Significance
The global population is aging, driving up age-related disease morbidity. Antiaging interventions are needed to reduce the burden of disease and protect population productivity. Young people are the most attractive targets for therapies to extend healthspan (because it is still possible to prevent disease in the young). However, there is skepticism about whether aging processes can be detected in young adults who do not yet have chronic diseases. Our findings indicate that aging processes can be quantified in people still young enough for prevention of age-related disease, opening a new door for antiaging therapies. The science of healthspan extension may be focused on the wrong end of the lifespan; rather than only studying old humans, geroscience should also study the young.
Keywords: biological aging, cognitive aging, aging, healthspan, geroscience
Abstract
Antiaging therapies show promise in model organism research. Translation to humans is needed to address the challenges of an aging global population. Interventions to slow human aging will need to be applied to still-young individuals. However, most human aging research examines older adults, many with chronic disease. As a result, little is known about aging in young humans. We studied aging in 954 young humans, the Dunedin Study birth cohort, tracking multiple biomarkers across three time points spanning their third and fourth decades of life. We developed and validated two methods by which aging can be measured in young adults, one cross-sectional and one longitudinal. Our longitudinal measure allows quantification of the pace of coordinated physiological deterioration across multiple organ systems (e.g., pulmonary, periodontal, cardiovascular, renal, hepatic, and immune function). We applied these methods to assess biological aging in young humans who had not yet developed age-related diseases. Young individuals of the same chronological age varied in their “biological aging” (declining integrity of multiple organ systems). Already, before midlife, individuals who were aging more rapidly were less physically able, showed cognitive decline and brain aging, self-reported worse health, and looked older. Measured biological aging in young adults can be used to identify causes of aging and evaluate rejuvenation therapies.
By 2050, the world population aged 80 y and above will more than triple, approaching 400 million individuals (1, 2). As the population ages, the global burden of disease and disability is rising (3). From the fifth decade of life, advancing age is associated with an exponential increase in burden from many different chronic conditions (Fig. 1). The most effective means to reduce disease burden and control costs is to delay this progression by extending healthspan, years of life lived free of disease and disability (4). A key to extending healthspan is addressing the problem of aging itself (5–8).
Fig. 1.
Burden of chronic disease rises exponentially with age. To examine the association between age and disease burden, we accessed data from the Institute for Health Metrics and Evaluation Global Burden of Disease database (www.healthdata.org/gbd) (43). Data graph (A) disability-adjusted life years (DALYs) and (B) deaths per 100,000 population by age. Bars, from bottom to top, reflect the burden of cardiovascular disease (navy), type-2 diabetes (light blue), stroke (lavender), chronic respiratory disease (red), and neurological disorders (purple).
At present, much research on aging is being carried out with animals and older humans. Paradoxically, these seemingly sensible strategies pose translational difficulties. The difficulty with studying aging in old humans is that many of them already have age-related diseases (9–11). Age-related changes to physiology accumulate from early life, affecting organ systems years before disease diagnosis (12–15). Thus, intervention to reverse or delay the march toward age-related diseases must be scheduled while people are still young (16). Early interventions to slow aging can be tested in model organisms (17, 18). The difficulty with these nonhuman models is that they do not typically capture the complex multifactorial risks and exposures that shape human aging. Moreover, whereas animals’ brief lives make it feasible to study animal aging in the laboratory, humans’ lives span many years. A solution is to study human aging in the first half of the life course, when individuals are starting to diverge in their aging trajectories, before most diseases (and regimens to manage them) become established. The main obstacle to studying aging before old age—and before the onset of age-related diseases—is the absence of methods to quantify the Pace of Aging in young humans.
We studied aging in a population-representative 1972–1973 birth cohort of 1,037 young adults followed from birth to age 38 y with 95% retention: the Dunedin Study (SI Appendix). When they were 38 y old, we examined their physiologies to test whether this young population would show evidence of individual variation in aging despite remaining free of age-related disease. We next tested the hypothesis that cohort members with “older” physiologies at age 38 had actually been aging faster than their same chronologically aged peers who retained “younger” physiologies; specifically, we tested whether indicators of the integrity of their cardiovascular, metabolic, and immune systems, their kidneys, livers, gums, and lungs, and their DNA had deteriorated more rapidly according to measurements taken repeatedly since a baseline 12 y earlier at age 26. We further tested whether, by midlife, young adults who were aging more rapidly already exhibited deficits in their physical functioning, showed signs of early cognitive decline, and looked older to independent observers.
Results
Are Young Adults Aging at Different Rates?
Measuring the aging process is controversial. Candidate biomarkers of aging are numerous, but findings are mixed (19–22). Multibiomarker algorithms have been suggested as a more reliable alternative to single-marker aging indicators (23–25). A promising algorithm is the 10-biomarker US National Health and Nutrition Survey (NHANES)-based measure of “Biological Age.” In more than 9,000 NHANES participants aged 30–75 y at baseline, Biological Age outperformed chronological age in predicting mortality over a two-decade follow-up (26). Because NHANES participants were all surveyed at one time point, age differences in biomarker levels were not independent of cohort effects; measured aging also included secular trends in environmental and behavioral influences on biomarkers. Similarly, most deaths observed during follow-up occurred to the oldest NHANES participants, leaving the algorithm’s utility for quantification of aging in younger persons uncertain. We therefore applied this algorithm to calculate the Biological Age of Dunedin Study members, who all shared the same birth year and birthplace, and were all chronologically 38 y old at the last assessment (SI Appendix). Even though the Dunedin cohort remained largely free of chronic disease, Biological Age took on a normal distribution, ranging from 28 y to 61 y (M = 38 y, SD = 3.23; Fig. 2). This distribution was consistent with the hypothesis that some 38-y-old cohort members were biologically older than others.
