Significance
Proofreading exonucleases contributing to replication fidelity in DNA viruses and cellular organisms are well known; however, proofreading in RNA viruses was unknown until recently. Coronavirus nonstructural protein 14 (nsp14) has been shown to function as a proofreading exoribonuclease. Additionally, nsp14 shows (guanine-N7) methyl transferase activity for viral mRNA capping. Both roles are important for viral replication and transcription. Here, we report the structures of severe acute respiratory syndrome-coronavirus nsp14 in complex with its activator nonstructural protein 10 (nsp10) and functional ligands. Structural observations coupled with mutagenesis and functional assays provide a better understanding of the function of nsp14. Furthermore, the structures of the nsp14–nsp10 complex demonstrate several unique niches that could be targeted for development of potent antiviral drugs.
Keywords: CoV, nsp14, proofreading, exoribonuclease, methyltransferase
Abstract
Nonstructural protein 14 (nsp14) of coronaviruses (CoV) is important for viral replication and transcription. The N-terminal exoribonuclease (ExoN) domain plays a proofreading role for prevention of lethal mutagenesis, and the C-terminal domain functions as a (guanine-N7) methyl transferase (N7-MTase) for mRNA capping. The molecular basis of both these functions is unknown. Here, we describe crystal structures of severe acute respiratory syndrome (SARS)-CoV nsp14 in complex with its activator nonstructural protein10 (nsp10) and functional ligands. One molecule of nsp10 interacts with ExoN of nsp14 to stabilize it and stimulate its activity. Although the catalytic core of nsp14 ExoN is reminiscent of proofreading exonucleases, the presence of two zinc fingers sets it apart from homologs. Mutagenesis studies indicate that both these zinc fingers are essential for the function of nsp14. We show that a DEEDh (the five catalytic amino acids) motif drives nucleotide excision. The N7-MTase domain exhibits a noncanonical MTase fold with a rare β-sheet insertion and a peripheral zinc finger. The cap-precursor guanosine-P3-adenosine-5′,5′-triphosphate and S-adenosyl methionine bind in proximity in a highly constricted pocket between two β-sheets to accomplish methyl transfer. Our studies provide the first glimpses, to our knowledge, into the architecture of the nsp14–nsp10 complex involved in RNA viral proofreading.
Coronaviruses (CoV), belonging to the Coronaviridae family in the order Nidovirales (1), are one of the major threats to public health. The most notable infections are the severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) caused by the SARS-CoV and MERS-CoV, respectively (2, 3). SARS-CoV brought about more than 8,000 infections and 800 deaths, and MERS-CoV has caused 1,139 cases of infections and 431 deaths till May 25, 2015 (www.who.int/en/).
CoVs have the largest genomes among RNA viruses (4). There are 14 ORFs in the genome of SARS-CoV. Among these, ORF1a and ORF1b encode 16 nonstructural proteins (nsp) that predominantly play a role in replication and transcription (5). Within these nsps, nsp12 functions as a RNA-dependent RNA polymerase (RdRp), and nsp8 together with nsp7 functions as a primase and confers processivity to polymerization by nsp12 (6–8). More importantly, the nsp7–nsp8–nsp12 complex can associate with nsp14 without impacting RNA synthesis (8). This interaction is crucial, because nsp14 has been shown to play a pivotal role in decreasing the incidence of mismatched nucleotides through its exoribonuclease domain (ExoN) (9–11), a role akin to a proofreading ExoN associated with a polymerase. Abrogation of the nsp14 ExoN activity results in enhanced sensitivity to the RNA mutagen 5-fluorouracil (12, 13). The nsp14–nsp10 complex can exquisitely excise 3′ mismatched nucleotides from dsRNA (14). Disturbance of the interaction between nsp14 and nsp10 has been shown to result in a decrease in replication fidelity (15). Consequently, in contrast to the general replication fidelity of RNA viruses (10−3–10−5), the low mutation rate (10−6–10−7) of SARS-CoV is tied to the ExoN activity. A DEDDh (the five catalytic amino acids) motif drives catalysis by nsp14 that is important for the viral replication and transcription (14, 16). In vivo studies using mouse models have demonstrated a role for this ExoN activity in viral virulence and pathogenesis (17). Nsp14 is highly conserved within the Coronaviridae family. Intriguingly, ExoNs also are encoded by RNA viruses belonging to the order Nidovirales with genomes larger than 20 kb (4, 18, 19).
In addition, nsp14 is also known to function as an S-adenosyl methionine (SAM)-dependent (guanine-N7) methyl transferase (N7-MTase) (20). Assembly of a cap1 structure at the 5′ end of viral mRNA assists in translation and evading host defense (21–23). Formation of this cap in SARS-CoV requires four sequential reactions. First, nsp13 RNA triphosphatase (RTPase) hydrolyzes nascent RNA to yield pp-RNA (24). Then an unknown guanylyltransferase (GTase) hydrolyzes GTP, transfers the product GMP to pp-RNA, and creates Gppp-RNA. Then nsp14 methylates the 5′ guanine of the Gppp-RNA at the N7 position, followed by methylation of the ribose of the first nucleotide at the 2′-O position by nsp16 (20, 25). Nsp10 has been shown to activate the 2′-O–MTase activity of nsp16 by stabilizing the SAM-binding pocket and extending the substrate RNA-binding groove of nsp16 (26, 27). Similarly, the ExoN activity of nsp14 is fully unleashed only in the presence of nsp10 (14). However, the molecular basis for this activation is poorly understood.
Here, we report the nsp14–nsp10 complex structures and show how nsp14 is activated by nsp10. Structural observations coupled with mutagenesis and functional assays unveil several previously unknown features of nsp14 and provide a better understanding of its participation in proofreading and mRNA capping.
Results and Discussion
Overall Structure of the nsp14–nsp10 Complex.
