H5 strains, and H5.3 forms highly sequence-specific interactions with two sites of polymorphism: Lys193 and Lys144. These sites are critical for dictating binding specificity of H5.3, which shows a strong preference for Lys at these positions (Fig. 4). The combination of CDRH3 inserting into the RBS and binding invariant residues on H5, and sequence-specific interactions with the variable periphery of the RBS, combine to produce an extremely potent and specific neutralizing antibody.
Fig. 4.
H5.3 forms critical interactions with polymorphic residues on the extreme edges of the interface. H5hd is shown in gold, the H5.3 light chain is shown in purple, and the H5.3 heavy chain is shown in teal. (A) Lys144 and Lys193 form two extreme edges of the H5.3-H5hd interface and are recognized exclusively by heavy chain and light chain residues, respectively. (B) Lys144 and Gly143 are recognized by Glu51 and Trp34 of the H5.3 heavy chain. (C) Lys193 makes three specific interactions with residues in the H5.3 light chain, two with the backbone carbonyls of Gly27 and Lys29 and a salt bridge to Asp49. (D) A sequence alignment of multiple H5N1 strains with ELISA EC50 values (taken from ref. 30) reveals the preference for Lys or Arg at 144 and the requirement for Lys at position 193. Strains shown in D are as follows: VN (A/Vietnam/1203/2004), rdt_Vn (VN/1203 N158D, N224K, Q226L), rdt_In (VN/1203 T160A, Q226L, G228A), Indo (A/Indonesia/05/2005), In rdt (Indo T160A, Q226L, G228S), Anhui (A/Anhui/1/2005), Egypt (A/Egypt/3300-NAMRU3/2008), BHG (A/bar-headed goose/Qinghai/1A/2005), HK 156 (A/Hong Kong/156/1997), and HK 213 (A/HK/213/2003).
Conformational Flexibility in H5.3 CDRs.
H5.3 was isolated from an H5N1 vaccine trial participant (∼3 mo after immunization), rather than a repeatedly infected or immunized volunteer, and is not highly mutated. H5.3 differs at 11 sites (4 in the heavy chain and 7 in the light chain) from its UCA. The low number of somatic mutations is consistent with the response expected after initial exposure to a new antigen and is representative of the antibodies we have isolated from an H5N1 vaccine trial. Six of the nine H5 antibodies we isolated and characterized from this trial were specific for only H5 (30). The H5-specific antibodies have significantly fewer somatic mutations than bnAbs (P = 0.008), which are reactive against strains to which most people have numerous exposures. The H5-specific antibodies have an average of 12.3 ± 5 mutations from the UCA compared with 21.6 ± 7 mutations for bnAbs (Table 1). Of the 11 mutations in H5.3, none contact H5hd (Fig. S3).
Table 1.
Somatic mutations in H5-specific Abs and bnAbs
| Antibody | Epitope* | No. of variable domain mutations | ||
| Heavy chain | Light chain | Total | ||
| H5-specific Abs | ||||
| H5.2 | Head | 10 | 8 | 18 |
| H5.3 | Head | 4 | 7 | 11 |
| H5.7 | Stem | 5 | 2 | 7 |
| H5.13 | Head | 4 | 2 | 6 |
| H5.16 | n.d. | 9 | 7 | 16 |
| H5.24 | n.d. | 6 | 10 | 16 |
| Avg | 12.3 | |||
| SD | 5.1 | |||
| bnAbs | ||||
| CH65 | Head | 12 | 6 | 18 |
| CH67 | Head | 10 | 9 | 19 |
| C05 | Head | 22 | 14 | 36 |
| CR6261 | Stem | 15 | 3 | 18 |
| CR8020 | Stem | 13 | 8 | 21 |
| FI6 | Stem | 14 | 9 | 23 |
| CR9114 | Stem | 17 | 12 | 29 |
| F045-092 | Head | 13 | 9 | 22 |
| H5.9 | n.d. | 16 | 11 | 27 |
| H5.31 | Head | 9 | 7 | 16 |
| H5.36 | n.d. | 2 | 7 | 9 |
| Avg | 21.6 | |||
| SD | 7.2 | |||
Fig. S3.
