Significance
Because of its selective expression on the surface of a variety of different cancer cells, but not on their normal counterparts, nucleolin (NCL) represents an attractive target for antineoplastic treatments. However, previously described NCL-targeting molecules, although promising, still suffer from intrinsic limitations. Here, we describe the identification of the first fully human anti-NCL immune-based agent displaying antineoplastic activity against solid tumors, both in vitro and in vivo. This molecule could represent the prototype of a novel class of NCL-targeting drugs with enormous clinical potential as tools for the diagnosis and therapy of a wide range of human cancers.
Keywords: nucleolin, microRNA, scFv, cancer immunotherapy, phage-display
Abstract
Nucleolin (NCL) is a nucleocytoplasmic protein involved in many biological processes, such as ribosomal assembly, rRNA processing, and mRNA stabilization. NCL also regulates the biogenesis of specific microRNAs (miRNAs) involved in tumor development and aggressiveness. Interestingly, NCL is expressed on the surface of actively proliferating cancer cells, but not on their normal counterparts. Therefore, NCL is an attractive target for antineoplastic treatments. Taking advantage of phage-display technology, we engineered a fully human single-chain fragment variable, named 4LB5. This immunoagent binds NCL on the cell surface, it is translocated into the cytoplasm of target cells, and it abrogates the biogenesis of NCL-dependent miRNAs. Binding of 4LB5 to NCL on the cell surface of a variety of breast cancer and hepatocellular carcinoma cell lines, but not to normal-like MCF-10a breast cells, dramatically reduces cancer cell viability and proliferation. Finally, in orthotopic breast cancer mouse models, 4LB5 administration results in a significant reduction of the tumor volume without evident side effects. In summary, here we describe, to our knowledge, the first anti-NCL single-chain fragment variable displaying antineoplastic activity against established solid tumors, which could represent the prototype of novel immune-based NCL-targeting drugs with clinical potential as diagnostic and therapeutic tools in a wide variety of human cancers.
Nucleolin (NCL) is one of the most abundant nonribosomal proteins in the nucleolus (1), first identified in ribosomal RNA processing (2). Additional studies have demonstrated that NCL is a multifunctional nucleocytoplasmic protein, involved in ribosomal assembly, chromatin decondensation, transcription, nucleo-cytoplasmic import/export, and chromatin remodeling (3, 4). NCL is frequently up-regulated in cancer and in cancer-associated endothelial cells compared with normal tissues (5, 6), where it is also present on the cell surface (7, 8). Altered NCL expression and localization results in oncogenic effects, such as stabilization of AKT, Bcl-2, Bcl-XL, and IL-2 mRNAs (9–11). Moreover, surface-NCL acts as a receptor for several oncogenic ligands (12–15) and viruses (16). Recently, we reported that NCL has a critical protumorigenic function regulating the biogenesis of selected microRNAs (miRNAs), a class of noncoding single-stranded RNAs 19–22 nt in length (17) that regulate gene expression at the posttranscriptional level by targeting mRNAs in a sequence-specific manner (18). In fact, NCL enhances the maturation of specific miRNAs (including miR-21, miR-221, and miR-222) causally involved in cancer pathogenesis and resistance to several antineoplastic treatments (19–23). Our findings demonstrated that NCL modulates the biogenesis of these miRNAs at the posttranscriptional level, enhancing their maturation from pri- to premiRNAs, identifying a novel NCL-dependent oncogenic mechanism (19).
Because of its oncogenic role and specific expression on cancer cells surface, NCL represents an attractive target for antineoplastic therapies (24). Several groups have attempted to develop molecules, such as aptamers (AS1411) or peptides (HB-19, V3 loop-mimicking pseudopeptide, N6L, and F3) (24–28), to bind and to inhibit NCL in cancer cells. These compounds have also been suggested as potential carriers for the targeted delivery into cancer cells of several antineoplastic agents (25, 29). Although promising, aptamers and peptides targeting NCL suffer from intrinsic limitations, such as extremely short half-life, undesired immunostimulatory actions, and still unknown toxicological effects (15).
Here, we report the isolation by phage-display of a fully human single-chain Fragment variable (scFv) antibody, named 4LB5, which binds with high affinity to the RNA binding domain (RBD) of NCL. 4LB5 impairs the interaction between NCL and its regulated miRNAs, affecting the biogenesis of miR-21, miR-221, and miR-222. NCL targeting with 4LB5 in vitro results in a marked decrease of cancer cell viability and proliferation, and administration of 4LB5 in vivo causes dramatic reduction of growth of established orthotopic breast cancer xenografts. To our knowledge, 4LB5 is the first scFv displaying antineoplastic activity in vivo. These results indicate that targeting NCL-RBD is a promising effective approach for cancer therapy, and indicate 4LB5 as the prototype of a novel class of antitumoral compounds.
Results
Selection, Purification, and Characterization of Anti-NCL scFv.
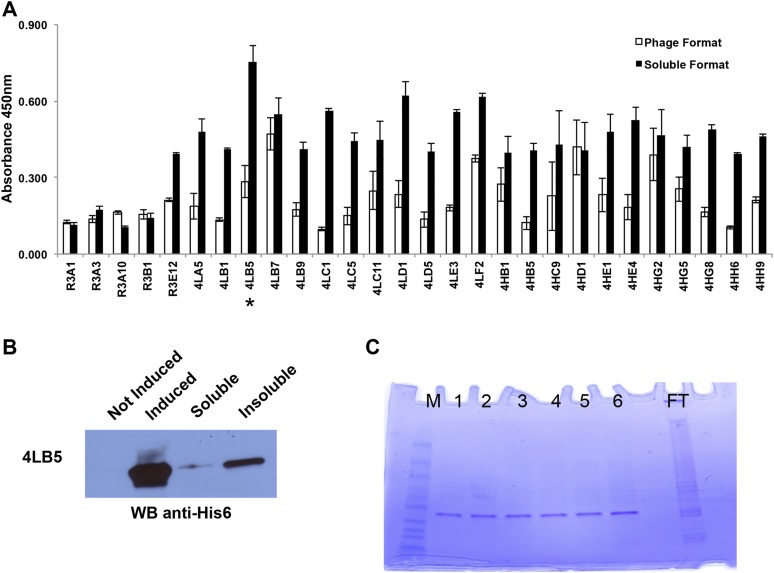
Using purified recombinant NCL-RBD (20 μg/mL) as bait, we performed four rounds of selection of scFvs from the Griffin.1 library (30). A fifth round of selection, using a lower amount (10 μg/mL) of recombinant protein, was also carried out to select phages with higher affinity for NCL-RBD. Ninety-six clones (from the third, fourth, and fifth round of selection) were analyzed by ELISA to select the best binders for recombinant NCL-RBD. Five, 11, and 12 phage clones were selected for further analysis from each round of selection, respectively. Results confirmed the binding of selected phages to recombinant NCL-RBD (Fig. S1A, white columns).
Fig. S1.
Selection and purification of human anti-NCL scFvs by phage display. (A) Binding of selected phage clones or soluble scFvs to NCL was assessed by ELISA using NCL-coated plates incubated with the indicated clones. Clone 4LB5 used for further experiments is indicated with an asterisk (*). The assay was performed three times in triplicate using different preparation of phages and scFvs, and mean ± SD is reported. (B) Selected clone named 4LB5 was subcloned in pET22b E. coli expression vector and transformed in BL21-DE3 bacterial cells. (C) IPTG-induced scFv was extracted from inclusion bodies, then refolded using urea gradient, and finally purified using Ni-NTA columns. FT, flow through; M, molecular standard; 1–6, Different elutions of the scFV.
Selected anti-NCL scFv clones were then analyzed in their soluble form by transforming bacterial SF110 cells with the pHEN2 phagemid vector (31) extracted from these clones. Periplasmic extracts, obtained following isopropyl-1-thio-β-d-galactopyranoside (IPTG) induction, were analyzed by ELISA (Fig. S1A, black columns), and clone 4LB5, identified in the fifth round of selection, exhibited the greatest affinity in the soluble form. Sequence analysis (www.vbase2.org) of clone 4LB5 indicated that the scFv VH belongs to the VH4 family (derived from the VH germ-line gene DP-71), whereas the VL belongs to the VL3 family (derived from the VL germ-line gene DPL-16).
