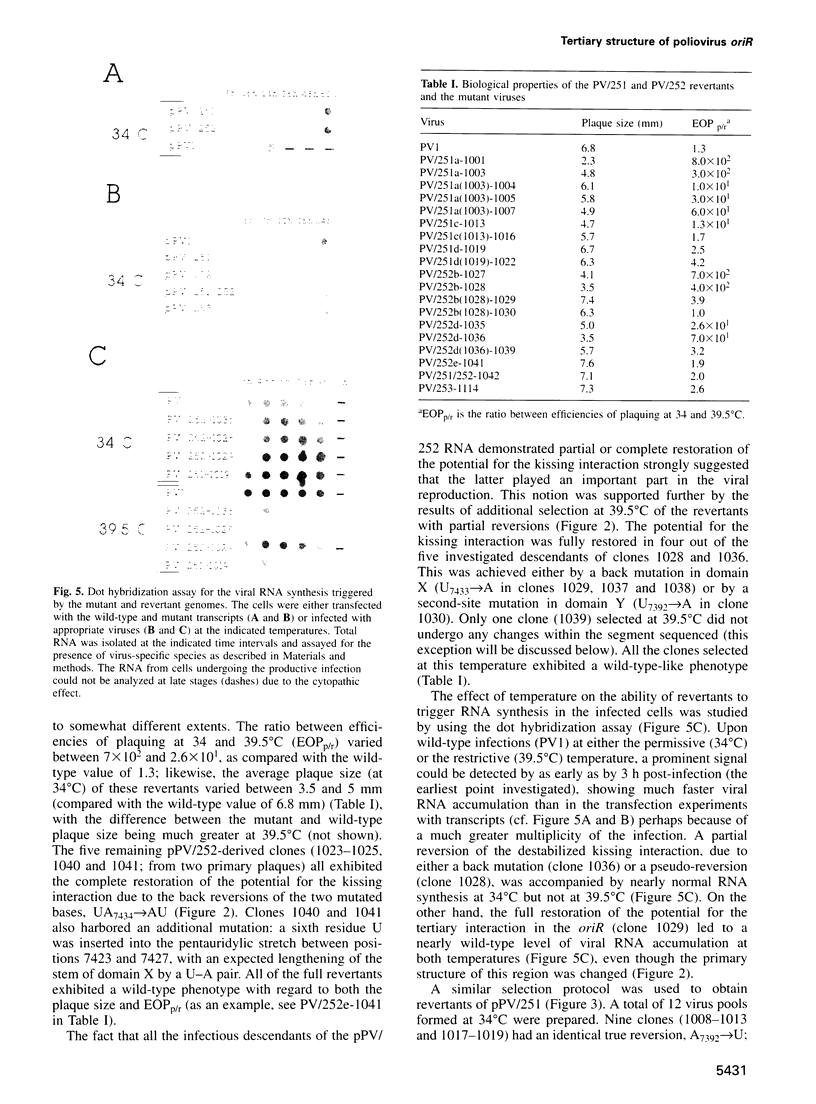
Abstract
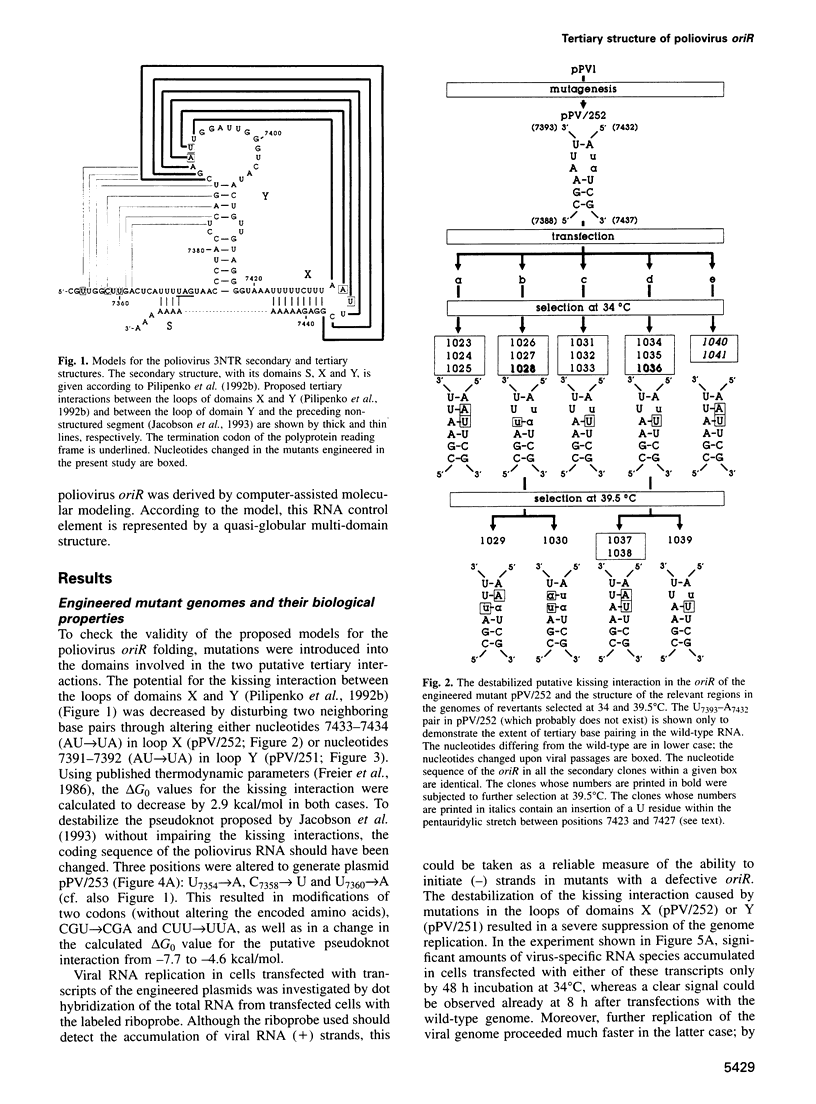
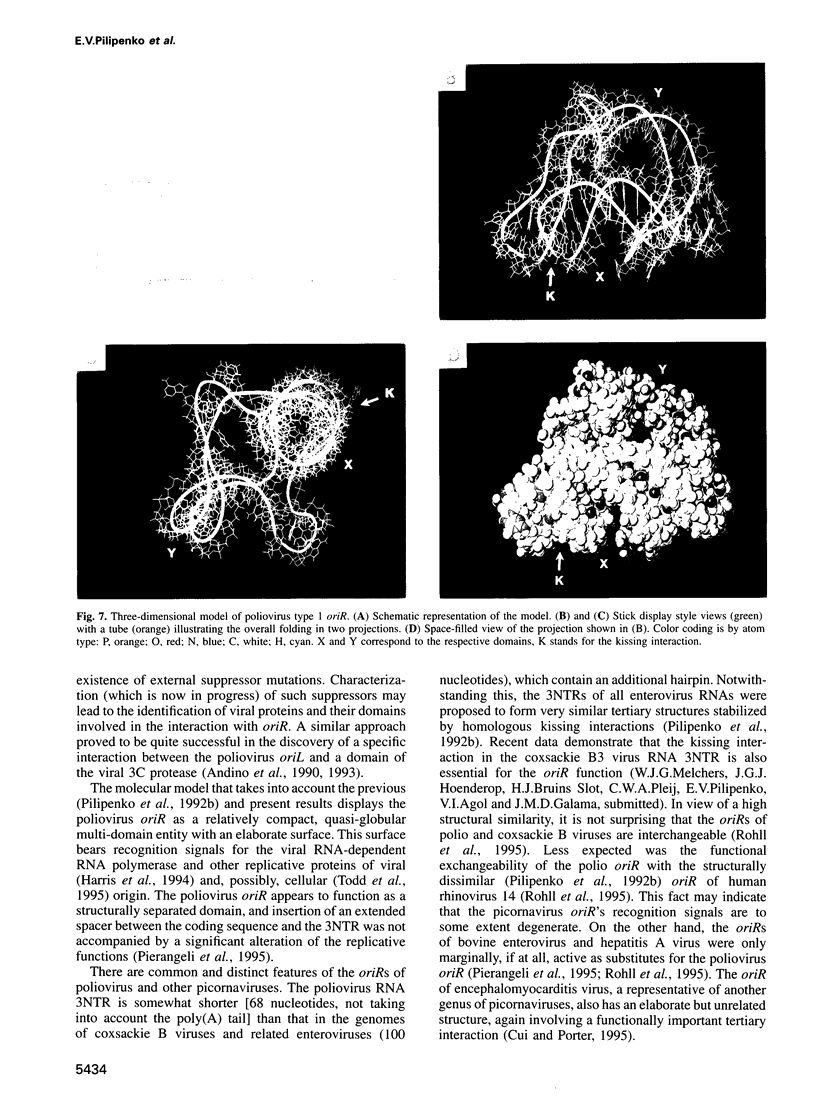
The key steps in the replication of the poliovirus genome, initiation of (-) and (+) strands, require two different cis-acting elements, oriR and oriL, respectively. It has been proposed that the spatial organization of these elements is maintained by tertiary ('kissing') interactions between the loops of two constituent hairpins. Here, the putative partners of the kissing interaction within the oriR of the full-length poliovirus RNA were modified by site-directed mutagenesis. The destabilization of this interaction resulted in a severe suppression of the viral RNA synthesis, but the mutant transcripts proved to be infectious. With a single exception, the potential for the kissing interaction within the oriR of the recovered viruses was partially or completely restored due to either true reversions or second-site compensatory mutations. There was a good correlation between the restoration of this potential and the phenotypic properties of the viruses. It was concluded that the kissing interaction in the poliovirus oriR is functionally important. Using the above experimental data, a three-dimensional structure was derived by molecular modeling techniques, which demonstrated the overall feasibility of the proposed interactions and displayed the poliovirus oriR as a quasi-globular multi-domain structure.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agol V. I. The 5'-untranslated region of picornaviral genomes. Adv Virus Res. 1991;40:103–180. doi: 10.1016/S0065-3527(08)60278-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino R., Rieckhof G. E., Achacoso P. L., Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5'-end of viral RNA. EMBO J. 1993 Sep;12(9):3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino R., Rieckhof G. E., Trono D., Baltimore D. Substitutions in the protease (3Cpro) gene of poliovirus can suppress a mutation in the 5' noncoding region. J Virol. 1990 Feb;64(2):607–612. doi: 10.1128/jvi.64.2.607-612.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K. Y., Tinoco I., Jr Characterization of a "kissing" hairpin complex derived from the human immunodeficiency virus genome. Proc Natl Acad Sci U S A. 1994 Aug 30;91(18):8705–8709. doi: 10.1073/pnas.91.18.