Affinity maturation mutations in H5.3 do not contact H5. There are 11 mutations in H5.3, shown in stick representation (4 in the heavy chain and 7 in the light chain), compared with its UCA, none of which are involved in the H5.3-H5 interface. H5.3 light chain is in purple, heavy chain is in teal, residues mutated from the H5.3 UCA are shown as blue sticks, H5hd is in gold, and H5 residues involved in the interface are shown as gray sticks.
The low number of somatic mutations in H5.3 results in an antibody that is not optimally configured for antigen binding. Comparison of the previously published structure of the H5.3 Fab alone (PDB ID code 4GSD) (30) with the conformation of H5.3 observed in the H5hd complexes reveals large conformational changes upon binding (Fig. 5). Most critically, the tip of CDRH3 is rotated ∼90° compared with the unliganded structure. This reorientation of CDRH3 is required to position the H5.3 H-bond donors such that they recapitulate the H-bonding pattern of sialic acid. The liganded structure of CDRH3 would clash with the unliganded structure of CDRL3, causing CDRL3 to shift away from its unliganded position by ∼5 Å. This shift causes CDRL3 to pack against the Cʹ strand of the heavy chain. Although this reorganization shifts CDRL3 away from H5, CDRL1 and L2 shift ∼1 Å closer to H5.
Fig. 5.
H5.3 retains germ-line flexibility. (A) An overlay of the free and bound structures of H5.3 shows the conformational reorganization associated with antigen binding. The bound H5.3 heavy chain is shown in teal, the free heavy chain is shown in gray, the bound light chain is shown in purple, the free light chain is shown in light pink, the tip of CDRL3 of the free light chain is highlighted in red, and the head domain is shown in gold. (B) View from the receptor-binding site. The heavy chain CDR3 is rotated ∼90°, the light chain CDR1 is shifted ∼5 Å. The free CDRH3 is highlighted in yellow.
Discussion
H5N1 influenza A viruses are not currently transmissible between humans, in part due to the receptor specificity of the virus. However, when humans are infected through contact with infected birds, the mortality rate is ∼60%, which is unusually high for an influenza infection (13). In this study, we show that a highly potent human antibody against H5 that also recognizes rdt variants shares many structural features with other anti-HA antibodies. H5.3 is an RBS-directed antibody that inserts CDRH3 into the RBS and recapitulates the H-bonding pattern between HA and the sialic acid receptor. H5.3 is able to bind H5 rdt variants, indicating that mutations that alter receptor preference are not necessarily escape mutants for RBS targeted antibodies.
Unlike HA-specific bnAbs, H5.3 forms critical interactions with residues on the extreme edges of the HA head domain. These interactions are formed between H5.3 heavy chain residues and H5 Lys144, as well as H5.3 light chain residues and H5 Lys193. Lys144 and Lys193 are sites of diversity within H5 strains and are critical for the high potency and specificity of H5.3. The combination of RBS-directed CDRH3 and peripheral interactions to polymorphic residues enables H5.3 to tightly bind H5 rdt variants, and indicates that antibodies produced in response to an appropriate H5 vaccine challenge can reasonably be expected to neutralize human transmissible influenza strains.
H5.3 undergoes a conformational change upon binding to H5 that is, to our knowledge, unprecedented among affinity-matured antibodies (Fig. 5). The critical binding determinants on CDRH3 rotate ∼90° from their positions in the unliganded structure (Fig. 5). Additionally, CDRL3 moves ∼5 Å to avoid steric clashes with H5hd. The rotation of CDRH3 emphasizes the difficulty of predicting binding mode, even within a family of structurally similar proteins. Conformational flexibility is typically a characteristic of unmutated, rather than affinity-matured, antibodies, yet the lack of a preformed combining site in H5.3 and large conformational change associated with binding indicate that H5.3 has retained some of the flexibility present in germ-line gene-encoded antibodies (34, 39–41). H5.3 differs from its UCA by only 11 mutations and is therefore substantially less mutated than most influenza bnAbs (Table 1), and, of these mutations, none directly contact H5 (Fig. S3).