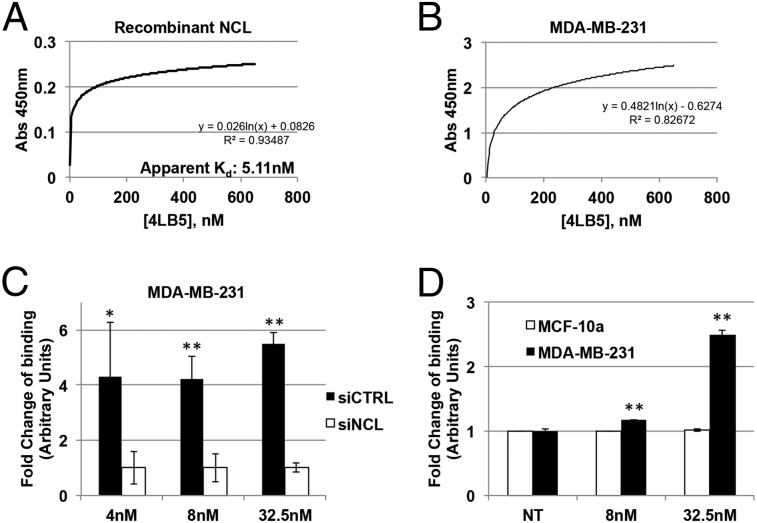
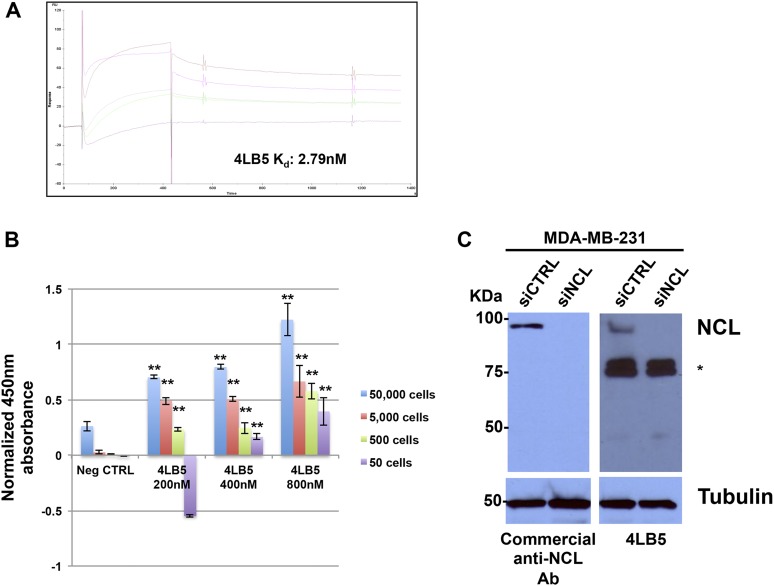
To obtain higher amounts of scFv, we subcloned 4LB5 cDNA into the pET22b(+) prokaryotic expression vector, fused with a C-terminal hexahistidine (His6) tag for its purification, and transformed in the Escherichia coli BL21(DE3) strain. However, fractionation of soluble and insoluble bacterial proteins revealed that 4LB5 scFv was mainly expressed in the insoluble form (Fig. S1B). For this reason, 4LB5 required denaturation before purification. SDS/PAGE analysis showed purified 4LB5 scFv to be ∼27 kDa in size and about 90% pure (Fig. S1C). To determine its binding affinity to NCL-RBD, we performed ELISA using increasing concentrations (0.1–600 nM) of purified 4LB5. As shown in Fig. 1A, significant binding to NCL-RBD was observed, with an apparent Kd of 5.11 nM. To further quantify the binding properties of 4LB5, surface plasmon resonance analysis was performed with different concentrations of 4LB5 (Fig. S2A). In our experimental settings, 4LB5 displayed a dissociation rate constant (kd) of 3.25 × 10−4 s−1, with equilibrium KD value of 2.79 nM.
Fig. 1.
Anti-NCL scFV 4LB5 specifically binds NCL in vitro and on the cell surface. (A) 4LB5 affinity for recombinant NCL was assessed by ELISA using different amounts of scFv. Apparent Kd is also indicated. Curve equation and R2 are also reported. (B) ELISA performed on MDA-MB-231 using different amounts of 4LB5. Curve equation and R2 are also reported. (C) ELISA performed using different amounts of 4LB5 on MDA-MB-231 cells following control (siCTRL) or anti-NCL (siNCL) siRNA transfection. *P < 0.05, **P < 0.01. (D) ELISA on MCF-10a (surface-NCL−) and MDA-MB-231 (surface-NCL+) breast cancer cells. **P < 0.01. All of the experiments are representative of three independent experiments performed in triplicate. Mean ± SD is reported.
Fig. S2.
Kinetic evaluation of 4LB5 binding to recombinant NCL and 4LB5 specific binding to NCL. (A) 4LB5 affinity for recombinant NCL-RBD was assessed by SPR using increasing concentrations (1–50 nM) of scFv. Resulting Kd is also indicated. (B) The detection limit of ELISA using 4LB5 (see Fig. 1) was assessed using different amounts of scFv and indicated numbers of MDA-MB-231 cells. Data (normalized for background levels) are representative of two independent experiments performed in triplicate, ± SD, **P < 0.01, compared with the corresponding negative control. (C) Specific binding of 4LB5 to NCL assessed by Western Blot on MDA-MB-231 cells transfected with control (siCTRL) or anti-NCL (siNCL) siRNA. 4LB5 was used as a primary antibody and HRP-conjugated anti-His6 as secondary antibody. Bands indicated by the asterisk are caused by the secondary anti-His6 antibody used for the detection of the NCL-bound scFv. Tubulin was used as a loading control.
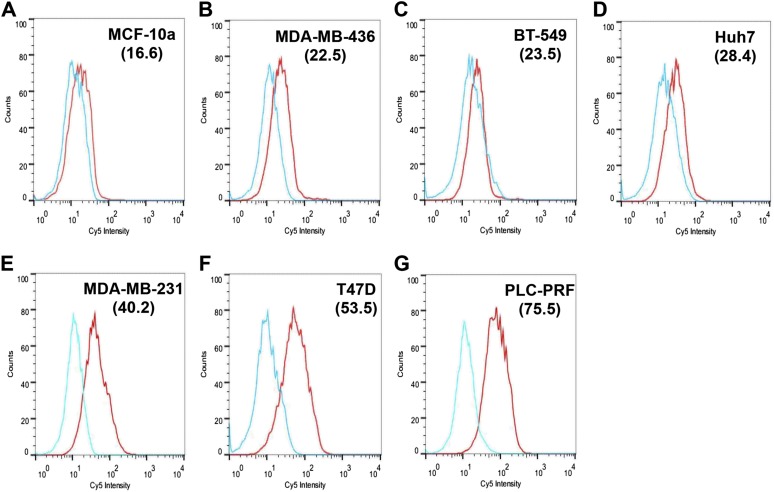
The binding of 4LB5 to surface-NCL was then assessed by ELISA using MDA-MB-231 breast cancer cells, which express high levels of surface-NCL (19). Fig. 1B shows the efficient binding of 4LB5 to the surface of these cells. To evaluate the detection limit of the ELISA performed using our scFv, the assay was performed using different amounts of MDA-MB-231 cells and different concentrations of 4LB5. As shown in Fig. S2B, at concentrations ranging from 400 to 600 nM, 4LB5 was able to detect as low as 50 cells, compared with the negative control. However, when used at 200 nM and 50 cells were plated, 4LB5 resulted in a signal that was not significantly different from the background. To confirm that the observed binding was caused by the specific interaction between 4LB5 and NCL, MDA-MB-231 cells were transfected with a control (siCTRL) or anti-NCL–specific siRNAs (siNCL) and analyzed by ELISA using different concentrations of 4LB5. Abrogation of NCL expression resulted in a significant reduction of 4LB5 binding (Fig. 1C). Western blot analysis of total MDA-MB-231 cell extracts using 4LB5 as a primary antibody further confirmed that 4LB5 was able to discriminate between siCTRL- and siNCL-transfected cells like the commercial anti-NCL antibody (Fig. S2C). We also observed, by ELISA, the differential binding of 4LB5 to MDA-MB-231 cells compared with normal-like MCF-10a breast cells expressing low levels of surface-NCL (19, 32) (Fig. 1D). Finally, we investigated the ability of 4LB5 to bind cancer cells by flow cytometry using a Cy5-labeled 4LB5 and different breast or hepatocellular carcinoma (HCC) cell lines (Fig. S3). In line with data shown in Fig. 1D, we did not detect binding of Cy5-4LB5 to surface-NCL− MCF-10a, compared with the Cy5 label alone (Fig. S3). Conversely, we detected strong binding of 4LB5 to MDA-MB-231 and T47D (breast cancer cells) and to PLC-PRF (HCC). Reduced binding was detected on breast cancer cells BT-549 and MDA-MB-436, but also on Huh7 HCC cells (Fig. S3). These data demonstrate that 4LB5 specifically binds to various cancer cell lines but not to normal-like breast cells or following abrogation of NCL expression.
Fig. S3.
4LB5 binds NCL on the surface of different cancer cell lines. Indicated cell lines—(A) MCF-10a normal-like breast, (B) MDA-MB-436 basal B TNBC, (C) BT-549 basal B TNBC, (D) Huh7 HCC, (E) MDA-MB-231 basal B TNBC, (F) T47D luminal breast cancer, (G) PLC-PRF, HCC—were stained (red line) or not (light blue line) for 1 h with 2 μg/mL Cy5.5-labeled 4LB5 and analyzed by flow cytometry. Mean fluorescent index is also reported in parenthesis. Data are representative of three independent experiments performed in duplicate.
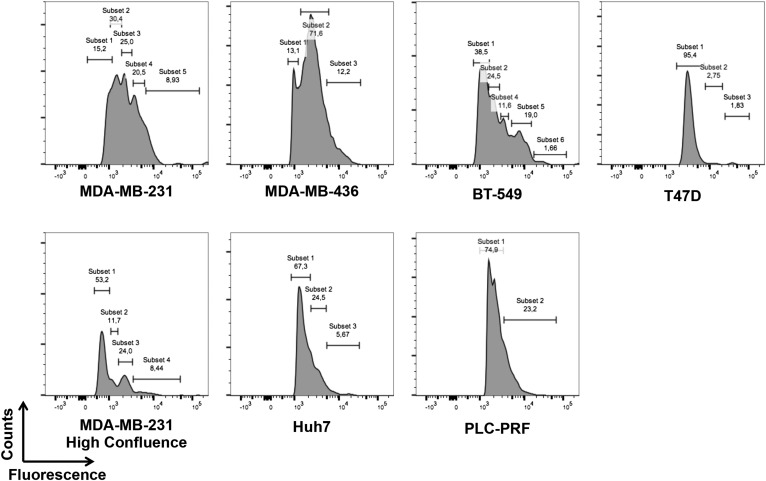
We also wanted to evaluate whether the cell lines used in this study might display heterogeneity in the levels of surface NCL. To this end, we performed an analysis by flow cytometry using a commercially available antibody against NCL (Fig. S4). Interestingly, most of the cancer cell lines that were used display a significant heterogeneity in surface-NCL expression levels and we observed the presence of different subpopulations in the majority of them. Of note, when MDA-MB-231 cells were cultured until they reached the maximum confluence, the amount of surface-NCL+ cells was reduced, especially those populations expressing higher levels of surface NCL. These observations are in line with previous findings indicating NCL as a useful marker of cell proliferation (5, 33).
