Abstract
Replication of human cytomegalovirus is regulated in part by cellular kinases and the single viral Ser/Thr kinase, pUL97. The virus-coded kinase augments the replication of human cytomegalovirus (HCMV) by enabling nuclear egress and altering cell cycle progression. These roles are accomplished through direct phosphorylation of nuclear lamins and the retinoblastoma protein, respectively. In an effort to identify additional pUL97 substrates, we analyzed the phosphoproteome of SILAC-labeled human fibroblasts during infection with either wild-type HCMV or a pUL97 kinase-dead mutant virus. Phosphopeptides were enriched over a titanium dioxide matrix and analyzed by high resolution mass spectrometry. We identified 157 unambiguous phosphosites from 106 cellular and 17 viral proteins whose phosphorylation required UL97. Analysis of peptides containing these sites allowed the identification of several candidate pUL97 phosphorylation motifs, including a completely novel phosphorylation motif, LxSP. Substrates harboring the LxSP motif were enriched in nucleocytoplasmic transport functions, including a number of components of the nuclear pore complex. These results extend the known functions of pUL97 and suggest that modulation of nuclear pore function may be important during HCMV replication.
Keywords: Human cytomegalovirus, herpesvirus, viral kinase, kinase motif, nuclear pore complex, TREX complex
1 Introduction
Human cytomegalovirus (HCMV) modulates its host cell to promote viral replication and subvert antiviral responses. This includes critical changes to intracellular trafficking, metabolism, and signaling (reviewed in [1]). Altered cellular kinase activity contributes to virus-mediated changes in each of these areas, and HCMV-encoded proteins modulate many cellular kinases. HCMV infection initiates major changes in total cellular phosphorylation patterns [2] and many HCMV proteins are themselves phosphorylated [1]. HCMV infection alters or requires the activity of a number of cellular kinases, including MAPK, PI3K, PKC, CDK1, and mTOR [3–12].
The HCMV-encoded kinase, pUL97, alters the phosphorylation dynamics of infected cells. Its activity is directed towards both small molecules and protein serine and threonine residues. Importantly, pUL97 phosphorylates ganciclovir (or the related pro-drug valganciclovir) leading to production of an active nucleoside analog which competitively inhibits the HCMV viral polymerase [1]. Ganciclovir is the standard of care for acute HCMV infection and prophylaxis. Resistance to ganciclovir occurs in some 5–30% of patients who undergo prolonged anti-HCMV therapy, and of these approximately 80% are mutations in pUL97 that inhibit its ability to phosphorylate ganciclovir [13]. Thus, pUL97 is an important player in HCMV therapeutics.
The pUL97 kinase is known to phosphorylate multiple cell-coded protein targets, including nuclear lamins to facilitate nuclear egress of virus capsids [3, 14, 15], the retinoblastoma protein to promote changes in cell cycle [16, 17], elongation factor 1 delta [18], and the C-terminus of RNA polymerase II [19]. Furthermore, pUL97 is incorporated into virions [20], possibly in association with the HCMV pUL83 protein [21]. Its virion localization suggests that it may phosphorylate virion components as well as cellular targets upon entry into a newly infected cell. pUL97 is classified as “augmenting” for laboratory growth of HCMV [22, 23]. That is, deletion or inactivation of pUL97 results in decreased yield of virus produced, but the gene or its product is not strictly essential. The primary defect appears to lie in nuclear egress; a kinase-dead point mutant, UL97-K355Q [24], accumulates viral capsids in the nucleus [25]. However, pUL97 also auto-phosphorylates [26] and phosphorylates UL44, the vDNA polymerase [25, 27]. Additionally, pUL97 interacts with and may phosphorylate pUL69, a viral factor involved in mRNA export from the nucleus to cytoplasm [28]. Thus pUL97 contributes to several aspects of the HCMV replication cycle.
A recent study identified dozens of additional cellular substrates for pUL97 using an in vitro phosphorylation assay [29]. Clearly, then, the full range of pUL97 functions during HCMV infection have not been fully characterized. To more comprehensively define pUL97 substrates and downstream effects during active infection, we used SILAC labeling and phosphopeptide enrichment strategies coupled with high-resolution tandem mass spectrometry to globally quantify relative changes in phosphopeptides and phosphosite occupancy over a time course of infection. Our results identify signatures of UL97-dependent signaling pathways, novel substrates and phosphosites, and several candidate kinase motifs which may mediate the effects of UL97 during HCMV replication.
2 Materials and methods
2.1 Cell Culture, SILAC Labeling, and HCMV Infection
MRC5 human lung fibroblasts (ATCC #CCL-171) were maintained in DMEM with 4.5 g/L glucose (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS) in a humidified incubator at 37°C with 5% CO2. Cells were cultured for five passages in SILAC medium supplemented with 10% dialyzed FBS to obtain near-uniform (>99%) labeling in accordance with published methods [30]. SILAC medium was prepared by dissolving the components of DMEM except for lysine and arginine, and then supplementing with either “light” isotope lysine and arginine (Sigma-Aldrich) or uniformly 13C and 15N-labeled lysine (U-13C6, 15N2; Δ8 Da) and arginine (U-13C6, 15N4; Δ10 Da) (Cambridge Isotope Laboratories) for the “heavy” labeling condition. Light- and heavy-SILAC labeled cells were infected with pUL97-expressing or pUL97-deficient viruses. The HCMV strain AD169 derivatives, UL97-K355Q and its revertant UL97-Q355K, were the generous gift of Dr. Donald Coen (Harvard Medical School), and have been previously described [16]. Stocks of HCMV were grown in MRC5 cells in roller bottles and viral titers were determined by 50% tissue culture infectious dose (TCID50) assay.
2.2 Sample Harvest
Following the temporal cascade of HCMV gene expression and modulation of host cell biology [1], samples were harvested at 24 h, 48 h, and 72 h time points post infection. Culture medium was aspirated and cells were washed quickly in ice-cold phosphate-buffered saline. Cells were lysed in situ in 4% SDS, 100 mM Tris pH 8, 5 mM DTT containing Complete™ protease inhibitor cocktail (Roche), 1mM sodium fluoride, 10 mM sodium pyrophosphate, 1mM sodium orthovanadate, and 1mM beta-glycerophosphate phosphatase inhibitors, followed by boiling for 10 min and clarification by centrifugation at 14,000 g for 10 min. Total protein content of each sample was determined using the BCA protein assay kit (ThermoFisher Scientific). Equal amounts of total protein from the heavy and light samples of each point were mixed, flash-frozen in liquid nitrogen, and stored at −80°C until further processing.
2.3 Peptide Preparation
Pooled heavy and light protein samples were subjected to buffer exchange, thiol reduction, alkylation, and trypsin digestion using the FASP procedure [31]. To achieve 2D and 3D LC-MS, resultant peptide samples were subjected to up-front fractionation either by strong cation exchange (SCX) chromatography (for the 24 h sample) or by isoelectric focusing electrophoresis (for the 48 h and 72 h samples). SCX chromatography was accomplished using STAGE-tips [32] employing Empore™ SCX-functionalized disks (3M,Saint Paul, MN) and a step-gradient elution of increasing potassium phosphate salt (increasing ratios of Buffer B to Buffer A; Buffer A: 7mM KH2PO4, pH 2.65, 30% ACN; Buffer B: 7mM KH2PO4, 350mM KCl, pH 2.65, 30% ACN). IEF was achieved by dilution of FASP peptides directly into IEF buffer and subjecting them to electrophoresis on an OffGel apparatus (Agilent) according to manufacturer’s protocols, using 24cm pH 3-10 IPG strips (GE Healthcare), followed by the collection of 12 peptide pools of differing pI. Peptide fractions were desalted using C18 reversed phase STAGE-tips and subjected directly to MS analysis. Additionally, peptide fractions were subjected to phosphopeptide enrichment by titanium dioxide affinity solid-phase extraction [33], followed by C18 reversed phase STAGE-tip cleanup and MS analysis.
2.4 Mass Spectrometry Data Acquisition
Peptide fractions were subjected to reverse-phase nano-LC-MS and MS/MS performed on a nano-flow capillary high-pressure UPLC system (Nano Ultra 2D plus, Eksigent) coupled to an LTQ-Orbitrap™ hybrid mass spectrometer (ThermoFisher Scientific), outfitted with a NanoMate ion source robot (Advion). Sample concentration and desalting were performed online using a trapping capillary column (150 μm x 4 cm, packed with 3 μm 100 Å Magic AQ C18 resin, Michrom, Auburn, CA) at 4μL/min for 7 min, while separation was conducted on an analytical capillary column (75μm x ca. 25cm, packed with 1.7μm, 130 Å C18 BEH resin; Waters, Billerica, MA) under a linear gradient of A and B mobile phase solvents (Solvent A: 3% acetonitrile/0.1% formic acid/0.1% acetic acid; Solvent B: 97% acetonitrile/0.1% formic acid/0.1% acetic acid; gradient from 8%–25% B) over 70, 180, or 300 min at a flow rate of ca. 0.4μL/min. Electrospray ionization was carried out using the NanoMate ion source at 1.74 kV, with the LTQ heated capillary at 200°C. Full-scan mass spectra were acquired in the Orbitrap in positive-ion mode over the range of m/z 335–1800 at a resolution of 60,000. MS/MS spectra were simultaneously acquired using the LTQ for the seven most abundant multiply charged species in the full-scan spectrum having signal intensities >2000 NL. Dynamic exclusion was set such that MS/MS was acquired only once for each species over 120 sec. All spectra were recorded in profile mode. Mass accuracy of the data remained typically within 0–6ppm of the external calibrants for the duration of the analysis. All raw LC-MS data generated in this study has been made publicly available through the Chorus project (chorus.org), as project number 745, accessible for download through the following URL: https://chorusproject.org/anonymous/download/experiment/653aeefe3fbb401c9dd240814c3d6d8f.
