Abstract
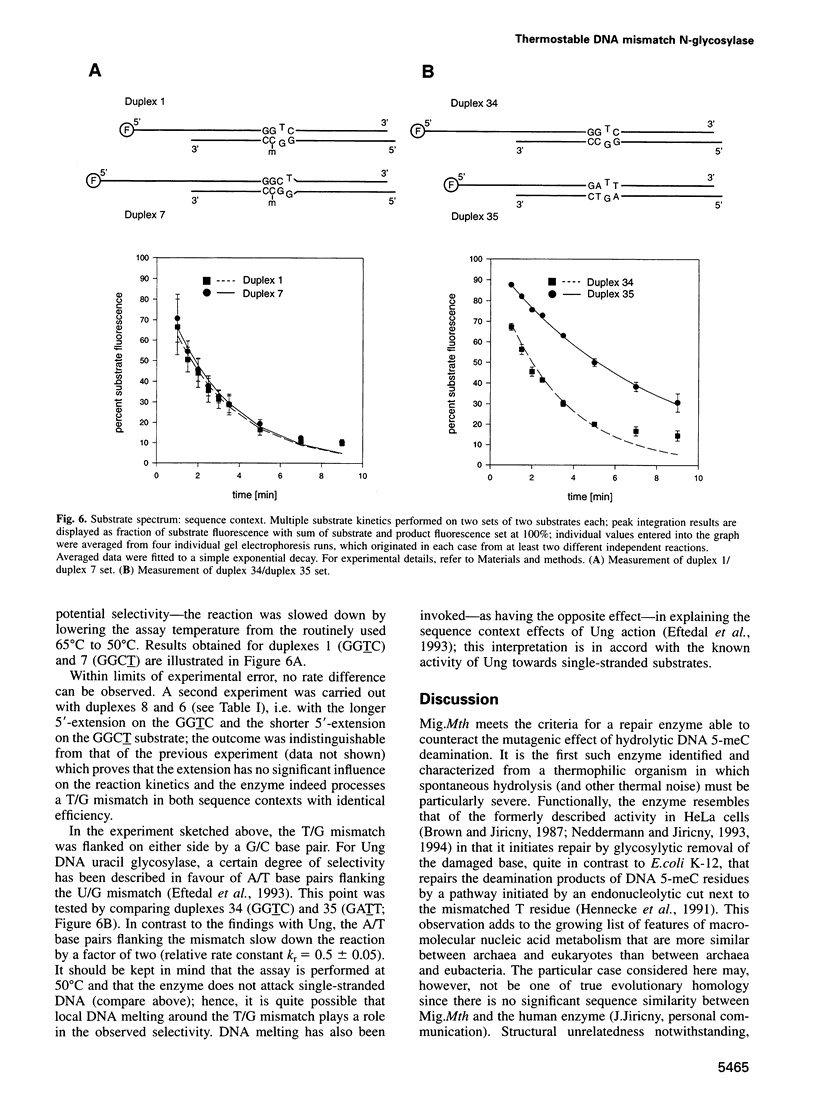
Spontaneous hydrolytic deamination of DNA 5-methylcytosine residues gives rise to T/G mismatches which are pre-mutagenic lesions requiring DNA repair. For fundamental reasons, the significance of this and other processes lowering genetic fidelity must be accentuated at elevated temperatures, making thermophilic organisms attractive objects for studying how cells cope with thermal noise threatening the integrity of their genetic information. Gene mig of Methanobacterium thermoautotrophicum THF, an anaerobic archaeon with an optimal growth temperature of 65 degrees C, was isolated and its product (Mig.Mth; EC3.2.2-) shown to be a T/G-selective DNA thymine N-glycosylase with the properties required for counteracting the mutagenic effect of hydrolytic 5-meC deamination. The enzyme acts on T/G and U/G oppositions with similar efficiency; G/G, A/G, T/C and U/C are minor substrates; no other opposition of common nucleobases is attacked and no removal of U from single-stranded DNA is observed. Substrate preferences are modulated by sequence context. Together with the results presented here, one example of an enzyme directed against the hydrolytic deamination damage of 5-meC is known from each of the three phylogenetic kingdoms; entry into the repair pathway is glycosylytic in the eukaryotic and the archaeal case, whereas the eubacterial repair starts with an endonucleolytic DNA incision.