Fig. S4.
Heterogeneous levels of surface NCL on the cancer cell lines used in the study. Indicated cell lines were stained using a commercially available anti-NCL antibody and analyzed by flow cytometry using the FlowJo software. The relative abundance of different subpopulations, expressing different levels of surface NCL, is reported.
Internalization of 4LB5 scFv.
NCL is able to shuttle between the cell surface and the cytoplasm of cancer cells (32, 34), a property that makes NCL an attractive target for the selective delivery of antineoplastic drugs or prodrugs, leaving normal cells unaffected (25).
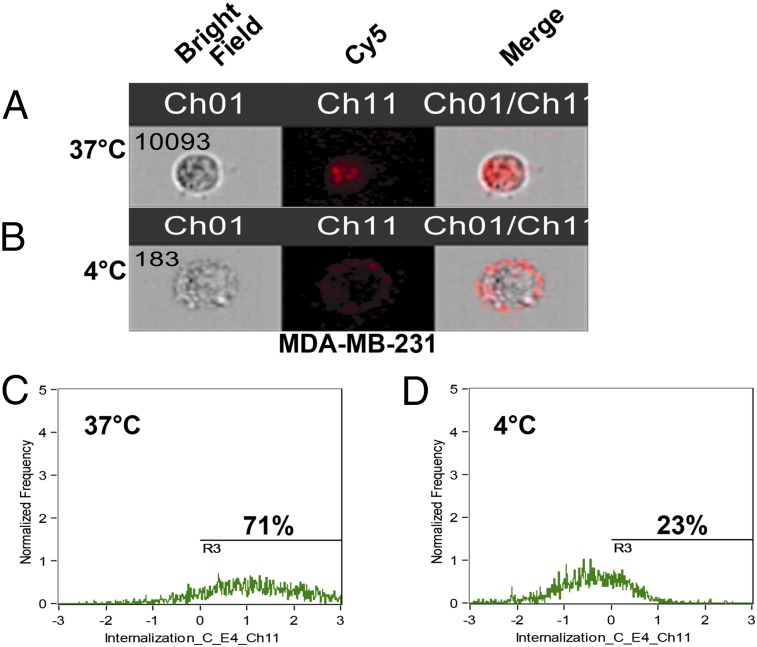
To test the ability of 4LB5 to undergo NCL-mediated endocytosis, MDA-MB-231 cells were incubated with Cy5-labeled 4LB5 (Cy5-4LB5) at 37 °C or at 4 °C for 6 h. Cells were extensively washed with PBS, harvested, and analyzed by Amnis FlowSight. Fig. 2 A and B shows representative bright field (Fig. 2 A and B, Left), Cy5 fluorescence (Fig. 2 A and B, Center), and merged (Fig. 2 A and B, Right) images of the analyzed cells. At 37 °C the majority of Cy5-4LB5 was mainly found in the cytoplasm, whereas at 4 °C, when the active mechanisms of NCL intracytoplasmic relocalization are slowed, Cy5-4LB5 remained on the cell surface. Quantitative analysis (Fig. 2 C and D) confirmed higher fluorescent internalization score for the cells incubated at 37 °C vs. 4 °C, when shuttling mechanisms are slowed.
Fig. 2.
4LB5 is internalized by target cells. MDA-MB-231 cells were incubated for 6 h at 37 °C (A) or 4 °C (B) with Cy5-labeled 4LB5. Cells were then harvested and analyzed using a FlowSight instrument (AMNIS) to acquire bright field (Ch01), Cy5 (Ch11), and merged images. At least 10.000 cells were acquired for each experimental point. (C and D) Internalization analysis was performed using the FlowSight Internalization wizard and quantification of cells internalizing 4LB5 in the two conditions is reported.
4LB5 scFv Affects miRNA Biogenesis.
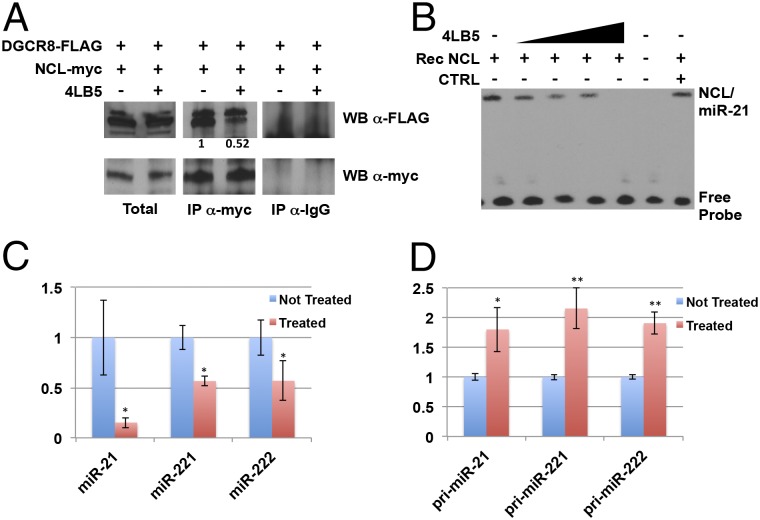
Because NCL has been shown to associate with DGCR8, one of the components of the microRNA microprocessor complex (19, 35, 36), we evaluated the effects of 4LB5 on this interaction in HeLa cells expressing FLAG-tagged DGCR8 and myc-tagged NCL. Fig. 3A shows that 4LB5 reduced the amount of coimmunoprecipitated NCL-myc and DGCR8-FLAG (fold-change 0.51).
Fig. 3.
Anti-NCL 4LB5 scFV inhibits microRNA biogenesis. (A) HeLa cells, transfected as indicated, were incubated for 24 h with or without 120 nM 4LB5. The interaction between myc-tagged NCL and FLAG-tagged DGCR8 was assessed by coimmunoprecipitation followed by SDS/PAGE and Western blot. Control IgG was used as negative control to evaluate the specificity of the anti-Myc antibody for the immunoprecipitation. (B) REMSA performed by incubating recombinant NCL and biotin-labeled miR-21 in the presence of increasing amounts (80–650 nM) of 4LB5. Control IgG was used as negative control (CTRL). (C and D) NCL-dependent microRNA levels were analyzed by real-time PCR 72 h following 15 nM 4LB5 treatment. Both mature (C) and primary (D) miRNA levels were analyzed. Data are the average of three independent experiments performed in triplicate. *P < 0.05, **P < 0.01.
NCL enhances the maturation of a subset of miRNAs (including miR-21, -221, and -222), and its inhibition by siRNAs or anti-NCL aptamers leads to down-regulation of these mature miRNAs and accumulation of their primary forms (19). Therefore, we assessed the ability of NCL to bind its target miRNAs in the presence of 4LB5 by RNA-EMSA (REMSA). As shown in Fig. 3B, 4LB5 reduced or completely abrogated the formation of the NCL/miR-21 complex, but no effect was observed when an unrelated control IgG was incubated with the complex.
Finally, MDA-MB-231 breast cancer cells were treated with 4LB5 or left untreated, and RNA was extracted after 72 h. In agreement with our previous studies, real-time analysis revealed that the mature forms of miR-21, miR-221, and miR-222 were significantly reduced by treatment with 4LB5 (Fig. 3C), but their primary forms (pri-miRNA) accumulated after the treatment (Fig. 3D).
Taken together, these data indicate that 4LB5 inhibits the interaction between NCL and the microprocessor complex, impairing the maturation of NCL-associated miRNAs.
4LB5 scFv Affects Cancer Cell Viability, Proliferation, and Migration in Vitro.
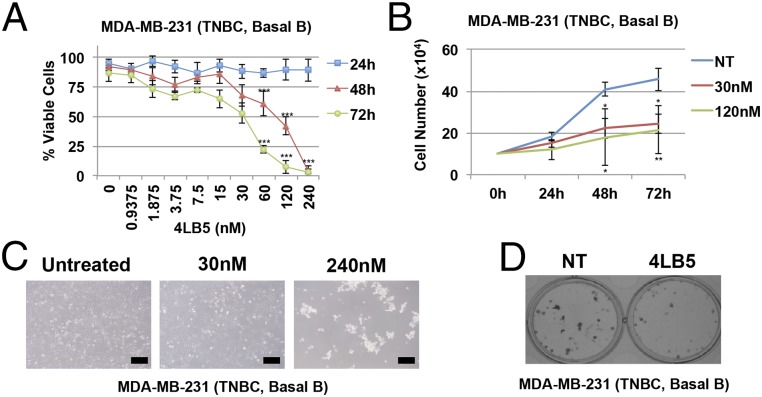
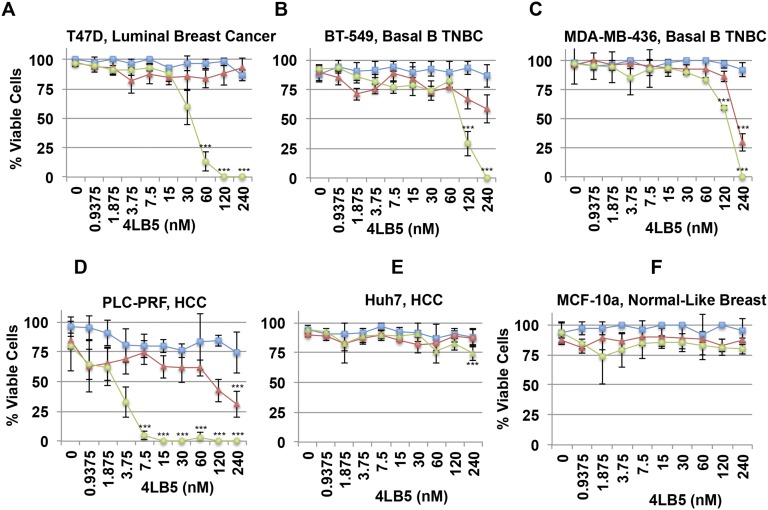
Several reports have shown that NCL inhibition by siRNAs or anti-NCL aptamers affects cell viability, proliferation, and migration (14, 19, 37–43). To assess the effects of 4LB5 on cell viability and proliferation, we performed a dose–response (0.9–240 nM) experiment using the scFv on MDA-MB-231 triple-negative breast cancer (TNBC) cells at different time points (24, 48, and 72 h). As shown in Fig. 4A, a significant reduction of cell viability was observed after 48 and 72 h of treatment, with an IC50 of ∼30 nM at 72 h. Growth curves and colony assays (Fig. 4 B–D) also indicated a significant reduction in cell proliferation at 48 and 72 h of treatment. Similar results were also observed on T47D (ER+, PgR+ luminal breast cancer) (IC50 ∼20 nM), BT549 (Basal B TNBC) (IC50 ∼58 nM), MDA-MB-436 (Basal B TNBC) (IC50 ∼50 nM), and PLC-PRF (HCC) (IC50 ∼3 nM) cell lines (Fig. S5 A–D), but no effect was observed on Huh7 (HCC) (Fig. S5E) or MCF-10a (normal-like breast) cells (Fig. S5F). The different response displayed by the cancer cell lines might be dependent on several factors, including, but not limited to, the relative abundance of subpopulations expressing different levels of surface NCL (Fig. S4), the different expression levels of NCL-dependent miRNAs and the different oncogenic pathways modulated by NCL in each different cellular context.