Among the structurally characterized antibodies that use CDRH3 to recapitulate the H-bonding pattern of sialic acid, all except H5.3 use an aspartic acid from CDRH3 to mimic the carboxylate group of sialic acid. In H5.3, a main chain carbonyl makes interactions with HA that are similar to those made by the carboxylate group of sialic acid, illustrating an additional way to recognize a conserved feature of the RBS.
The potency of H5.3 comes from binding energy derived from interactions between the RBS and CDRH3, as well from key interactions outside the RBS (to Lys144 and Lys193). In H5.3, potency comes at the expense of breadth because Lys144 and Lys193 are polymorphic sites in H5N1 strains. H5.3 was isolated from the blood of a healthy donor who had participated, ∼3 mo earlier, in a clinical trial of an experimental H5N1 vaccine (25, 30). The structural and sequence analysis presented here indicate that H5.3 is lightly mutated and flexible, consistent with an antibody that is not optimally matured. Of the nine human monoclonal antibodies we have isolated against H5, the six that are specific for only H5 have, on average, fewer mutations than bnAbs (Table 1). BnAbs have significantly more mutations and likely arise through stimulation of memory B cells. In general, the presence of reactive memory B cells correlates with the ability to produce a bnAb response (42–45). It seems that, for both influenza and HIV, bnAbs require significantly more somatic mutation than seems common in vaccine-induced antibodies in otherwise antigen-naive humans (46, 47), suggesting that bnAbs might be elicited by repeated immunization against multiple strains, rather than fortuitous gene use. H5.3 is illustrative of the lack of breadth typical of vaccine-induced antibodies, and our results indicate that breadth and potency directly conflict in H5.3 interactions. The H5.3 interactions with Lys144 and Lys193 prevent tight binding to variant H5 strains that do not contain these residues, yet H5.3 is extremely potent against VN/1203 (30), indicating it was likely strongly selected for, and expanded, in germinal centers during affinity maturation.
H5.3 was elicited by a VN/1203 H5N1 experimental vaccine and is representative of the type of antibodies an H5-naive person is likely to produce. Subsequent infection or immunization with a variant strain would likely be an effective route to generate broadly reactive anti-H5 antibodies. Such strategies are known to be effective within influenza subtypes (25, 48, 49). Our structural understanding of H5.3 supports the hypothesis that repeated challenge through immunization or infection is an effective strategy to enhance universal binding determinants, such as a CDRH3 loop that effectively binds conserved features of the RBS, similar to the natural receptor, while minimizing strain-specific interactions. Achieving a universal flu vaccine may require methods to develop and maintain a memory B-cell population that has already expanded multiple times against diverse antigens.
Methods
Expression and Purification of H5.3 Fab.
The H5.3 hybridoma cell line was generated as previously reported (30), and additional details are given in SI Methods.
Expression and Purification of HA.
A/Vietnam/1203/2004 H5 head domain (HA1 residues 57–264) was cloned into the pET28a vector, containing an N-terminal 6x-His tag and a thrombin cleavage site. The H5 rdt variant HA molecules were made in pET28a using PCR-based mutagenesis and were verified by nucleotide sequencing. These proteins were expressed and purified as described in SI Methods.
Crystallization.
Experimental details are given in SI Methods. H5.3-H5hd crystals were grown from a reservoir containing 100 mM Hepes, pH 7.5, 20% PEG 10,000. H5.3-H5hd_rdt_In crystals were grown from 200 mM ammonium sulfate, 100 mM Hepes, pH 7.5, 25% PEG 3350. H5.3-H5hd_rdt_Vn crystals were grown from 200 mM sodium malonate, pH 7, 20% PEG 3350.