2.5 Peptide Identification and Quantification
SILAC MS peak pair quantitation and peptide assignments to tandem MS were accomplished using ProteomeDiscoverer™ software (v. 1.4, ThermoFisher Scientific), employing a Mascot™ database search node (v. 2.5, Matrix Science, London, UK) and the Percolator node [34] for empirical peptide false discovery rate (FDR) minimization. MS/MS data were searched against a database consisting of the SwissProt human proteome (ed. Nov 12, 2014) concatenated to the UniProt HCMV AD169 proteome. Accommodation for both light and heavy (KΔ8 Da, RΔ10 Da) peptide assignments was enabled as a quantitation parameter in the Mascot™ node. The analysis of both non-enriched and phosphopeptide-enriched samples enabled the generation of a database of high confidence protein assignments for each time point, which we further employed as the templates for the assignment of phosphopeptides. Thus, to determine the proteins present with high confidence in each time point sample, searches were initially conducted using the parameters of trypsinolysis with ≤ 2 missed cleavages, a precursor mass error window of ±10ppm, a fragment mass error window of 1.2 Da, fixed cysteine carbamidomethylation with variable methionine oxidation and N-terminal protein acetylation. Proteins defined by all unique peptides identified within an FDR of 1% (through reversed database search via the Percolator node) were ascribed as high confidence, and new databases containing only these proteins were generated for each time point. Phosphopeptide assignment was conducted using these time point sample-specific high-confidence protein databases as templates, and the altered parameters of trypsinolysis with ≤ 3 missed cleavages, variable phosphorylation on serine, threonine, and tyrosine, along with variable methionine oxidation and N-terminal protein acetylation, and fixed cysteine carbamidomethylation. Phosphopeptide assignments were similarly constrained to a 1% FDR using Percolator. Positional confidence scoring for phosphosite assignments was accomplished using a PhosphoRS node [35]. SILAC ratio quantification was achieved using the Precursor Ion Quantifier node tailored to detect light and heavy (KΔ8 Da, RΔ10 Da) peak pairs. Baseline signal values were employed in place of missing peaks to capture single peak species in peptide SILAC ratios, with a limit on the H/L of a maximum of 100 and minimum of 0.01.
2.6 Statistical analyses, motif analyses, and functional/interaction network annotation
To correct for any initial inequalities in heavy and light sample mixing, all SILAC ratios were normalized within each time point by setting the median ratio to 1. Protein SILAC ratios were calculated as the median values of the ratios for the unmodified, unique peptides ascribed to those proteins. SILAC ratios for key peptides of interest were confirmed by manual inspection of summed mass spectra spanning the peaks of peptide chromatographic elution using Xcalibur Software (ThermoFisher Scientific). Statistical modeling of SILAC ratios was performed using R [36] and the fitdistrplus [37] and gamlss [38] packages. Peptide modifications other than phosphorylation were stripped from all peptides in order to facilitate comparison of phosphosite occupancy. Redundant peptides caused by modification stripping were averaged geometrically. Standard deviations and samples numbers for these cases are listed in Supporting Information Table S2. Phosphopeptide ratios were log2 transformed and corrected by subtracting the log2 protein ratio from the total protein SILAC ratios [39, 40], and 4-parameter Johnson parameters were determined by maximum likelihood estimation. The fitted parameters were used to determine the Johnson cumulative distribution function and extract p-values. Peptides containing phosphosites with a PhosphoRS probability < 0.70 and ≥ 0.11 were considered ambiguously localized, whereas phosphosites with a PhosphoRS probability ≥ 0.70 were considered unambiguous. Peptides and their parent proteins containing ambiguous phosphosites were used for ontology analysis, since such proteins can be considered putative UL97 targets although their precise phosphorylation sites cannot be determined. However, only unambiguous phosphorylation sites were used for phosphorylation motif analysis in order to eliminate false positives.
Motif analysis was done with Motif-X [41, 42] and MMFPh [43]. In both cases the “foreground” dataset consisted of pooled, corrected, phosphopeptide ratios greater than 2.0 from all three time points sampled. Relevant Motif-X parameters were: width=13, occurrences=20, significance=0.000001,background=IPI Human Proteome. Relevant MMFPh parameters were: analysis type= complete, width=13, min occurrences=10–30%, significance=10−6, background=SWISS Human Proteome.
Interaction network analysis was performed using Cytoscape [44], Cluego [45] and Cluepedia [46].
3 Results
3.1 Experimental design
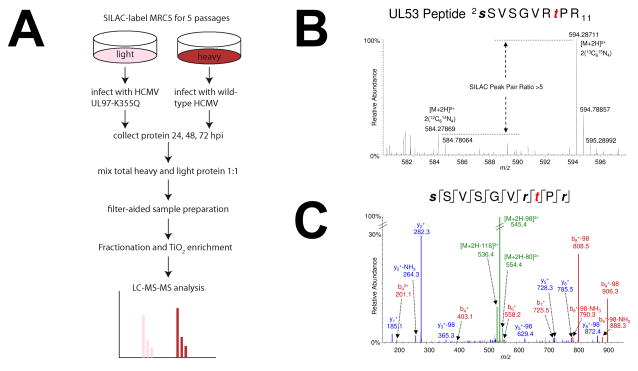
We used SILAC to facilitate quantitative comparisons of protein and phosphopeptide abundances by mass spectrometry (Fig. 1A). This method has been shown to yield accurate peptide ratios over a broad dynamic range, although some ratio compression occurs at extreme ratios. MRC-5 primary human fibroblasts exhibited >99% labeling of lysine and arginine residues after five passages in SILAC media, consistent with previously reported labeling efficiencies [30]. Light- or heavy-labeled MRC-5 cells were infected with HCMV at a multiplicity of 3 TCID50/cell, which is projected to infect >95% of cells. Cells received either the kinase-dead UL97 mutant, UL97-K355Q, or a wild-type revertant, UL97-Q355K [16], (hereafter referred to as UL97mut and UL97wt, respectively). At selected time points following infection, tryptic peptides were prepared from pooled protein isolated from heavy and light populations of infected cells, fractionated by strong cation exchange chromatography or isoelectric focusing, and subjected to phosphopeptide-enrichment using TiO2 chromatography, prior to tandem mass-spectrometry analysis. Relative expression ratios in the UL97wt vs. UL97mut samples were determined by measuring the relative intensities of SILAC peak pairs in the MS1 signals, for each positively identified peptide (Fig.1B; shown here for a pUL53 peptide, one of the substrates identified as strongly positively influenced by pUL97). All ratios are expressed as UL97wt/UL97mut throughout the analysis. Peptides were identified by their MS2 fragmentation and peptide spectral matching (see Materials and Methods). A representative MS2 spectral match, from the pUL53 SILAC pear pair shown in Fig. 1B, demonstrating unambiguous identification and phosphosite localization is shown in Fig. 1C.
Figure 1.
Experimental design and validation. (A) SILAC workflow for enriching phosphopeptides. (B) Representative MS1 spectrum of a pUL53 phosphopeptide showing relative abundances of heavy and light SILAC precursor ions. (C) Corresponding MS2 spectrum showing b- and y- fragment ions used for precursor ion sequence identification and phosphosite mapping.
HCMV, like many other viruses, expresses genes and modulates various aspects of cell biology in a temporal cascade [1]. In order to capture the temporal component of pUL97 phosphorylation we sampled the pUL97 phosphoproteome at 24, 48, and 72 hours post-infection (hpi), corresponding to the phases of early gene expression, initiation of late gene expression, and initiation of virion release, respectively. Though pUL97 is incorporated into virions [20], we anticipated that most of the pUL97-dependent changes would occur at the later time points post-infection because virion-associated pUL97 is likely to be much less abundant than newly synthesized pUL97. It is expressed with early kinetics [47], so we anticipated that the key pUL97-inuduced changes in phosphorylation would begin to occur starting at ~24hpi.
In order to identify differentially regulated phosphopeptides in our study, we employed a two-tier search strategy on our combined phospho-enriched and non-enriched sample datasets: following the assumption that authentic phosphopeptdes can be expected to be derived from proteins identified via their non-phosphorylated peptides, we first identified the proteins present in our samples at each time point with high confidence (≤1% FDR at the peptide level) by their non-phosphorylated peptide matches, and at the same time, derived their SILAC ratios. Then, we employed these high-confidence empirical proteomes as templates for assigning phosphopeptides authentically present in our samples. This strategy allowed a comprehensive characterization of phosphopeptides, quantification of their SILAC ratios, and quantification of the median SILAC ratios of their underlying unmodified protein templates.
