Abstract
Telomeric DNA represents a novel target for the development of anticancer drugs. By application of a catalytic metallodrug strategy, a copper–acridine–ATCUN complex (CuGGHK-Acr) has been designed that targets G-quadruplex telomeric DNA. Both fluorescence solution assays and gel sequencing demonstrate the CuGGHK-Acr catalyst to selectively bind and cleave the G-quadruplex telomere sequence. The cleavage pathway has been mapped by matrix assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) experiments. CuGGHK-Acr promotes significant inhibition of cancer cell proliferation and shortening of telomere length. Both senescence and apoptosis are induced in the breast cancer cell line MCF7.
Keywords: anticancer agents, bioinorganic chemistry, carbon, DNA cleavage, peptides
Essential to DNA replication and cell division,[1] telomeric DNA has the potential to form a type of nucleic acid secondary structure known as the G-quadruplex (G4).[2] This, and other structured nucleic acid motifs are a current focus of drug discovery efforts.[3] The replicative capacity of cells is determined by telomere length since cellular senescence occurs following its reduction to a critical level (Hayflick limit).[4] Telomere length is maintained by telomerase in most cancer cells, but is shorter as a consequence of frequent cell division.[5] Accordingly, there is increasing interest in the development of G4 ligands as anticancer drugs.[6]
G4 ligands can interfere with telomere maintenance by stabilizing G-quadruplex telomere structure.[7] However, a significant change of a cancer cell’s telomere length typically requires long-term treatment, because only about 50 to 200 nt of telomere length is lost during each round of cell division.[8] In that regard, a CeIVEDTP–DNA (EDTP =ethylenedi-aminetetra(methylenephosphonic acid)) conjugate has recently been reported to induce sequence-specific cleavage of telomeric DNA by assembling a (3 +1) intermolecular G-quadruplex. However, a combination of low cellular uptake, instability to natural nucleases, and self-cleavage are unfavorable to further application.[9] Thus far, no DNA-cleaving agents have been reported that exhibit selective cleavage of intramolecular G-quadruplex telomeric DNA.
In our previous studies, an amino-terminal copper/nickel binding motif (ATCUN) has been incorporated into a variety of peptide frameworks to develop antiviral metallopeptides that cleave HIV and HCV (hepatitis C virus) RNA.[10] The ATCUN motif coordinates a copper ion with a high affinity, is redox active in the 3 +/2 + states, and can promote DNA cleavage under physiologically relevant conditions.[11] Herein, we develop a G4-cleaving agent by coupling GGHK, an ATCUN peptide, to an acridine-based G4 ligand that has the capability of positioning a CuGGH moiety in close proximity to the G4 telomeric DNA, and promoting selective cleavage (Scheme 1). In addition, recent studies suggest that more G4 DNA is formed during DNA replication than the G0/G1 phase of cell cycle division; therefore, cancer cells should be more vulnerable to G4-targeting drugs as a result of their frequent cell division.[12]
Scheme 1.
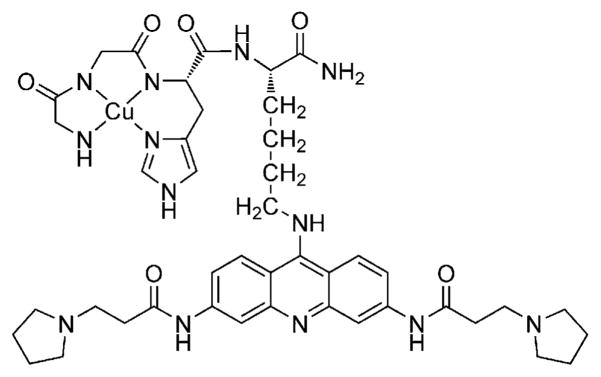
Chemical structure of CuGGHK-Acr.