Fig. 2.
Biological Age is normally distributed in a cohort of adults aged 38 y.
Biological Age is assumed to reflect ongoing longitudinal change within a person. However, it is a cross-sectional measure taken at a single point in time. Therefore, we next tested the hypothesis that young adults with older Biological Age at age 38 y were actually aging faster. To quantify the pace at which an individual is aging, longitudinal repeated measures are needed that track change over time. The Dunedin Study contains longitudinal data on 18 biomarkers established as risk factors or correlates of chronic disease and mortality. Our selection of 18 biomarkers was constrained by measures available 15 y ago at time one, that can be assayed with high throughput, and that are scalable to epidemiologic studies. Still, these biomarkers track the physiological integrity of study members’ cardiovascular, metabolic, and immune systems, their kidneys, livers, and lungs, their dental health, and their DNA (SI Appendix). We analyzed within-individual longitudinal change in these 18 biomarkers across chronological ages 26 y, 32 y, and 38 y to quantify each study member’s personal rate of physiological deterioration, their “Pace of Aging.”
The Pace of Aging was calculated from longitudinal analysis of the 18 biomarkers in three steps (SI Appendix). First, all biomarkers were standardized to have the same scale (mean = 0, SD = 1 based on their distributions when study members were 26 y old) and coded so that higher values corresponded to older levels (i.e., scores were reversed for cardiorespiratory fitness, lung function, leukocyte telomere length, creatinine clearance, and high density lipoprotein cholesterol, for which values are expected to decline with increasing chronological age). Even in our cohort of young adults, biomarkers showed a pattern of age-dependent decline in the functioning of multiple organ system over the 12-y follow-up period (Fig. 3). Second, we used mixed-effects growth models to calculate each study member’s personal slope for each of the 18 biomarkers; 954 individuals with repeated measures of biomarkers contributed data to this analysis. Of the 51,516 potential observations (n = 954 study members × 18 biomarkers × 3 time points), 44,475 (86.3%) were present in the database and used to estimate longitudinal growth curves modeling the Pace of Aging. The models took the form , where Bit is a biomarker measured for individual i at time t, γ0 and γ1 are the fixed intercept and slope estimated for the cohort, and μ0i and μ1i are the random intercepts and slopes estimated for each individual i. Finally, we calculated each study member’s Pace of Aging as the sum of these 18 slopes: . We calculated Pace of Aging from slopes because our goal was to quantify change over time. We summed slopes across biomarkers because our goal was to quantify change across organ systems. An additive model reduces the influence of temporary change isolated to any specific organ system, e.g., as might arise from a transient infection. The resulting Pace of Aging measure was normally distributed in the cohort, consistent with the hypothesis that some cohort members were aging faster than others.
Fig. 3.
Healthy adults exhibit biological aging of multiple organ systems over 12 y of follow-up. Biomarker values were standardized to have mean = 0 and SD = 1 across the 12 y of follow-up (Z scores). Z scores were coded so that higher values corresponded to older levels of the biomarkers; i.e., Z scores for cardiorespiratory fitness, lung function (FEV1 and FEV1/FVC), leukocyte telomere length, creatinine clearance, and HDL cholesterol, which decline with age, were reverse coded so that higher Z scores correspond to lower levels.
The Pace of Aging can be scaled to reflect physiological change relative to the passage of time. Because the intact birth cohort represents variation in the population, it provides its own norms. We scaled the Pace of Aging so that the central tendency in the cohort indicates 1 y of physiological change for every one chronological year. On this scale, cohort members ranged in their Pace of Aging from near 0 y of physiological change per chronological year to nearly 3 y of physiological change per chronological year.
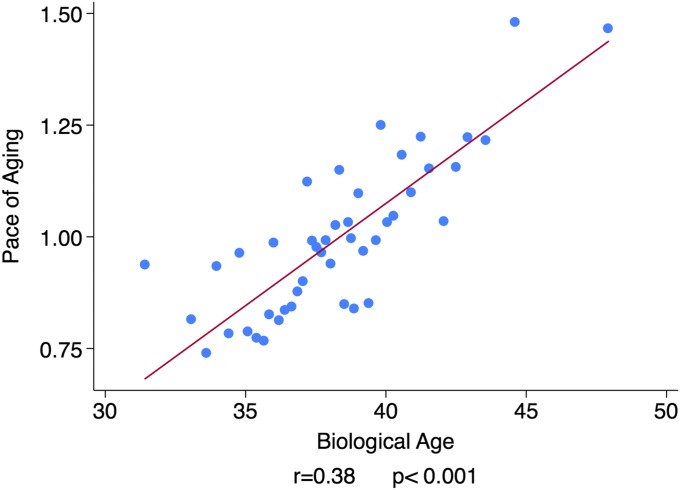
Study members with advanced Biological Age had experienced a more rapid Pace of Aging over the past 12 y compared with their biologically younger age peers (r = 0.38, P < 0.001; Fig. 4). Each year increase in Biological Age was associated with a 0.05-y increase in the Pace of Aging relative to the population norm. Thus, a 38-y-old with a Biological Age of 40 y was estimated to have aged 1.2 y faster over the course of the 12-y follow-up period compared with a peer whose chronological age and Biological Age were 38. This estimate suggests that a substantial component of individual differences in Biological Age at midlife emerges during adulthood.