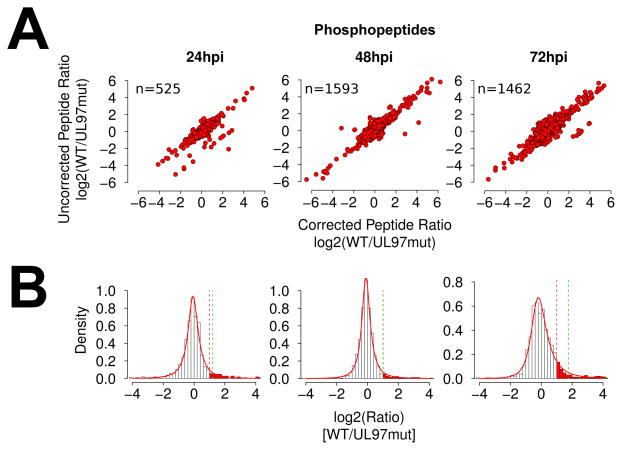
At each time point, phosphopeptide ratios were log transformed and corrected for cognate protein level changes in order for phosphosite occupancy to be accurately determined [39]. Phosphopeptides without corresponding experimental protein ratios were omitted from further analysis. This correction modified the apparent phosphopeptide ratios of a small, but substantial, number of phosphopeptides (Fig. 2A). The resulting dataset yielded 525 phosphopeptides at 24 hpi, 1593 at 48 hpi, and 1462 at 72 hpi (Fig. 2A and Supporting Information Table S1).
Figure 2.
Phosphopeptide data and ratio modeling. (A) Scatterplots comparing corrected and uncorrected phosphopeptide ratios. (B) Phosphopeptide ratio distributions and fitting. Solid red line; density plot of Johnson distribution fitted by maximum likelihood estimation. Dashed red line; 2-fold significance threshold. Dashed green line; p0.95 significance threshold.
To assess the data quality, the corrected ratio distributions were then modeled by distribution fitting. Ratio distributions were narrow, symmetrical, and found to fit extremely well to a 4-parameter Johnson distribution (Fig. 2B). The modeled Johnson parameters and their corresponding cumulative distribution function were then used to assign p-values to each variate. At 24 and 48hpi, a right-sided p-value of 0.05 corresponded to roughly 2-fold change in peptide abundance, indicating that 95% percent of the peptides showed less than 50% reduction in phosphorylation levels upon inhibition of pUL97 activity. However, at 72 hpi the 2-fold and p0.05 thresholds diverged, with ~13% of peptides showing ≤ 50% reduction in phosphorylation levels. This suggests a pulse of increased pUL97 activity occurs late during infection, which is consistent with the known function of pUL97 in nuclear breakdown during capsid egress commonly observed between 48 and 72 hpi [48–50].
3.2 Comparison of the current data set to previously identified pUL97 substrates
We identified regulated phosphopeptides from the known pUL97 substrates RB1 [16], LMNA [51, 52], UL44 [52], pUL53 [53], and UL69 [54]. Each of these was identified via at least one phosphopeptide that was ≥ 2-fold less abundant in the kinase-dead virus infection than in the wild-type infection. We also observed the known pUL97 auto-phosphorylation site S180 [26] at all three times after infection that were assayed. This pUL97 phosphosite, as well as T177 and a site at the N-terminus likely to be S13 (PhosphoRS probabilities for S11, S13, and T16 are 16.9%, 59.1%, and 16.9%, respectively), showed > 10-fold lower abundances in UL97mut-infected cells, suggesting T177 and S13 may be novel auto-phosphorylation sites. These results validate our SILAC approach and lend confidence to the newly identified pUL97 substrates. We did not, however, identify regulated phosphopeptides from RNA polymerase II or HDAC1, also known pUL97 substrates [55, 56]. Therefore, the list of pUL97 substrates we have generated is by no means complete. Importantly, we now assign specific residues as candidate pUL97 target sites on approximately 100 previously unidentified cellular and viral substrate proteins, as well as the known substrates mentioned above (Tables 1 & 2; Supporting Information Table 2).
Table 1.
Upregulated Cellular Phosphopeptides
| Protein_p-sites1 | Peptide Sequence2 | Motif | UL97wt/UL97mut | ||||
|---|---|---|---|---|---|---|---|
| LxSP | SP | SxxK | 24 hpi | 48 hpi | 72 hpi | ||
| ACD11_S762|T766 | VLRLADGPDEVHLS#AIAT#MELR | NA | NA | NA | NA | 73.87 | NA |
| AHNK_S1158 | ADVVVS#GPK | No | No | Yes | NA | NA | 27.49 |
| AHNK_S177 | DIDISS#PEFK | No | Yes | No | 2.36 | NA | 2.11 |
| AHNK_S511 | ISMQDVDLSLGS#PK | Yes | Yes | No | 4.07 | 2.23 | 2.85 |
| AHNK_T490 | VKT#PEMIIQKPK | No | No | No | NA | NA | 2.09 |
| ARHG2_S174 | ILSQS#TDSLNMR | No | No | No | 3.59 | NA | NA |
| ARHG7_S252|T254|S257 | S#GT#LKS#PPKGFDTTAINK | NA | NA | NA | NA | NA | 7.36 |
| ARMX2_T185|T191|T194|T203 | AAVPPGTVVPT#EAAAPT#EVT#EGPGVAAPT#K | NA | NA | NA | NA | 36.03 | NA |
| ATX2_S784 | GIS#PVVSEHR | No | Yes | No | NA | NA | 2.22 |
| ATX2L_S111 | GPPQS#PVFEGVYNNSR | No | Yes | No | 2.19 | NA | NA |
| ATX2L_S558 | LQPSS#SPENSLDPFPPR | No | No | No | 7.93 | NA | NA |
| BASP1_S194|S195|T196 | ETPAATEAPS#S#T#PK | NA | NA | NA | NA | 10.49 | NA |
| BASP1_T196 | ETPAATEAPSST#PK | No | No | No | NA | NA | 18.40 |
| BCLF1_S578 | LLASTLVHS#VK | No | No | Yes | NA | NA | 12.11 |
| CALD1_S789 | VTS#PTKV | No | Yes | Yes | NA | NA | 2.42 |
| CDK1_Y15 | IGEGTY#GVVYK | No | No | No | NA | NA | 6.97 |
| CH10_S51 | VLQATVVAVGS#GSK | No | No | Yes | 16.70 | 31.16 | 25.15 |
| CLIP1_T182|S186 | EPSAT#PPIS#NLTK | NA | NA | NA | NA | NA | 2.20 |
| COG4_S6 | ADLDS#PPKLSGVQQPSEGVGGGR | Yes | Yes | Yes | NA | NA | 2.62 |
| COIL_S301|T303 | LGFS#LT#PSK | NA | NA | NA | NA | NA | 2.49 |
| COIL_T303 | LGFSLT#PSK | No | No | No | NA | NA | 2.57 |
| CT2NL_S488 | DLS#PTLIDNSAAK | No | Yes | No | NA | NA | 7.87 |
| CT2NL_S556|S557 | VS#S#PLSPLSPGIK | NA | NA | NA | NA | NA | 8.72 |
| CT2NL_S560|S563 | VSSPLS#PLS#PGIK | NA | NA | NA | NA | NA | 6.72 |
| CTNB1_S196 | HAIMRSPQMVS#AIVRTMQNTNDVETAR | No | No | No | NA | NA | 2.73 |
| DC1L1_S421 | SVSSNVASVS#PIPAGSK | No | Yes | No | NA | NA | 2.37 |
| DC1L2_S405|S406|S407|T410 | GGPASVPS#S#S#PGT#SVK | NA | NA | NA | NA | NA | 2.19 |
| DDB1_T118 | IGRPSET#GIIGIIDPECR | No | No | No | 19.22 | NA | NA |
| DEST_S3 | AS#GVQVADEVCR | No | No | No | 9.29 | NA | NA |
| DHX29_T198 | FQSPQIQAT#ISPPLQPK | No | No | No | NA | NA | 3.29 |
| EFHD2_S74 | ADLNQGIGEPQS#PSR | No | Yes | No | NA | NA | 8.97 |
| EFHD2_S74|S76 | RADLNQGIGEPQS#PS#R | NA | NA | NA | NA | NA | 9.15 |
| EFHD2_S76 | RADLNQGIGEPQSPS#RR | No | No | No | NA | NA | 7.70 |
| ELAV1_S202 | NVALLSQLYHS#PAR | No | Yes | No | NA | NA | 3.29 |
| ELYS_S2226 | LISPLAS#PADGVK | Yes | Yes | No | NA | 3.56 | NA |
| ELYS_S528 | CLVAGLLS#PR | Yes | Yes | No | NA | 17.15 | NA |
| EP15R_S146 | FDGIFESLLPINGLLS#GDK | No | No | Yes | NA | NA | 2.79 |
| EP400_S736 | ALS#PVTSR | No | Yes | No | NA | 17.77 | NA |
| FAKD5_S95 | ASTLQLGS#PR | Yes | Yes | No | NA | 9.82 | 12.31 |
| FIP1_S259 | AEFTS#PPSLFK | No | Yes | No | NA | NA | 2.56 |
| FLNA_T1385,S1396 | GAGT#GGLGLAVEGPS#EAK | No | No | Yes | NA | NA | 2.30 |
| FLNB_S1433 | IAGPGLGS#GVR | No | No | No | NA | NA | 17.37 |
| FOCAD_S764,T766 | LLS#LT#PPLVLPALEEFFTSLVK | No | No | No | NA | 14.23 | NA |