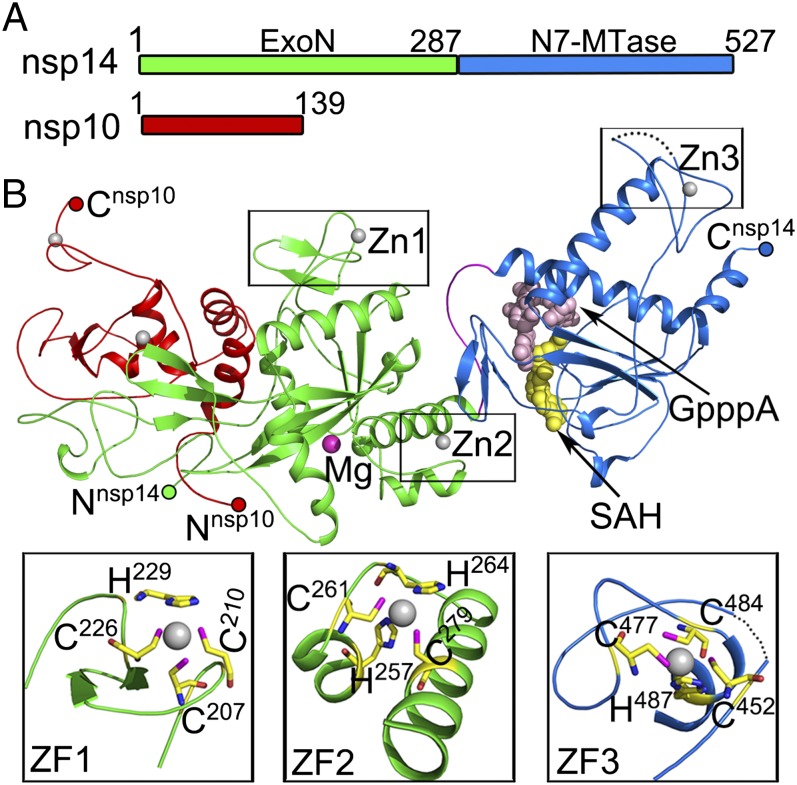
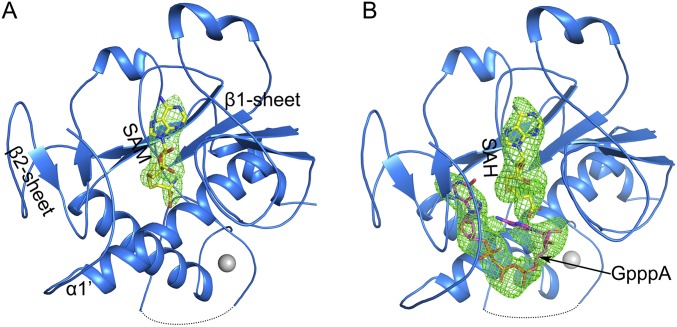
Full-length nsp14 was coexpressed with nsp10 in Escherichia coli and purified as a preformed complex (Fig. 1A and Fig. S1). The unliganded, SAM-bound, and S-adenosyl homocysteine (SAH)–guanosine-P3-adenosine-5′,5′-triphosphate (GpppA)–bound nsp14–nsp10 complex structures were refined to 3.4 Å, 3.2 Å, and 3.3 Å resolutions, respectively (Table S1). With the exception of the region encompassing amino acids 454–464 of nsp14 and the C terminus of nsp10 (residues 132–139), all residues of nsp14 and nsp10 could be built in the final model (Fig. 1B). The complex structure reveals that one molecule of nsp10 binds one molecule of nsp14 (Fig. 1B). Furthermore, nsp14 is bimodular; amino acids 1–287 fold into the ExoN domain, and amino acids 288–527 form the N7-MTase domain. A convoluted loop consists of amino acids 288–301, and a break in it could result in abolishment of the N7-MTase activity (28). Electron density maps of the two active centers are shown in Fig. S2. Nsp10 interacts exclusively with the ExoN domain of nsp14 (Fig. 1B), as is consistent with previous biochemical results showing that nsp10 stimulates the ExoN activity without perturbing the N7-MTase activity (14, 25).
Fig. 1.
Overall structure of the nsp14–nsp10 complex. (A) Domain organization of nsp14 and nsp10. Domain boundaries are marked with residue numbers. (B) Cartoon representation of the structure of the nsp14–nsp10 heterodimer. Nsp10, the ExoN domain, the N7-MTase domain, and the loop at the N terminus of N7-MTase are marked red, green, marine, and pink, respectively. Invisible residues from 454 to 464 of nsp14 are shown by a dashed line. Magnesium, zinc ions, and the N7-MTase substrates SAH and GpppA are shown as spheres and are colored magenta, gray, yellow, and light pink, respectively. Three zinc fingers (ZF) of nsp14 are highlighted with residues shown as sticks.
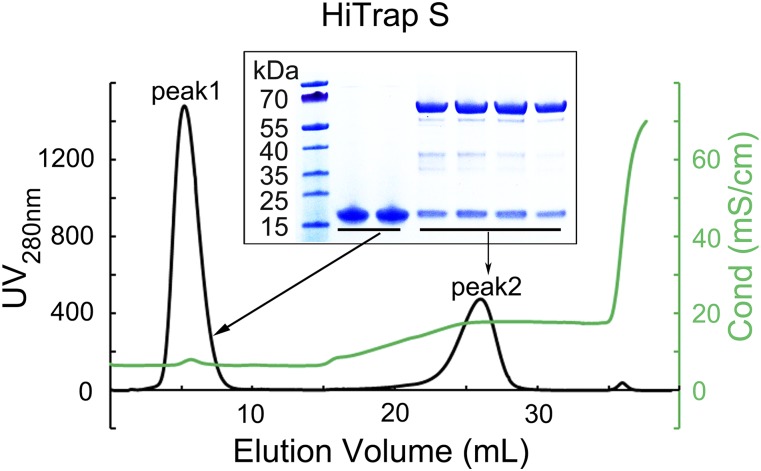
Fig. S1.
Purification of the nsp14–nsp10 complex. During coexpression in E. coli, nsp10 was expressed in excess of nsp14. The nsp14–nsp10 complex was separated from the excess nsp10 by HiTrap S ion-exchange chromatography. Nsp10 did not bind to the column (Peak 1). Peak 2 eluted from the column using NaCl contained the complex of nsp14 with nsp10. (Inset) SDS/PAGE.
Table S1.
Data collection and refinement statistics
| Datasets | nsp14–nsp10 | nsp14–nsp10–SAM | nsp14–nsp10–SAH–GpppA |
| Data collection statistics | |||
| Cell parameters | |||
| a, b, c, Å | 189.9, 195.0, 179.8 | 189.0, 196.9, 180.1 | 187.0, 195.9, 181.0 |
| α, β, γ, ° | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Space group | C2 2 21 | C2 2 21 | C2 2 21 |
| Wavelength, Å | 0.984 | 0.979 | 0.979 |
| Resolution, Å* | 50.0 (3.46) −3.4 | 50.0 (3.31) −3.2 | 50.0 (3.36) −3.30 |
| No. of all reflections | 346,535 | 747,221 | 339,184 |
| No. of unique reflections | 46,052 | 56,140 | 50,763 |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) |
| Average I/σ (I) | 26.8 (2.3) | 24.0 (2.44) | 16.5 (1.4) |
| Rmerge† (%) | 8.6 (91.7) | 9.9 (93.0) | 7.6 (87.7) |
| Refinement statistics | |||
| No. of reflections used [σ(F) > 0] | 45,999 | 55,716 | 48,424 |
| Rwork‡, % | 21.8 | 24.3 | 21.7 |
| Rfree‡, % | 26.6 | 26.5 | 26.0 |
| rmsd bond distance, Å | 0.024 | 0.027 | 0.025 |
| rmsd bond angle, ° | 1.343 | 1.179 | 1.445 |
| Average B-value, Å | 151.6 | 107.7 | 125.0 |
| No. of protein atoms | 10,172 | 10,172 | 10,172 |
| No. of ligand atoms | 0 | 52 | 152 |
| No. of solvent atoms | 0 | 0 | 0 |
| Ramachandran plot | |||
| Residues in favored regions, % | 96.02 | 97.35 | 95.63 |
| Residues in allowed regions, % | 3.67 | 2.50 | 4.21 |
| Residues in disallowed regions, % | 0.31 | 0.15 | 0.16 |
Numbers in parentheses are corresponding values for the highest-resolution shell.
Rmerge = ΣhΣl | Iih-<Ih> |/ΣhΣI <Ih>, where <Ih> is the mean of the observations Iih of reflection h.
Rwork = Σ(||Fp(obs)|-|Fp(calc)||)/Σ|Fp(obs)|; Rfree is an R factor for a selected subset (5%) of the reflections that was not included in prior refinement calculations.
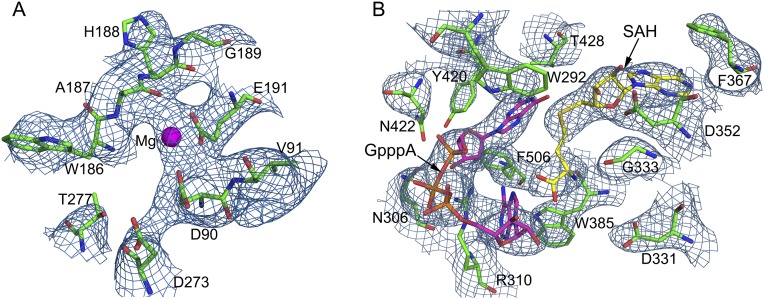
Fig. S2.