Fig. 4.
4LB5 affects cancer cell proliferation and survival. (A) Basal B TNBC cells MDA-MB-231 were treated with increasing amounts of 4LB5. Viable cells were counted using Trypan blue staining at different time points (light blue squares, 24 h; red triangles 48 h; green circles, 72 h). All of the experiments are representative of four independent experiments performed in triplicate. Mean ± SD is reported. *P < 0.05, **P < 0.01, ***P < 0.001. (B) Growth curves performed on MDA-MB-231 cells left untreated or treated with 30 nM or 120 nM 4LB5. Total cells were counted at the indicated time points. All of the experiments are representative of three independent experiments performed in triplicate. Mean ± SD is reported. *P < 0.05, **P < 0.01, ***P < 0.001. (C) Representative images of the cells shown in A, treated for 72 h, as indicated. (Scale bars, 100 μm.) (D) Colony assay on MDA-MB-231 cells treated with or without 30 nM 4LB5, and stained after 10 d with Crystal violet. All of the experiments are representative of three independent experiments performed in triplicate.
Fig. S5.
4LB5 affects cancer cell survival. Indicated panels were treated with increasing amount of 4LB5: (A) T47D, (B) BT-549, (C) MDA-MB-436, (D) PLC-PRF, (E) Huh7; (F) MCF-10a. Viable cells were counted using trypan blue staining at different time points (24, 48, and 72 h). All of the experiments are representative of three independent experiments performed in quadruplicate. Mean ± SD is reported. ***P < 0.001.
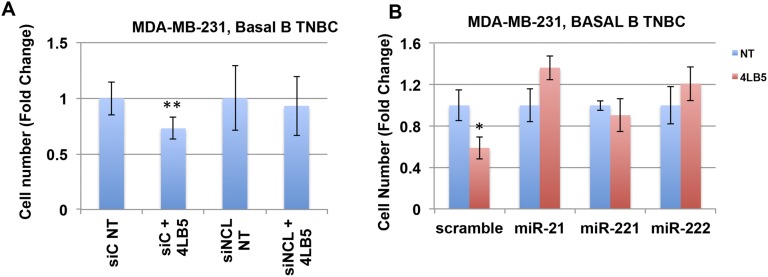
To confirm that the cytotoxic effect of 4LB5 was dependent on the specific binding of the scFv to NCL, MDA-MB-231 cells were transfected with anti-NCL siRNAs (siNCL) and treated with 4LB5. Fig. S6A shows that 4LB5 treatment failed to inhibit cell proliferation of MDA-MB-231 cells with abolished NCL expression compared with cells transfected with siNCL and not treated with the scFv. Moreover, we also assessed whether the cytotoxic effect of NCL inhibition could be rescued by the overexpression of mature miRNAs, whose biological activity is not dependent on NCL. Fig. S6B shows that overexpression of NCL-regulated miRs, such as mature miR-21, miR-221, and miR-222, prevented 4LB5 mediated inhibition of cell proliferation.
Fig. S6.
4LB5 cytotoxic effect depends on surface-NCL expression and is prevented by overexpression of specific miRNAs. (A) MDA-MB-231 cells were transfected with control (siC) or anti-NCL (siNCL) siRNAs for 24 h, and then untreated or treated with 30 nM 4LB5 for 48 h. Total cells were counted. Data are representative of three independent experiments performed in quadruplicate. Mean ± SD is reported. **P < 0.01. (B) MDA-MB-231 cells were transfected with scramble RNA or indicated mature miRNAs for 24 h, and treated or not with 50 nM 4LB5 for 48 h. Total cells were counted. Data are representative of two independent experiments performed in quadruplicate. Mean ± SE is reported. *P < 0.05.
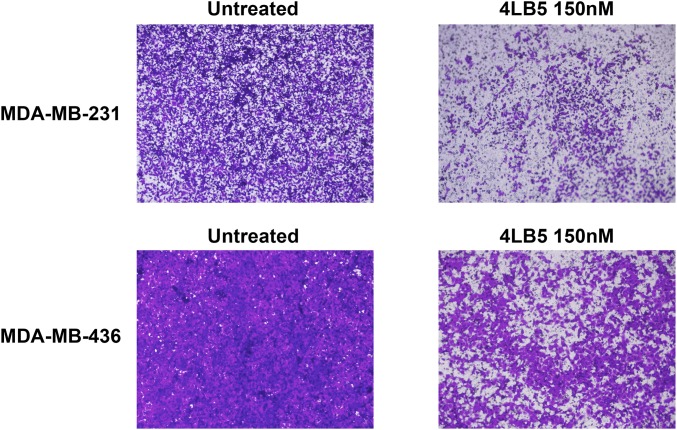
Because miR-21, -221, and -222 are extensively associated with an invasive phenotype of breast cancer (44–46) and NCL inhibition affects breast cancer cell migration (19), we also tested whether 4LB5 was able to inhibit this process in vitro. MDA-MB-231 and MDA-MB-436 cells were treated for 24 h with 4LB5 and then counted and reseeded into transwell plates for additional 24 h. Compared with untreated cells, Crystal violet staining revealed that 4LB5 treatment impaired cell migration in both cell lines (Fig. S7).
Fig. S7.
4LB5 inhibits cancer cell migration. Indicated cell lines were treated or left untreated for 24 h with 150 nM 4LB5, then counted and 5 × 104 viable cells were plated in the presence or in the absence of the scFv in transwell chambers for additional 24 h. Following migration, cells were stained with Crystal violet and acquired using a phase-contrast microscope. Data are representative of two independent experiments performed in triplicate. (Magnification, 4×.)
These observations indicate that NCL inhibition by 4LB5 significantly reduces cell viability, proliferation, and migration in vitro.
4LB5 scFv Induces Apoptosis in Cancer Cells.
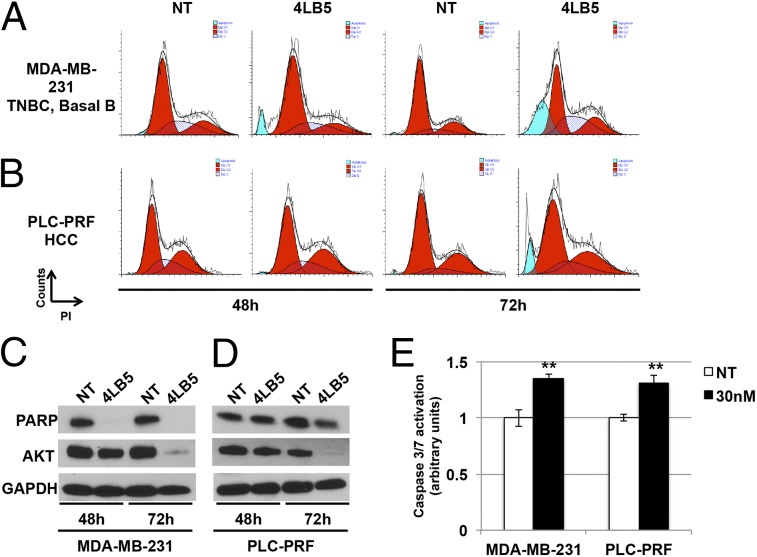
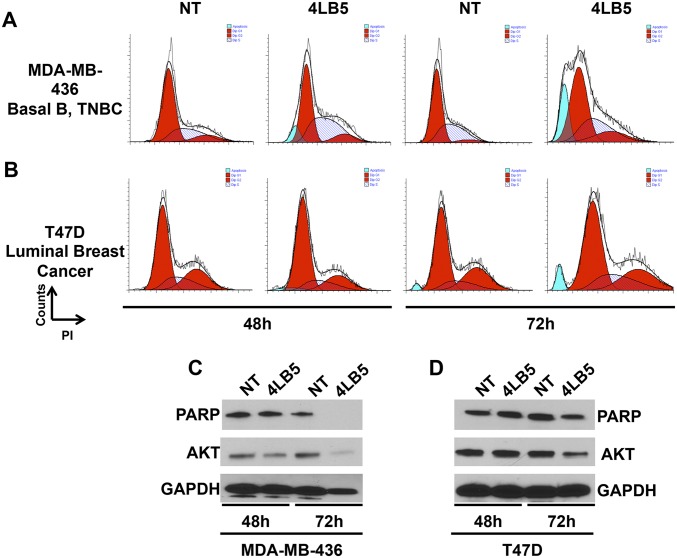
The reduced cell viability and proliferation observed following NCL inhibition by 4LB5 treatment led us to hypothesize that 4LB5 might also be able to induce apoptosis. We first performed a flow-cytometric analysis of different cell lines treated with 4LB5 for 48 or 72 h (Fig. 5 A and B and Fig. S8 A and B) and stained with propidium iodide. In all analyzed cell lines, a sub-G1 peak, compatible with the accumulation of dead cells, was observed at 72 h of treatment with 4LB5. Western blot analysis of poly(ADP-ribose) polymerase (PARP) levels confirmed the activation of apoptosis following 4LB5 treatment, resulting in inactive-PARP degradation (Fig. 5 C and D and Fig. S8 C and D). In the same experiment, we also measured the expression levels of AKT, a previously described antiapoptotic factor whose expression is dependent on NCL (11). Interestingly, NCL inhibition upon 4LB5 treatment reduced AKT levels. To further demonstrate the activation of apoptotic pathways following scFv treatment, we also measured caspase 3/7 activation in MDA-MB-231 and PLC-PRF cells treated with 4LB5 or left untreated. Fig. 5E shows a significant caspase 3/7 cleavage upon 4LB5 treatment.