A fluorescein-labeled G4 oligonucleotide of human telomeric DNA (22G4: 5′-FAM-d(AGGGTTAGGGT-TAGGGTTAGGG)) was employed as a model for binding and cleavage reactivity studies. Binding of CuGGHK-Acr to 22G4 DNA yielded a KD ~ 0.51 μM by monitoring the quenching of FAM (fluorescein) emission at 520 nm (λex = 494 nm), which indicates a significant affinity of CuGGHK-Acr toward 22G4 in K+ solution (Figure S1). By contrast, titration of the metal-binding motif CuGGH to a solution of 22G4 DNA resulted in a negligible change of emission (data not shown), suggesting Fçrster resonance energy transfer from FAM to the acridine ring of CuGGHK-Acr. The affinity of CuGGHK-Acr is consistent with the affinity of other reported acridine analogues.[13] In fact, prior structural studies suggest the acridine ring to stack on the surface of the G-tetrad, with the positively charged nitrogen atom of the pyrrolidine ring interacting with a water molecule near the phosphate backbone.[14]
The addition of calf thymus DNA (CT-DNA) to a solution containing both CuGGHK-Acr and 22G4 recovered the emission of 22G4 due to the nonselective binding of G4 ligand to CT-DNA. However, the addition of 120-fold equivalents of CT-DNA only yielded a 32% recovery of emission (Figure S2). In addition, titration of CuGGHK-Acr to a 22mer self-complementary duplex telomeric DNA (ds22Telo: 5′-FAM-d(TTAGGGTTAGG)-(CH2CH2O)6-d(CCTAACCC-TAA)) resulted in a KD ~ 12.0 μM (Figure S3). Both the competition assay with CT-DNA and binding affinity with ds22telo indicate CuGGHK-Acr to have a significant preference for binding to 22G4 DNA over duplex DNA.
The Michaelis–Menten kinetics of CuGGHK-Acr was measured from the change of emission following cleavage, which may result either from the change of microenvironment or the dissociation of catalyst from cleavage product. No significant cleavage was observed in the absence of ascorbate or H2O2. A plot of initial velocity versus substrate concentration yielded a KM of 1.43 μM and kcat of 0.0578 s−1 (Figure S4). The KM is approximately 3 times the value of KD, yielding a k1 of 0.063 μM−1s−1 and k−1 of 0.032 s−1. In addition, CuGGH exhibited a similar kcat (0.0215 s−1) as