Fig. 4.
Dunedin Study members with older Biological Age at 38 y exhibited an accelerated Pace of Aging from age 26–38 y. The figure shows a binned scatterplot and regression line. Plotted points show means for bins of data from 20 Dunedin Study members. Effect size and regression line were calculated from the raw data.
We next tested whether individual variation in Biological Age and the Pace of Aging related to differences in the functioning of study members’ bodies and brains, measured with instruments commonly used in clinical settings (SI Appendix).
Does Accelerated Aging in Young Adults Influence Indicators of Physical Function?
In gerontology, diminished physical capability is an important indication of aging-related health decline that cuts across disease categories (27, 28). Study members with advanced Biological Age performed less well on objective tests of physical functioning at age 38 than biologically younger peers (Fig. 5). They had more difficulty with balance and motor tests (for unipedal stance test of balance, r = −0.22, P < 0.001; for grooved pegboard test of fine motor coordination, r = −0.13, P < 0.001), and they were not as strong (grip strength test, r = −0.19, P < 0.001). Study members’ Biological Ages were also related to their subjective experiences of physical limitation. Biologically older study members reported having more difficulties with physical functioning than did biologically younger age peers (SF-36 physical functioning scale, r = 0.13, P < 0.012). We repeated these analyses using the Pace of Aging measure. Consistent with findings for Biological Age, study members with a more rapid Pace of Aging exhibited diminished capacity on the four measures of physical functioning relative to more slowly aging age peers.
Fig. 5.
Healthy adults who were aging faster exhibited deficits in physical functioning relative to slower-aging peers. The figure shows binned scatter plots of the associations of Biological Age and Pace of Aging with tests of physical functioning (unipedal stance test, grooved pegboard test, grip strength) and study members’ reports of their physical limitations. In each graph, Biological Age associations are plotted on the left in blue (red regression line) and Pace of Aging associations are plotted on the right in green (navy regression line). Plotted points show means for bins of data from 20 Dunedin Study members. Effect size and regression line were calculated from the raw data.
Does Accelerated Aging in Young Adults Influence Indicators of Brain Aging?
In neurology, cognitive testing is used to evaluate age-related decline in brain integrity. The Dunedin Study conducted cognitive testing when study members were children and repeated this testing at the age-38 assessment (29). Study members with older Biological Ages had poorer cognitive functioning at midlife (r = −0.17, P < 0.001). Moreover, this difference in cognitive functioning reflected actual cognitive decline over the years. When we compared age-38 IQ test scores to baseline test scores from childhood, study members with older Biological Age showed a decline in cognitive performance net of their baseline level (r = −0.09, P = 0.010). Results were similar for the Pace of Aging (Fig. 6). The literature on cognitive aging divides the composite IQ into “crystallized versus fluid” constituents (30, 31). Crystallized abilities (such as the Information subtest) peak in the fifties and show little age-related decline thereafter. In contrast, fluid abilities (such as the digit symbol coding subtest) peak in the twenties and show clear decline thereafter (31). The overall IQ aggregates these age trends. This aggregation makes it a highly reliable measure, albeit a conservative choice as a correlate of Biological Age and Pace of Aging. Therefore, we also report results for individual subtests in SI Appendix. As expected, the largest declines in cognitive functioning, and the largest correlations between decline and Pace of Aging, were observed for tests of fluid intelligence, in particular the digit symbol coding test (r = −0.15, P < 0.001).
Fig. 6.
Healthy adults who were aging faster showed evidence of cognitive decline and increased risk for stroke and dementia relative to slower-aging peers. The figure shows binned scatter plots of the associations of Biological Age and Pace of Aging with cognitive functioning and cognitive decline (Top) and with the calibers of retinal arterioles and venules (Bottom). The y axes in the graphs of cognitive functioning and cognitive decline are denominated in IQ points. The y axes in the graphs of arteriolar and venular caliber are denominated in SD units. In each graph, Biological Age associations are plotted on the left in blue (red regression line) and Pace of Aging associations are plotted on the right in green (navy regression line). Plotted points show means for bins of data from 20 Dunedin Study members. Effect size and regression line were calculated from the raw data.
Neurologists have also begun to use high-resolution 2D photographs of the retina to evaluate age-related loss of integrity of blood vessels within the brain. Retinal and cerebral small vessels share embryological origin and physiological features, making retinal vasculature a noninvasive indicator of the state of the brain’s microvasculature (32). Retinal microvascular abnormalities are associated with age-related brain pathology, including stroke and dementia (33–35). Two measurements of interest are the relative diameters of retinal arterioles and venules. Narrower arterioles are associated with stroke risk (36). Wider venules are associated with hypoxia and dementia risk (37, 38). We calculated the average caliber of study members’ retinal arterioles and venules from images taken at the age-38 assessment. Consistent with the cognitive testing findings, study members with advanced Biological Age had older retinal vessels (narrower arterioles, r = −0.20, P < 0.001; wider venules, r = 0.17, P < 0.001). Results were similar for the Pace of Aging measure (Fig. 6).
Do Young Adults Who Are Aging Faster Feel and Look Older?