Data Collection and Structure Determination.
Diffraction data were collected from single crystals at 100 K at the Life Sciences Collaborative Access Team (LS-CAT) Sector 21 at the Advance Photon Source (Argonne, IL). Details describing the data collection and structure determination are given in SI Methods and in Table S1. The structures have been deposited in the Protein Data Bank under accession codes 4XNM (H5.3-H5hd), 4XNQ (H5.3-H5hd_rdt_Vn), and 4XRC (H5.3-H5hd_rdt_In).
SI Methods
Expression and Purification of H5.3 Fab.
The H5.3 hybridoma cell line was generated as previously reported (30). Briefly, the H5.3 expressing hybridoma cell line was grown in serum-free medium (Life Tech), and H5.3 IgG was purified from the supernatant using Protein G resin. Purified full-length antibody was digested with papain, and H5.3 Fab fragments were purified using Protein A resin. The Fab was concentrated and stored at −80 °C until needed.
Expression and Purification of HA.
The proteins were expressed in BL21 (DE3) cells grown in LB medium at 37 °C to an optical density (at 600 nm) of 0.6 and were induced with 0.1 mM isopropyl β-d-thiogalactopyranoside for 12 h at 20 °C. Bacteria were harvested by centrifugation and lysed with a French press in PBS with ∼1 µg/mL hen egg white lysozyme, ∼1 µg/mL DNase I, 10 µg/mL leupeptin, 1 mM PMSF, and 0.7 µg/mL pepstatin. The lysate was clarified by centrifugation, and the insoluble fraction was resuspended in 8 M urea, 50 mM Tris, pH 8, and purified by Co-NTA affinity using TALON resin. The protein was refolded overnight by rapid dilution (10 mL into 200 mL) into 1 M arginine, 10 mM Tris, pH 7.5, 0.5 mM reduced glutathione, 0.05 mM oxidized glutathione, followed by dialysis against 150 mM NaCl, 10 mM Tris, pH 7.5, for 24 h, and centrifugal concentration. The 6x-His tag was removed using thrombin, followed by gel filtration on a 24-mL Superdex 200 (GE Healthcare) column. Biotinylated H5hd was prepared the same as H5hd, with the following modifications: a BirA tag (GGGLNDIFEAQKIEQHE) was introduced at the carboxyl terminus by PCR, and the purified, refolded protein was labeled with Escherichia coli biotin ligase following standard protocols (50).
Crystallization.
The H5.3 Fab was mixed with excess H5_hd, and the complex was isolated by gel filtration on a Superdex 200 column maintained in 150 mM NaCl, 10 mM Tris, pH 7.5. The complex was concentrated to ∼10 mg/mL and immediately used for crystallization trials in sitting drop vapor diffusion trays. H5.3-H5hd crystals were grown from a reservoir containing 100 mM Hepes, pH 7.5, 20% (wt/vol) PEG 10,000. H5.3-H5hd_rdt_In crystals were grown from 200 mM ammonium sulfate, 100 mM Hepes, pH 7.5, 25% (wt/vol) PEG 3350. H5.3-H5hd_rdt_Vn crystals were grown from 200 mM sodium malonate, pH 7, 20% (wt/vol) PEG 3350. After 3–10 d, crystals were cryoprotected with a solution containing the reservoir solution with 2% (wt/vol) additional PEG and 15% (wt/vol) glycerol, harvested onto nylon loops, and flash cooled in liquid nitrogen.
Data Collection and Structure Determination.