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui T., Porter A. G. Localization of binding site for encephalomyocarditis virus RNA polymerase in the 3'-noncoding region of the viral RNA. Nucleic Acids Res. 1995 Feb 11;23(3):377–382. doi: 10.1093/nar/23.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gmyl A. P., Pilipenko E. V., Maslova S. V., Belov G. A., Agol V. I. Functional and genetic plasticities of the poliovirus genome: quasi-infectious RNAs modified in the 5'-untranslated region yield a variety of pseudorevertants. J Virol. 1993 Oct;67(10):6309–6316. doi: 10.1128/jvi.67.10.6309-6316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon K. H., Johnson K. N., Hanzlik T. N. The larger genomic RNA of Helicoverpa armigera stunt tetravirus encodes the viral RNA polymerase and has a novel 3'-terminal tRNA-like structure. Virology. 1995 Apr 1;208(1):84–98. doi: 10.1006/viro.1995.1132. [DOI] [PubMed] [Google Scholar]
- Gregorian R. S., Jr, Crothers D. M. Determinants of RNA hairpin loop-loop complex stability. J Mol Biol. 1995 May 19;248(5):968–984. doi: 10.1006/jmbi.1995.0275. [DOI] [PubMed] [Google Scholar]
- Harris K. S., Xiang W., Alexander L., Lane W. S., Paul A. V., Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5' and 3' termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994 Oct 28;269(43):27004–27014. [PubMed] [Google Scholar]
- Jacobson S. J., Konings D. A., Sarnow P. Biochemical and genetic evidence for a pseudoknot structure at the 3' terminus of the poliovirus RNA genome and its role in viral RNA amplification. J Virol. 1993 Jun;67(6):2961–2971. doi: 10.1128/jvi.67.6.2961-2971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahser F. C., Marsh L. E., Hall T. C. Contributions of the brome mosaic virus RNA-3 3'-nontranslated region to replication and translation. J Virol. 1993 Jun;67(6):3295–3303. doi: 10.1128/jvi.67.6.3295-3303.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S. Y., Sonenberg N., Maizel J. V., Jr Unusual folding regions and ribosome landing pad within hepatitis C virus and pestivirus RNAs. Gene. 1995 Mar 10;154(2):137–143. doi: 10.1016/0378-1119(94)00859-q. [DOI] [PubMed] [Google Scholar]
- Mans R. M., Pleij C. W., Bosch L. tRNA-like structures. Structure, function and evolutionary significance. Eur J Biochem. 1991 Oct 15;201(2):303–324. doi: 10.1111/j.1432-1033.1991.tb16288.x. [DOI] [PubMed] [Google Scholar]