2Fo-Fc maps of the active centers of nsp14. The electron density for the ExoN catalytic center (A) and the N7-MTase catalytic center (B) are contoured at 2σ. Amino acids surrounding the catalytic centers and substrates SAH and GpppA are shown as sticks, and Mg2+ is shown as a magenta sphere.
Structure of the nsp14 ExoN Domain.
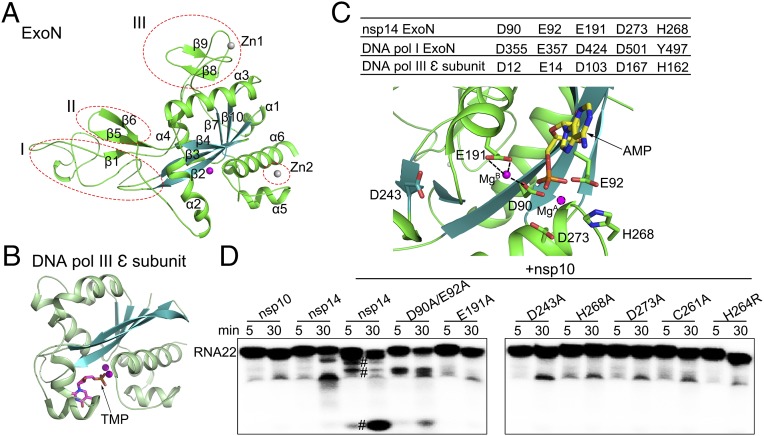
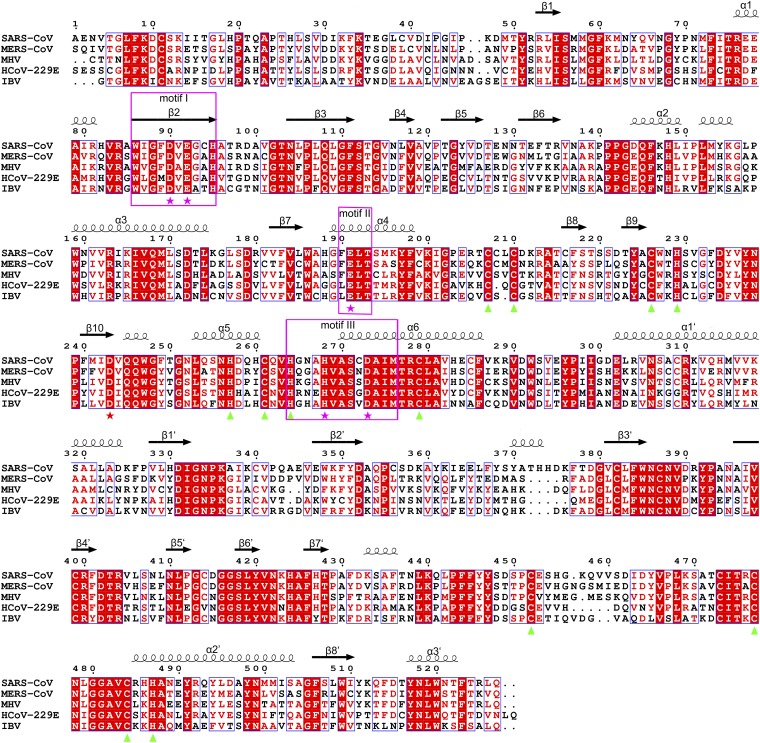
The ExoN domain contains a central, twisted β-sheet made up of five β-strands (Fig. 2A). The strands form a parallel β-sheet with the exception of β3 and are flanked by α-helices on either side. Such an arrangement of the core structural elements is reminiscent of the structures of the DEDD superfamily exonucleases, such as the ε subunit of E. coli DNA polymerase III [Protein Data Bank (PDB) code 1J53; Z score of 11.7, rmsd of 3.0 Å over 174 aligned Cα atoms as calculated by DALI server (Fig. 2B)] (29). One Mg2+ ion is observed at its active center (Fig. 2A). However, three major structural differences set nsp14 apart from typical DEDD family exonucleases (Fig. 2A). First, the N-terminal Ala1-Arg76 forms a long, flexible region that interacts with nsp10. Second, a β-hairpin structure containing β5 and β6 (Ala119-Asp145) also is observed to interact with nsp10. Last, the most striking difference is the presence of a zinc finger on either side of the β-sheet. The first zinc finger, comprising of Cys207, Cys210, Cys226, and His229, is located between α4 and β10, and the second zinc finger comprising, His257, Cys261, His264, and Cys279, is located between α5 and α6 (Figs. 1B and 2A).
Fig. 2.
Comparison of the structure and catalytic residues of nsp14 ExoN domain with proofreading homologs. (A) Cartoon representation of the ExoN domain marked with secondary structural elements. The three different regions from other DEDD superfamily exonucleases are indicated by red dashed ellipses. (B) The structure of the E. coli ε subunit of polymerase III (Pol III) is shown in the same orientation as nsp14 for comparison. Metal ions are shown as spheres, and bound ligands are shown as sticks. (C) The active center of the ExoN domain of nsp14. (Upper) Catalytic residues of the ExoN domain of nsp14, the exonuclease domain of DNA polymerase I, and the ε subunit of DNA polymerase III of E. coli are listed in the table. (Lower) Catalytic residues, the modeled substrate AMP, and the mistaken D243 are shown as sticks. MgB observed in the structure and MgA modeled are shown as spheres. Dashed lines indicate the hydrogen bonds between MgB and D90 and E191. (D) Exoribonuclease assays for nsp10, nsp14 alone, and nsp14 or nsp14 mutants in complex with nsp10 on 5′-labeled ssRNA of 22 nucleosides (RNA22). The symbol “#” indicates cleavage products.
Comparison of nsp14 ExoN Active Sites with Proofreading Homologs.
Although the overall structure of the nsp14 ExoN domain has diverged significantly from other proteins, the architecture of the catalytic core and active sites resembles those used by DEDD-type exonucleases, such as the proofreading ExoN domain of DNA polymerase I Klenow fragment (PDB ID code 1KLN) and the ε subunit of DNA polymerase III (PDB ID code 1J53) of E. coli, suggesting a conserved mechanism for catalysis. These proteins and nsp14 share a similar two-metal-ion–assisted mechanism for removal of misincorporated nucleotides (30–32). In the nsp14–nsp10 complex structure, only one Mg2+ coordinated by Asp90 and Glu191 is observed. The absence of the second metal ion could be attributed to the lack of the substrate or product binding (30). Ingredients such as the 3′ end of DNA, metal ions, and the solvent molecule required for catalysis are positioned in place by side chains of D355, E357, D424, Y497, and D501 of the Klenow fragment (Fig. 2C) (33). The ExoN domain of nsp14 exhibits a similar constellation of acidic amino acids, with one notable difference. A conserved aspartate of motif II (34, 35), for instance Asp424 of the Klenow fragment and Asp103 of the ε subunit of polymerase III, is replaced by Glu191 in nsp14 (Fig. 2C and Fig. S3) (31, 33). In addition, His268 categorizes nsp14 as a DEDDh-type exoribonuclease. The position of these catalytic amino acids around a nucleotide modeled in the active site of ExoN is shown in Fig. 2C. The MgA activates one molecule of water to initiate the nucleophilic attack on the phosphorous of the substrate, whereas MgB facilitates the leaving of the product (Fig. 2C). Simultaneously mutating Asp90 and Glu92 to alanine impaired the ExoN activity significantly, whereas E191A, H268A, or D273A mutants were severely deficient in their ability to degrade RNA, confirming the importance of these amino acids in the excision of nucleotides (Fig. 2D). Intriguingly, Asp243, previously mistaken as a catalytic residue (5, 16), is the fifth highly conserved acidic amino acid located between motifs II and III (Fig. 2C and Fig. S3) (34). The ExoN activity of D243A mutant is completely lost (Fig. 2D). In contrast to reported results, the activity of D90A/E92A, D243A, H268A, and D273A complexed with nsp10 or not on different substrates has been shown to be nearly identical (14, 16, 20).