that observed for CuGGHK-Acr, consistent with CuGGH attacking DNA from various orientations, whereas the position of the Cu center in CuGGHK-Acr is restricted by the interaction between the G-tetrad and acridine groups. CuGGH, lacking the binding moiety for G4 substrate, has a significantly higher KM (20.5 μM) than that observed for CuGGHK-Acr, resulting in a ~ 38-fold smaller kcat/KM for CuGGH relative to CuGGHK-Acr.
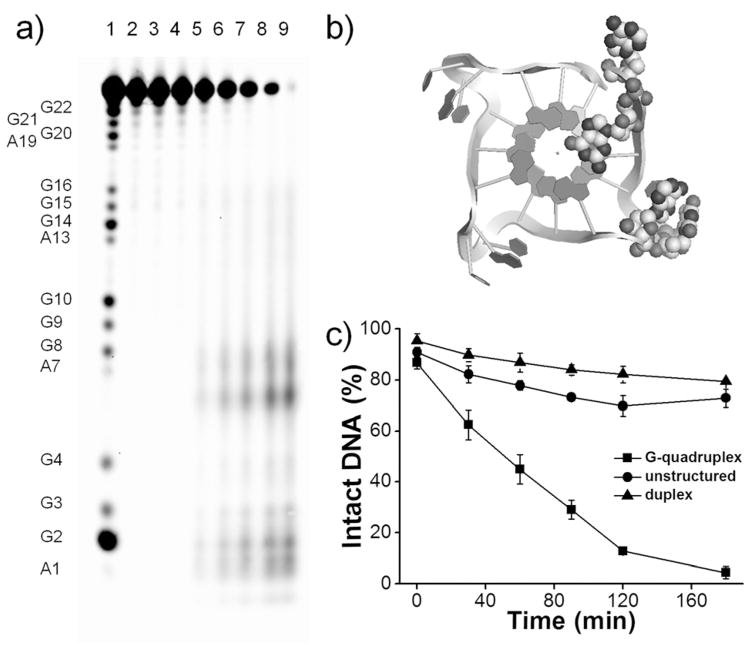
To characterize the 22G4 cleavage site(s) promoted by CuGGHK-Acr, and confirm its specific binding pattern, products from the reaction mixture were separated by denaturing PAGE. Time-dependent cleavage by CuGGHK-Acr was observed (Figure 1a). Especially, in lane 9, the major cleavage sites were located at A1 (12.8%), G2 (10.2%), T6 (13.7%), A7 (7.6%), and other nearby nucleotides, demonstrating the Cu center to be located near the groove formed by the stacked G2–G4 trinucleotides and the T5-A7 loop (Figure 1b). Minor cleavage events at other nucleotides near this groove most likely reflect the flexibility of the lysine side chain carrying the reactive CuGGH moiety, as well as the short-range local diffusion of reactive oxygen species. By contrast, DNA fragments from the cleavage by CuGGH are manifested as an average distribution indicating the metal-binding motif to exhibit a random cleavage pattern (Figure S5).
Figure 1.
a) Denaturing polyacrylamide gel electrophoresis (15% PAGE) showing the sequence-specific cleavage of 22G4 by CuGGHK-Acr in 10 mM Tris-HCl and 100 mM KCl at 37°C. Lane 1: A +G DNA Ladder; Lane 2: DNA only; Lane 3: DNA +ascorbate+H2O2 ; Lanes 4 to 9: CuGGHK-Acr +DNA +ascorbate+H2O2 (0, 30, 60, 90, 120, and 180 min). [22G4] =10 μM (strand concentration), [CuGGHK-Acr] =2 μM, [ascorbate] =1 mM, [H2O2] = 1 mM. b) Summary of major cleavage sites (highlighted by CPK atoms) identified in parallel G4 telomeric DNA (PDB: 1KF1). c) Selective cleavage by CuGGHK-Acr on three substrates at 37°C. G-quadruplex DNA (■): 22G4 in 10 mM Tris-HCl (pH 7.4), 100 mM KCl; unstructured DNA (●): 22G4 in 10 mM Tris-HCl (pH 7.4); duplex DNA (▲): ds12Telo in 10 mM Tris-HCl (pH 7.4), 100 mM LiCl. Each of the above reactions used 220 μM (nucleotide concentration) of DNA, 2 μM CuGGHK-Acr, 1 mM of ascorbate, and 1 mM of H2O2.