| GTF2I_S210 | SILS#PGGSCGPIK | No | Yes | No | NA | 2.26 | NA |
| HACD3_S114 | WLDES#DAEMELR | No | No | No | NA | 2.38 | NA |
| HDGR2_S490 | FALKVDS#PDVKR | No | Yes | No | NA | NA | 2.63 |
| HDGR2_S490 | FALKVDS#PDVK | No | Yes | No | NA | NA | 2.73 |
| HNRPK_S284 | DYDDMS#PR | No | Yes | No | NA | 12.95 | NA |
| HUWE1_S3752 | LGS#SGLGSASSIQAAVR | No | No | No | NA | NA | 3.53 |
| HUWE1_S3752|S3753|S3757|S3759 | LGS#S#GLGS#AS#SIQAAVR | NA | NA | NA | NA | 3.56 | NA |
| HUWE1_S3757|S3759|S3760 | LGSSGLGS#AS#S#IQAAVR | NA | NA | NA | 3.67 | NA | 3.69 |
| IF4G1_S1231 | EAALPPVS#PLK | No | Yes | Yes | 3.02 | 2.82 | 4.21 |
| IF4G1_S1231 | EAALPPVS#PLKAALSEEELEK | No | Yes | Yes | NA | 3.88 | 6.35 |
| IF5_S410 | AAS#VPKVETVK | No | No | Yes | NA | 2.16 | NA |
| KI67_S579,S584 | AQSLVIS#PPAPS#PR | No | Yes | Yes | NA | NA | 2.62 |
| KPYM_S37 | LDIDS#PPITAR | No | Yes | No | NA | NA | 2.15 |
| LASP1_T68 | QSFTMVADT#PENLR | No | No | No | NA | NA | 2.19 |
| LCLT1_Y365,S368 | LLSILYWTLFSPAMCLLIY#LYS#LVK | No | No | Yes | NA | NA | 14.31 |
| LMNA_S632 | SVGGS#GGGSFGDNLVTR | No | No | No | 3.25 | NA | NA |
| LMNB1_S23 | AGGPTTPLS#PTR | No | Yes | No | NA | NA | 2.19 |
| LR16A_T1228 | FGLGT#PEK | No | No | No | NA | NA | 2.28 |
| LR16A_T1228 | FGLGT#PEKNTK | No | No | No | NA | NA | 2.21 |
| LRBA_S2201 | ISLAS#PR | Yes | Yes | No | NA | NA | 13.39 |
| LRCH4_S457 | AVVGGAAAVSTQAMHNGS#PK | No | Yes | No | NA | NA | 2.30 |
| LTOR1_S26|S27|T30 | KLLLDPS#S#PPT#K | NA | NA | NA | NA | NA | 2.06 |
| LTOR1_S26|T30 | KLLLDPS#SPPT#K | NA | NA | NA | NA | NA | 2.15 |
| LTOR1_S27 | LLLDPSS#PPTK | No | Yes | No | NA | NA | 2.13 |
| MAGD2_S247 | ALLSLRS#PK | Yes | Yes | No | NA | 3.86 | NA |
| MAP1B_S1793,S1797 | ESS#PLYS#PTFSDSTSAVK | Yes | Yes | No | NA | NA | 2.61 |
| MAP1B_S1797 | ESSPLYS#PTFSDSTSAVK | Yes | Yes | No | NA | NA | 3.72 |
| MAP4_S280|T282 | DMES#PT#KLDVTLAK | NA | NA | NA | NA | NA | 10.19 |
| MARCS_S170 | LSGFS#FKK | No | No | Yes | NA | 10.66 | 9.21 |
| MARK2_S569 | VPVAS#PSAHNISSSGGAPDR | No | Yes | No | NA | 3.36 | NA |
| MBB1A_S1290 | GVLGKS#PLSALAR | No | Yes | No | NA | NA | 3.11 |
| MBB1A_S1308 | S#PSLLQSGAK | No | Yes | No | NA | NA | 2.52 |
| MCM2_S13 | AESSESFTMASS#PAQR | No | Yes | No | NA | 7.26 | NA |
| MSH6_S830 | IHNVGS#PLK | No | Yes | Yes | NA | 3.41 | NA |
| MYO9B_S716|S717|S722 | AAGMS#S#PGAQS#HPEELPR | NA | NA | NA | NA | 2.42 | NA |
| MYO9B_S717 | AAGMSS#PGAQSHPEELPR | No | Yes | No | NA | NA | 2.79 |
| MYO9B_T1348 | ATGAALTPT#EER | No | No | No | NA | NA | 2.23 |
| NAV1_S1000 | GQLTNIVS#PTAATTPR | No | Yes | No | NA | NA | 15.21 |
| NAV1_T647 | KT#SLDVSNSAEPGFLAPGAR | No | No | No | NA | NA | 5.29 |
| NCOR2_S1487 | SLIGS#PGR | No | Yes | No | NA | 2.00 | NA |
| NELFA_S233 | SPTAPSVFS#PTGNR | No | Yes | No | NA | NA | 3.43 |
| NELFA_T157 | NALTTLAGPLT#PPVK | No | No | No | NA | NA | 3.54 |
| NHRF1_S280 | EALAEAALES#PRPALVR | Yes | Yes | No | NA | NA | 2.05 |
| NOP2_T663|S666 | GADSELST#VPS#VTK | NA | NA | NA | NA | 10.46 | NA |
| NPM_S243 | GPSS#VEDIK | No | No | No | NA | 2.23 | 4.01 |
| NU133_S50 | GLPLGSAVSSPVLFS#PVGR | Yes | Yes | No | NA | 8.44 | NA |
| NU153_S334 | IPSIVSS#PLNSPLDR | No | Yes | No | NA | NA | 2.16 |
| NU153_S338 | RIPSIVSSPLNS#PLDR | Yes | Yes | No | NA | 3.23 | NA |
| NU153_S518|S519 | VQMTSPS#S#TGSPMFK | NA | NA | NA | NA | 3.11 | NA |
| NU153_S518|S519|T520|S522 | VQMTSPS#S#T#GS#PMFK | NA | NA | NA | NA | NA | 2.80 |
| NU153_S522 | VQMTSPSSTGS#PMFK | No | Yes | No | NA | 12.76 | 10.97 |
| NUP53_S259 | CALSS#PSLAFTPPIK | Yes | Yes | No | 2.28 | NA | NA |
| NUP98_S612 | NLNNSNLFS#PVNR | Yes | Yes | No | NA | 2.46 | 2.82 |
| ODPA_S232 | YGMGTS#VER | No | No | No | NA | 3.62 | NA |
| P3C2A_S108 | LLLDDS#FETK | No | No | No | NA | NA | 2.83 |
| P4K2A_S9 | MDETSPLVS#PER | Yes | Yes | No | NA | NA | 2.37 |
| P66B_S486 | LQQQAALS#PTTAPAVSSVSK | No | Yes | No | NA | NA | 2.10 |
| PAK2_S141 | YLS#FTPPEK | No | No | No | 2.87 | NA | NA |
| PDS5A_S1266 | VDESGPPAPS#KPR | No | No | No | NA | 5.26 | NA |
| PDXD1_T691|S693 | AQGAGVTLPPT#PS#GSR | NA | NA | NA | NA | NA | 6.57 |
| PGRC1_S57 | IVRGDQPAASGDS#DDDEPPPLPRLK | No | No | No | NA | 30.13 | NA |
| RANB3_S333 | VLS#PPKLNEVSSDANR | No | Yes | Yes | 2.24 | NA | NA |
| RB_T823|T826 | ISEGLPTPT#KMT#PR | NA | NA | NA | NA | 2.15 | NA |
| RBBP6_S516 | LGYLVS#PPQQIR | Yes | Yes | No | NA | NA | 2.53 |
| RBM14_T206 | QPT#PPFFGR | No | No | No | NA | 9.14 | NA |
| RBP2_S2510 | AVVS#PPKFVFGSESVK | No | Yes | Yes | NA | 6.99 | 13.33 |
| RIF1_S2161 | ELDPSLVSANDS#PSGMQTR | No | Yes | No | NA | 9.54 | NA |
| RL12_S38 | IGPLGLS#PK | No | Yes | Yes | 2.58 | NA | 2.34 |
| RL18_S130 | ILTFDQLALDS#PK | Yes | Yes | No | NA | 3.44 | 6.16 |
| RL4_S295 | ILKS#PEIQR | Yes | Yes | No | NA | 2.14 | 2.37 |
| RNBP6_T4 | MAAT#ASAGVPATVSEK | No | No | No | NA | NA | 2.66 |
| RNZ2_S736 | VPLFS#PNFSEK | Yes | Yes | No | NA | NA | 15.20 |
| RRP1B_S706 | SILVS#PTGPSR | Yes | Yes | No | NA | 2.28 | NA |
| RRP1B_S706|S711 | SILVS#PTGPS#R | NA | NA | NA | NA | NA | 4.00 |
| SARNP_S162|S163 | FGLNVS#S#ISR | NA | NA | NA | 2.96 | NA | NA |
| SCAM3_T130 | ELQHAALGGTAT#R | No | No | No | 2.71 | NA | NA |
| SF3B2_S307 | SSLGQS#ASETEEDTVSVSK | No | No | No | 2.33 | NA | NA |
| SMAG2_S592 | MGLLS#PSGIGGVSPR | Yes | Yes | No | NA | NA | 4.96 |
| SMCA2_S1377 | GRPPAEKLS#PNPPK | No | Yes | No | NA | NA | 7.58 |
| SMTN_S503 | APPTLLSTSS#GGK | No | No | Yes | NA | 3.80 | 7.70 |
| SRC_Y419 | LIEDNEY#TAR | No | No | No | NA | 2.10 | NA |
| SRRM2_S1179 | MALPPQEDATAS#PPR | No | Yes | No | NA | 2.14 | NA |
| SRRM2_S2272 | TPAAAAAMNLAS#PR | Yes | Yes | No | 2.01 | NA | 2.12 |
| SRSF9_S211,S216 | GS#PHYFS#PFRPY | No | Yes | No | NA | 2.31 | NA |
| STMN1_S38 | ESVPEFPLS#PPK | No | Yes | Yes | 2.14 | NA | NA |
| STRUM_S917 | TLMNAVS#PLK | No | Yes | Yes | NA | NA | 2.24 |
| TALDO_S237 | TIVMGAS#FR | No | No | No | NA | 2.58 | NA |
| TB182_S1029 | GSGGLFS#PSTAHVPDGALGQR | Yes | Yes | No | NA | 2.30 | NA |
| TB182_S435 | FSEGVLQS#PSQDQEK | Yes | Yes | No | NA | NA | 2.52 |
| TCOF_S1378 | LGAGEGGEASVS#PEKTSTTSK | No | Yes | Yes | 2.79 | NA | NA |
| TCOF_S1386 | LGAGEGGEASVSPEKTSTTS#K | No | No | Yes | 3.06 | NA | NA |
| TCOF_T1384 | KLGAGEGGEASVSPEKTST#TSK | No | No | No | 2.46 | NA | NA |
| TENA_T2134|S2139|Y2140 | DTDSAIT#NCALS#Y#K | NA | NA | NA | 3.85 | NA | NA |
| TFP11_S128 | LKTGGNFKPS#QKGFAGGTK | No | No | No | NA | NA | 16.86 |