Full text
PDF

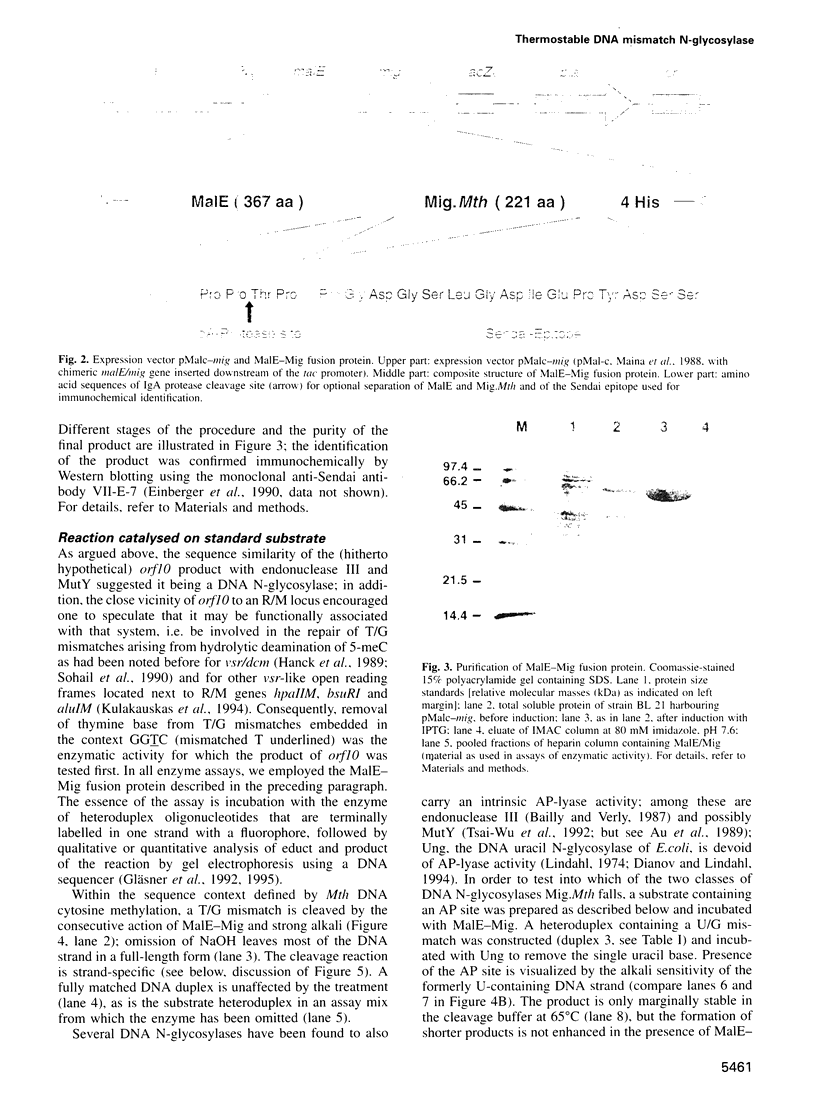
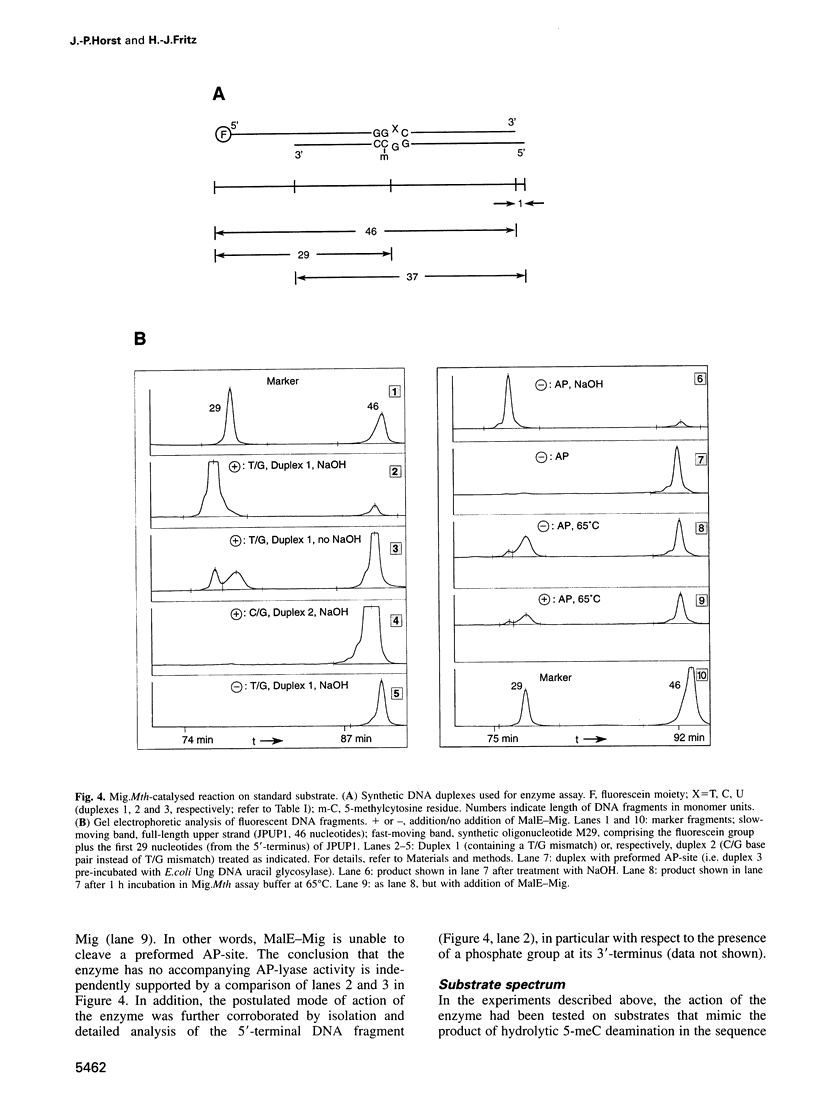
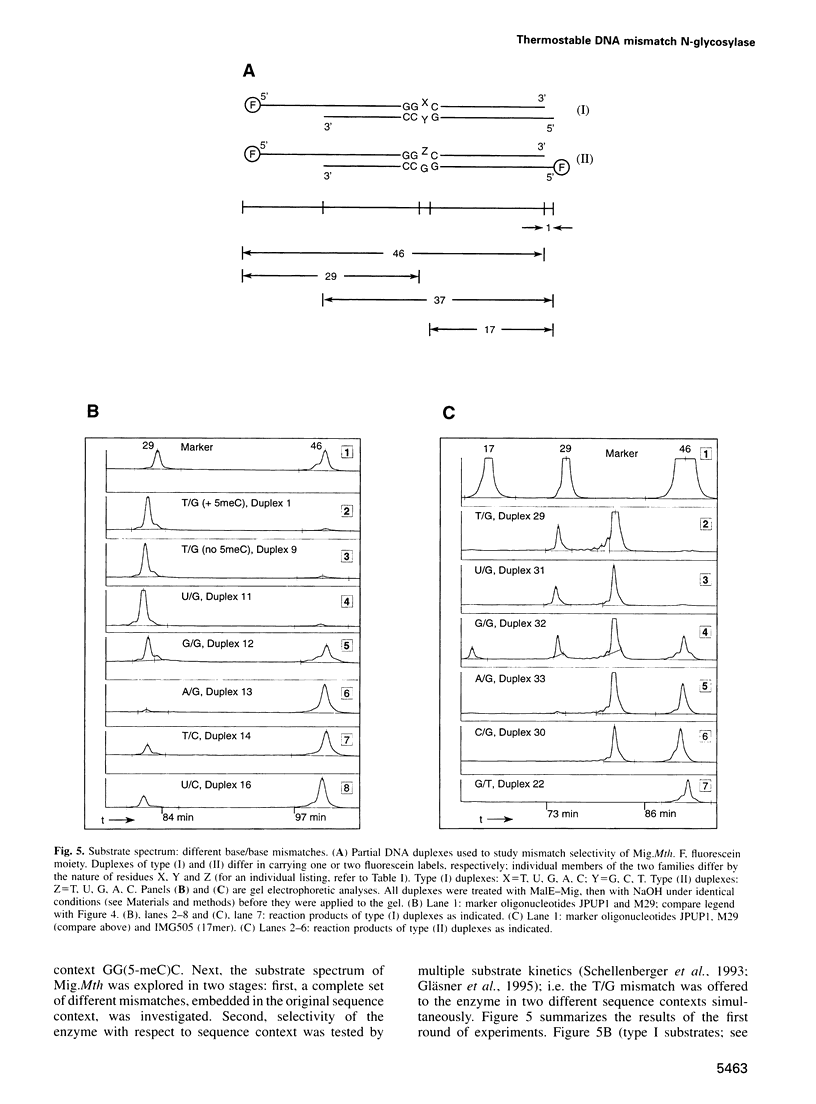





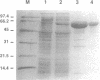
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ansorge W., Sproat B., Stegemann J., Schwager C., Zenke M. Automated DNA sequencing: ultrasensitive detection of fluorescent bands during electrophoresis. Nucleic Acids Res. 1987 Jun 11;15(11):4593–4602. doi: 10.1093/nar/15.11.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahara H., Wistort P. M., Bank J. F., Bakerian R. H., Cunningham R. P. Purification and characterization of Escherichia coli endonuclease III from the cloned nth gene. Biochemistry. 1989 May 16;28(10):4444–4449. doi: 10.1021/bi00436a048. [DOI] [PubMed] [Google Scholar]
- Au K. G., Clark S., Miller J. H., Modrich P. Escherichia coli mutY gene encodes an adenine glycosylase active on G-A mispairs. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8877–8881. doi: 10.1073/pnas.86.22.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly V., Verly W. G. Escherichia coli endonuclease III is not an endonuclease but a beta-elimination catalyst. Biochem J. 1987 Mar 1;242(2):565–572. doi: 10.1042/bj2420565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breimer L. H., Lindahl T. DNA glycosylase activities for thymine residues damaged by ring saturation, fragmentation, or ring contraction are functions of endonuclease III in Escherichia coli. J Biol Chem. 1984 May 10;259(9):5543–5548. [PubMed] [Google Scholar]
- Brown T. C., Jiricny J. A specific mismatch repair event protects mammalian cells from loss of 5-methylcytosine. Cell. 1987 Sep 11;50(6):945–950. doi: 10.1016/0092-8674(87)90521-6. [DOI] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H., Farabaugh P. J., Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978 Aug 24;274(5673):775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Dianov G., Lindahl T. Reconstitution of the DNA base excision-repair pathway. Curr Biol. 1994 Dec 1;4(12):1069–1076. doi: 10.1016/s0960-9822(00)00245-1. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M., Laval J., Boiteux S. Substrate specificity of the Escherichia coli endonuclease III: excision of thymine- and cytosine-derived lesions in DNA produced by radiation-generated free radicals. Biochemistry. 1993 Nov 16;32(45):12105–12111. doi: 10.1021/bi00096a022. [DOI] [PubMed] [Google Scholar]
- Duncan B. K., Rockstroh P. A., Warner H. R. Escherichia coli K-12 mutants deficient in uracil-DNA glycosylase. J Bacteriol. 1978 Jun;134(3):1039–1045. doi: 10.1128/jb.134.3.1039-1045.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplay P., Bedouelle H., Fowler A., Zabin I., Saurin W., Hofnung M. Sequences of the malE gene and of its product, the maltose-binding protein of Escherichia coli K12. J Biol Chem. 1984 Aug 25;259(16):10606–10613. [PubMed] [Google Scholar]
- Eftedal I., Guddal P. H., Slupphaug G., Volden G., Krokan H. E. Consensus sequences for good and poor removal of uracil from double stranded DNA by uracil-DNA glycosylase. Nucleic Acids Res. 1993 May 11;21(9):2095–2101. doi: 10.1093/nar/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einberger H., Mertz R., Hofschneider P. H., Neubert W. J. Purification, renaturation, and reconstituted protein kinase activity of the Sendai virus large (L) protein: L protein phosphorylates the NP and P proteins in vitro. J Virol. 1990 Sep;64(9):4274–4280. doi: 10.1128/jvi.64.9.4274-4280.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gläsner W., Merkl R., Schellenberger V., Fritz H. J. Substrate preferences of Vsr DNA mismatch endonuclease and their consequences for the evolution of the Escherichia coli K-12 genome. J Mol Biol. 1995 Jan 6;245(1):1–7. doi: 10.1016/s0022-2836(95)80033-6. [DOI] [PubMed] [Google Scholar]