Fig. S3.
Primary sequence alignment of nsp14 homologs by ClustalW (53) and rendered using ESPript version 3.0 (54). The secondary structural elements and numbering of SARS-CoV nsp14 are displayed above the alignment. Identical residues in all sequences are boxed with red background; those conserved are marked in red. Catalytic residues, D243, and residues involved in formation of zinc fingers are marked with magenta stars, red stars, and green triangles, respectively. Sequences of nsp14 in SARS-CoV (accession number ADC35510), MERS-CoV (AIZ48758), mouse hepatitis virus (MHV; NP_068668), human coronavirus [(HCoV)-229E; (AGT21366.1], and infectious bronchitis virus (IBV; P0C6Y2) were used for the analysis.
Further, we mutated residues of the two zinc fingers of the ExoN domain of nsp14 to decipher their functions. Zinc finger 2 is in proximity to the catalytic residues. Disruption of this zinc finger via a C261A or H264R mutation abolished the enzymatic activity, suggesting this zinc finger has a role in catalysis (Fig. 2D). In stark contrast to mutants of zinc finger 2, none of the mutants of zinc finger 1 could be expressed as soluble proteins. Inspection of the structure reveals that zinc finger 1 contributes the structural stability of nsp14.
Thus, Asp90, Glu92, Glu191, His268, and Asp273 of nsp14 are likely to provide the electrophilic environment necessary for ligand binding and catalysis. In addition, both the zinc fingers of the ExoN domain are essential for the function of nsp14.
Stimulation of ExoN Activity of nsp14 by nsp10.
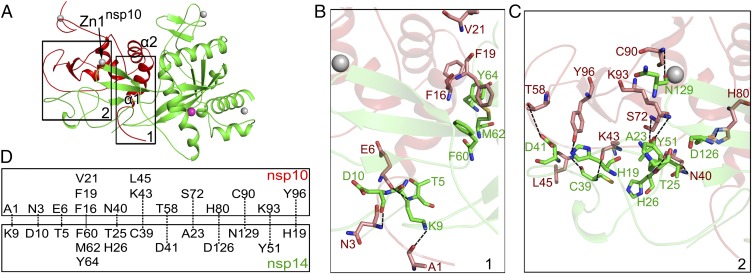
In the absence of nsp10, nsp14 cannot catalyze nucleotide excision efficiently (Fig. 2D). The structure of the nsp14–nsp10 complex explains this requirement of nsp10 for enzymatic activity of nsp14. Known structures of nsp10 could be superimposed over the nsp14–nsp10 complex with an rmsd of <0.8 Å between matching Cα atoms over the entire length of nsp10. Two regions of nsp10 contribute the major residues for nsp14–nsp10 interaction (Fig. 3A). The first contact area involves the entire N-terminal loop and helix α1 (Pro1–Leu24) of nsp10, which has led to interpretable electron density for these first nine residues of nsp10 that were not observed in previous structures (26, 27, 36, 37). The residues Ala1, Asn3, and Glu6 of nsp10 stabilize the N terminus of nsp14 by forming hydrogen bonds with Lys9, Asp10, and Thr5, respectively, whereas Phe16, Phe19, and Val21 of nsp10 form van der Waals interactions with Phe60, Met62, and Tyr64 of nsp14 (Fig. 3 B and D). The second region of intermolecular interactions is extensive and encompasses residues from the loop region following helix α2 and residues around zinc finger 1. Here, a number of complementary hydrogen bonds are observed; Asn40, Lys43, Leu45, Thr58, Ser72, Lys93, and Tyr96 of nsp10 interact with Thr25, His26, Cys39, Asp41, Ala23, Tyr51, and His19 of the N terminus of nsp14. A salt bridge formed between His80 of nsp10 and Asp126 of nsp14 and a hydrogen bond between Cys90 of nsp10 and Asn129 of nsp14 stabilize the structural elements between β5 and β6 of nsp14 (Fig. 3 C and D). The extensive interaction of nsp10 with nsp14 suggests that nsp10 might be necessary to maintain the structural stability of the ExoN domain and fully unleash the ExoN activity of nsp14 (14).
Fig. 3.
Intermolecular interactions between nsp14 and nsp10. Zinc ions are represented as gray spheres in A–C. (A) The Nsp14 ExoN domain (green) is stabilized by nsp10 (red). Two regions of nsp10 (boxed) contribute major interactions with nsp14. Zn1nsp10, the first zinc ion of nsp10. (B and C) Interaction details by regions 1 (B) and 2 (C). Hydrogen bonds between residues are shown by dashed lines. Residues of nsp14 and nsp10 involved in interaction are displayed as green and red sticks, respectively. (D) Schematic representation of the contacts between nsp10 and nsp14.
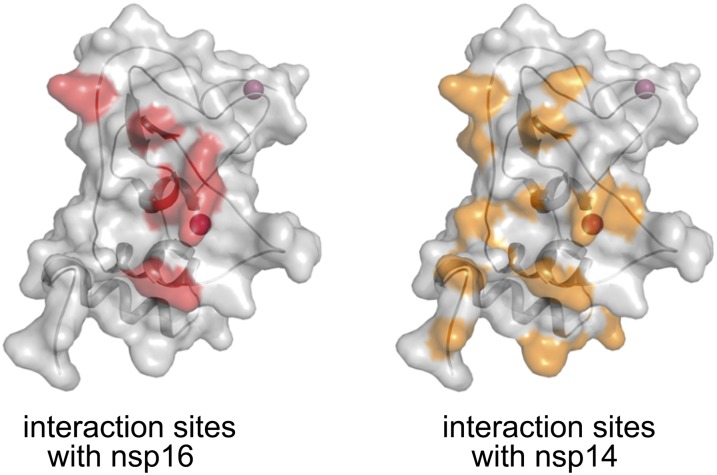
Previous mutagenesis studies had shown that regions of nsp10 contacting nsp14 and nsp16 overlap substantially (38, 39). Comparison of the nsp14–nsp10 complex with the nsp16–nsp10 complex surprisingly reveals that a significantly larger surface area of nsp10 contacts with nsp14 (Fig. S4). The buried solvent-accessible areas of nsp10 for contacting nsp14 and nsp16 are 2,236 Å2 and 938 Å2, respectively. Because nsp10 is encoded in about three- to sixfold excess over nsp14 and nsp16, the nsp14–nsp10 complex and nsp16–nsp10 complex can exist simultaneously.
Fig. S4.
Comparison of the nsp16 and nsp14 interaction sites on the nsp10 surface. Nsp10 of the nsp14–nsp10–SAM complex is shown as a gray surface and cartoon. The interaction sites for nsp10 with nsp16 and nsp14 are colored in salmon and orange, respectively.
Structure of the nsp14 N7-MTase Domain.