Diffraction data were collected from single crystals at 100 K at LS-CAT Sector 21 at the Advance Photon Source (Argonne, IL). Data were indexed, integrated, and scaled with either HKL2000 (H5.3-H5hd and H5.3-H5hd_rdt_Vn) or XDS and Scala as called from xia2 (H5.3-H5hd_rdt_In) (51–56). Data collection statistics are given in Table S1. Molecular replacement was performed with Molrep (57) by iteratively searching a library of Fab fragments. A solution, obtained with PDB ID code 1IGI, was used in Phaser (58) to locate the head domain of 2FK0. The variable and constant domains of H5.3 (PDB ID code 4GSD) were aligned individually to the molecular replacement solution, and residues that differed between the recombinant H5.3 structure (the initial structure of H5.3 was determined using a recombinant protein) and the hybridoma-derived H5.3 were mutated to alanine, and the resulting model was refined in Phenix (59). The sequence of H5.3 variable regions has been described (30). Germ-line gene-encoded antibody constant region sequences IGLC1*02 and IGHG1*03 were used for the light or heavy chain constant regions, respectively. An additional two residues, observed at the amino terminus of the light chain in all structures, were modeled as Y and E. Refinement included rigid body refinement of the individual domains, simulated annealing, positional refinement, individual B-factor refinement, and translation/libration/screw (TLS) refinement. Loops were rebuilt and side chains added in COOT (60) using simulated annealing composite omit maps (61) generated by Phenix (59). TLS refinement (62) was incorporated in the final rounds of refinement using TLS groups identified using Phenix. N-linked glycans were placed in the final rounds of refinement. Data and model statistics are given in Table 1. The structures have been deposited in the Protein Data Bank under accession codes 4XNM (H5.3-H5hd), 4XNQ (H5.3-H5hd_rdt_Vn), and 4XRC (H5.3-H5hd_rdt_Vn).
Biosensor Assays.
Affinity measurements were performed using Fab fragments and biotinylated H5hd on an Octet interferometry instrument (ForteBio) with streptavidin biosensors (Fortebio 18-5019). HA and Fab were diluted in 1× kinetics buffer (Forte bio, 18-5032) with PBS. Fabs were diluted to 100 nM, 50 nM, 25 nM, 12.5 nm, 7.5 nM, and 3.6 nM. Tips were equilibrated in 1× kinetics buffer for 60 s, HA was immobilized on biosensors for 120 s, baselines were established in 1× kinetics buffer for 60 s, and dilutions of H5.3 Fab were used for 180-s association curves, and 180-s 1× kinetic buffer dissociation curves. The data were analyzed with ForteBio data analysis software version 7.1. Briefly, the y axis was aligned to baseline, and data were processed with Savitzky–Galay filtering. Affinities were calculated by doing curve fits of association and dissociations with a 1:1 binding model and local fitting of the full curves. All R2 were greater than 0.98 and X2 were less than 0.25.
Acknowledgments
We thank Frances Smith-House and David Blum for technical support. This work was supported by NIH Grant R01 AI106002, NIH Grant R21 AI092268NIH, contract HHSN272200900047C, and Defense Threat Reduction Agency Grant HDTRA1-10-1-0067. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. Use of the Life Sciences Collaborative Access Team at the Advanced Photon Source was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817). K.L.W. was supported by a Molecular Biophysics training grant at Vanderbilt (NIH Grant T32 GM008320). A portion of the work described here used the Vanderbilt robotic crystallization facility, which was supported by NIH Grant RR026915.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4XNM, 4XNQ, and 4XRC).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502762112/-/DCSupplemental.