Duplex telomeric DNA and unstructured telomeric DNA (22G4 in the absence of monovalent metal cation) served as controls to evaluate the cleavage selectivity. Sequencing gels demonstrate CuGGHK-Acr to exhibit weaker cleavage activity with unstructured 22G4 relative to the G-quadruplex (Figures 1c and S6). Unfortunately, ds22telo cannot be completely denatured under the PAGE conditions used; therefore a shorter self-complementary duplex telomeric DNA ds12Telo (5′-FAM-d(TTAGGG)-(CH2CH2O)6-d-(CCCTAA)) was designed to evaluate the cleavage selectivity. Cleavage of ds12Telo was performed in the presence of Li+, which destabilizes G-quadruplex DNA without influencing duplex DNA.[15] In fact, cleavage of ds12Telo was observed to be even weaker than for unstructured 22G4, reflecting preferential binding of CuGGHK-Acr to G-quad-ruplex DNA (Figures S6 and S7). In comparison to the duplex telomeric DNA, unstructured 22G4 may be more accessible. Moreover, the double filter mechanism for metallodrug activity can also contribute to this selective cleavage.[16]
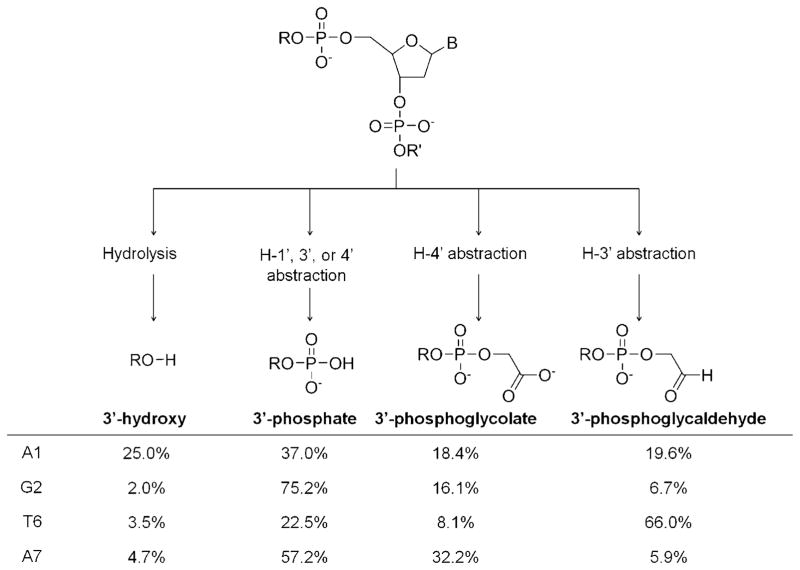
Mass spectrometry was used to characterize the 3′ overhang fragments from specific degradation paths at each site. Major 3′ overhang products were identified as 3′-hydroxy, 3′-phosphate, 3′-phosphoglycolate, and 3′-phosphoglycaldehyde (Figure 2), and confirmed by the broadening of gel bands in PAGE due to the distinct negative charges of these fragments. Unlike the oxidative path initiated by hydrogen abstraction from 1′, 3′, and 4′ CH positions of the deoxyribose ring,[17] the 3′-hydroxy product should reflect hydrolysis of phosphate backbone by transient formation of the strong Lewis acid Cu3+ species.[18] Since oxidative cleavage products are not generally accepted as typical substrates of telomerase,[19] the advantage of this cleavage pattern is that telomeric DNA will be more difficult to elongate by telomerase following cleavage, unless these DNA lesions are first repaired by other DNA repair mechanisms.
Figure 2.