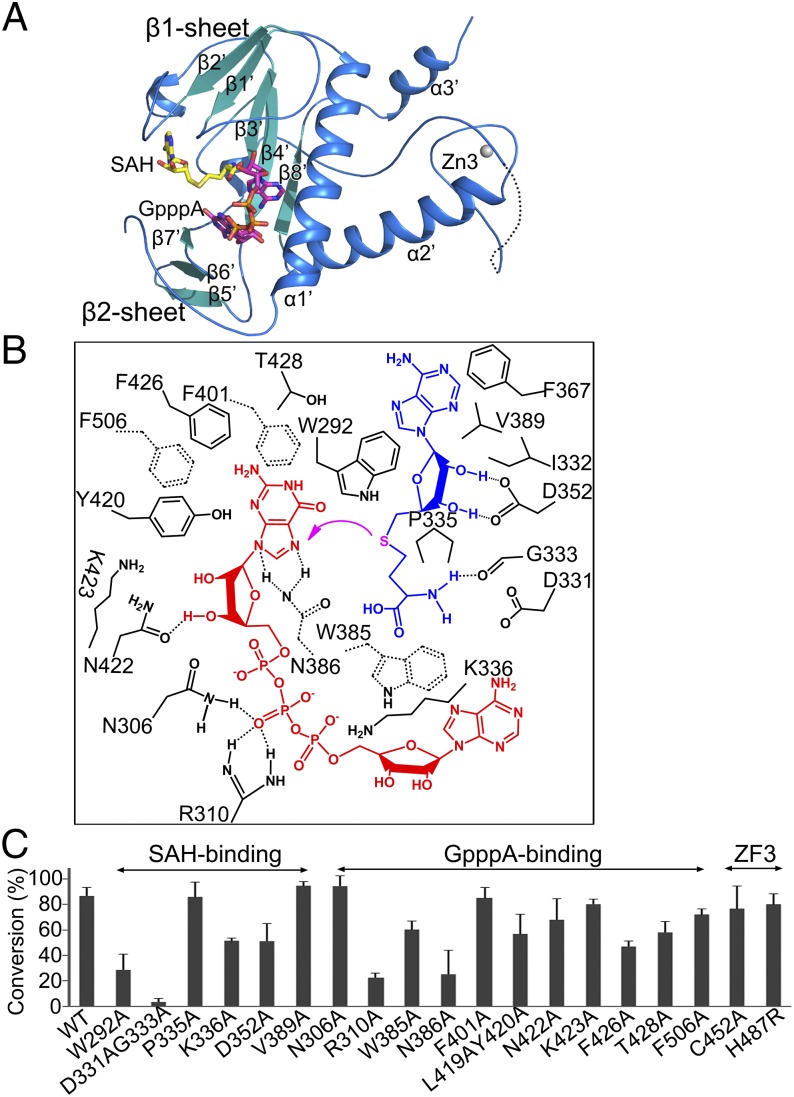
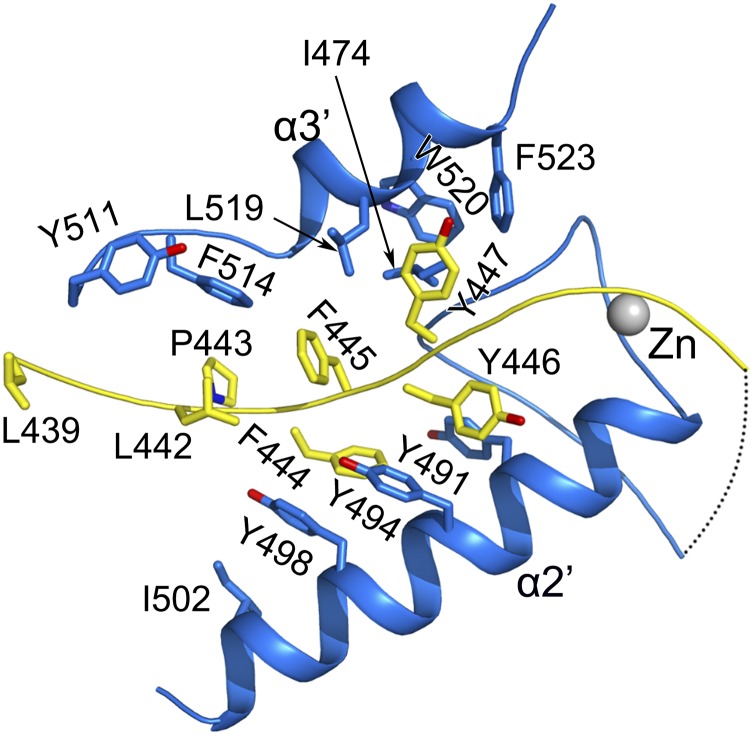
The N7-MTase domain of nsp14 contains an atypical MTase fold (Fig. 4A). The central β-sheet is made up of five β-strands instead of seven; the canonical β3 and β6 stands are missing. Among these strands, β2′, β1′, β3′, and β4′ are positioned parallel, whereas β8′ runs antiparallel. Contrarily, a seldom-observed insertion of a three-stranded antiparallel β-sheet between canonical strands β5 and β6 of the central sheet is observed in nsp14 (Fig. 4A). The presence of such a sheet has been noted in the structure of the N7-MTase from Vaccinia virus (PDB ID code 2VDW). This small β2-sheet is positioned almost perpendicular to the central β1-sheet. A cavity between sheets β1 and β2 functions as a ligand-binding pocket. Two small helices are embedded in the connecting loops of the β1-sheet, and a lone α-helix, α1′, is stacked against the opposite face of the central β1-sheet. Behind this α-helix lies another long α-helix, α2′. The third zinc finger of nsp14 formed by Cys452, Cys477, Cys484, and His487 is located at the tip of this helix and protrudes from the protein at its C terminal (Figs. 1B and 4A). Intriguingly, an additional α-helix, α3′ spanning the last 12 amino acids (Thr516–Gly527) is observed stabilizing the local hydrophobic environment (Fig. S5). Such a modification at the C terminus has been observed previously in the nucleic acid-binding SAM-dependent MTases (40). Truncation of this region has been shown to attenuate greatly or abolish the N7-MTase activity of nsp14 (20).
Fig. 4.
Structure and methyl transfer mechanism of the nsp14 N7-MTase domain. (A) Cartoon representation of the N7-MTase domain marked with secondary structural elements. (B) Amino acids within 4 Å of the ligands SAH (blue) and GpppA (red) are labeled and numbered. The magenta arrow indicates the methyl transfer. Dashed lines between residues indicate hydrogen bonds. Trp385, Asn386, Phe401, and Phe506 are shown by dashed bonds to depict their position below the plane of ligand GpppA. (C) The ability of nsp14 to methylate N7 of guanine of GpppA-RNA was measured. The results depict the efficiency of the conversion of substrate to product (%) and are plotted as a bar graph. WT nsp14 and its mutants were complexed with nsp10.
Fig. S5.
C-terminal hydrophobic interaction network of the N7-MTase domain of nsp14. The less stable region between Leu439 and Glu453 (shown in yellow) interacts with α2′ and the last α-helix α3′ via hydrophobic interactions. Key residues involved in formation of the hydrophobic network are shown as sticks and are labeled.
Structural Basis for Methylation of N7 of Guanine by nsp14.
In comparing the structures of unliganded, SAM-bound, and SAH–GpppA–bound nsp14–nsp10 complexes, no significant structure movement is observed, suggesting that the ligand-binding sites are preformed. The ligands bind in a pocket surrounded by sheets β1 and β2 and helix α1′ (Fig. 4A and Fig. S6).
Fig. S6.
Ligand-binding sites of the nsp14 N7-MTase domain. Substrates SAM (A) and GpppA and SAH (B) bind in pockets formed by the β1-sheet, β2-sheet, and α1′. Fo-Fc electron density maps (omit map) for SAM, GpppA, and SAH contoured at 2σ are shown.