- Marc D., Drugeon G., Haenni A. L., Girard M., van der Werf S. Role of myristoylation of poliovirus capsid protein VP4 as determined by site-directed mutagenesis of its N-terminal sequence. EMBO J. 1989 Sep;8(9):2661–2668. doi: 10.1002/j.1460-2075.1989.tb08406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe C., Eyermann F., Bénard L., Portier C., Ehresmann B., Ehresmann C. Ribosomal protein S15 from Escherichia coli modulates its own translation by trapping the ribosome on the mRNA initiation loading site. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4394–4398. doi: 10.1073/pnas.90.10.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierangeli A., Bucci M., Pagnotti P., Degener A. M., Perez Bercoff R. Mutational analysis of the 3'-terminal extra-cistronic region of poliovirus RNA: secondary structure is not the only requirement for minus strand RNA replication. FEBS Lett. 1995 Nov 6;374(3):327–332. doi: 10.1016/0014-5793(95)01127-z. [DOI] [PubMed] [Google Scholar]
- Pilipenko E. V., Blinov V. M., Romanova L. I., Sinyakov A. N., Maslova S. V., Agol V. I. Conserved structural domains in the 5'-untranslated region of picornaviral genomes: an analysis of the segment controlling translation and neurovirulence. Virology. 1989 Feb;168(2):201–209. doi: 10.1016/0042-6822(89)90259-6. [DOI] [PubMed] [Google Scholar]
- Pilipenko E. V., Gmyl A. P., Maslova S. V., Belov G. A., Sinyakov A. N., Huang M., Brown T. D., Agol V. I. Starting window, a distinct element in the cap-independent internal initiation of translation on picornaviral RNA. J Mol Biol. 1994 Aug 19;241(3):398–414. doi: 10.1006/jmbi.1994.1516. [DOI] [PubMed] [Google Scholar]
- Pilipenko E. V., Gmyl A. P., Maslova S. V., Khitrina E. V., Agol V. I. Attenuation of Theiler's murine encephalomyelitis virus by modifications of the oligopyrimidine/AUG tandem, a host-dependent translational cis element. J Virol. 1995 Feb;69(2):864–870. doi: 10.1128/jvi.69.2.864-870.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko E. V., Gmyl A. P., Maslova S. V., Svitkin Y. V., Sinyakov A. N., Agol V. I. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell. 1992 Jan 10;68(1):119–131. doi: 10.1016/0092-8674(92)90211-t. [DOI] [PubMed] [Google Scholar]