The 3′ overhang cleavage products generated from both hydrolysis and oxidative cleavage mechanisms. The relative intensities of the cleavage products characterized by MALDI-MS are shown. DNA cleavage was performed with the following reagents: [22G4]=10 μM (strand concentration), [CuGGHK-Acr] =2 μM, [ascorbate] =1 mM, [H2O2] = 1 mM, in 10 mM Tris-HCl, and 100 mM KCl at 37 °C for 180 min.
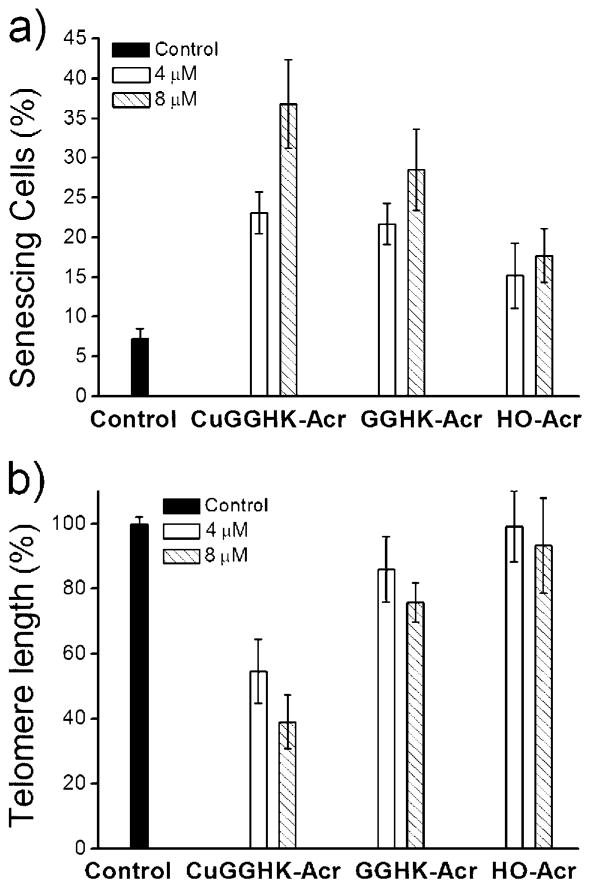
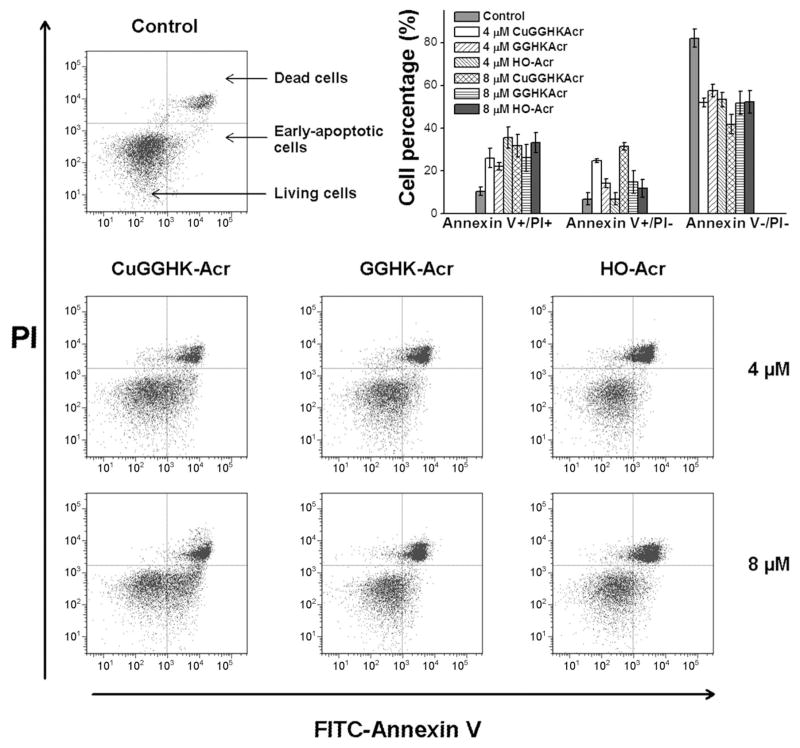
The effects of CuGGHK-Acr on cancer cell proliferation have also been studied. CuGGHK-Acr shows more significant anticancer activity against both MCF7 and HuH7 cell lines (Table 1) than the metal-free ligand GGHK-Acr, or the prescursor (HO-Acr) lacking the ATCUN motif, indicating the importance of Cu. Caco2 cells are growing slower and as a result the influence is less clear. To further confirm the effect on MCF7 breast carcinoma cells, cellular senescence and apoptosis studies were performed. In fact, 8 μM of CuGGHK-Acr can induce more senescent cells (~ 37%) and apoptotic cells (~ 32%) than the same amount of either GGHK-Acr or HO-Acr (Figures 3a, 4, and S8). Despite the apoptotic resistance often observed in many types of senescent cell,[20] both apoptosis and senescence are actually p53-dependent.[21] In addition, the telomere length in MCF7 cells measured by real-time PCR suggests that CuGGHK-Acr can shorten the telomere by 50–60%, relative to that of the untreated MCF7 cells, whereas HO-Acr alone decreases the length by <7% (Figure 3b). We hypothesized that CuGGHK-Acr could cleave the telomeric DNA of cancer cells by targeting the G-quadruplex, with persistent telomere shortening eventually activating p53. Relative to HO-Acr, both a more extensive shortening of telomeric DNA, and a larger number of senescent cells were observed after treatment of copper-free GGHK-Acr, which most likely reflects the recruitment of copper from serum to form the active CuGGHK-Acr complex. Interestingly, the percentage of dead cells (~ 33%) caused by HO-Acr approximates to that of CuGGHK-Acr (~ 32%). Considering that HO-Acr does not induce obvious early apoptosis, the dead cells (annexin V+/PI +) caused by HO-Acr should mainly arise from necrosis.