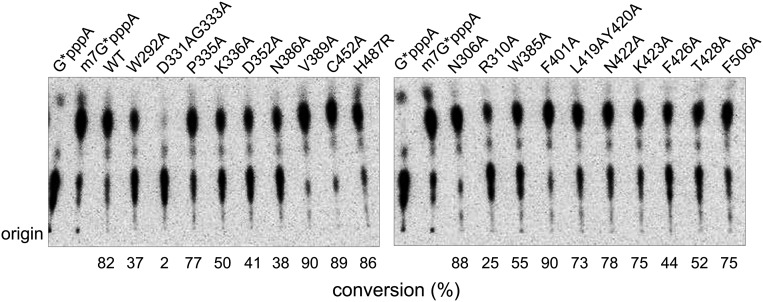
The methyl donor SAM sits above the central β1 sheet in the space between the loops connecting strands β2′ with β3′ and β3′ with β4′. The purine ring is bound in a hydrophobic environment created by amino acids such as Ile332, Phe367, and Val389 (Fig. 4B). The Oδ1 and Oδ2 of Asp352 form hydrogen bonds with the O3′ and O2′ atoms of the ribose, respectively (Fig. 4B). Substitution of Asp352 with alanine reduces the N7-MTase activity of nsp14 by 20% (Fig. 4C). Asp331 and Gly333, at the end of β1′, which previously were shown to be essential for the N7-MTase activity of nsp14 (20), are observed in proximity to the carboxyl group of SAM. The carbonyl oxygen of Gly333 forms a hydrogen bond with the amino nitrogen of methionine of SAM. Not surprisingly, a D331A/G333A double mutation completely abolished the N7-MTase activity (Fig. 4C). The Trp292 at the N-terminal loop of N7-MTase (Lys288–Asp301) stacks against the ribose for optimal positioning of SAM for catalysis. Disruption of the interaction of Trp292 with ribose reduced the N7-MTase activity by more than 50% (Fig. 4 B and C). These results are consistent with those reported for the methyltransferase from the Encephalitozoon cuniculi (Ecm1), in which mutations disrupting the interaction of the N7-MTase with ribose and methionine moieties of SAM impair activity greatly or completely, but those affecting interactions with adenine are not important for activity (41).
Methyl receptor GpppA binds near SAM. Side chains of Phe401, Tyr420, Phe426, Thr428, and Phe506 entrench and hold the purine moiety of guanosine in position (Fig. 4B). Among these amino acids, Phe426 showed the largest influence on the N7-MTase activity, and F426A mutation reduced MTase activity by 50% (Fig. 4C). Asn386 is located in immediate proximity to the atoms involved in methyl transfer and forms two hydrogen bonds with the guanine moiety to help orient it during catalysis. An N386A mutant abolished more than 50% of the N7-MTase activity (Fig. 4C). The Oδ atom of Asn422 is observed to form hydrogen bonds with the O3′ atom of the guanosine ribose moiety. Mutating it to alanine does not significantly impact the N7-MTase activity. Asn306, Arg310, and Lys336 contribute the positive potential for binding the triphosphate moiety. In particular, Nδ2 of Asn306 as well as Nη1 and Nη2 of Arg310 interact with the second phosphate group (Fig. 4B). The importance of these side chains in catalysis is underscored by the fact that the R310A mutant retained only 20% of the activity, whereas the K336A mutant retained 50% (Fig. 4C). Last, Trp385 stacks against the adenosine moiety of GpppA, and a W385A mutation reduced the activity by more than one third (Fig. 4 B and C).
The mutations R310A, P335A, K336A, D352A, and Y420A have been reported; the changes in the activity of these mutants are consistent with our results, except that Y420A was reported to be inactive (28), but we found that the L419A/Y420A double mutant retained 70% activity. We believe our results to be reasonable, based on the location of these two residues.
Thus, the ligands are held in a highly constricted pocket, ensuring that the methyl donor SAM and acceptor GpppA are held in close proximity. By enforcing charge and shape complementarity, the ligands are oriented in the pocket so that the methyl group of SAM is brought in the vicinity of the N7 of guanine to realize the methyl transfer using an in-line mechanism, as previously proposed for Ecm1 (41).
Intriguingly, zinc finger 3 of nsp14 located in the N7-MTase domain is spatially isolated from the active site, ruling out a direct role for this motif in catalysis (Fig. 4A). Consistent with this observation, C452A and H487R mutations of zinc finger 3 had only a marginal effect on the MTase activity (Fig. 4C). The peripheral location of zinc finger 3 may be more suited to forging protein–protein interactions, for example binding with the nsp16–nsp10 complex to accomplish the second methyl transfer reaction essential for formation of cap1 structure.
Raw TLC data of the N7-MTase activity assays are included as Fig. S7.
Fig. S7.
N7-MTase activity of nsp14 and its mutants. Nsp14 and its mutants were tested for their ability to methylate 32P-labeled G*pppA-RNA. After incubation for 6 min at 37 °C, the reaction products were digested by nuclease P1 into single nucleotides and methylated m7G*pppA or unmethylated G*pppA. They were analyzed by separating the mixture using TLC. The position of origin is marked on the left. The percentage of conversion of G*pppA-RNA to m7G*pppA-RNA is listed at the bottom.
The Interactions Between the Two Naturally Fused Domains.
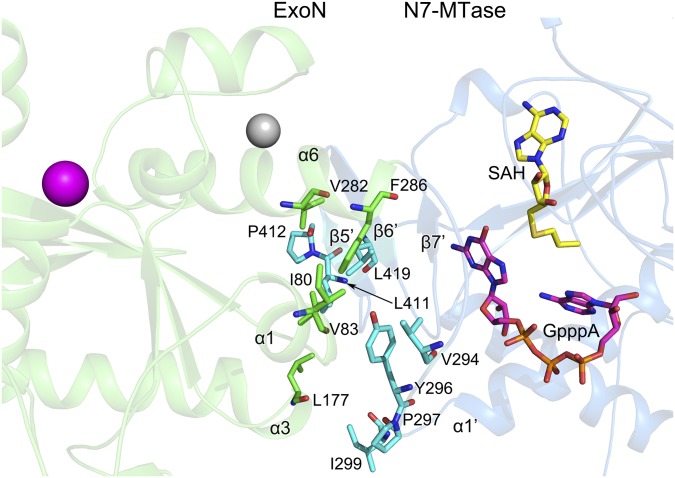
The ExoN and MTase domains of nsp14 are interlinked, and they interact using hydrophobic interactions (Fig. S8). Three α-helices of ExoN stabilize the N-terminal loop (Lys288–Asp301) and the β2-sheet of N7-MTase domain, the base of the substrate binding pocket of N7-MTase. Ile80 and Val83 of α1, Leu177 of α3, and Val282 and Phe286 of α6 form hydrophobic interactions with Val294, Tyr296, Pro297, Ile299, Leu411, Pro412, and Leu419. All these amino acids are highly conserved within members of the coronavirus genus (Fig. S3), highlighting an important conserved role for these residues (28).
Fig. S8.
The hydrophobic interactions between the ExoN domain and N7-MTase domain. Interfacing residues between the ExoN and N7-MTase domains are displayed as green and cyan sticks, respectively. Secondary structural elements involved in the interactions are labeled.
Model for the Role of nsp14 in Proofreading and mRNA Capping.
