Abstract
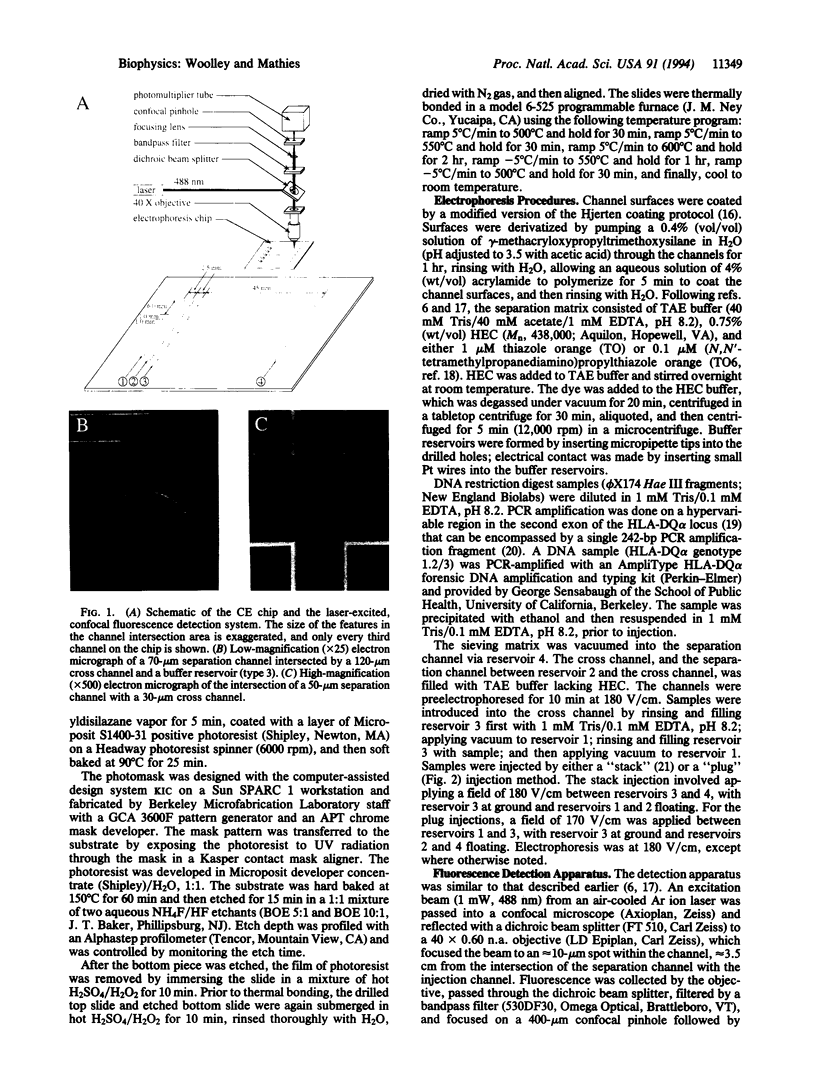
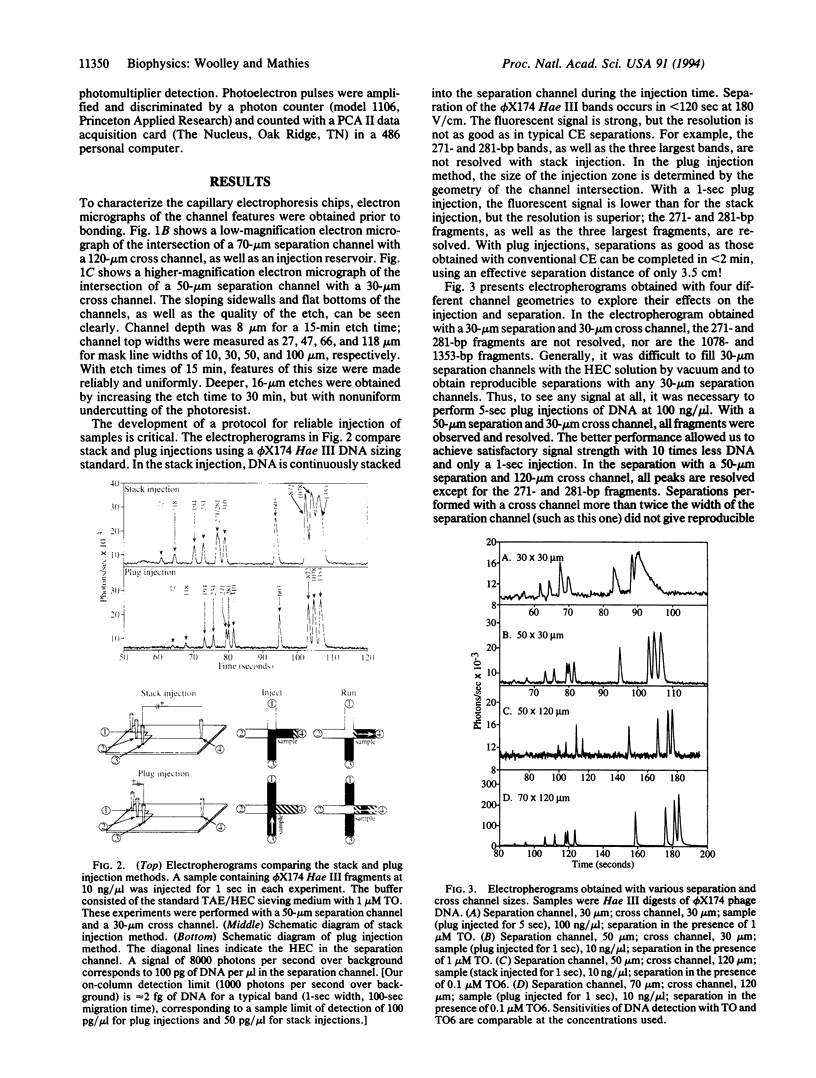
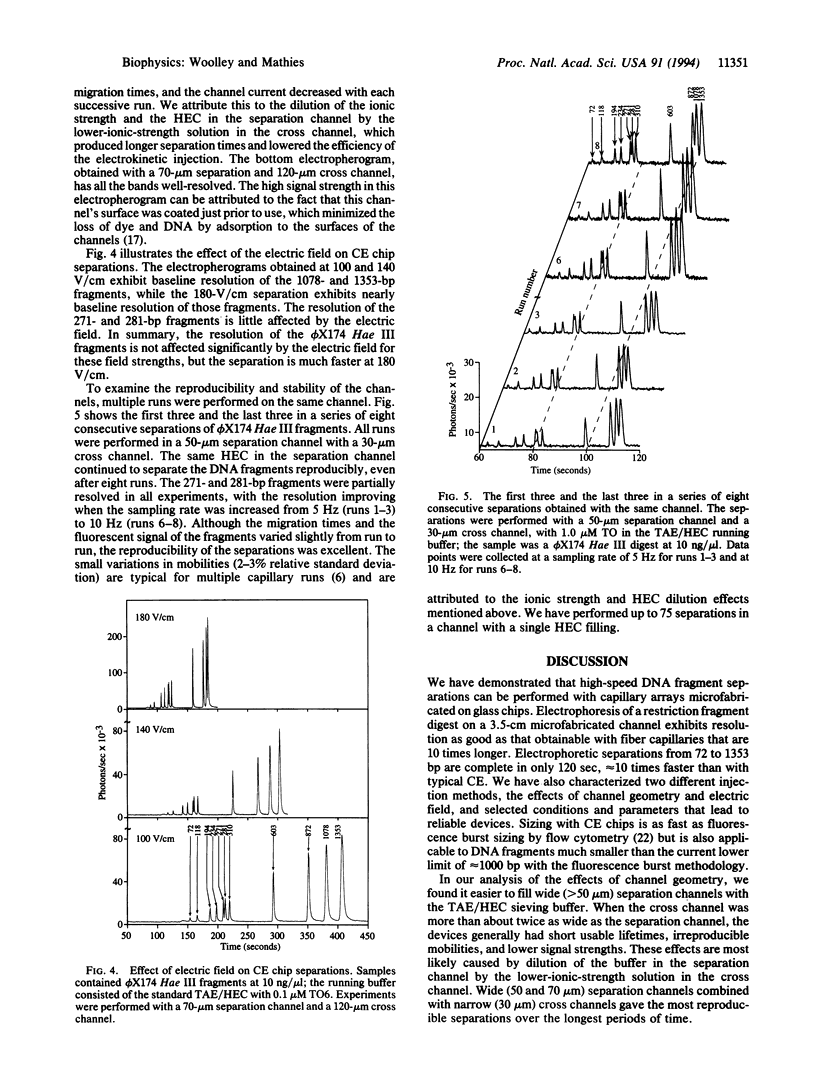
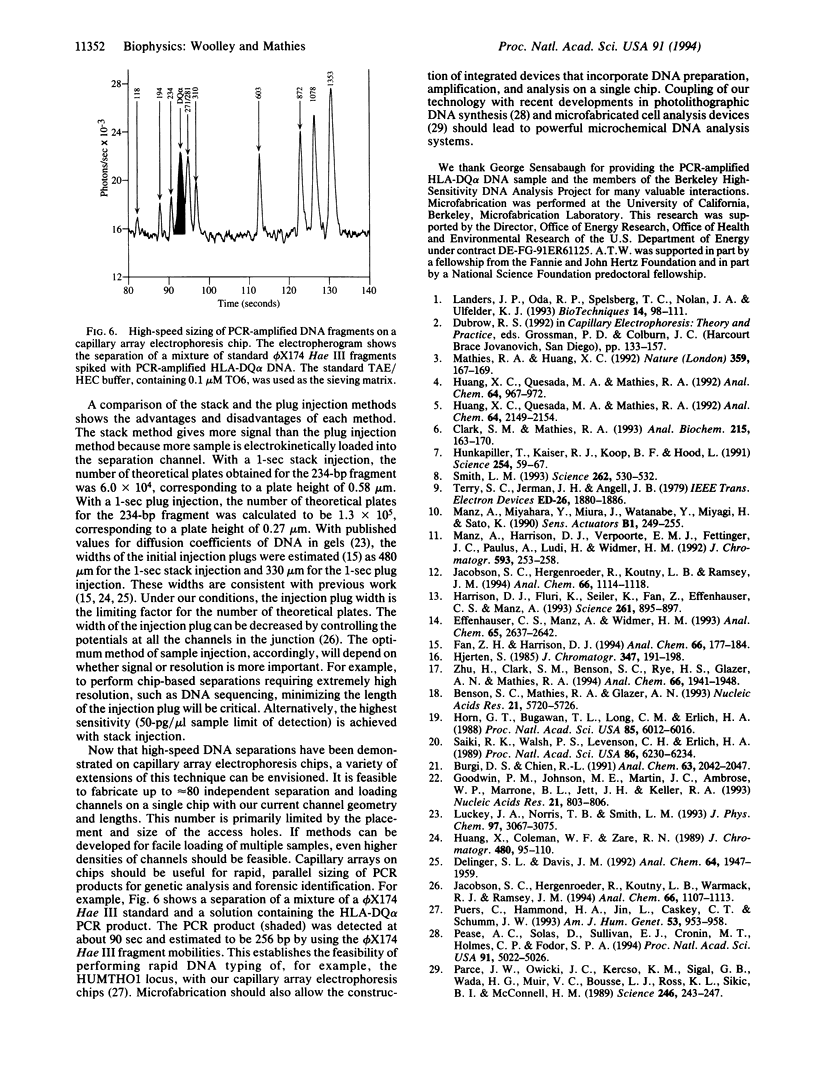
Capillary electrophoresis arrays have been fabricated on planar glass substrates by photolithographic masking and chemical etching techniques. The photolithographically defined channel patterns were etched in a glass substrate, and then capillaries were formed by thermally bonding the etched substrate to a second glass slide. High-resolution electrophoretic separations of phi X174 Hae III DNA restriction fragments have been performed with these chips using a hydroxyethyl cellulose sieving matrix in the channels. DNA fragments were fluorescently labeled with dye in the running buffer and detected with a laser-excited, confocal fluorescence system. The effects of variations in the electric field, procedures for injection, and sizes of separation and injection channels (ranging from 30 to 120 microns) have been explored. By use of channels with an effective length of only 3.5 cm, separations of phi X174 Hae II DNA fragments from approximately 70 to 1000 bp are complete in only 120 sec. We have also demonstrated high-speed sizing of PCR-amplified HLA-DQ alpha alleles. This work establishes methods for high-speed, high-throughput DNA separations on capillary array electrophoresis chips.