| THOC2_S1393 | TPVSGSLKS#PVPR | Yes | Yes | No | NA | 2.46 | NA |
| TIF1B_S19 | AASAAAASAAAASAASGS#PGPGEGSAGGEKR | No | Yes | No | 27.69 | NA | NA |
| TM263_S44|T46 | VTGGIFS#VT#K | NA | NA | NA | NA | NA | 11.31 |
| TP53B_S1618 | AADIS#LDNLVEGK | No | No | No | NA | NA | 2.19 |
| TPD54_S12 | MDSAGQDINLNS#PNK | Yes | Yes | Yes | NA | 4.74 | 5.23 |
| TRI33_S862 | QSGLSSLVNGKS#PIR | No | Yes | No | NA | NA | 2.19 |
| TRIPC_S936|S938|T941|S942 | HLAES#ES#LLT#S#PPK | NA | NA | NA | NA | NA | 4.01 |
| TRIPC_S942 | HLAESESLLTS#PPK | Yes | Yes | Yes | NA | 3.66 | 3.77 |
| TRIPC_S991 | DDSLDLS#PQGR | No | Yes | No | NA | NA | 2.34 |
| TSC2_T659|S660|S664 | T#S#GPLS#PPTGPPGPAPAGPAVR | NA | NA | NA | 4.01 | NA | NA |
| TSSC1_T19,T22 | ALT#PQT#AETDAIR | No | No | No | NA | 2.70 | NA |
| TUT4_S155|S156 | VPS#S#PAEAEKGPSLLLK | NA | NA | NA | NA | 7.72 | NA |
| UFD1_S245|S247 | GVEPS#PS#PIKPGDIKR | NA | NA | NA | NA | NA | 12.71 |
| UFD1_S247 | GVEPSPS#PIKPGDIKR | No | Yes | Yes | NA | 4.40 | NA |
| UFD1_S247 | GVEPSPS#PIKPGDIK | No | Yes | Yes | NA | 4.25 | 4.53 |
| UIF_S282 | GVPLQFDINS#VGK | No | No | Yes | NA | 20.55 | NA |
| WDR11_S645|T647|S650|S654|S655|S658 | QTVVS#DT#ELS#IVES#S#VIS#LLQEAESK | NA | NA | NA | NA | NA | 17.48 |
| ZC12A_S344 | ANALLS#PPR | Yes | Yes | No | NA | NA | 2.82 |
| ZC3H4_S907|S908 | ALPTSKPEGSLHS#S#PVGPSSSK | NA | NA | NA | NA | NA | 2.26 |
| ZC3H4_S907|S908|S913|S914|S915 | ALPTSKPEGSLHS#S#PVGPS#S#S#K | NA | NA | NA | NA | 2.24 | NA |
| ZFR_S1054 | RRDS#DGVDGFEAEGK | No | No | No | NA | 7.27 | NA |
| ZO2_S130 | KVQVAALQAS#PPLDQDDR | No | Yes | No | NA | NA | 2.05 |
| ZYX_S344 | S#PGAPGPLTLK | No | Yes | No | NA | NA | 2.02 |
| = “logical or” and indicates ambiguous localization. A phosphorylation site is considered ambiguously localized if 0.11 ≤ phosphoRS probability < 0.7. Fully annotated versions of Table 1 and Table 2 are appended in Supporting Information Table S2.
”#” indicates phosphorylation site on prior amino acid.
Table 2.
Upregulated Viral Phosphopeptides
| Gene | Protein_p-sites1 | Peptide Sequence 2 | Motif | UL97wt/UL97mut | ||||
|---|---|---|---|---|---|---|---|---|
| LxSP | SP | SxxK | 24 hpi | 48 hpi | 72 hpi | |||
| UL24 | VP22_S51 | SVVCDGPPGS#PTDSAR | No | Yes | No | NA | NA | 2.47 |
| UL25 | PP85_Y61 | MEAGLSPY#SVSSDAPSSFELVR | No | No | No | NA | NA | 2.30 |
| UL32 | PP150_S1008 | TVAFDLSS#PQK | Yes | Yes | Yes | NA | 3.85 | 2.07 |
| UL32 | PP150_S335 | HVPTQPLDGWSWIAS#PWK | No | Yes | Yes | NA | NA | 2.62 |
| UL32 | PP150_S504 | HAAFSLVSPQVTKAS#PGR | No | Yes | No | NA | 3.16 | NA |
| UL32 | PP150_S923 | AVVGRPPSVPVSGS#APGR | No | No | No | 6.66 | 5.71 | NA |
| UL32 | PP150_S991 | STGTAAVGS#PVK | No | Yes | Yes | NA | 3.47 | NA |
| UL32 | PP150_T675 | MPTTSTASQNTVSTT#PR | No | No | No | NA | NA | 2.10 |
| UL32 | PP150_T797 | AQTVTST#PVQGR | No | No | No | NA | NA | 2.03 |
| UL32 | PP150_T984,S991 | ST#GTAAVGS#PVK | No | Yes | Yes | NA | NA | 3.53 |
| UL34 | UL34_S377 | HRADEEDGPLAS#QTAVR | No | No | No | NA | NA | 7.73 |
| UL44 | VPAP_S286 | GDPFDKNYVGNS#GK | No | No | No | NA | 2.57 | 11.71 |
| UL44 | VPAP_S354|S355 | KMS#S#GGGGGDHDHGLSSK | NA | NA | NA | NA | 2.77 | NA |
| UL45 | RIR1_S123|S124 | GGRVS#S#GLSTFNPAGATR | NA | NA | NA | NA | NA | 3.44 |
| UL46 | VP19_T137,S139 | T#VS#LGITSLLTCVMR | No | No | No | NA | NA | 28.40 |
| UL53 | UL53_S2,T9 | S#SVSGVRT#PR | No | No | No | NA | NA | 4.49 |
| UL57 | DNBI_S806 | GNAGS#VFFR | No | No | No | NA | NA | 3.68 |
| UL57 | DNBI_S991 | SLVTVPVS#VEK | No | No | Yes | NA | 21.60 | 40.43 |
| UL69 | ICP27_Y151 | VAASASVPLNPHY#GK | No | No | No | NA | NA | 16.23 |
| UL71 | UL71_S360 | KKPTAAALLSS#A | No | No | No | NA | NA | 2.24 |
| UL80 | SCAF_T354 | SHPLSAAVPAATAPPGAT#VAGASPAVSSLAWPHDGVYLPK | No | No | No | 5.89 | NA | NA |
| UL82 | PP71_S7,S13 | SQASSS#PGEGPS#SEAAAISEAEAASGSFGR | No | Yes | No | NA | NA | 2.24 |
| UL83 | PP65_S430 | VTGGGAMAGAS#TSAGR | No | No | No | NA | NA | 2.63 |
| UL83 | PP65_S430|T431|S432 | VTGGGAMAGAS#T#S#AGRK | NA | NA | NA | NA | NA | 3.39 |
| UL83 | PP65_S432 | VTGGGAMAGASTS#AGR | No | No | No | 3.94 | 2.06 | 2.09 |
| UL83 | PP65_S432 | VTGGGAMAGASTS#AGRK | No | No | No | NA | NA | 4.89 |
| UL83 | PP65_S439 | S#ASSATACTSGVMTR | No | No | No | 2.25 | NA | NA |
| UL83 | PP65_S554 | HRQDALPGPCIAS#TPK | No | No | Yes | 3.46 | NA | NA |
| UL83 | PP65_S554|T555 | QDALPGPCIAS#T#PK | NA | NA | NA | NA | 8.16 | 15.06 |
| UL83 | PP65_S554|T555 | QDALPGPCIAS#T#PKK | NA | NA | NA | NA | NA | 34.52 |
| UL83 | PP65_S554|T555 | QDALPGPCIAS#T#PKK | NA | NA | NA | NA | 5.94 | NA |
| UL83 | PP65_S554|T555 | HRQDALPGPCIAS#T#PKK | NA | NA | NA | 4.21 | NA | NA |
| UL83 | PP65_T431|S432 | VTGGGAMAGAST#S#AGRK | NA | NA | NA | NA | NA | 3.45 |
| UL83 | PP65_T555 | HRQDALPGPCIAST#PKK | No | No | No | NA | NA | 29.50 |
| UL95 | UL95_T218 | NDAAT#PSFLR | No | No | No | NA | 2.72 | 2.60 |
| UL97 | UL97_S11|S13|T16 | S#AS#LGT#TTQGWDPPPLR | NA | NA | NA | NA | NA | 11.30 |
| UL97 | UL97_S180 | ALFTGGS#DPSDSVSGVR | No | No | No | 16.22 | 31.36 | 12.33 |
| UL97 | UL97_S180,S185 | ALFTGGS#DPSDS#VSGVR | No | No | No | NA | NA | 10.17 |
| UL97 | UL97_T134|S135|S136 | RPVVPST#S#S#R | NA | NA | NA | NA | NA | 4.79 |
| UL97 | UL97_T177 | RALFT#GGSDPSDSVSGVR | No | No | No | NA | 42.24 | NA |
| UL97 | UL97_T177|S180|S183 | RALFT#GGS#DPS#DSVSGVR | NA | NA | NA | NA | NA | 12.24 |
| UL122 | VIE2_S331 | RIS#ELDNEKVR | No | No | No | NA | NA | 3.55 |
| UL122 | VIE2_T351 | NTPFCT#PNVQTR | No | No | No | NA | NA | 2.28 |
| UL122 | VIE2_T351 | DKNTPFCT#PNVQTR | No | No | No | NA | NA | 2.38 |
| UL123 | VIE1_S365 | VCS#PSVDDLR | No | Yes | No | NA | 6.81 | 7.44 |
We also compared our data to the list of putative pUL97 substrates identified by in vitro protein chip analysis [29]. Of the 276 proteins from this study that we could convert to UniProt accessions, only 29 were identified in our SILAC dataset, and of those only 4 (coded by NUP133, EP400, PDS5A, and ZYX) showed a 50% or greater decrease in phospho occupancy upon mutation of pUL97. The source of this incongruity between the earlier in vitro data and our in vivo data is presently unclear, potentially attributable to numerous factors. However, we would argue that in vivo data is likely to represent a more accurate landscape of relevant biological pUL97 activity, since it reflects the combinatorial effects of all of the cellular and viral signaling pathways modulated during infection.