- Gläsner W., Merkl R., Schmidt S., Cech D., Fritz H. J. Fast quantitative assay of sequence-specific endonuclease activity based on DNA sequencer technology. Biol Chem Hoppe Seyler. 1992 Dec;373(12):1223–1225. doi: 10.1515/bchm3.1992.373.2.1223. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hanck T., Gerwin N., Fritz H. J. Nucleotide sequence of the dcm locus of Escherichia coli K12. Nucleic Acids Res. 1989 Jul 25;17(14):5844–5844. doi: 10.1093/nar/17.14.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennecke F., Kolmar H., Bründl K., Fritz H. J. The vsr gene product of E. coli K-12 is a strand- and sequence-specific DNA mismatch endonuclease. Nature. 1991 Oct 24;353(6346):776–778. doi: 10.1038/353776a0. [DOI] [PubMed] [Google Scholar]
- Hergersberg M. Biological aspects of cytosine methylation in eukaryotic cells. Experientia. 1991 Dec 1;47(11-12):1171–1185. doi: 10.1007/BF01918381. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989 Apr;5(2):151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- Jones P. A., Rideout W. M., 3rd, Shen J. C., Spruck C. H., Tsai Y. C. Methylation, mutation and cancer. Bioessays. 1992 Jan;14(1):33–36. doi: 10.1002/bies.950140107. [DOI] [PubMed] [Google Scholar]
- Kulakauskas S., Barsomian J. M., Lubys A., Roberts R. J., Wilson G. G. Organization and sequence of the HpaII restriction-modification system and adjacent genes. Gene. 1994 May 3;142(1):9–15. doi: 10.1016/0378-1119(94)90348-4. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kuo C. F., McRee D. E., Fisher C. L., O'Handley S. F., Cunningham R. P., Tainer J. A. Atomic structure of the DNA repair [4Fe-4S] enzyme endonuclease III. Science. 1992 Oct 16;258(5081):434–440. doi: 10.1126/science.1411536. [DOI] [PubMed] [Google Scholar]
- Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974 Jul 30;13(16):3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Tsai-Wu J. J., Cillo J. DNA determinants and substrate specificities of Escherichia coli MutY. J Biol Chem. 1995 Oct 6;270(40):23582–23588. doi: 10.1074/jbc.270.40.23582. [DOI] [PubMed] [Google Scholar]