Fig. 5.
4LB5 induces apoptosis. (A and B) Cell cycle analysis of MDA-MB-231 (A) and PLC-PRF (B) cells by propidium iodide staining, treated or not (NT, not treated) with 240 nM 4LB5 for 48 h and 72 h. Red peaks indicate cells in G1 and G2 phase of the cell cycle. Stripes indicate cells in S phase. Blue peaks indicate sub-G1 apoptotic cells. Data are representative of three independent experiments. (C and D) Western blot analysis of total lysates from cells treated as in A and B to evaluate inactive-PARP cleaveage and AKT levels. GAPDH was used as loading control. (E) Caspase 3/7 activation assay performed on MDA-MB-231 and PLC-PRF cells 24 h following the treatment with 30 nM 4LB5 or control vector (NT). **P < 0.01. Data are representative of three independent experiments performed in triplicate.
Fig. S8.
4LB5 induces apoptosis. (A and B) Cell cycle analysis of MDA-MB-436 (A) and T47D (B) cells by propidium iodide staining, treated or not (NT, not treated) with 240 nM 4LB5 for 48 h and 72 h. Red peaks indicate cells in G1 and G2 phase of the cell cycle. Stripes indicate cells in S phase. Blue peaks indicate sub-G1 apoptotic cells. (C and D) Western blot analysis of total lysates from cells treated as in A and B to evaluate inactive-PARP cleavage and AKT levels. GAPDH was used as loading control.
Overall, these data indicate that NCL inhibition by 4LB5 treatment results in decreased cell viability and activation of programmed cell death.
4LB5 Displays Potent Antitumor Activity in Vivo.
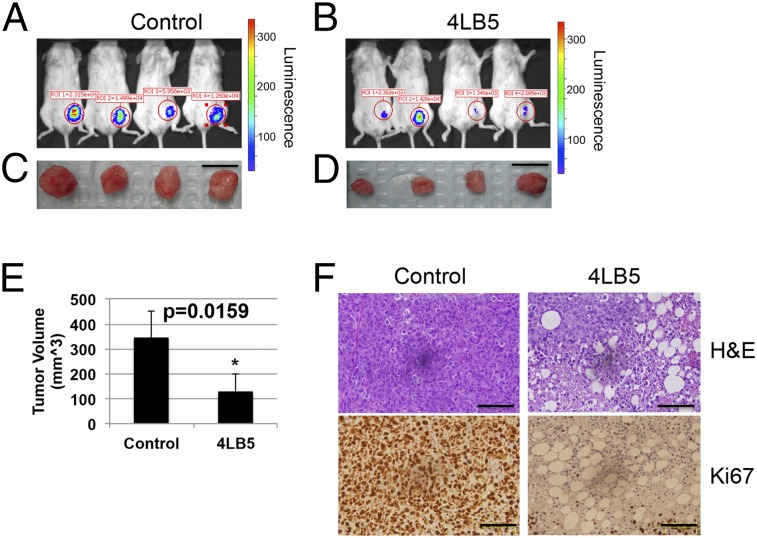
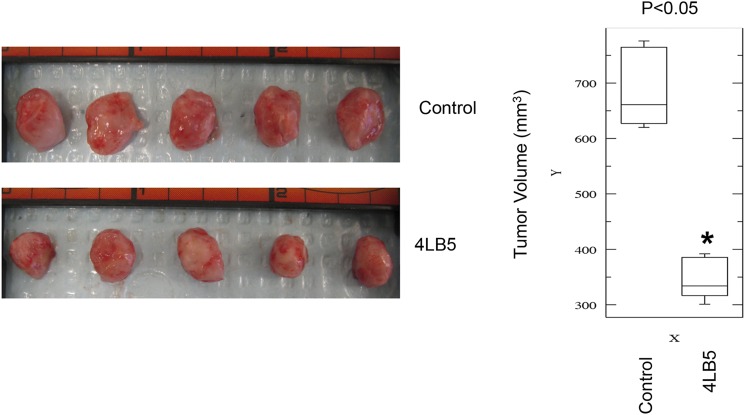
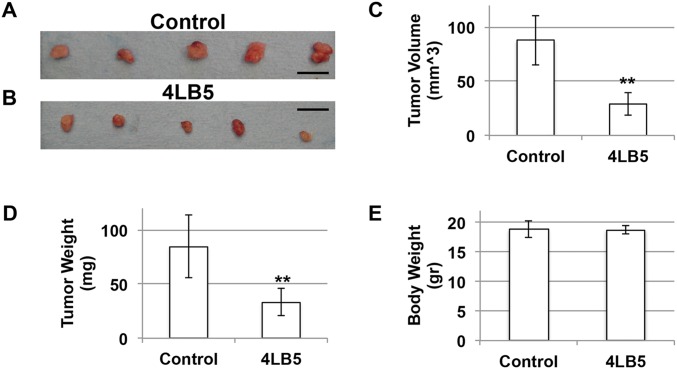
To verify the potential anticancer activity of our anti-NCL scFv 4LB5 in vivo, we used an orthotopic xenograft mouse model in which Luc-MDA-MB-231 (19) were injected into the mammary fat pad of nonobese diabetic (NOD)-SCID mice. Two weeks after the injection, mice bearing tumors of comparable size received intraperitoneal injections of vehicle (n = 4 in Fig. 6; n = 5 in Fig. S9) or 2 mg/kg of 4LB5 (n = 4 in Fig. 6; n = 5 in Fig. S9) twice weekly. Two weeks after the first treatment, we observed a clear reduction of tumor size in 4LB5-treated mice, in comparison with the control-treated ones, by an in vivo imaging system (IVIS) (Fig. 6 A and B). Mice were then killed and tumors removed for further analysis (Fig. 6 C–E and Fig. S9). Compared with controls, 4LB5 treatment significantly reduced the tumor volume (P = 0.0159). Interestingly, H&E staining (Fig. 6F, Upper) showed reduced cellularity and several areas of necrosis following treatment with the scFv. In addition, Ki-67 IHC staining of treated tumors indicated a reduced proliferation compared with controls (Fig. 6F, Lower). In a different experiment, the treatment with 2 mg/kg of 4LB5 (n = 5) or vehicle (n = 5), twice a week for 4 wk, begun at 3 d after the orthotopic implantation of MDA-MB-231 cells. Excised tumors (Fig. S10 A–D) displayed a significant reduction in the tumor volume and weight in 4LB5-treated mice compared with controls, but we did not observe alteration of health conditions and body weight (Fig. S10E) in scFv-treated mice, suggesting that 4LB5 was not toxic for normal cells.
Fig. 6.
4LB5 inhibits breast cancer cell growth in vivo. (A–D) NOD-SCID (n = 8) mice were injected with 2 × 106 MDA-MB-231-Luc cells into the mammary fat pad. After 2 wk, mice were treated with control solution (n = 4) (A and C) or 2 mg/kg of 4LB5 (n = 4) (B and D), twice a week. Mice were monitored by IVIS weekly. At 4 wk from injection (2 wk of treatment), mice were killed. Tumors were excised and measured. (Scale bars, 1 cm.) (E) Average volume for the tumors reported in A–D (L × W × H) is reported. *P < 0.05. (F) Representative images of H&E and Ki-67 staining of tumors shown in (C and D); 20× magnification is reported. (Scale bars, 50 μm.) See also Fig. S9, where a different batch of 4LB5 was used in a separate experimental setting.
Fig. S9.
4LB5 inhibits breast cancer cell growth in vivo (i). NOD-SCID (n = 10) mice were injected with 2 × 106 MDA-MB-231-Luc cells into the mammary fat pad. After 2 wk, mice were treated with control solution (n = 5) (Left) or 2 mg/kg of 4LB5 (n = 5) (B and D), twice a week. Mice were monitored by IVIS weekly. At 4 wk from injection (2 wk of treatment) mice were killed. Tumors were excised and measured. Distribution of the tumor volumes (L × W × H) is reported (Right). *P < 0.05. See also Fig. 6, where a different batch of 4LB5 was used in a separate experimental setting.
Fig. S10.
4LB5 inhibits breast cancer cell growth in vivo (ii). (A and B) NOD-SCID mice (n = 10) were injected with 2 × 106 MDA-MB-231-Luc cells into the mammary fat pad. After 2 d, mice were treated with 2 mg/kg (n = 5) of 4LB5 or with control solution (n = 5), twice a week. At 4 wk of treatment, mice were killed. Tumors were excised and measured. (C and D) Average tumor volume (L × W × H) (C) and weight (D) of tumors in A and B is reported. **P < 0.01. (E) Following euthanization, body weight was measured to evaluate potential toxic effects of the treatment.
Our in vivo observations clearly indicate that 4LB5 is effective in reducing the viability and proliferation of aggressive breast cancer cells in the absence of detectable side effects.