3.3 Phosphomotif analysis
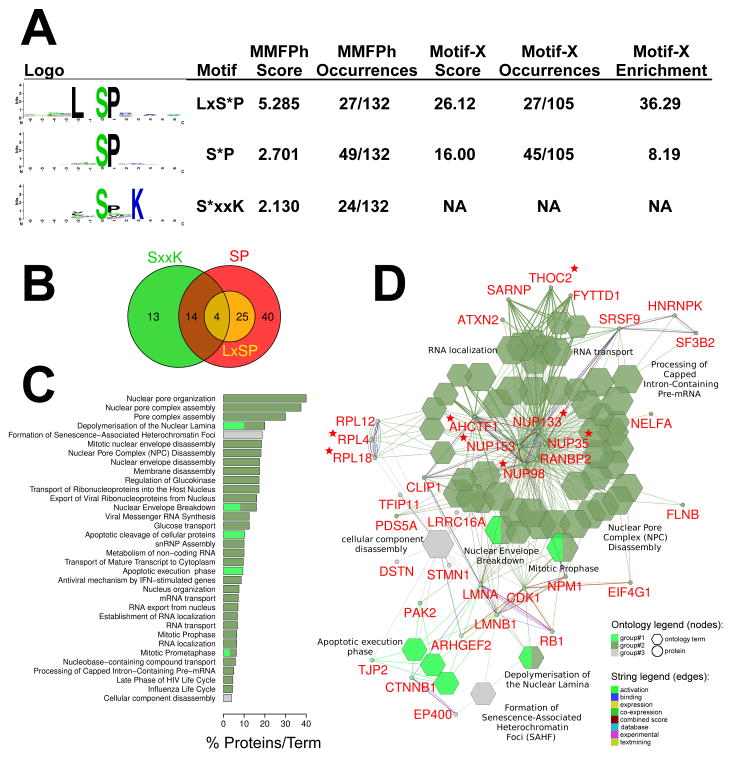
pUL97 has previously been shown to recognize substrates containing arginine or lysine in the +5 position after a serine or threonine [57]. This S/TxxxxK/R motif was deduced more than a decade ago by performing in vitro phosphorylation experiments with purified pUL97 and candidate substrate proteins, most significantly histone H2B, which is not known to be a physiological pUL97 substrate. We hypothesized that peptides from our SILAC phospho-proteome with high UL97wt/UL97mut ratios might provide a more robust and unbiased dataset for in silico phospho motif analysis, and thereby provide complementary evidence for the S/TxxxxK/R motif and/or identify novel kinase motifs specific to pUL97. To test this hypothesis, we extracted all quantified phosphopeptides showing 2-fold or greater unambiguous phosphosite occupancy reduction in UL97mut-infected cells, across all three time points assayed (Tables 1 and 2; unambiguous sites only). These peptides were input into Motif-X [41] and MMFPh [58] as the “foreground” dataset and analyzed for motif enrichment versus peptides from the entire human genome (“background”). Both tools were able to identify two common enriched motifs, SP and LxSP, while MMFPh also identified SxxK with a slight preference for Pro in the +1 position (Fig. 3A). Both tools scored the LxSP motif highest, which had a Motif-X enrichment of >30-fold, followed by the lower complexity SP motif, whose score was penalized due to its low complexity (and thus higher frequency in the background dataset), but was present in the highest number of peptides (~30–40% depending on the engine used). Four peptides from the proteins COG4, TPD54, TRIPC, and pp150 contained both an LxSP and SxxK motif (and necessarily an SP motif; Fig. 3B, Tables 1 & 2). Neither tool identified the previously identified S/TxxxxK/R motif, although the SxxK motif is conspicuously similar.
Figure 3.
Putative UL97 kinase motifs and pathway analysis. (A) Enriched phosphorylation motifs and statistics. All significantly regulated peptides (≥2-fold [UL97wt/UL97mut]) across all time points were analyzed non-redundantly using Motif-X [41, 90] and MMFPH [43]. (B) Euler diagram displaying overlap of enriched phosphorylation motifs in all phosphopeptides showing 2-fold or greater occupancy decrease upon mutation of pUL97. Note that the numbers of peptides in each class differ from those identified by Motif-X and MMFPH as these tools discard all peptides absent from their background databases. (C) Selected ontology terms enriched in putative pUL97 substrates. This list is a subset of the full list of ontology terms used to construct the network in panel D. All enriched ontology terms, their p-values, and constituent proteins are listed in Supporting Information Table S4. (D) Cluego/Clupedia ontology and STRING network. Hexagonal nodes represent significantly enriched gene ontology terms (p>0.005). Circular nodes represent proteins. Connecting lines (edges) indicate interrelatedness. Multicolored lines connecting proteins highlight different STRING protein-protein interaction evidence codes. Stars indicate that a protein contains an LxSP phosphorylation motif.
Interestingly, the minimal pUL97 kinase motif, SP, is the same as the minimal motif for the CDK1-coded protein, cyclin-dependent kinase 1 (Cdk1) [59], and the SxxK motif, especially considering the preference for Pro in the +1 position, closely resembles the full Cdk1 motif [S/T]Px[RK]. However, to our knowledge, the LxSP motif appears to be novel, having no significant similarity to any motif in the Human Protein Reference Database [60].
Our foreground dataset contained several peptides with phospho-Tyrosine (pY; see Table 1) sites that changed upon removal of pUL97 function. This set of peptides lacks the complexity required to extract enriched motifs. Nonetheless, since there is no evidence for pUL97 Tyr kinase activity, we interpret changes in pY occupancy as an indirect consequence of removing pUL97 activity.
3.4 Substrate functional classification
To identify related components and pathways that were affected by pUL97 kinase activity, we performed gene ontology (GO) enrichment and protein association network analysis on the same peptides used for motif analysis. For this analysis we used the programs ClueGO [45] and CluePedia [46] which first create an association network of enriched GO terms, and then overlay a STRING [61] protein-protein interaction network in order visualize functional relatedness. ClueGO analysis of all ≥2-fold upregulated peptides revealed one dominant enrichment node containing GO terms related to nuclear envelope and lamina breakdown, RNA transport, and mitosis (Figs. 3C & 3D). Tightly grouped within the STRING sub-network is a cluster of nuclear pore proteins (encoded by NUP133, NUP35, NUP98, NUP153, RANBP2, and AHCTF1), with smaller peripheral clusters of nuclear lamina proteins (LMNA, LMNB1), cell cycle related proteins (CDK1, RB1, ARHGEF2, PAK2), ribosomal proteins (RPL12, RPL4, RPL18), splicing factors (SRSF9, SF3B2) and RNA transport proteins (FYTTD1/UIF, THOC2, SARNP). Interestingly, the novel LxSP motif was contained in 5 of 6 nuclear pore related proteins (Nup133, Nup35, Nup98, Nup153, AHCTF1) and 2 of 3 ribosomal proteins (RPL4, RPL18) analyzed in this ontology network.