- Pilipenko E. V., Maslova S. V., Sinyakov A. N., Agol V. I. Towards identification of cis-acting elements involved in the replication of enterovirus and rhinovirus RNAs: a proposal for the existence of tRNA-like terminal structures. Nucleic Acids Res. 1992 Apr 11;20(7):1739–1745. doi: 10.1093/nar/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole T. L., Wang C., Popp R. A., Potgieter L. N., Siddiqui A., Collett M. S. Pestivirus translation initiation occurs by internal ribosome entry. Virology. 1995 Jan 10;206(1):750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- Reynolds J. E., Kaminski A., Kettinen H. J., Grace K., Clarke B. E., Carroll A. R., Rowlands D. J., Jackson R. J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995 Dec 1;14(23):6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld K., Linschooten K., Pleij C. W., Bosch L. The three-dimensional folding of the tRNA-like structure of tobacco mosaic virus RNA. A new building principle applied twice. EMBO J. 1984 Nov;3(11):2613–2619. doi: 10.1002/j.1460-2075.1984.tb02182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohll J. B., Moon D. H., Evans D. J., Almond J. W. The 3' untranslated region of picornavirus RNA: features required for efficient genome replication. J Virol. 1995 Dec;69(12):7835–7844. doi: 10.1128/jvi.69.12.7835-7844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spedding G., Draper D. E. Allosteric mechanism for translational repression in the Escherichia coli alpha operon. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4399–4403. doi: 10.1073/pnas.90.10.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spedding G., Gluick T. C., Draper D. E. Ribosome initiation complex formation with the pseudoknotted alpha operon messenger RNA. J Mol Biol. 1993 Feb 5;229(3):609–622. doi: 10.1006/jmbi.1993.1067. [DOI] [PubMed] [Google Scholar]
- Todd S., Nguyen J. H., Semler B. L. RNA-protein interactions directed by the 3' end of human rhinovirus genomic RNA. J Virol. 1995 Jun;69(6):3605–3614. doi: 10.1128/jvi.69.6.3605-3614.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K., Iizuka N., Kohara M., Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992 Mar;66(3):1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Le S. Y., Ali N., Siddiqui A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5' noncoding region. RNA. 1995 Jul;1(5):526–537. [PMC free article] [PubMed] [Google Scholar]
- Wang C., Sarnow P., Siddiqui A. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J Virol. 1994 Nov;68(11):7301–7307. doi: 10.1128/jvi.68.11.7301-7307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer E., Hellen C. U., Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]