Discussion
The widely demonstrated role of NCL in tumorigenesis suggests that inhibition of its oncogenic actions reduces tumor aggressiveness (9, 13, 19, 24, 32, 34, 47), and several studies have proposed NCL as an ideal target for antineoplastic therapies in different solid and hematological malignancies (9, 19, 25, 34, 37, 38, 41, 42, 48, 49). Given the selective presence of NCL on cancer cells and cancer-associated endothelial cells, but not on normal cells, molecules targeting NCL might represent an effective approach for the selective delivery of drugs or toxins to tumors while minimizing side effects (25, 41). In addition, NCL-ligands have the potential to be modified to develop novel cancer imaging and diagnostic tools (25).
The aim of this study was to identify a novel anti-NCL molecule with a strong relevance in terms of efficacy and clinical pertinence for cancer therapy. Taking advantage of phage-display technology, we selected a fully human recombinant scFv, named 4LB5, which specifically binds NCL on the cell surface of cancer cells. This molecule displayed a significant ability to discriminate between cancer and normal-like breast cells.
In accordance with our previous study (19), we demonstrate that 4LB5 treatment affects the expression of mature miR-21, -221, and -222, affecting NCL interaction with DGCR8 and halting the maturation of the primary forms of these miRNAs. Further studies will be carried out to evaluate whether NCL inhibition using this scFv might directly or indirectly affect the expression of miRNAs other than those regulated by previously described anti-NCL approaches.
Furthermore, 4LB5 treatment of breast cancer cells, but not of normal-like breast cells, significantly reduces cell viability, proliferation, and migration, and induces apoptosis, in agreement with previous in vitro studies using other anti-NCL molecules (19, 32). Similar results were obtained using HCC cells expressing high levels of surface-NCL (50), suggesting that NCL inhibition might represent a valid approach for the treatment of different types of tumors. However, factors other than surface-NCL levels, might determine the specific cellular sensitivity to NCL inhibition. Further studies analyzing the relative expression levels of miRNAs and mRNAs regulated by NCL in different tumor systems will be needed.
Here, we also demonstrate that 4LB5 treatment reduces tumor growth in vivo in an orthotopic xenograft mouse model of breast cancer (19), in the absence of any evident side effect. Additional studies will be needed to evaluate the pharmacokinetic and pharmacodynamic parameters of 4LB5, to better characterize the therapeutic potential of this molecule and to establish more effective treatment protocols.
Despite its already evident efficacy, 4LB5 may be further improved as a novel tool for cancer diagnosis and therapy. Specifically, scFvs can be easily modified to obtain a compact (51) or a full-length human Ig with the same specificity, but with a prolonged in vivo half-life and the ability to activate CDC and ADCC, combining the antitumoral activity of NCL inhibition with an immune response against cancer cells.
We also show that 4LB5 translocates into the cytoplasm following NCL binding, suggesting its use to vehicle antitumoral molecules (prodrugs, enzymes, toxins, and radionuclides) directly into cancer cells, enhancing their therapeutic activity while reducing their adverse effects.
In summary, here we describe a novel NCL-targeting scFv with potent antineoplastic activity that could represent the prototype of a novel class of immune-based drugs with enormous clinical potential as tools for the diagnosis and therapy of a wide range of human cancers.
Materials and Methods
Cell Cultures and Transfections.
MDA-MB-231, MDA-MB-436, BT549, T47D, Huh7, and PLC-PRF cells were cultured in RPMI with 10% (vol/vol) FBS, l-glutamine, and antibiotics. HeLa cells were cultured in DMEM with 10% FBS, l-glutamine, and antibiotics (Sigma). MCF10a cells were cultured in Mammary Epithelial Cell Growth Medium (MEGM, Lonza) supplemented with 10% FBS, bovine pituitary extract, hydrocortisone, hEGF, and insulin (BulletKit, Lonza). Cell lines were purchased from the American Type Culture Collection (ATCC) and cultured in humidified atmosphere containing 5% CO2 at 37 °C. Transfection was performed by using Lipofectamine 2000 (Life Technologies), as suggested by the manufacturer.
Plasmids and siRNAs.
pET15b and pET22b(+) prokaryotic expression vectors were purchased from Novagen. pHEN2 phagemid vector was described previously (31). pET-15b-NCL-RBD-His6, FLAG-DGCR8, and NCL-Myc were kindly provided by F.P. and described elsewhere (19). NCL siRNA (sc29230) and control siRNA (sc37007) were from Santa Cruz Biotechnology.
Internalization Experiments.
Subconfluent MDA-MB-231 cells were treated with 1 μg/mL of Cy5-4LB5 diluted in complete medium and cultured at 37 °C or at 4 °C for 6 h to allow the internalization of the scFv. Cells were then extensively washed with PBS, gently scraped and acquired by ImageStream (Amnis) to determine the extent of internalization. Bright field and Cy5 images were acquired and analyzed using the built-in Amnis internalization wizard.
REMSA.
REMSA was performed using the LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. In brief, 1 nmol of biotinylated miR-21 [kindly provided by F.P. and described elsewhere (19)] were incubated with 50 ng of recombinant NCL-RBD-His6 for 30 min at room temperature. For competition experiments, recombinant proteins were preincubated with increasing concentrations of 4LB5 (80–650 nM) or with control IgG. Binding reactions were run on a native 7% (wt/vol) polyacrylamide–1× TBE gel. Transfer of binding reactions to nylon membranes and detection were performed according to the manufacturer’s instructions.
Quantitative Real-Time PCR.
Quantitative real-time PCR (qRT-PCR) were performed using the TaqMan Fast-PCR kit (Applied Biosystems) according to the manufacturer’s instructions, using the appropriate TaqMan probes for miRNA and pri-miRNA quantification, followed by detection with the 7900HT Sequence Detection System (Applied Biosystems). All reactions were performed in triplicate. Simultaneous quantification of RNU6 was used as reference for miRNA quantification. Simultaneous quantification of GAPDH mRNAs was used as reference for pri-mRNA quantification. The comparative cycle threshold (Ct) method for relative quantification of gene and miRNA expression (User Bulletin #2; Applied Biosystems) was used to determine miRNA and pri-miRNA, expression levels.
Cell Viability and Growth Assays.
For viability assays, 1 × 105 cells were plated in 12-well plates and treated with the indicated amounts (1–240 nM) of 4LB5. At the indicated time points, cells were harvested, mixed 1:1 with Trypan blue and counted using a hemacytometer. The percentage of viable cells is reported. IC50 was evaluated using the Prism 6.0 software (Graphpad software).
For cell growth curves, 1 × 105 cells were plated in 12-well plates and treated with the indicated amounts (30–120 nM) of 4LB5. Cells were harvested every 24 h for 3 d and counted as described above. Total cell numbers were reported.
Colony Assay.
For colony assay experiments, 200 MDA-MB-231 cells were plated in 12-well plates and treated with 30 nM 4LB5 in complete medium for 72 h. Then, cells were replenished with complete medium without 4LB5 and allowed to grow for 7 additional days, to allow the formation of the colonies. Cells were then fixed with 1% glutaraldehyde in PBS and stained with Crystal violet.
Migration Assay.
Transwell insert chambers with 8-μm porous membrane (Greiner-Bio-One) were used for migration assay. MDA-MB-231 and MDA-MB-436 cells were treated with 150 nM 4LB5 for 24 h, harvested and 5 × 104 viable cells were added to the top chamber in serum-free media plus 150 nM 4LB5. The lower chamber was filled with complete media. Chambers were incubated for 24 h at 37 °C in humidified atmosphere. Cells on the top of the chamber were then removed using a cotton swab, and migrated cells were fixed in 1% glutaraldehyde-PBS, stained with Crystal violet and visualized under a phase-contrast microscope (E200, Nikon).
In Vivo Experiments.
Animal experiments were performed according to the Ohio State University institutional guidelines after review by an institutional review board. For the establishment of xenograft models, 2 × 106 viable Luc+ MDA-MB-231 cells were injected into the fourth left-side mammary fat pad of female NOD-SCID mice (NOD/ShiLtSz; Charles River). Three days or 2 wk after tumor cell inoculation, mice were treated twice a week with intraperitoneal injections of 4LB5 (2 mg/kg) or control buffer (25 mM imidazole in PBS) diluted in 100 µL PBS. Tumor size was assessed every 7 d by bioluminescence imaging, as described below. After 4 wk of treatment, mice were analyzed by bioluminescence images and then killed. For in vivo bioluminescence analysis, mice were injected with 75 mg/kg Luciferin (Xenogen), and tumor growth was detected by bioluminescence at 20 min after the injection. The home-built bioluminescence system used an electron multiplying charge-coupled device (IVIS-200, Perkin-Elmer) with an exposure time of 30 s and an electron multiplication gain of 500 voltage gain × 200, 5-by-5 binning, and with background subtraction. The tumor size was measured using a caliper, and the volume was calculated in cubed millimeters using the formula L × W × H.
Statistical Analysis.
Student’s t test was used to determine the statistical significance (indicated as P value) for each experiment. All error bars represent the SD of the mean. Data were considered statistically significant for P < 0.05, at least.
SI Materials and Methods
Selection of scFv Phage Clones.