Beyond clinical indicators, a person’s experience of aging is structured by their own perceptions about their well-being and by the perceptions of others. Consistent with tests of aging indicators, study members with older Biological Age perceived themselves to be in poorer health compared with biologically younger peers (r = −0.22, P < 0.001). In parallel, these biologically older study members were perceived to be older by independent observers. We took a frontal photograph of each study member’s face at age 38, and showed these to a panel of Duke University undergraduates who were kept blind to all other information about the study members, including their age. Based on the facial images alone, student raters scored study members with advanced Biological Age as looking older than their biologically younger peers (r = 0.21, P < 0.001). Results for self-perceived well-being and facial age were similar when analyses were conducted using the Pace of Aging measure (Fig. 7).
Fig. 7.
Healthy adults who were aging faster felt less healthy and were rated as looking older by independent observers. The figure shows binned scatter plots of the associations of Biological Age and the Pace of Aging with self-rated health (Top) and with facial aging (Bottom). The y axes are denominated in SD units. In each graph, Biological Age associations are plotted on the left in blue (red regression line) and Pace of Aging associations are plotted on the right in green (navy regression line). Plotted points show means for bins of data from 20 Dunedin Study members. Effect size and regression line were calculated from the raw data.
Discussion
Aging is now understood as a gradual and progressive deterioration of integrity across multiple organ systems (7, 39). Here we show that this process can be quantified already in young adults. We followed a birth cohort of young adults over 12 y, from ages 26–38, and observed systematic change in 18 biomarkers of risk for age-related chronic diseases that was consistent with age-dependent decline. We were able to measure these changes even though the typical age of onset for the related diseases was still one to two decades in the future and just 1.1% of the cohort members had been diagnosed with an age-related chronic disease.
Measuring aging remains controversial. We measured aging in two ways. First, we used a biomarker scoring algorithm previously calibrated on a large, mixed-age sample. We applied this algorithm to cross-sectional biomarker data collected when our study members were all chronologically aged 38 y to calculate their Biological Age. Second, we conducted longitudinal analysis of 18 biomarkers in our population-representative birth cohort when they were aged 26 y, 32 y, and 38 y. We used this longitudinal panel dataset to model how each individual changed over the 12-y period to calculate their personal Pace of Aging.
Pace of Aging and Biological Age represent two different approaches to quantifying aging. Pace of Aging captures real-time longitudinal change in human physiology across multiple systems and is suitable for use in studies of within-individual change. For this analysis, we examined all 18 biomarkers with available longitudinal data in the Dunedin Study biobank. The other approach, Biological Age, provides a point-in-time snapshot of physiological integrity in cross-sectional samples. For this analysis, we used the published 10-biomarker set developed from the NHANES. These two approaches yielded consistent results. Study members with older Biological Age had evidenced faster Pace of Aging over the preceding 12 y. Based on Pace of Aging analysis, we estimate that roughly 1/2 of the difference in Biological Age observed at chronological age 38 had accumulated over the past 12 y. Our analysis shows that Biological Age can provide a summary of accumulated aging in cases where only cross-sectional data are available. For purposes of measuring the effects of risk exposures and antiaging treatments on the aging process, Pace-of-Aging-type longitudinal measures provide a means to test within individual change.
Biological measures of study members’ aging were mirrored in their functional status, brain health, self-awareness of their own physical well-being, and their facial appearance. Study members who had older Biological Age and who experienced a faster Pace of Aging scored lower on tests of balance, strength, and motor coordination, and reported more physical limitations. Study members who had an older Biological Age and who experienced a faster Pace of Aging also scored lower on IQ tests when they were aged 38 y, showed actual decline in full-scale IQ score from childhood to age-38 follow-up, and exhibited signs of elevated risk for stroke and for dementia measured from images of microvessels in their eyes. Further, study members who had an older Biological Age and who experienced a faster Pace of Aging reported feeling in worse health. Undergraduate student raters who did not know the study members beyond a facial photograph were able to perceive differences in the aging of their faces.
Together, these findings constitute proof of principle for the measures of Biological Age and Pace of Aging studied here to serve as technology to measure aging in young people. Further research is needed to refine and elaborate this technology. Here we identify several future directions that can build on our initial proof-of-principle for measuring accelerated aging up to midlife.
First, our analysis was limited to a single cohort, and one that lacked ethnic minority populations. Replication in other cohorts is needed, in particular in samples including sufficient numbers of ethnic minority individuals to test the “weathering hypothesis” that the stresses of ethnic minority status accelerate aging (40, 41). Larger samples can also help with closer study of relatively rare aging trajectories. Three Dunedin Study members had Pace of Aging less than zero, appearing to grow physiologically younger during their thirties. In larger cohorts, study of such individuals may reveal molecular and behavioral pathways to rejuvenation.
Second, data were right censored (follow-up extended only to age 38); aging trajectories may change at older ages. Some cohort members experienced negligible aging per year, a pace that cannot be sustained throughout their lives. Future waves of data collection in the Dunedin cohort will allow us to model these nonlinear patterns of change. A further issue with right censoring is that we lack follow-up data on disability and mortality with which to evaluate the precision of the Pace of Aging measure. Continued follow-up of the Dunedin cohort and analysis of other cohorts with longer-range follow-up can be used to conduct, e.g., receiver operating characteristic curve and related analyses (42) to evaluate how well Pace of Aging forecasts healthspan and lifespan.