References
- 1.WHO . Influenza (Seasonal): Fact Sheet 211. World Health Organization; Geneva: 2014. [Google Scholar]
- 2.Lambert LC, Fauci AS. Influenza vaccines for the future. N Engl J Med. 2010;363(21):2036–2044. doi: 10.1056/NEJMra1002842. [DOI] [PubMed] [Google Scholar]
- 3.Garten RJ, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325(5937):197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kilbourne ED. Influenza pandemics of the 20th century. Emerg Infect Dis. 2006;12(1):9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 6.Knossow M, Skehel JJ. Variation and infectivity neutralization in influenza. Immunology. 2006;119(1):1–7. doi: 10.1111/j.1365-2567.2006.02421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nobusawa E, et al. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182(2):475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 8.Laursen NS, Wilson IA. Broadly neutralizing antibodies against influenza viruses. Antiviral Res. 2013;98(3):476–483. doi: 10.1016/j.antiviral.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong S, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9(10):e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shinya K, et al. Avian flu: Influenza virus receptors in the human airway. Nature. 2006;440(7083):435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 11.Yamada S, et al. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature. 2006;444(7117):378–382. doi: 10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- 12.Couceiro JN, Paulson JC, Baum LG. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium: The role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993;29(2):155–165. doi: 10.1016/0168-1702(93)90056-s. [DOI] [PubMed] [Google Scholar]
- 13.WHO . Influenza at the Human-Animal Interface. World Health Organization; Geneva: 2014. [Google Scholar]
- 14.Weis W, et al. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333(6172):426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 15.Xu R, et al. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol. 2013;20(3):363–370. doi: 10.1038/nsmb.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogers GN, et al. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304(5921):76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 17.Xiong X, et al. Enhanced human receptor binding by H5 haemagglutinins. Virology. 2014;456-457:179–187. doi: 10.1016/j.virol.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong X, et al. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature. 2013;497(7449):392–396. doi: 10.1038/nature12144. [DOI] [PubMed] [Google Scholar]
- 19.Matrosovich M, et al. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000;74(18):8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205(1):17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 21.Imai M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herfst S, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336(6088):1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, et al. An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level. Science. 2013;340(6139):1463–1467. doi: 10.1126/science.1236787. [DOI] [PubMed] [Google Scholar]
- 24.Treanor JJ. Expanding the options for confronting pandemic influenza. JAMA. 2014;312(14):1401–1402. doi: 10.1001/jama.2014.12558. [DOI] [PubMed] [Google Scholar]
- 25.Belshe RB, et al. National Institute of Allergy and Infectious Diseases–Funded Vaccine and Treatment Evaluation Units Immunogenicity of avian influenza A/Anhui/01/2005(H5N1) vaccine with MF59 adjuvant: a randomized clinical trial. JAMA. 2014;312(14):1420–1428. doi: 10.1001/jama.2014.12609. [DOI] [PubMed] [Google Scholar]
- 26.Nicholson KG, et al. Safety and antigenicity of non-adjuvanted and MF59-adjuvanted influenza A/Duck/Singapore/97 (H5N3) vaccine: A randomised trial of two potential vaccines against H5N1 influenza. Lancet. 2001;357(9272):1937–1943. doi: 10.1016/S0140-6736(00)05066-2. [DOI] [PubMed] [Google Scholar]
- 27.Treanor JJ, et al. Safety and immunogenicity of a recombinant hemagglutinin vaccine for H5 influenza in humans. Vaccine. 2001;19(13-14):1732–1737. doi: 10.1016/s0264-410x(00)00395-9. [DOI] [PubMed] [Google Scholar]
- 28.Treanor JJ, Campbell JD, Zangwill KM, Rowe T, Wolff M. Safety and immunogenicity of an inactivated subvirion influenza A (H5N1) vaccine. N Engl J Med. 2006;354(13):1343–1351. doi: 10.1056/NEJMoa055778. [DOI] [PubMed] [Google Scholar]