Phagemid particles were rescued with M13-K07 (Life Technologies) from the Griffin.1 library, as previously described (51). For each round of selection, phages (1013 cfu) were blocked with 5% (wt/vol) Nonfat Dry Milk (Bio-Rad) in PBS for 15 min. Polypropylene tubes were coated with recombinant NCL-RBD in PBS at a concentration of 20 μg/mL in the first four rounds of selection and at a concentration of 10 μg/mL in the fifth round. Blocked phages were incubated for 16 h at 4 °C in rotation in the coated tubes, and then elutes with 50 mM citric acid (pH 2.5) in PBS for 5 min, and then neutralized with 1 M Tris⋅HCl (pH 7.4). Recovered phages were amplified by infecting Escherichia coli TG1 bacterial strain to prepare phages for the next round of selection. Phage screening was carried out by ELISA as described below. Cultures of E. coli SF110 bacterial strain, previously infected with selected phage clones, were grown at 37 °C in 2xTY medium containing 100 μg/mL ampicillin and 1% glucose, until OD = 600 nm was reached. Cells were centrifuged at 4,000 × g for 15 min and resuspended in glucose-free medium. The expression of soluble scFv was induced by the addition of IPTG (Calbiochem) to a final concentration of 1 mM in the cell culture, which was then grown at room temperature overnight. Cells were harvested by centrifugation at 4,000 × g for 15 min, and a periplasmic extract was obtained by resuspending cells in B-PER buffer (Thermo Pierce), according to the manufacturer’s recommendations.
Purification of Recombinant Proteins.
pET15b-NCL-RBD-His6 was expressed in E. coli BL21(DE3) (Agilent Technologies) bacterial cells as soluble protein following IPTG induction and purified using nickel affinity chromatography (Qiagen), according to the manufacturer’s instructions.
Recombinant 4LB5 scFv showed reduced solubility and for this reason was extracted from the insoluble fraction of pET22b(+)-4LB5–transformed BL21(DE3) bacterial cells. The fraction was solubilized with 7 M urea, 2 M thiourea, 20 mM Tris, pH 8.0, and 50 mM NaCl and mixed overnight at room temperature. The solution was centrifuged at 20,000 × g for 1 h to pellet. Supernatant was removed and the resulting pellet was resolubilized with the same buffer with the addition of 0.5 mM aminosulfobetaine-14 and 0.1% IGEPAL for 48 h and centrifuged at 20,000 × g rpm for 1 h. The supernatant was diluted to one-half concentration and applied to a pretreated tandem Q-HiTrap/S-HiTrap at 0.5 mL/min. The flow-through was collected and the column washed with 10 column volumes of 3.5 M urea, 2 M thiourea, 10 mM Tris, pH 8.0, and 25 mM NaCl. The columns were eluted with wash buffer with the addition of 1 M NaCl. Flow through was applied to a 5-mL Hitrap column charged with nickel sulfate solution and prepared with 3.5 M urea, 2 M thiourea, 10 mM Tris, pH 8.0, 100 mM NaCl, and 20 mM of imidazole. The protein solution, diluted with 3.5 mM urea buffer, was applied to the column and washed with 10 column volumes of buffer. The column was then washed slowly with decreasing urea concentrations to promote folding in 10 mM Tris pH 8.0, 100 mM NaCl and 10% (vol/vol) glycerol to support the solubility. The column was then washed in PBS and eluted with PBS with 250 mM imidazole. Protein purity and quantification were assessed by SDS/PAGE and Coomassie blue staining.
ELISA.
Phages and soluble scFvs, prepared as described above, were evaluated for their affinity to bind NCL-RBD by ELISA. Flat-bottom 96-well plates were coated with 20 μg/mL of recombinant NCL-RBD in 2% (wt/vol) Nonfat Dry Milk (NFDM) in PBS. Phages or soluble scFvs were added to the plates in 2% (wt/vol) NFDM and incubated for 2 h at room temperature. Plates were washed with PBS and incubated with HRP-conjugated anti-M13 antibody (Amersham) for 1 h, washed again and incubated with TMB reagent (Sigma) for 10 min before quenching with an equal volume of 1 M HCl.
For cell ELISA, 1 × 104 MDA-MB-231 or MCF10a cells were incubated in round-bottom 96-well plates with different concentrations (0–600 nM) of 4LB5 in 2% (wt/vol) NFDM for 2 h at room temperature with gentle agitation. Plates were then centrifuged and cell pellets were washed with PBS and incubated with HRP-conjugated antipenta-His antibody (Qiagen) for 1 h at room temperature. Following additional washes, TMB reagent (Sigma) was added for 10 min before quenching with an equal volume of 1 M HCl. ELISA plates were read (A450) using a Spectramax 340 microtiter plate reader (Molecular Devices).
Surface Plasmon Resonance.
The surface plasmon resonance (SPR) analyses were performed at 25 °C on a BIAcore 3000 instrument (Biacore), equipped with research-grade CM5 sensor chips (Biacore). The running buffer was HBS-EP (10 mm Hepes, 0.15 m NaCl, 3.4 mm EDTA, and 0.005% surfactant P20 at pH 7.4). Coupling reagents, N-hydroxysuccinimide, 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride, ethanolamine hydrochloride, and HBS-EP running buffer were purchased from Biacore. To measure the binding properties of 4LB5 to the NCL-RBD, recombinant NCL-RBD was immobilized onto the surface of sensor chip CM5 using the standard amine coupling chemistry. Typically, 350 and 700 RU of NCL-RBD were immobilized onto the sensor surface. Binding curves were recorded by injecting 4LB5 (5–500 nM) over the immobilized NCL-RBD at a constant flow rate of 50 μL/min−1. Association and dissociation phases were recorded for 300 and 1,200 s, respectively. The rate constants of the interactions described above were calculated by nonlinear analysis of the association and dissociation curves using SPR kinetic evaluation software BIAevaluation (Biacore), fitting data to the 1:1 Langmuir binding model. The equilibrium dissociation constants (KD) were calculated from the values of the association rate constant ka and dissociation rate constant kd, according to the thermodynamic relationship KD = kd/ka.
Western Blot, Coimmunoprecipitation Experiments, and Antibodies.
Periplasmic extracts were obtained as previously described (51) using the B-PER extraction buffer (Thermo Scientific) supplemented with protease inhibitors (Calbiochem). Insoluble fractions were resuspended in 8 M urea before SDS/PAGE and Coomassie blue staining or Western blot using standard procedures. Eukaryotic protein extracts were obtained using 1% Nonidet P-40, 1 mm EDTA, 50 mm Tris⋅HCl, pH 7.5, and 150 mm NaCl, supplemented with complete protease and phosphatase inhibitors (Calbiochem). Protein extracts were subjected to SDS/PAGE, electroblotted onto nitrocellulose membranes, and probed with antibodies as indicated, according to standard procedures. For immunoprecipitation experiments, subconfluent HeLa cells were transfected with the FLAG-DGCR8 and NCL-Myc expression vectors. Twenty-four hours after transfection, cells were treated with 120 nM 4LB5 or control buffer (12.5 mM imidazole in PBS) for an additional 24 h. Proteins were extracted as described above, and 2 mg of total protein extracts were immunoprecipitated with anti–FLAG-M2 resin (Sigma) overnight at 4 °C with rotation. Immunoprecipitates were washed as suggested by the manufacturer, subjected to SDS/PAGE, electroblotted onto nitrocellulose membranes, and probed with antibodies as indicated. Used antibodies were antipenta-His antibody (Qiagen), anti-PARP, anti-AKT, anti-GAPDH (Cell Signaling), anti-NCL, anti–Myc-tag, and anti-Tubulin (Santa Cruz Biotechnology).
Cy5 Labeling and Flow-Cytometry Analysis.
4LB5 was Cy5 labeled using the LYNX Rapid Cy5 Antibody conjugation kit (AbD Serotec) according to the manufacturer’s instructions. Briefly, 1 mg of 4LB5 was incubated with modifier reagent and the LYNX lyophilized mix overnight at room temperature. The reaction was then stopped using the quencher reagent.
For flow-cytometry analysis, cells were harvested and incubated with 1 μg/mL of Cy5-4LB5 diluted in PBS containing 2% FBS, on ice. Cells were then washed and analyzed with a FACS-Calibur flow cytometer (Becton Dickinson).
Cell Cycle Analysis.
For cell cycle analysis, 24 h following the seeding, cells were treated with 240 nM 4LB5. Cells were harvested at different time-points, fixed and permeabilized with ice-cold 70% ethanol, treated with RNase-A (Invitrogen), and stained with 10 mg/mL of propidium iodide. Cells were sorted on a FACS-Calibur flow cytometer (Becton Dickinson), and the results were analyzed with ModFit software, v3.2 (Verity Software House).
Caspase Activation Assay.
Apoptosis activation was quantified by measuring caspase 3 and 7 activation 48 h following 4LB5 treatment, using Caspase-Glo 3/7 assay (Promega) according to the manufacturer’s instructions on a Bio-Tek Synergy HT multidetection microplate reader.
Paraffin-Embedded Tissue Staining.
Xenograft tumor samples were fixed in 10% neutral-buffered formalin, processed, embedded in paraffin, and sectioned at 4 µm. H&E staining was performed according to standard procedures. For immunohistochemical staining, slides with specimens were placed in a 60 °C oven for 1 h, cooled, deparaffinized, and rehydrated through xylene and graded ethanol solutions to water. All slides were placed for 5 min in a 3% hydrogen peroxide solution in water to block the endogenous peroxidase. Antigen retrieval was performed by HIER, in which the slides were placed in a citric acid solution, pH 6.1, for 25 min at 96 °C and cooled for 15 min in solution. Sections were then treated with primary antibodies for Ki-67, followed by biotinylated secondary antibodies and the DAB chromogen.
Acknowledgments
This work was supported by National Institutes of Health Grants U01CA166905 and U01CA152758 (to C.M.C. and D.P.) and the Pelotonia Intramural Research Program (D.P.). T.R. and T.S. are recipients of a Medical Student Research Scholarship (The Ohio State University, College of Medicine).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507087112/-/DCSupplemental.