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benson S. C., Mathies R. A., Glazer A. N. Heterodimeric DNA-binding dyes designed for energy transfer: stability and applications of the DNA complexes. Nucleic Acids Res. 1993 Dec 11;21(24):5720–5726. doi: 10.1093/nar/21.24.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. M., Mathies R. A. High-speed parallel separation of DNA restriction fragments using capillary array electrophoresis. Anal Biochem. 1993 Dec;215(2):163–170. doi: 10.1006/abio.1993.1571. [DOI] [PubMed] [Google Scholar]
- Goodwin P. M., Johnson M. E., Martin J. C., Ambrose W. P., Marrone B. L., Jett J. H., Keller R. A. Rapid sizing of individual fluorescently stained DNA fragments by flow cytometry. Nucleic Acids Res. 1993 Feb 25;21(4):803–806. doi: 10.1093/nar/21.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison D. J., Fluri K., Seiler K., Fan Z., Effenhauser C. S., Manz A. Micromachining a miniaturized capillary electrophoresis-based chemical analysis system on a chip. Science. 1993 Aug 13;261(5123):895–897. doi: 10.1126/science.261.5123.895. [DOI] [PubMed] [Google Scholar]
- Horn G. T., Bugawan T. L., Long C. M., Erlich H. A. Allelic sequence variation of the HLA-DQ loci: relationship to serology and to insulin-dependent diabetes susceptibility. Proc Natl Acad Sci U S A. 1988 Aug;85(16):6012–6016. doi: 10.1073/pnas.85.16.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. C., Quesada M. A., Mathies R. A. DNA sequencing using capillary array electrophoresis. Anal Chem. 1992 Sep 15;64(18):2149–2154. doi: 10.1021/ac00042a021. [DOI] [PubMed] [Google Scholar]
- Hunkapiller T., Kaiser R. J., Koop B. F., Hood L. Large-scale and automated DNA sequence determination. Science. 1991 Oct 4;254(5028):59–67. doi: 10.1126/science.1925562. [DOI] [PubMed] [Google Scholar]
- Landers J. P., Oda R. P., Spelsberg T. C., Nolan J. A., Ulfelder K. J. Capillary electrophoresis: a powerful microanalytical technique for biologically active molecules. Biotechniques. 1993 Jan;14(1):98–111. [PubMed] [Google Scholar]
- Parce J. W., Owicki J. C., Kercso K. M., Sigal G. B., Wada H. G., Muir V. C., Bousse L. J., Ross K. L., Sikic B. I., McConnell H. M. Detection of cell-affecting agents with a silicon biosensor. Science. 1989 Oct 13;246(4927):243–247. doi: 10.1126/science.2799384. [DOI] [PubMed] [Google Scholar]