- Maina C. V., Riggs P. D., Grandea A. G., 3rd, Slatko B. E., Moran L. S., Tagliamonte J. A., McReynolds L. A., Guan C. D. An Escherichia coli vector to express and purify foreign proteins by fusion to and separation from maltose-binding protein. Gene. 1988 Dec 30;74(2):365–373. doi: 10.1016/0378-1119(88)90170-9. [DOI] [PubMed] [Google Scholar]
- Michaels M. L., Pham L., Nghiem Y., Cruz C., Miller J. H. MutY, an adenine glycosylase active on G-A mispairs, has homology to endonuclease III. Nucleic Acids Res. 1990 Jul 11;18(13):3841–3845. doi: 10.1093/nar/18.13.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels M. L., Tchou J., Grollman A. P., Miller J. H. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 1992 Nov 17;31(45):10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- Neddermann P., Jiricny J. Efficient removal of uracil from G.U mispairs by the mismatch-specific thymine DNA glycosylase from HeLa cells. Proc Natl Acad Sci U S A. 1994 Mar 1;91(5):1642–1646. doi: 10.1073/pnas.91.5.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neddermann P., Jiricny J. The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells. J Biol Chem. 1993 Oct 5;268(28):21218–21224. [PubMed] [Google Scholar]
- Nölling J., de Vos W. M. Characterization of the archaeal, plasmid-encoded type II restriction-modification system MthTI from Methanobacterium thermoformicicum THF: homology to the bacterial NgoPII system from Neisseria gonorrhoeae. J Bacteriol. 1992 Sep;174(17):5719–5726. doi: 10.1128/jb.174.17.5719-5726.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nölling J., van Eeden F. J., Eggen R. I., de Vos W. M. Modular organization of related Archaeal plasmids encoding different restriction-modification systems in Methanobacterium thermoformicicum. Nucleic Acids Res. 1992 Dec 25;20(24):6501–6507. doi: 10.1093/nar/20.24.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L. C., Aasland R., Wittwer C. U., Krokan H. E., Helland D. E. Molecular cloning of human uracil-DNA glycosylase, a highly conserved DNA repair enzyme. EMBO J. 1989 Oct;8(10):3121–3125. doi: 10.1002/j.1460-2075.1989.tb08464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival K. J., Klein M. B., Burgers P. M. Molecular cloning and primary structure of the uracil-DNA-glycosylase gene from Saccharomyces cerevisiae. J Biol Chem. 1989 Feb 15;264(5):2593–2598. [PubMed] [Google Scholar]