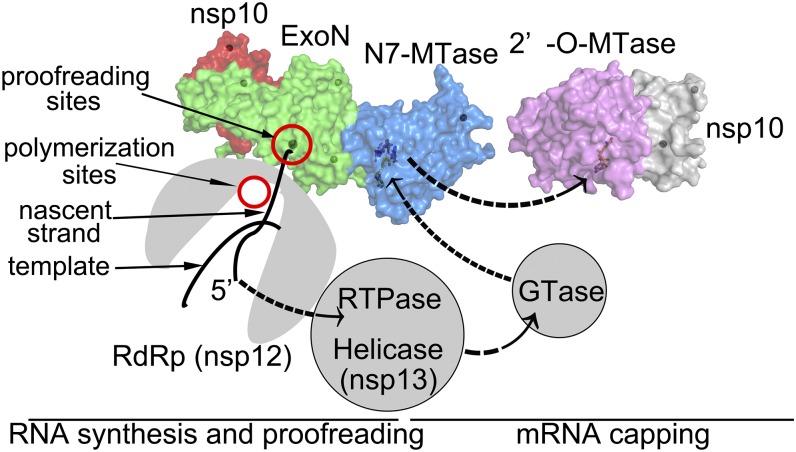
To envision how nsp14 could participate in the functioning of the replication and transcription complex (RTC), we attempted to infer the interaction mode between nsp14 and nsp12, the RdRp. The polymerization and proofreading sites of DNA polymerases are located on different domains or subunits that are typically separated by a distance of 30 Å or more (42). In contrast, during transcription, RNA polymerases carry out both these functions using the same active sites (43). In this context, nsp14 is functionally and structurally more similar to the proofreading exonuclease domain of DNA polymerases. Using the known structure of the Klenow fragment of DNA polymerase I (PDB ID code 1KLN) as a guide, the ExoN domain of nsp14 was placed next to the polymerization site (Fig. 5). During instances of mismatch of nucleotides, the nascent strand is moved to the ExoN site for excision. As the newly synthesized strand is extended, a cap0 structure is first assembled at the 5′ end. This assembly would require the enzymatic activities of a RTPase contributed by nsp13 (24), a guanylyl transferase, which is yet to be identified, and the N7-MTase activity of nsp14. Last, the 2′-O-MTase activity of nsp16 would result in the formation of a cap1 structure at the 5′ end of the newly synthesized RNA. Such a model accounts for nsp13 functioning as a helicase for unwinding double-stranded nucleic acids and the direct nsp13–nsp12 interactions (44, 45). Further experiments are necessary to shed more light on the relative orientation of the various active sites of the RTC and how the RNA is maneuvered for formation of the cap1 structure.
Fig. 5.
Model for the role of nsp14 in proofreading and mRNA capping. Nascent RNA is synthesized at the polymerization sites (red circle) of the nsp12 RdRp domain (gray) or is mismatch excised at the proofreading site (red circles) of nsp14 ExoN (green surface). The relative orientation of the polymerization and proofreading domains is built based on the Klenow fragment. The 5′ end of newly synthesized mRNA is modified by sequential activities contributed by RTPase of nsp13, GTase (currently unknown), N7-MTase of nsp14, and 2′-O–MTase of nsp16 (light pink surface, PDB ID code 3R24) for formation of a cap1 structure.
Conclusions
The structures of SARS-CoV nsp14 in complex with nsp10 described here reveal that the core structural elements and catalytic residues of the proofreading ExoN domain of nsp14 are highly similar to those of its eukaryotic and prokaryotic predecessors. Furthermore, nsp14 of SARS-CoV has gained some striking and remarkable structural features to accomplish additional tasks such as the methylation of N7 of guanine of cap precursors. Our studies unveil several of these nsp14-specific features. First, nsp10 contacts the nsp14 ExoN domain to provide structural support and facilitate its ExoN activity for proofreading. Second, two zinc fingers are located on the ExoN domain. Our mutagenesis studies show that both zinc fingers are essential for the nsp14 function. Third, the N7-MTase domain of nsp14 adopts an atypical MTase fold and ropes in a second β-sheet to bind the substrates in a highly constricted cavity to accomplish methyl transfer. This N7-MTase domain carries a third zinc finger that is located remote from the active sites. Combined with previous functional data, this work provides evidence for the likely mechanism underlying the involvement of nsp14 in viral replication and transcription.
Materials and Methods
Protein Production.
Full-length nsp10 and nsp14 of SARS-CoV isolate Tor2/FP1-10851 were synthesized by GENEWIZ Inc. and cloned into pGEX-6p-1 and pRSFDuet-1, respectively. Nsp10 with an N-terminal GST tag and nsp14 with a C-terminal His tag were coexpressed in E. coli BL21 (DE3). Cells were grown at 37 °C and induced with 0.1 mM isopropyl β-d-thiogalactoside when OD600nm reached 0.8. After induction at 16 °C for 18 h, cells were harvested, resuspended, and lysed by sonication in 20 mM Hepes (pH 7.0), 150 mM NaCl, 4 mM MgCl2, and 5% (vol/vol) glycerol. After centrifugation, recombinant protein was purified by GST-affinity chromatography. The GST tag of nsp10 was removed by PreScission protease. Excess nsp10 was separated from the nsp14–nsp10 complex by HiTrap S ion-exchange chromatography (GE Healthcare).
Mutagenesis was performed using the Easy Mutagenesis System (Transgen Biotech). All nsp14 mutants were coexpressed with nsp10 and purified as described above.
Nsp14 alone was purified using Ni-nitrilotriacetate affinity resin (GE Healthcare) with a gradient of imidazole for washing and elution.
Crystallization.
Crystallization was performed by the hanging-drop vapor-diffusion method at 16 °C. Each crystallization drop consisted of 1 μL of nsp14–nsp10 complex (10 mg/mL) mixed with 10 mM DTT and 0.25 mM SAM and 1 μL of the mother liquor equilibrated over 200 μL of reservoir solution. Diffraction-quality crystals grew in 15% (vol/vol) Tacsimate (pH 7.0), 0.1 M Hepes (pH 7.0), and 2% (wt/vol) polyethylene glycol 3,350 after optimization by microseeding. Diffraction was further improved by dehydration in 25% glycerol for 3 min. To obtain a complex of nsp14–nsp10 with SAM and GpppA, crystals were soaked with 50 mM GpppA (New England Biolabs).
Data Collection and Structure Determination.
X-ray diffraction data were collected at Shanghai Synchrotron Radiation Facility beamlines BL19U and 17B. All data were processed with the program HKL3000 (46). The initial phases were calculated by the single-wavelength anomalous dispersion method using the anomalous signal of zinc atoms with the program PHENIX.autosol (47). A total of 10 zinc atoms were found using HYSS (48). After phasing, twofold noncrystallographic symmetry was identified and used in subsequent density modification, dramatically improving the phases of diffraction data. Two molecules of nsp10 (PDB ID code 2G9T) were docked into the map using MOLREP (49). Other parts of the complex were manually built in COOT (50) and iteratively refined in PHENIX.refine (51). To solve the structure of the unliganded nsp14–nsp10 complex or nsp14–nsp10–SAH–GpppA complex, molecular replacement was done first in MOLREP using the structure of the nsp14–nsp10–SAM complex as the search model, and then the structures were refined in PHENIX.refine. Data collection and refinement statistics are listed in Table S1.
Exoribonuclease Activity Assay.
ssRNA made up of 22 nucleotides (RNA22, 5′-GGGCGAUUAGGAGCUAACUGCG-3′) was used as a substrate for activity assays (16). To obtain 5′-labeled RNA22, it was incubated with T4 polynucleotide kinase (New England Biolabs) and γ-[32P]-ATP (PerkinElmer). MicroSpin G-25 columns (GE Healthcare) were used to remove excess γ-[32P]-ATP. Later, RNA22 was extracted with phenol-chloroform and precipitated with ethanol.