- Pease A. C., Solas D., Sullivan E. J., Cronin M. T., Holmes C. P., Fodor S. P. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci U S A. 1994 May 24;91(11):5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puers C., Hammond H. A., Jin L., Caskey C. T., Schumm J. W. Identification of repeat sequence heterogeneity at the polymorphic short tandem repeat locus HUMTH01[AATG]n and reassignment of alleles in population analysis by using a locus-specific allelic ladder. Am J Hum Genet. 1993 Oct;53(4):953–958. [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Walsh P. S., Levenson C. H., Erlich H. A. Genetic analysis of amplified DNA with immobilized sequence-specific oligonucleotide probes. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6230–6234. doi: 10.1073/pnas.86.16.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. M. The future of DNA sequencing. Science. 1993 Oct 22;262(5133):530–532. doi: 10.1126/science.8211178. [DOI] [PubMed] [Google Scholar]
- Zhu H., Clark S. M., Benson S. C., Rye H. S., Glazer A. N., Mathies R. A. High-sensitivity capillary electrophoresis of double-stranded DNA fragments using monomeric and dimeric fluorescent intercalating dyes. Anal Chem. 1994 Jul 1;66(13):1941–1948. doi: 10.1021/ac00085a004. [DOI] [PubMed] [Google Scholar]