- Pohlner J., Klauser T., Kuttler E., Halter R. Sequence-specific cleavage of protein fusions using a recombinant Neisseria type 2 IgA protease. Biotechnology (N Y) 1992 Jul;10(7):799–804. doi: 10.1038/nbt0792-799. [DOI] [PubMed] [Google Scholar]
- Schellenberger V., Siegel R. A., Rutter W. J. Analysis of enzyme specificity by multiple substrate kinetics. Biochemistry. 1993 Apr 27;32(16):4344–4348. doi: 10.1021/bi00067a025. [DOI] [PubMed] [Google Scholar]
- Schlagman S., Hattman S., May M. S., Berger L. In vivo methylation by Escherichia coli K-12 mec+ deoxyribonucleic acid-cytosine methylase protects against in vitro cleavage by the RII restriction endonuclease (R. Eco RII). J Bacteriol. 1976 May;126(2):990–996. doi: 10.1128/jb.126.2.990-996.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail A., Lieb M., Dar M., Bhagwat A. S. A gene required for very short patch repair in Escherichia coli is adjacent to the DNA cytosine methylase gene. J Bacteriol. 1990 Aug;172(8):4214–4221. doi: 10.1128/jb.172.8.4214-4221.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Thayer M. M., Ahern H., Xing D., Cunningham R. P., Tainer J. A. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995 Aug 15;14(16):4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai-Wu J. J., Liu H. F., Lu A. L. Escherichia coli MutY protein has both N-glycosylase and apurinic/apyrimidinic endonuclease activities on A.C and A.G mispairs. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8779–8783. doi: 10.1073/pnas.89.18.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin E. A., Wolfe R. S., Wolin M. J. Viologen dye inhibition of methane formation by Methanobacillus omelianskii. J Bacteriol. 1964 May;87(5):993–998. doi: 10.1128/jb.87.5.993-998.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]