Third, data were left censored (biomarker follow-up began at age 26); when and how aging trajectories began to diverge was not observed. Studies tracking Pace of Aging earlier in adulthood and studies of children are needed.
Fourth, measurements were taken only once every 6 y. Contiguous annual measurements would provide better resolution to measure aging, but neither our funders nor our research participants favored this approach. Medical record datasets comprising primary care health screenings may provide annual follow-up intervals (although patients seeking annual physicals will not fully represent population aging).
Fifth, Pace of Aging analyses applied a unit weighting scheme to all biomarkers. We weighted all biomarkers equally to transparently avoid assumptions, and to avoid sample-specific findings. Nonetheless, aging is likely to affect different bodily systems to differing degrees at different points in the lifespan. Further study is needed to refine weightings of biomarker contributions to Pace of Aging measurement. For example, longitudinal data tracking biomarkers could be linked with follow-up records of disability and mortality to estimate weights for biomarker change.
Sixth, biomarkers used to measure aging in our study were restricted to those scalable to a cohort based on technology available during the measurement period (1998–2012). They necessarily provide an incomplete picture of age-related changes to physiology. Similarly, it is possible that not every biomarker in our set of 18 is essential to measure aging processes. We used all of the biomarkers that were repeatedly measured in the Dunedin Study, some of which may become more (or less) important for modeling Pace of Aging as our cohort grows older. Our leave-one-out analysis showed that associations between Pace of Aging and measures of physical and cognitive functioning and subjective aging did not depend on any one biomarker. A next step is add-one-in-type analysis to test the relative performance of biomarker subsets with the aim of identifying a “short form” of the Pace of Aging. This analysis will require multiple datasets so that an optimal short-form Pace of Aging identified in a training dataset can be evaluated in an independent test dataset.
Seventh, methods are not available to estimate confidence intervals for a person’s Pace of Aging score. Datasets with repeated measures of multiple biomarkers are becoming available. Our findings suggest that future studies of aging incorporate longitudinal repeated measures of biomarkers to track change. They also suggest that these studies of aging incorporate multiple biomarkers to track change across different organ systems. Such studies will require new statistical methods to calculate confidence intervals around Pace of Aging-type scores.
Within the bounds of these limitations, the implication of the present study is that it is possible to quantify individual differences in aging in young humans. This development breaks through two blockades separating model organism research from human translational studies. One blockade is that animals age quickly enough that whole lifespans can be observed whereas, in humans, lifespan studies outlast the researchers. A second blockade is that humans are subject to a range of complex social and genomic exposures impossible to completely simulate in animal experiments. If aging can be measured in free-living humans early in their lifespans, there are new scientific opportunities. These include testing the fetal programming of accelerated aging (e.g., does intrauterine growth restriction predispose to faster aging in young adulthood?); testing the effects of early-life adversity (e.g., does child maltreatment accelerate aging in the decades before chronic diseases develop?); testing social gradients in health (e.g., do children born into poor households age more rapidly than their age-peers born into rich ones and can such accelerated aging be slowed by childhood interventions?); and searching for genetic regulators of aging processes (e.g., interrogating biological aging using high throughput genomics). There are also potential clinical applications. Early identification of accelerated aging before chronic disease becomes established may offer opportunities for prevention. Above all, measures of aging in young humans allow for testing the effectiveness of antiaging therapies (e.g., caloric restriction) without waiting for participants to complete their lifespans.
Materials and Methods
A more detailed description of study measures, design, and analysis is provided in SI Appendix.
Sample.
Participants are members of the Dunedin Multidisciplinary Health and Development Study, which tracks the development of 1,037 individuals born in 1972–1973 in Dunedin, New Zealand.
Measuring Biological Age.
We calculated each Dunedin Study member’s Biological Age at age 38 y using the Klemera–Doubal equation (23) and parameters estimated from the NHANES-III dataset (26) for 10 biomarkers. Biological Age took on a normal distribution, ranging from 28 y to 61 y (M = 38 y, SD = 3.23).
Measuring the Pace of Aging.
We measured Pace of Aging from repeated assessments of a panel of 18 biomarkers, 7 of which overlapped with the Biological Age algorithm. We modeled change over time in each biomarker and composited results within each individual to calculate their Pace of Aging. Study members ranged in their Pace of Aging from near 0 y of physiological change per chronological year to nearly 3 y of physiological change per chronological year.
Measuring Diminished Physical Capacity.
We measured physical capacity as balance, strength, motor coordination, and freedom from physical limitations when study members were aged 38 y.
Measuring Cognitive Aging.
We measured cognitive aging using neuropsychological tests in childhood and at age 38 y and images of retinal microvessels.
Measuring Subjective Perceptions of Aging.
We measured subjective perceptions of aging using study members’ self reports and evaluations of facial photographs of the study members made by independent raters.
Supplementary Material
Acknowledgments
We thank Dunedin Study members, their families, unit research staff, and Dunedin Study founder Phil Silva. The Dunedin Multidisciplinary Health and Development Research Unit is supported by the New Zealand Health Research Council. This research received support from US National Institute on Aging (NIA) Grants AG032282 and AG048895 and UK Medical Research Council Grant MR/K00381X. Additional support was provided by the Jacobs Foundation. D.W.B. received support from NIA through a postdoctoral fellowship T32 AG000029 and P30 AG028716-08. S.I. was supported by a Rothschild Fellowship from the Yad Hanadiv Rothschild Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506264112/-/DCSupplemental.