Phosphorylation of nuclear pore and lamina proteins is thought to contribute to breakdown of the nucleus during mitosis (reviewed in [62]). pUL97 has been shown to mimic this process late during HCMV infection in order to facilitate capsid egress from the nucleus [48–50]. The effect of pUL97 on the ribosomal proteins identified is undocumented and unclear, although it is possible that they regulate translation of a select subset of mRNAs beneficial to the virus. As discussed below, removal of pUL97 activity had a fairly modest effect on global steady state protein levels. This suggests that pUL97 phosphorylation of ribosomal proteins may be involved in a very specific form of translational regulation. The identification of enriched RNA transport and splicing functions in the list of putative pUL97 substrates suggests a direct role in RNA post-transcriptional processing and translation. In particular, identification of putative phosphosites in several members of the TRanscription-EXport (TREX) [63] complex (SARNP, THOC2, FYTTD1/UIF) suggests an expanded role for pUL97 in coupled mRNA transport beyond its known ability to regulate pUL69/ICP27 (see “Discussion”).
Overall, the features of the pUL97 functional network suggest that pUL97 mediates pleiotropic activities that affect mRNA splicing and transport, translation, nuclear organization, and the cell cycle.
3.5 Analysis of viral phosphopeptides
Peptides from 17 HCMV-coded proteins exhibited a ≥2-fold increase in S/T phosphorylation (Table 2). The SP phosphorylation motif was present in peptides corresponding to four viral proteins (pUL24, pUL32, pUL82 and pUL123), the SxxK motif was present in three viral proteins (pUL32, pUL57and pUL83) and the LxSP motif was not found in any viral protein. The direct interaction of pUL97 with pUL83 [21] provides support for the view that SxxK is a target of the viral kinase. Several of the viral proteins identified in our analysis have been identified as pUL97 targets previously, including pUL44 [52, 64], pUL53 [48], pUL69[54], and pUL97 itself [26, 65, 66]. To the best of our knowledge, the remaining 14 viral proteins that identified here have not been shown previously to be phosphorylated either directly or indirectly by pUL97. The role of these phosphorylation events in the viral lifecycle remains to be investigated.
3.6 Cdk1
One critical question arising from our motif analysis is whether the phospho sites identified in our proteomics experiments are direct pUL97 substrates or indirect substrates of one or more secondary kinases regulated downstream of pUL97. The novelty of the LxSP motif supports the idea that peptides containing these sites are directly phosphorylated by pUL97, as no other known kinase has been shown to have this specificity for LxSP substrates. On the other hand, the overlap between the SP and SxxR motifs with the known CDK1 motifs raises the possibility that pUL97 might be exclusively or partially working through Cdk1. The majority of proteins in our foreground motif search dataset are not known to be targets of Cdk1, although several are, such as the proteins encoded by RB1, NUP153, LMNA, and LMNB1 [59]. Furthermore, three phospho peptides from Cdk1 itself containing the sites pT14+pY15, pY15, and pT161 were present in our SILAC data (Table 3). None were identified at 24 hpi, all three were present at 48 hpi, and one was identified at 72 hpi. Interestingly, at 48 hpi phospho occupancy at all three peptides remained unchanged in the UL97mut compared to UL97wt sample. However, at 72 hpi, occupancy at Y15 decreased dramatically (~7-fold) in the UL97mut sample. Phosphorylation of Cdk1 at Y15 has been shown to inhibit Cdk1 kinase activity, as well as entry into mitosis [67, 68]. Phosphorylation at this site is likely indirect since there is no evidence that pUL97 has intrinsic tyrosine-kinase activity. Therefore, Cdk1 may be inactivated by pUL97 late after infection, which is consistent recent results showing that Cdk1 inhibitors do not prevent nuclear lamina disruption and viral egress during HCMV infection [49].
Table 3.
CDK1 Phosphopeptides
| Protein_p-sites1 | Peptide Sequence 2 | Motif | UL97wt/UL97mut | ||||
|---|---|---|---|---|---|---|---|
| LxSP | SP | SxxK | 24 hpi | 48 hpi | 72 hpi | ||
| CDK1_T14,Y15 | IGEGT#Y#GVVYK | No | No | No | NA | 1.05 | NA |
| CDK1_T161 | VYT#HEVVTLWYR | No | No | No | NA | 1.15 | NA |
| CDK1_Y15 | IGEGTY#GVVYK | No | No | No | NA | 1.27 | 6.97 |
3.7 Protein level changes induced by pUL97
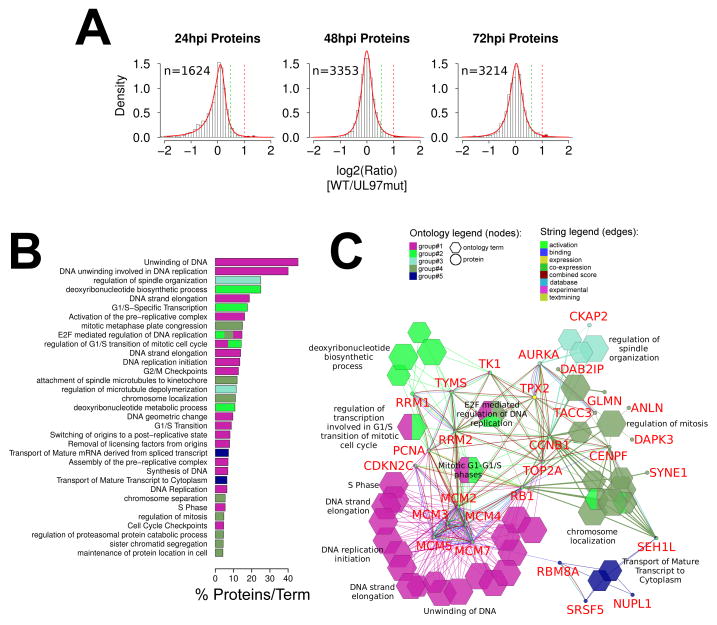
In order to uncover addition pathways regulated by pUL97, we analyzed total protein changes induced by pUL97mut in our SILAC dataset. Like the phosphopeptides, protein ratios at each time point were median normalized and modeled as Johnson distributions (Fig. 4A). The 24 hpi distribution showed a significant negative skew which was not apparent at 48 and 72 hpi. One interpretation of the skewedness is that pUL97 globally destabilizes proteins and therefore when UL97 activity is removed protein abundances increase. However, since there is no other evidence that this is the case, we interpret this result as an unexplainable systematic bias in the 24h dataset. We therefore performed a functional analysis only on upregulated ratios, across all time points. At each time point the protein ratio distributions were narrower than the phosphopeptide distributions, with ~95% of the ratios being ≤ 1.5-fold. Therefore, rather than using a right-sided p-value of 0.05 we opted to use an empirical 2-fold significance cutoff. We then extracted all of the proteins with UL97wt/UL97mut ratios of 2-fold or greater (Supporting Information Table S3), pooled the proteins from all time points and performed ClueGO/CluePedia analysis.
Figure 4.
Protein level ontology network analysis. (A) SILAC protein ratios modeled as in Fig. 2B. (B) Selected ontology terms enriched in pUL97 substrates at the protein level. This list is a subset of the full list of ontology terms used to construct the network in panel C. All enriched ontology terms, their p-values, and constituent proteins are listed in Supporting Information Table S5. (C) Cluego network analysis of all proteins with a 2-fold or greater SILAC ratio.
The resulting ontology network showed one primary enrichment node, with four smaller peripheral nodes (Figs. 4B and C). The primary node contained a cluster of five mini-chromosome maintenance (MCM) proteins. The peripheral nodes were enriched in several G1/S and M-phase cell-cycle related proteins (coded by CCNB1, CDKN2C, CENFP, RB1), and functions including spindle organization (TPX2, AURKA), DNA replication (TOP2A, PCNA), and nucleotide biosynthesis (RRM1, RRM2, TYMS, TK1). MCM proteins form a hexameric DNA helicase complex that is essential for initiation of cellular DNA pre-replication complexes (reviewed in [69]). All of the five MCM proteins identified showed ~2-fold steady-state protein increases in the presence of functional pUL97 at all time points. This result is curious and difficult to interpret since several recent studies have all indicated that MCM proteins are inhibited during HCMV replication in order to shut-down cellular DNA replication and redirect resources towards viral DNA replication, which does not require MCM-helicase activity [70–72]. Given these results it is unclear why pUL97 appears to stabilize MCM proteins, and we conclude that this stabilization is likely an indirect effect of pUL97’s Cdk-like activity. Most of the other increases in pUL97-mediated cellular proteins identified in this network are relatively modest (2–3 fold) and occur late in infection, also echoing pUL97’s Cdk-like activities. Only 12 viral proteins changed more than 2-fold at one of the sampled time points (Supporting Information Table S3), and no viral protein changed more than 3-fold at any time. Additionally, no viral protein showed elevated ratios at more than one time point.
These data suggest that UL97 has little effect on cellular and viral steady-state protein levels and what little effect it does have is likely an indirect consequence of its cell-cycle regulatory activity.