- 29.Belshe RB, et al. National Institute of Allergy and Infectious Diseases-Funded Vaccine and Treatment Evaluation Units Safety and immunogenicity of influenza A H5 subunit vaccines: Effect of vaccine schedule and antigenic variant. J Infect Dis. 2011;203(5):666–673. doi: 10.1093/infdis/jiq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thornburg NJ, et al. Human antibodies that neutralize respiratory droplet transmissible H5N1 influenza viruses. J Clin Invest. 2013;123(10):4405–4409. doi: 10.1172/JCI69377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stevens J, et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312(5772):404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 32.Jones S, Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci USA. 1996;93(1):13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whittle JRR, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci USA. 2011;108(34):14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt AG, et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci USA. 2013;110(1):264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong M, et al. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J Virol. 2013;87(22):12471–12480. doi: 10.1128/JVI.01388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee PS, et al. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun. 2014;5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bizebard T, et al. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature. 1995;376(6535):92–94. doi: 10.1038/376092a0. [DOI] [PubMed] [Google Scholar]
- 38.Barbey-Martin C, et al. An antibody that prevents the hemagglutinin low pH fusogenic transition. Virology. 2002;294(1):70–74. doi: 10.1006/viro.2001.1320. [DOI] [PubMed] [Google Scholar]
- 39.Wedemayer GJ, Patten PA, Wang LH, Schultz PG, Stevens RC. Structural insights into the evolution of an antibody combining site. Science. 1997;276(5319):1665–1669. doi: 10.1126/science.276.5319.1665. [DOI] [PubMed] [Google Scholar]
- 40.Thorpe IF, Brooks CL., 3rd Molecular evolution of affinity and flexibility in the immune system. Proc Natl Acad Sci USA. 2007;104(21):8821–8826. doi: 10.1073/pnas.0610064104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willis JR, Briney BS, DeLuca SL, Crowe JE, Jr, Meiler J. Human germline antibody gene segments encode polyspecific antibodies. PLOS Comput Biol. 2013;9(4):e1003045. doi: 10.1371/journal.pcbi.1003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dogan I, et al. Multiple layers of B cell memory with different effector functions. Nat Immunol. 2009;10(12):1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- 43.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41(4):529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bentebibel S-E, et al. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci Transl Med. 2013;5:176ra32. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pallikkuth S, et al. Impaired peripheral blood T-follicular helper cell function in HIV-infected nonresponders to the 2009 H1N1/09 vaccine. Blood. 2012;120(5):985–993. doi: 10.1182/blood-2011-12-396648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kepler TB, et al. Immunoglobulin gene insertions and deletions in the affinity maturation of HIV-1 broadly reactive neutralizing antibodies. Cell Host Microbe. 2014;16(3):304–313. doi: 10.1016/j.chom.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klein F, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153(1):126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suguitan AL, Jr, et al. Influenza H5 hemagglutinin DNA primes the antibody response elicited by the live attenuated influenza A/Vietnam/1203/2004 vaccine in ferrets. PLoS ONE. 2011;6(7):e21942. doi: 10.1371/journal.pone.0021942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mulligan MJ, et al. DMID 13-0032 H7N9 Vaccine Study Group Serological responses to an avian influenza A/H7N9 vaccine mixed at the point-of-use with MF59 adjuvant: a randomized clinical trial. JAMA. 2014;312(14):1409–1419. doi: 10.1001/jama.2014.12854. [DOI] [PubMed] [Google Scholar]
- 50.Howarth M, Ting AY. Imaging proteins in live mammalian cells with biotin ligase and monovalent streptavidin. Nat Protoc. 2008;3(3):534–545. doi: 10.1038/nprot.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276A:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 52.Winter G. xia2: An expert system for macromolecular crystallography data reduction. J Appl Cryst. 2010;43:186–190. [Google Scholar]
- 53.Sauter NK, Grosse-Kunstleve RW, Adams PD. Robust indexing for automatic data collection. J Appl Cryst. 2004;37(Pt 3):399–409. doi: 10.1107/S0021889804005874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 55.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 58.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 4):458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 59.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 60.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 61.Brünger AT, Adams PD, Rice LM. New applications of simulated annealing in X-ray crystallography and solution NMR. Structure. 1997;5(3):325–336. doi: 10.1016/s0969-2126(97)00190-1. [DOI] [PubMed] [Google Scholar]
- 62.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 4):439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]