Reaction mixtures contained 300 nM nsp14–nsp10 complex (or nsp14 mutant–nsp10 complex or nsp14 alone), 1,000-cpm labeled RNA22, and 300 nM unlabeled RNA22 in a buffer made up of 50 mM Hepes (pH 7.0), 50 mM NaCl, 5 mM MgCl2, and 1 mM DTT. After incubation at 37 °C for 5 or 30 min, the reactions were stopped by the addition of an equal volume of loading buffer (96% formamide with 10 mM EDTA). Products were separated on 20% 7-M urea-containing polyacrylamide gels and visualized through PhosphorImager.
MTase Activity Assay.
DNA fragments including the optimized T7 class II promoter Ф2.5 with ATP as initial nucleotide (52) and the 5′-terminal 259 nucleotides of the SARS-CoV genome (20) were used as a template for in vitro transcription. We then used the Vaccinia capping system (except for SAM) (New England Biolabs) and α-[32P]-GTP to label the 5′ terminus of the RNA to G*pppA-RNA. To improve the efficiency of the reaction, 0.05 U inorganic pyrophosphatase (New England Biolabs) was added. G-50 Sephadex columns (Roche) were used to remove unused α-[32P]-GTP. RNA was extracted with phenol-chloroform and precipitated with ethanol.
MTase activity was tested as follows: 0.1 µg nsp14–nsp10 complex or nsp14 mutant–nsp10 complex was mixed with 1,000-cpm labeled RNA in a buffer made up of 50 mM Hepes (pH 7.0), 6 mM KCl, 5 mM DTT, 1 mM MgCl2, and 0.2 mM SAM. After incubation at 37 °C for 6 min, 5 µg nuclease P1 (Sigma) and 1 mM ZnCl2 were added to digest the RNA. Reaction products were spotted on polyethylenimine cellulose plates (Merck) to separate G*pppA from capped m7G*pppA and were visualized using a PhosphorImager.
The marker m7G*pppA was prepared as above, except that inorganic pyrophosphatase was replaced by 0.2 mM SAM.
Acknowledgments
We thank Deyin Guo, Jun Li, Cong Zeng, Zhenhua Ming, Xiangxi Wang, Wei Wang, Xiuna Yang, and Yu Dong for technical assistance and valuable advice. This work was supported by Ministry of Science and Technology of China Project 973 Grants 2014CB542800 and 2014CBA02003, National Natural Science Foundation of China Grant 81330036, and Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB08020200.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.K.A. is a guest editor invited by the Editorial Board.
Data deposition: The structures reported in this article have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5C8S, 5C8T, and 5C8U).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508686112/-/DCSupplemental.
References
- 1.De Groot R, et al. Order nidovirales. In: King AM, Adams MJ, Lefkowitz EJ, editors. Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; London: 2011. pp. 785–795. [Google Scholar]
- 2.Peiris JS, et al. SARS study group Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 4.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. Nidovirales: Evolving the largest RNA virus genome. Virus Res. 2006;117(1):17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snijder EJ, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhai Y, et al. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat Struct Mol Biol. 2005;12(11):980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imbert I, et al. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25(20):4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subissi L, et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci USA. 2014;111(37):E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckerle LD, Lu X, Sperry SM, Choi L, Denison MR. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J Virol. 2007;81(22):12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckerle LD, et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6(5):e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denison MR, Graham RL, Donaldson EF, Eckerle LD, Baric RS. Coronaviruses: An RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8(2):270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith EC, Blanc H, Vignuzzi M, Denison MR. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: Evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9(8):e1003565. doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith EC, Denison MR. Coronaviruses as DNA wannabes: A new model for the regulation of RNA virus replication fidelity. PLoS Pathog. 2013;9(12):e1003760. doi: 10.1371/journal.ppat.1003760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouvet M, et al. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc Natl Acad Sci USA. 2012;109(24):9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith EC, et al. Mutations in coronavirus nonstructural protein 10 decrease virus replication fidelity. J Virol. 2015;89(12):6418–6426. doi: 10.1128/JVI.00110-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minskaia E, et al. Discovery of an RNA virus 3′->5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci USA. 2006;103(13):5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham RL, et al. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat Med. 2012;18(12):1820–1826. doi: 10.1038/nm.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nga PT, et al. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 2011;7(9):e1002215. doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zirkel F, et al. An insect nidovirus emerging from a primary tropical rainforest. MBio. 2011;2(3):e00077–11. doi: 10.1128/mBio.00077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci USA. 2009;106(9):3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89(6):951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 22.Nallagatla SR, Toroney R, Bevilacqua PC. A brilliant disguise for self RNA: 5′-end and internal modifications of primary transcripts suppress elements of innate immunity. RNA Biol. 2008;5(3):140–144. doi: 10.4161/rna.5.3.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Decroly E, Ferron F, Lescar J, Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol. 2012;10(1):51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivanov KA, et al. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J Virol. 2004;78(11):5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouvet M, et al. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6(4):e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, et al. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7(10):e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Decroly E, et al. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7(5):e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, et al. Structure-function analysis of severe acute respiratory syndrome coronavirus RNA cap guanine-N7-methyltransferase. J Virol. 2013;87(11):6296–6305. doi: 10.1128/JVI.00061-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue) suppl 2:W545-9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beese LS, Steitz TA. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: A two metal ion mechanism. EMBO J. 1991;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamdan S, Carr PD, Brown SE, Ollis DL, Dixon NE. Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure. 2002;10(4):535–546. doi: 10.1016/s0969-2126(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 32.Chen P, et al. Biochemical characterization of exoribonuclease encoded by SARS coronavirus. J Biochem Mol Biol. 2007;40(5):649–655. doi: 10.5483/bmbrep.2007.40.5.649. [DOI] [PubMed] [Google Scholar]
- 33.Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985;313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 34.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: Structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29(5):1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernad A, Blanco L, Lázaro JM, Martín G, Salas M. A conserved 3′----5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59(1):219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- 36.Joseph JS, et al. Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs. J Virol. 2006;80(16):7894–7901. doi: 10.1128/JVI.00467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su D, et al. Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10. J Virol. 2006;80(16):7902–7908. doi: 10.1128/JVI.00483-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bouvet M, et al. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J Biol Chem. 2014;289(37):25783–25796. doi: 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lugari A, et al. Molecular mapping of the RNA Cap 2′-O-methyltransferase activation interface between severe acute respiratory syndrome coronavirus nsp10 and nsp16. J Biol Chem. 2010;285(43):33230–33241. doi: 10.1074/jbc.M110.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin JL, McMillan FM. SAM (dependent) I AM: The S-adenosylmethionine-dependent methyltransferase fold. Curr Opin Struct Biol. 2002;12(6):783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 41.Fabrega C, Hausmann S, Shen V, Shuman S, Lima CD. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol Cell. 2004;13(1):77–89. doi: 10.1016/s1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
- 42.Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 43.Sydow JF, Cramer P. RNA polymerase fidelity and transcriptional proofreading. Curr Opin Struct Biol. 2009;19(6):732–739. doi: 10.1016/j.sbi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 44.Tanner JA, et al. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J Biol Chem. 2003;278(41):39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adedeji AO, et al. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS One. 2012;7(5):e36521. doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otwinowski Z, Minor W. Processing of X-ray diffraction data. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 47.Terwilliger TC, et al. Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 6):582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grosse-Kunstleve RW, Adams PD. Substructure search procedures for macromolecular structures. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 11):1966–1973. doi: 10.1107/s0907444903018043. [DOI] [PubMed] [Google Scholar]
- 49.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Cryst. 1997;30(6):1022–1025. [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coleman TM, Wang G, Huang F. Superior 5′ homogeneity of RNA from ATP-initiated transcription under the T7 ϕ 2.5 promoter. Nucleic Acids Res. 2004;32(1):e14. doi: 10.1093/nar/gnh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 54.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(Web Server issue):W320-4. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]