References
- 1.Department of Economic and Social Affairs, Population Division . World Population Ageing 2013. United Nations; New York: 2013. [Google Scholar]
- 2.Harper S. Economic and social implications of aging societies. Science. 2014;346(6209):587–591. doi: 10.1126/science.1254405. [DOI] [PubMed] [Google Scholar]
- 3.Vos T, et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990-2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2163–2196. doi: 10.1016/S0140-6736(12)61729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burch JB, et al. Advances in geroscience: Impact on healthspan and chronic disease. J Gerontol A Biol Sci Med Sci. 2014;69(Suppl 1):S1–S3. doi: 10.1093/gerona/glu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayflick L. The future of ageing. Nature. 2000;408(6809):267–269. doi: 10.1038/35041709. [DOI] [PubMed] [Google Scholar]
- 6.Gavrilov LA, Gavrilova NS. The reliability theory of aging and longevity. J Theor Biol. 2001;213(4):527–545. doi: 10.1006/jtbi.2001.2430. [DOI] [PubMed] [Google Scholar]
- 7.Kaeberlein M. Longevity and aging. F1000Prime Rep. 2013;5:5. doi: 10.12703/P5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldman DP, et al. Substantial health and economic returns from delayed aging may warrant a new focus for medical research. Health Aff (Millwood) 2013;32(10):1698–1705. doi: 10.1377/hlthaff.2013.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yashin AI, et al. How genes influence life span: The biodemography of human survival. Rejuvenation Res. 2012;15(4):374–380. doi: 10.1089/rej.2011.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barzilai N, et al. The place of genetics in ageing research. Nat Rev Genet. 2012;13(8):589–594. doi: 10.1038/nrg3290. [DOI] [PubMed] [Google Scholar]
- 11.Heyn H, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci USA. 2012;109(26):10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barker DJP, Eriksson JG, Forsén T, Osmond C. Fetal origins of adult disease: Strength of effects and biological basis. Int J Epidemiol. 2002;31(6):1235–1239. doi: 10.1093/ije/31.6.1235. [DOI] [PubMed] [Google Scholar]
- 13.Gillman MW. Developmental origins of health and disease. N Engl J Med. 2005;353(17):1848–1850. doi: 10.1056/NEJMe058187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gavrilov LA, Gavrilova NS. Early-life programming of aging and longevity: The idea of high initial damage load (the HIDL hypothesis) Ann N Y Acad Sci. 2004;1019:496–501. doi: 10.1196/annals.1297.091. [DOI] [PubMed] [Google Scholar]
- 15.Danese A, McEwen BS. Adverse childhood experiences, allostasis, allostatic load, and age-related disease. Physiol Behav. 2012;106(1):29–39. doi: 10.1016/j.physbeh.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 16.Fontana L, Kennedy BK, Longo VD, Seals D, Melov S. Medical research: Treat ageing. Nature. 2014;511(7510):405–407. doi: 10.1038/511405a. [DOI] [PubMed] [Google Scholar]
- 17.de Cabo R, Carmona-Gutierrez D, Bernier M, Hall MN, Madeo F. The search for antiaging interventions: From elixirs to fasting regimens. Cell. 2014;157(7):1515–1526. doi: 10.1016/j.cell.2014.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longo VD, et al. Interventions to slow aging in humans: Are we ready? Aging Cell. April 22, 2015 doi: 10.1111/acel.12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simm A, et al. Potential biomarkers of ageing. Biol Chem. 2008;389(3):257–265. doi: 10.1515/BC.2008.034. [DOI] [PubMed] [Google Scholar]
- 20.Johnson TE. Recent results: Biomarkers of aging. Exp Gerontol. 2006;41(12):1243–1246. doi: 10.1016/j.exger.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Sprott RL. Biomarkers of aging and disease: Introduction and definitions. Exp Gerontol. 2010;45(1):2–4. doi: 10.1016/j.exger.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 22.Mather KA, Jorm AF, Parslow RA, Christensen H. Is telomere length a biomarker of aging? A review. J Gerontol A Biol Sci Med Sci. 2011;66(2):202–213. doi: 10.1093/gerona/glq180. [DOI] [PubMed] [Google Scholar]
- 23.Klemera P, Doubal S. A new approach to the concept and computation of biological age. Mech Ageing Dev. 2006;127(3):240–248. doi: 10.1016/j.mad.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Gruenewald TL, Seeman TE, Ryff CD, Karlamangla AS, Singer BH. Combinations of biomarkers predictive of later life mortality. Proc Natl Acad Sci USA. 2006;103(38):14158–14163. doi: 10.1073/pnas.0606215103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen AA, et al. Detection of a novel, integrative aging process suggests complex physiological integration. PLoS ONE. 2015;10(3):e0116489. doi: 10.1371/journal.pone.0116489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine ME. Modeling the rate of senescence: Can estimated biological age predict mortality more accurately than chronological age? J Gerontol A Biol Sci Med Sci. 2013;68(6):667–674. doi: 10.1093/gerona/gls233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clegg A, Young J, Iliffe S, Rikkert MO, Rockwood K. Frailty in elderly people. Lancet. 2013;381(9868):752–762. doi: 10.1016/S0140-6736(12)62167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Applegate WB, Blass JP, Williams TF. Instruments for the functional assessment of older patients. N Engl J Med. 1990;322(17):1207–1214. doi: 10.1056/NEJM199004263221707. [DOI] [PubMed] [Google Scholar]