Table 1.
Activity of cell proliferation inhibition expressed as IC50 value.
| Compound | Features of molecule | IC50 [μM][a] | |||
|---|---|---|---|---|---|
| ACTUN | Cu | HuH-7 | MCF7 | Caco2 | |
| CuGGHK-Acr | + | + | 9.8 ±2.3 | 8.9 ±1.1 | 17.2 ±2.9 |
| GGHK-Acr | + | − | 26.6 ±2.2 | 20.8 ±0.8 | 24.1 ±3.0 |
| HO-Acr | − | − | 16.7 ±1.6 | 18.3 ±1.8 | 24.6 ±3.5 |
IC50 values were measured with the MTT assay after 72 h exposure.
Figure 3.
a) Percentage of senescing MCF7 cells revealed by a senescence-associated beta-galactosidase assay after a 7 day treatment period with the indicated concentration of CuGGHK-Acr, GGHK-Acr, or HO-Acr. b) Relative telomere length of MCF7 cells measured by real-time PCR after a 7 day treatment period with the indicated compounds.
Figure 4.
Apoptosis analysis of MCF7 by flow cytometry (FITC-Annexin V and PI assay) after incubation with CuGGHK-Acr, GGHK-Acr, or HO-Acr for 7 days. The histogram insert shows the percentage of cells at different life stages: live cells (Annexin V−/PI−), early-apoptotic cells (Annexin V+/PI−), and dead cells (Annexin V+/PI+), respectively.
In summary, we have designed a novel DNA-cleaving agent CuGGHK-Acr that targets G4 telomeric DNA by application of a catalytic metallodrug. These results demonstrate that CuGGHK-Acr can selectively bind to G4 telo-meric DNA, relative to CT-DNA, and promote efficient irreversible cleavage of G4 telomeric DNA, relative to telomeric DNA in other structural states. Major selective cleavage sites were identified as A1-G2 and T6-A7 nucleotides from a model G-quadruplex oligonucleotide derived from telomeric DNA. Results from MALDI-MS suggest that both hydrolysis and oxidative cleavage mechanisms are involved in DNA cleavage, but both require redox co-reagents. Significant inhibition of cancer cell proliferation was also observed. As a result of the significant shortening of telomeric DNA, more effective cellular senescence and apoptosis are induced in MCF7 cells by CuGGHK-Acr than its analogues lacking the Cu center. Overall, this study represents an important first step in a novel approach to designing selective telomeric DNA-cleaving agents that have the potential to be developed into potent anticancer drugs.
Supplementary Material
Footnotes
This work was supported by grants from the National Institutes of Health (HL093446 and AA016712).
Contributor Information
Zhen Yu, Department of Chemistry and Biochemistry, The Ohio State University, 100 West 18th Avenue, Columbus, OH 43210 (USA).
Menglu Han, College of Pharmacy, the Ohio State University, 500 West 12th Avenue, Columbus, OH 43210 (USA).