References
- 1.Bugler B, Caizergues-Ferrer M, Bouche G, Bourbon H, Amalric F. Detection and localization of a class of proteins immunologically related to a 100-kDa nucleolar protein. Eur J Biochem. 1982;128(2-3):475–480. doi: 10.1111/j.1432-1033.1982.tb06989.x. [DOI] [PubMed] [Google Scholar]
- 2.Warner JR. The nucleolus and ribosome formation. Curr Opin Cell Biol. 1990;2(3):521–527. doi: 10.1016/0955-0674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- 3.Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56(3):379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 4.Mongelard F, Bouvet P. Nucleolin: A multiFACeTed protein. Trends Cell Biol. 2007;17(2):80–86. doi: 10.1016/j.tcb.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Srivastava M, Pollard HB. Molecular dissection of nucleolin’s role in growth and cell proliferation: New insights. FASEB J. 1999;13(14):1911–1922. [PubMed] [Google Scholar]
- 6.Ridley L, et al. Children’s Cancer and Leukaemia Group Biological Studies Committee Multifactorial analysis of predictors of outcome in pediatric intracranial ependymoma. Neuro-oncol. 2008;10(5):675–689. doi: 10.1215/15228517-2008-036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hovanessian AG, et al. The cell-surface-expressed nucleolin is associated with the actin cytoskeleton. Exp Cell Res. 2000;261(2):312–328. doi: 10.1006/excr.2000.5071. [DOI] [PubMed] [Google Scholar]
- 8.Christian S, et al. Nucleolin expressed at the cell surface is a marker of endothelial cells in angiogenic blood vessels. J Cell Biol. 2003;163(4):871–878. doi: 10.1083/jcb.200304132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otake Y, et al. Overexpression of nucleolin in chronic lymphocytic leukemia cells induces stabilization of bcl2 mRNA. Blood. 2007;109(7):3069–3075. doi: 10.1182/blood-2006-08-043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen CY, et al. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000;14(10):1236–1248. [PMC free article] [PubMed] [Google Scholar]
- 11.Abdelmohsen K, et al. Enhanced translation by nucleolin via G-rich elements in coding and non-coding regions of target mRNAs. Nucleic Acids Res. 2011;39(19):8513–8530. doi: 10.1093/nar/gkr488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reyes-Reyes EM, Akiyama SK. Cell-surface nucleolin is a signal transducing P-selectin binding protein for human colon carcinoma cells. Exp Cell Res. 2008;314(11-12):2212–2223. doi: 10.1016/j.yexcr.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tate A, et al. Met-independent hepatocyte growth factor-mediated regulation of cell adhesion in human prostate cancer cells. BMC Cancer. 2006;6:197. doi: 10.1186/1471-2407-6-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wise JF, et al. Nucleolin inhibits Fas ligand binding and suppresses Fas-mediated apoptosis in vivo via a surface nucleolin-Fas complex. Blood. 2013;121(23):4729–4739. doi: 10.1182/blood-2012-12-471094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdelmohsen K, Gorospe M. RNA-binding protein nucleolin in disease. RNA Biol. 2012;9(6):799–808. doi: 10.4161/rna.19718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tayyari F, et al. Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat Med. 2011;17(9):1132–1135. doi: 10.1038/nm.2444. [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 18.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007;17(3):118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Pichiorri F, et al. In vivo NCL targeting affects breast cancer aggressiveness through miRNA regulation. J Exp Med. 2013;210(5):951–968. doi: 10.1084/jem.20120950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao X, et al. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. 2011;30(9):1082–1097. doi: 10.1038/onc.2010.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pogribny IP, et al. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer. 2010;127(8):1785–1794. doi: 10.1002/ijc.25191. [DOI] [PubMed] [Google Scholar]
- 22.Anastasov N, et al. Radiation resistance due to high expression of miR-21 and G2/M checkpoint arrest in breast cancer cells. Radiat Oncol. 2012;7:206. doi: 10.1186/1748-717X-7-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mei M, et al. Downregulation of miR-21 enhances chemotherapeutic effect of taxol in breast carcinoma cells. Technol Cancer Res Treat. 2010;9(1):77–86. doi: 10.1177/153303461000900109. [DOI] [PubMed] [Google Scholar]
- 24.Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp Mol Pathol. 2009;86(3):151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koutsioumpa M, Papadimitriou E. Cell surface nucleolin as a target for anti-cancer therapies. Recent Pat Anticancer Drug Discov. 2014;9(2):137–152. doi: 10.2174/1574892808666131119095953. [DOI] [PubMed] [Google Scholar]
- 26.Destouches D, et al. Suppression of tumor growth and angiogenesis by a specific antagonist of the cell-surface expressed nucleolin. PLoS ONE. 2008;3(6):e2518. doi: 10.1371/journal.pone.0002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El Khoury D, et al. Targeting surface nucleolin with a multivalent pseudopeptide delays development of spontaneous melanoma in RET transgenic mice. BMC Cancer. 2010;10:325. doi: 10.1186/1471-2407-10-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krust B, El Khoury D, Nondier I, Soundaramourty C, Hovanessian AG. Targeting surface nucleolin with multivalent HB-19 and related Nucant pseudopeptides results in distinct inhibitory mechanisms depending on the malignant tumor cell type. BMC Cancer. 2011;11:333. doi: 10.1186/1471-2407-11-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Lorenzo C, D’Alessio G. From immunotoxins to immunoRNases. Curr Pharm Biotechnol. 2008;9(3):210–214. doi: 10.2174/138920108784567254. [DOI] [PubMed] [Google Scholar]
- 30.Marks JD, et al. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222(3):581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 31.Nissim A, et al. Antibody fragments from a ‘single pot’ phage display library as immunochemical reagents. EMBO J. 1994;13(3):692–698. doi: 10.1002/j.1460-2075.1994.tb06308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soundararajan S, Chen W, Spicer EK, Courtenay-Luck N, Fernandes DJ. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008;68(7):2358–2365. doi: 10.1158/0008-5472.CAN-07-5723. [DOI] [PubMed] [Google Scholar]
- 33.Derenzini M, Sirri V, Trerè D, Ochs RL. The quantity of nucleolar proteins nucleolin and protein B23 is related to cell doubling time in human cancer cells. Lab Invest. 1995;73(4):497–502. [PubMed] [Google Scholar]
- 34.Soundararajan S, et al. Plasma membrane nucleolin is a receptor for the anticancer aptamer AS1411 in MV4-11 leukemia cells. Mol Pharmacol. 2009;76(5):984–991. doi: 10.1124/mol.109.055947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N. Nucleolar localization of DGCR8 and identification of eleven DGCR8-associated proteins. Exp Cell Res. 2007;313(20):4196–4207. doi: 10.1016/j.yexcr.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 36.Pickering BF, Yu D, Van Dyke MW. Nucleolin protein interacts with microprocessor complex to affect biogenesis of microRNAs 15a and 16. J Biol Chem. 2011;286(51):44095–44103. doi: 10.1074/jbc.M111.265439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenberg JE, et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Invest New Drugs. 2014;32(1):178–187. doi: 10.1007/s10637-013-0045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamada T, Park CS, Shen Y, Rabin KR, Lacorazza HD. G0S2 inhibits the proliferation of K562 cells by interacting with nucleolin in the cytosol. Leuk Res. 2014;38(2):210–217. doi: 10.1016/j.leukres.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schokoroy S, Juster D, Kloog Y, Pinkas-Kramarski R. Disrupting the oncogenic synergism between nucleolin and Ras results in cell growth inhibition and cell death. PLoS ONE. 2013;8(9):e75269. doi: 10.1371/journal.pone.0075269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang X, et al. Cell surface nucleolin is crucial in the activation of the CXCL12/CXCR4 signaling pathway. Tumour Biol. 2014;35(1):333–338. doi: 10.1007/s13277-013-1044-0. [DOI] [PubMed] [Google Scholar]
- 41.Wu J, et al. Nucleolin targeting AS1411 modified protein nanoparticle for antitumor drugs delivery. Mol Pharm. 2013;10(10):3555–3563. doi: 10.1021/mp300686g. [DOI] [PubMed] [Google Scholar]
- 42.Birmpas C, Briand JP, Courty J, Katsoris P. The pseudopeptide HB-19 binds to cell surface nucleolin and inhibits angiogenesis. Vasc Cell. 2012;4(1):21. doi: 10.1186/2045-824X-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Z, et al. Knocking down nucleolin expression in gliomas inhibits tumor growth and induces cell cycle arrest. J Neurooncol. 2012;108(1):59–67. doi: 10.1007/s11060-012-0827-2. [DOI] [PubMed] [Google Scholar]
- 44.Yan LX, et al. Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast Cancer Res. 2011;13(1):R2. doi: 10.1186/bcr2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah MY, Calin GA. MicroRNAs miR-221 and miR-222: A new level of regulation in aggressive breast cancer. Genome Med. 2011;3(8):56. doi: 10.1186/gm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Leva G, et al. MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst. 2010;102(10):706–721. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishimaru D, et al. Mechanism of regulation of bcl-2 mRNA by nucleolin and A+U-rich element-binding factor 1 (AUF1) J Biol Chem. 2010;285(35):27182–27191. doi: 10.1074/jbc.M109.098830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, et al. Aptamer imaging with Cu-64 labeled AS1411: Preliminary assessment in lung cancer. Nucl Med Biol. 2014;41(2):179–185. doi: 10.1016/j.nucmedbio.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 49.Birmpas C, Briand JP, Courty J, Katsoris P. Nucleolin mediates the antiangiogenesis effect of the pseudopeptide N6L. BMC Cell Biol. 2012;13:32. doi: 10.1186/1471-2121-13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Semenkovich CF, Ostlund RE, Jr, Olson MO, Yang JW. A protein partially expressed on the surface of HepG2 cells that binds lipoproteins specifically is nucleolin. Biochemistry. 1990;29(41):9708–9713. doi: 10.1021/bi00493a028. [DOI] [PubMed] [Google Scholar]
- 51.De Lorenzo C, et al. A human, compact, fully functional anti-ErbB2 antibody as a novel antitumour agent. Br J Cancer. 2004;91(6):1200–1204. doi: 10.1038/sj.bjc.6602110. [DOI] [PMC free article] [PubMed] [Google Scholar]