- 29.Belsky DW, et al. Is obesity associated with a decline in intelligence quotient during the first half of the life course? Am J Epidemiol. 2013;178(9):1461–1468. doi: 10.1093/aje/kwt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salthouse TA. What and when of cognitive aging. Curr Dir Psychol Sci. 2004;13(4):140–144. [Google Scholar]
- 31.Hartshorne JK, Germine LT. When does cognitive functioning peak? The asynchronous rise and fall of different cognitive abilities across the life span. Psychol Sci. 2015;26(4):433–443. doi: 10.1177/0956797614567339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shalev I, et al. Retinal vessel caliber and lifelong neuropsychological functioning: Retinal imaging as an investigative tool for cognitive epidemiology. Psychol Sci. 2013;24(7):1198–1207. doi: 10.1177/0956797612470959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong TY. Is retinal photography useful in the measurement of stroke risk? Lancet Neurol. 2004;3(3):179–183. doi: 10.1016/s1474-4422(04)00682-9. [DOI] [PubMed] [Google Scholar]
- 34.Ikram MK, Cheung CY, Wong TY, Chen CPLH. Retinal pathology as biomarker for cognitive impairment and Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 2012;83(9):917–922. doi: 10.1136/jnnp-2011-301628. [DOI] [PubMed] [Google Scholar]
- 35.Heringa SM, et al. Associations between retinal microvascular changes and dementia, cognitive functioning, and brain imaging abnormalities: A systematic review. J Cereb Blood Flow Metab. 2013;33(7):983–995. doi: 10.1038/jcbfm.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong TY, et al. Retinal microvascular abnormalities and incident stroke: The Atherosclerosis Risk in Communities Study. Lancet. 2001;358(9288):1134–1140. doi: 10.1016/S0140-6736(01)06253-5. [DOI] [PubMed] [Google Scholar]
- 37.Liew G, et al. Retinal microvascular signs and cognitive impairment. J Am Geriatr Soc. 2009;57(10):1892–1896. doi: 10.1111/j.1532-5415.2009.02459.x. [DOI] [PubMed] [Google Scholar]
- 38.de Jong FJ, et al. Arteriolar oxygen saturation, cerebral blood flow, and retinal vessel diameters. The Rotterdam Study. Ophthalmology. 2008;115(5):887–892. doi: 10.1016/j.ophtha.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 39.Kirkwood TBL. Understanding the odd science of aging. Cell. 2005;120(4):437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 40.Geronimus AT, Hicken M, Keene D, Bound J. “Weathering” and age patterns of allostatic load scores among blacks and whites in the United States. Am J Public Health. 2006;96(5):826–833. doi: 10.2105/AJPH.2004.060749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levine ME, Crimmins EM. Evidence of accelerated aging among African Americans and its implications for mortality. Soc Sci Med. 2014;118(118):27–32. doi: 10.1016/j.socscimed.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pencina MJ, D’Agostino RB, Sr, D’Agostino RB, Jr, Vasan RS. Evaluating the added predictive ability of a new marker: From area under the ROC curve to reclassification and beyond. Stat Med. 2008;27(2):157–172. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 43. Institute for Health Metrics and Evaluation (2013) GBD Cause Patterns (Univ Washington, Seattle, WA). Available at vizhub.healthdata.org/gbd-cause-patterns/. Accessed September 20, 2014.
- 44.Poulton R, et al. The Dunedin Multidisciplinary Health and Development Study: Are its findings consistent with the overall New Zealand population? N Z Med J. 2006;119(1235):U2002. [PubMed] [Google Scholar]
- 45.Moffitt TE, Caspi A, Rutter M, Silva PA. Sex Differences in Antisocial Behavior: Conduct Disorder, Delinquency, and Violence in the Dunedin Longitudinal Study. Cambridge Univ Press; Cambridge, UK: 2001. [Google Scholar]
- 46.Jackson SHD, Weale MR, Weale RA. Biological age—What is it and can it be measured? Arch Gerontol Geriatr. 2003;36(2):103–115. doi: 10.1016/s0167-4943(02)00060-2. [DOI] [PubMed] [Google Scholar]
- 47.Mitnitski A, Rockwood K. Biological age revisited. J Gerontol A Biol Sci Med Sci. 2014;69(3):295–296. doi: 10.1093/gerona/glt137. [DOI] [PubMed] [Google Scholar]
- 48.Levine ME. Response to Dr. Mitnitski’s and Dr. Rockwood’s Letter to the Editor: Biological age revisited. J Gerontol A Biol Sci Med Sci. 2013;69A(3):297–298. doi: 10.1093/gerona/glt138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho IH, Park KS, Lim CJ. An empirical comparative study on biological age estimation algorithms with an application of Work Ability Index (WAI) Mech Ageing Dev. 2010;131(2):69–78. doi: 10.1016/j.mad.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 50.Singer JD, Willett JB. Applied Longitudinal Data Analysis. Oxford Univ Press; New York: 2003. [Google Scholar]