Dr. James A. Cowan, Email: cowan@chemistry.ohio-state.edu, Department of Chemistry and Biochemistry, The Ohio State University, 100 West 18th Avenue, Columbus, OH 43210 (USA). MetalloPharm, 1790 Riverstone Drive, Delaware, OH 43015 (USA)
References
- 1.a) O’Sullivan RJ, Karlseder J. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Blackburn EH. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 2.a) Henderson A, Wu Y, Huang YC, Chavez EA, Platt J, Johnson FB, Brosh RM, Jr, Sen D, Lansdorp PM. Nucleic Acids Res. 2014;42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lipps HJ, Rhodes D. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]; c) Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. J Am Chem Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Malina J, Hannon MJ, Brabec V. FEBS J. 2014;281:987–997. doi: 10.1111/febs.12696. [DOI] [PubMed] [Google Scholar]; b) Phongtongpasuk S, Paulus S, Schnabl J, Sigel RKO, Spingler B, Hannon MJ, Freisinger E. Angew Chem Int Ed. 2013;52:11513–11516. doi: 10.1002/anie.201305079. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2013;125:11727–11730. [Google Scholar]; c) del Mundo IMA, Siters KE, Fountain MA, Morrow JR. In org Chem. 2012;51:5444–5457. doi: 10.1021/ic3004245. [DOI] [PubMed] [Google Scholar]; d) Wu K, Liu S, Luo Q, Hu W, Li X, Wang F, Zheng R, Cui J, Sadler PJ, Xiang J, et al. Inorg Chem. 2013;52:11332–11342. doi: 10.1021/ic401606v. [DOI] [PubMed] [Google Scholar]
- 4.a) Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, Greider CW, Harley CB. Proc Natl Acad Sci USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chiu CP, Harley CB. Proc Soc Exp Biol Med. 1997;214:99–106. doi: 10.3181/00379727-214-44075. [DOI] [PubMed] [Google Scholar]; c) Allsopp RC, Harley CB. Exp Cell Res. 1995;219:130–136. doi: 10.1006/excr.1995.1213. [DOI] [PubMed] [Google Scholar]
- 5.a) Hiyama E, Hiyama K. Cancer Lett. 2003;194:221–233. doi: 10.1016/s0304-3835(02)00709-7. [DOI] [PubMed] [Google Scholar]; b) Neidle S, Parkinson G. Nat Rev Drug Discovery. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 6.a) Dixon IM, Lopez F, Tejera AM, Esteve JP, Blasco MA, Pratviel G, Meunier B. J Am Chem Soc. 2007;129:1502–1503. doi: 10.1021/ja065591t. [DOI] [PubMed] [Google Scholar]; b) Verga D, Hamon F, Poyer F, Bombard S, Teulade-Fichou MP. Angew Chem Int Ed. 2014;53:994–998. doi: 10.1002/anie.201307413. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2014;126:1012–1016. [Google Scholar]; c) McLuckie KIE, Di Antonio M, Zecchini H, Xian J, Caldas C, Krippendorff BF, Tannahill D, Lowe C, Balasubramanian S. J Am Chem Soc. 2013;135:9640–9643. doi: 10.1021/ja404868t. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Di Antonio M, Doria F, Richter SN, Bertipaglia C, Mella M, Sissi C, Palumbo M, Freccero M. J Am Chem Soc. 2009;131:13132–13141. doi: 10.1021/ja904876q. [DOI] [PubMed] [Google Scholar]; e) Dai JX, Carver M, Hurley LH, Yang DZ. J Am Chem Soc. 2011;133:17673–17680. doi: 10.1021/ja205646q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Tahara H, Shin-ya K, Seimiya H, Yamada H, Tsuruo T, Ide T. Oncogene. 2006;25:1955–1966. doi: 10.1038/sj.onc.1209217. [DOI] [PubMed] [Google Scholar]; b) Casa-grande V, Salvati E, Alvino A, Bianco A, Ciammaichella A, D’Angelo C, Ginnari-Satriani L, Serrilli AM, Iachettini S, Leonetti C, Neidle S, Ortaggi G, Porru M, Rizzo A, Franceschin M, Biroccio A. J Med Chem. 2011;54:1140–1156. doi: 10.1021/jm1013665. [DOI] [PubMed] [Google Scholar]; c) Yaku H, Fujimoto T, Murashima T, Miyoshi D, Sugimoto N. Chem Commun. 2012;48:6203–6216. doi: 10.1039/c2cc31037f. [DOI] [PubMed] [Google Scholar]
- 8.a) Greider CW. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]; b) Huffman KE, Levene SD, Tesmer VM, Shay JW, Wright WE. J Biol Chem. 2000;275:19719–19722. doi: 10.1074/jbc.M002843200. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Suzuki Y, Lonnberg T, Komiyama M. J Am Chem Soc. 2009;131:2871–2874. doi: 10.1021/ja807313x. [DOI] [PubMed] [Google Scholar]
- 10.a) Bradford S, Cowan JA. Chem Commun. 2012;48:3118–3120. doi: 10.1039/c2cc17377h. [DOI] [PubMed] [Google Scholar]; b) Joyner JC, Cowan JA. J Am Chem Soc. 2011;133:9912–9922. doi: 10.1021/ja203057z. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Jin Y, Cowan JA. J Am Chem Soc. 2006;128:410–411. doi: 10.1021/ja055272m. [DOI] [PubMed] [Google Scholar]
- 11.a) Joyner JC, Reichfield J, Cowan JA. J Am Chem Soc. 2011;133:15613–15626. doi: 10.1021/ja2052599. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Jin Y, Lewis MA, Gokhale NH, Long EC, Cowan JA. J Am Chem Soc. 2007;129:8353–8361. doi: 10.1021/ja0705083. [DOI] [PubMed] [Google Scholar]; c) Jin Y, Cowan JA. J Am Chem Soc. 2005;127:8408–8415. doi: 10.1021/ja0503985. [DOI] [PubMed] [Google Scholar]
- 12.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Nat Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Moore MJB, Schultes CM, Cuesta J, Cuenca F, Gunaratnam M, Tanious FA, Wilson WD, Neidle S. J Med Chem. 2006;49:582–599. doi: 10.1021/jm050555a. [DOI] [PubMed] [Google Scholar]; b) Cheng MK, Modi C, Cookson JC, Hutchinson I, Heald RA, McCarroll AJ, Missailidis S, Tanious F, Wilson WD, Mergny JL, Laughton CA, Stevens MFG. J Med Chem. 2008;51:963–975. doi: 10.1021/jm070587t. [DOI] [PubMed] [Google Scholar]
- 14.a) Collie GW, Sparapani S, Parkinson GN, Neidle S. J Am Chem Soc. 2011;133:2721–2728. doi: 10.1021/ja109767y. [DOI] [PubMed] [Google Scholar]; b) Campbell NH, Parkinson GN, Reszka AP, Neidle S. J Am Chem Soc. 2008;130:6722–6724. doi: 10.1021/ja8016973. [DOI] [PubMed] [Google Scholar]
- 15.a) Viglaský V, Bauer L, Tluckova K. Biochemistry. 2010;49:2110–2120. doi: 10.1021/bi902099u. [DOI] [PubMed] [Google Scholar]; b) Nakano S, Fujimoto M, Hara H, Sugimoto N. Nucleic Acids Res. 1999;27:2957–2965. doi: 10.1093/nar/27.14.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hocharoen L, Cowan JA. Chem Eur J. 2009;15:8670–8676. doi: 10.1002/chem.200900821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pogozelski WK, Tullius TD. Chem Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 18.Bradford SS, Ross MJ, Fidai I, Cowan JA. Chem Med Chem. 2014;9:1275–1285. doi: 10.1002/cmdc.201400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Strahl C, Blackburn EH. Nucleic Acids Res. 1994;22:893–900. doi: 10.1093/nar/22.6.893. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Strahl C, Blackburn EH. Mol Cell Biol Hum Dis. 1996;16:53–65. doi: 10.1128/mcb.16.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.a) Spaulding C, Guo W, Effros RB. Exp Gerontol. 1999;34:633–644. doi: 10.1016/s0531-5565(99)00033-9. [DOI] [PubMed] [Google Scholar]; b) Ryu SJ, Cho KA, Oh YS, Park SC. Apoptosis. 2006;11:303–313. doi: 10.1007/s10495-006-3978-9. [DOI] [PubMed] [Google Scholar]
- 21.a) Atadja P, Wong H, Garkavtsev I, Veillette C, Riabowol K. Proc Natl Acad Sci USA. 1995;92:8348–8352. doi: 10.1073/pnas.92.18.8348. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Harris SL, Levine AJ. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]; c) Lowe SW, Ruley HE, Jacks T, Housman DE. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.