- 51.Verbeke G, Fieuws S, Molenberghs G, Davidian M. The analysis of multivariate longitudinal data: A review. Stat Methods Med Res. 2014;23(1):42–59. doi: 10.1177/0962280212445834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bohannon RW, Larkin PA, Cook AC, Gear J, Singer J. Decrease in timed balance test scores with aging. Phys Ther. 1984;64(7):1067–1070. doi: 10.1093/ptj/64.7.1067. [DOI] [PubMed] [Google Scholar]
- 53.Vereeck L, Wuyts F, Truijen S, Van de Heyning P. Clinical assessment of balance: Normative data, and gender and age effects. Int J Audiol. 2008;47(2):67–75. doi: 10.1080/14992020701689688. [DOI] [PubMed] [Google Scholar]
- 54.Springer BA, Marin R, Cyhan T, Roberts H, Gill NW. Normative values for the unipedal stance test with eyes open and closed. J Geriatr Phys Ther. 2007;30(1):8–15. doi: 10.1519/00139143-200704000-00003. [DOI] [PubMed] [Google Scholar]
- 55.Rantanen T, et al. Midlife hand grip strength as a predictor of old age disability. JAMA. 1999;281(6):558–560. doi: 10.1001/jama.281.6.558. [DOI] [PubMed] [Google Scholar]
- 56.Mathiowetz V, et al. Grip and pinch strength: Normative data for adults. Arch Phys Med Rehabil. 1985;66(2):69–74. [PubMed] [Google Scholar]
- 57.Lezak DM, Howieson DB, Loring DW, Hannay HJ, Fischer JS. Neuropsychological Assessment. 4th Ed Oxford Univ Press; New York: 2004. [Google Scholar]
- 58.Ware JE, Jr, Sherbourne CD. The MOS 36-item short-form health survey (SF-36). I. Conceptual framework and item selection. Med Care. 1992;30(6):473–483. [PubMed] [Google Scholar]
- 59.Wechsler D. Wechsler Intelligence Scale for Children. 4th Ed Harcourt Assessment; San Antonio, TX: 2003. [Google Scholar]
- 60.Wechsler D. Wechsler Adult Intelligence Scale. 4th Ed Pearson Assessment; San Antonio, TX: 2008. [Google Scholar]
- 61.Cheung CY-L, et al. A new method to measure peripheral retinal vascular caliber over an extended area. Microcirculation. 2010;17(7):495–503. doi: 10.1111/j.1549-8719.2010.00048.x. [DOI] [PubMed] [Google Scholar]
- 62.Cheung CY, et al. Quantitative and qualitative retinal microvascular characteristics and blood pressure. J Hypertens. 2011;29(7):1380–1391. doi: 10.1097/HJH.0b013e328347266c. [DOI] [PubMed] [Google Scholar]
- 63.Knudtson MD, et al. Revised formulas for summarizing retinal vessel diameters. Curr Eye Res. 2003;27(3):143–149. doi: 10.1076/ceyr.27.3.143.16049. [DOI] [PubMed] [Google Scholar]
- 64.Sun C, Wang JJ, Mackey DA, Wong TY. Retinal vascular caliber: Systemic, environmental, and genetic associations. Surv Ophthalmol. 2009;54(1):74–95. doi: 10.1016/j.survophthal.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 65.Liew G, et al. Measurement of retinal vascular caliber: Issues and alternatives to using the arteriole to venule ratio. Invest Ophthalmol Vis Sci. 2007;48(1):52–57. doi: 10.1167/iovs.06-0672. [DOI] [PubMed] [Google Scholar]
- 66.Wong TY, Wang JJ, Rochtchina E, Klein R, Mitchell P. Does refractive error influence the association of blood pressure and retinal vessel diameters? The Blue Mountains Eye Study. Am J Ophthalmol. 2004;137(6):1050–1055. doi: 10.1016/j.ajo.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 67.Cullinane EM, Siconolfi S, Carleton RA, Thompson PD. Modification of the Astrand-Rhyming sub-maximal bicycle test for estimating VO2max of inactive men and women. Med Sci Sports Exerc. 1988;20(3):317–318. [PubMed] [Google Scholar]
- 68.Bowtell DDL. Rapid isolation of eukaryotic DNA. Anal Biochem. 1987;162(2):463–465. doi: 10.1016/0003-2697(87)90421-0. [DOI] [PubMed] [Google Scholar]
- 69.Jeanpierre M. A rapid method for the purification of DNA from blood. Nucleic Acids Res. 1987;15(22):9611. doi: 10.1093/nar/15.22.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30(10):e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shalev I, et al. Exposure to violence during childhood is associated with telomere erosion from 5 to 10 years of age: A longitudinal study. Mol Psychiatry. 2013;18(5):576–581. doi: 10.1038/mp.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cockcroft DW, Gault MH. Prediction of creatinine clearance from serum creatinine. Nephron. 1976;16(1):31–41. doi: 10.1159/000180580. [DOI] [PubMed] [Google Scholar]
- 73.Winter MA, Guhr KN, Berg GM. Impact of various body weights and serum creatinine concentrations on the bias and accuracy of the Cockcroft-Gault equation. Pharmacotherapy. 2012;32(7):604–612. doi: 10.1002/j.1875-9114.2012.01098.x. [DOI] [PubMed] [Google Scholar]
- 74.Brown DL, Masselink AJ, Lalla CD. Functional range of creatinine clearance for renal drug dosing: A practical solution to the controversy of which weight to use in the Cockcroft-Gault equation. Ann Pharmacother. 2013;47(7-8):1039–1044. doi: 10.1345/aph.1S176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.