4 Discussion
By mapping the phosphoproteome of cells infected with wild-type or UL97 kinase-dead virus, we have identified suites of changes that can be attributed to the catalytic activity of pUL97. pUL97 activity is required for global changes in the phosphorylation state of both cellular and viral proteins. The pUL97 phospho-proteome extends our previous knowledge about HCMV and its kinase by assigning specific residues to the phosphorylation of >100 pUL97 substrate proteins. Furthermore, it identifies several putative pUL97 substrate motifs and highlights the similarity between the pUL97 minimal consensus sequence and that of Cdk1.
As UL97 has been shown to functionally complement Cdk1 [16] and its phosphorylation motif overlaps with that of Cdk1, it is possible that the changes in phosphosite occupancy we observed within infected cells are largely the consequence of pUL97 activating Cdk1. Two facts argue against this. First, pUL97 phosphosites showed enrichment in a novel motif LxSP, which appears in most of the regulated nuclear pore peptides (Table 1). As far as we can tell this kinase specificity is novel and suggests that, minimally, changes in LxSP containing peptides reported here represent bona fide pUL97 targets. Second, pUL97 mediates phosphorylation of Cdk1 at Y15 late after infection, which is a known Cdk inactivating modification [67, 68]. These facts strongly suggest that at least some of the phosphosites we have identified are true pUL97 substrates. Nonetheless, it will be important to validate these new substrates in the presence of Cdk inhibitors, as has been done previously for pUL97-mediated phosphorylation of UL50 and UL53 during capsid egress [49]. Additionally, in vitro phosphorylation assays with recombinant pUL97 and LxSP-containing peptides will be necessary to confirm pUL97-mediated phosphorylation of the LxSP and SxxK motifs.
UL97-regulated LxSP-containing peptides were enriched in nuclear pore and ribosomal proteins. Nuclear pore complexes mediate transit of HCMV viral DNA into the nucleus as incoming capsids dock at the outer nuclear membrane. Viral DNA transits through the central channel of the nuclear pore. It is possible that virion associated pUL97, which enters with the virus, facilitates this process by phosphorylating nuclear pore proteins, however the early kinetics (i.e. not immediate-early) of de novo pUL97 expression suggest that the viral kinase is modifying nuclear pores for other reasons. Consistent with this idea, we observed significant increases in nuclear pore protein phosphosite occupancy in UL97wt samples up to 72hpi.
Since pUL97 has known roles in nuclear lamina dissolution during HCMV capsid egress, it is tempting to speculate that its activity towards nuclear pore complexes might facilitate egress. However, no evidence exists supporting the view that nuclear pores are involved in capsid egress. Rather, an overwhelming body of evidence from several different herpes viruses suggests that capsids transit through the perinuclear space via envelopment at the inner nuclear membrane and subsequent budding at the outer nuclear membrane (reviewed in [73] and [74]). pUL97 contributes to early steps of this process by phosphorylating and destabilizing nuclear lamina proteins [16, 51, 75], which normally occlude access to the inner nuclear membrane, and the viral proteins pUL53 and pUL50, which are pUL97 substrates thought to potentiate primary envelopment [50].
Importantly, a dominant negative Lamin A mutant has recently been shown to complement pUL97 deficiency [49], arguing strongly that lamins are the primary targets of pUL97 facilitating viral egress, not nuclear pores. Additionally, HCMV capsids are generally thought to be too large to be transported through the nuclear pore complex. Little is known about the function of nuclear pore proteins during the later stages of HCMV infection. The function of nuclear pore complexes has been investigated during herpes simplex virus type 1 (HSV-1) infection but is incompletely understood. During HSV-1 infection nuclear pore complexes swell and their numbers increase [76], yet transport and selectivity through these distended pores appears to remain intact [77]. Recent experiments with the HSV-1 immediate-early protein ICP27 suggest that direct binding of ICP27 to nuclear pore proteins, in particular NUP62, may be involved in selective viral mRNA export from the nucleus [78]. The HCMV ICP27 homolog, pUL69, mediates the selective export of HCMV mRNAs. pUL97 can phosphorylate pUL69 in vitro and increases its RNA transport efficiency [54]. One pUL97-regulated phosphosite was identified in pUL69 in our study, a phospho-Tyrosine at amino-acid 151. This site is unlikely to be a direct substrate of pUL97, which is not known to have pY activity, and we therefore conclude that either this phosphorylation is indirect or other that bona fide pUL97 sites in pUL69 were missed. Rather than binding of nuclear pore complexes, pUL69-mediated selective RNA transport is thought to occur through interaction with components of the TRanscription-EXport (TREX) complex, particularly the DEAD-Box RNA Helicase UAP56 [79, 80]. The mammalian TREX complex is composed of UAP56, the adaptor protein ALY, and several THO proteins referred which form the THO transcription elongation sub-complex [63, 81]. Although the molecular details are not completely understood, ICP27 homologs from several herpes viruses, including KSHV, EBV, HSV-1, and HCMV are thought to promote the selective export of intronless viral mRNAs by directly interacting with and subverting TREX complex proteins (for review see[82]).
Interestingly, we observed decreased phosphorylation of three TREX proteins when pUL97 activity was abrogated, the core THO component THOC2 [81], SARNP/CIP29 [83], and the UAP56 interacting protein FYTDD1/UIF1 [84]. Further examination of pUL97 mediated phosphorylation of TREX complex proteins during HCMV replication will be required to determine the exact role of TREX components in viral mRNA transport. However, our data suggest that pUL97 may have a broader role in mediating the selective translation of viral mRNAs than previously appreciated. Rather than simply phosphorylating the viral export factor pUL69, pUL97 may be a master regulator of selective viral RNA transport and translation, phosphorylating (and thereby regulating) a continuum of cellular and viral proteins from the transcription-transport machinery, to nuclear pore proteins, to specific ribosomal subunits. This is in addition to the known functions of pUL97 in cell cycle regulation and nuclear envelope remodeling, which we also confirmed as significantly regulated pathways in our dataset.
The specific role of nuclear pore phosphorylation by pUL97 during HCMV replication is unclear, but we speculate that it could be involved in the later steps of UL69/TREX mediated RNA transport. Alternatively, pUL97 phosphorylated nuclear pores might be involved in herpesvirus capsid egress. Indeed, it has been proposed that nuclear pores can serve as docking receptors for capsid fusion with the inner nuclear membrane during HSV-1 infection [85]. Further experiments will be required to test these possibilities, as well as define the exact function of each putative pUL97 phosphorylation site. Nonetheless, this work provides a robust framework upon which to initiate such studies and identifies TREX, nuclear pore, and ribosomal proteins as candidate pUL97-regulated pathways for further analysis.
To the best of our knowledge this work represents one of the first global phosphoproteome characterizations of signaling by a viral enzyme. This work extends our previous knowledge about HCMV and UL97 by assigning specific residues to the phosphorylation of known UL97 substrates. We view these sites as potential fertile ground for future functional characterization.
From the perspective of the virus, signaling is likely important to establish a host cell environment that is favorable to viral replication and evades immune surveillance. From a clinical perspective, we propose that these signaling signatures may be biomarkers and lend insight into signaling synergies that contribute to essential steps of the viral replication program. Identifying and disrupting such steps could suggest novel strategies for therapeutic intervention. For example, if pUL97 phosphorylation of a cellular factor is essential to the virus, a drug which blocks or serves as a decoy target might be useful clinically. Disrupting host-pathogen kinase interactions has been touted as a novel antiviral strategy (reviewed in [86] and [87]).
Other studies of viral phosphopeptides have focused on virions [88, 89], rather than on the complex milieu of infected cells. We project that expansion of this technology will identify signatures and essential signaling events required for replication of a range of viruses. Application of SILAC technology to the study of host-pathogen interactions is an exciting tool for uncovering patterns and actions of viruses.
Supplementary Material
Acknowledgments
This work was supported by a grant to T.S. from the National Institutes of Health (AI112951). A.O. was supported by a National Institutes of Health Postdoctoral Fellowship (F32AI106175-02), and L.J.T. was supported by an American Cancer Society Postdoctoral Fellowship (PF-11-087-01-MPC).
Abbreviations
- HCMV
human cytomegalovirus
- hpi
hours post infection
- TCID50
50% tissue culture infectious dose
- HSV-1
herpes simplex virus type 1
- MCM
mini-chromosome maintenance
- KSHV
Kaposi’s sarcoma-associated herpesvirus
- EBV
Epstein–Barr virus
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Mocarski ES, Shenk T, Pass RF. Cytomegaloviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams and Wilkins; Philadelphia: 2007. pp. 2701–2772. [Google Scholar]
- 2.Muganda-Ojiaku PM, Huang ES. Alteration of protein phosphorylation patterns in cell lines morphologically transformed by human cytomegalovirus. Cancer Biochem Biophys. 1987;9:179–189. [PubMed] [Google Scholar]
- 3.Muranyi W, Haas J, Wagner M, Krohne G, Koszinowski UH. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science. 2002;297:854–857. doi: 10.1126/science.1071506. [DOI] [PubMed] [Google Scholar]
- 4.Sun B, Harrowe G, Reinhard C, Yoshihara C, et al. Modulation of human cytomegalovirus immediate-early gene enhancer by mitogen-activated protein kinase kinase kinase-1. J Cell Biochem. 2001;83:563–573. doi: 10.1002/jcb.1251. [DOI] [PubMed] [Google Scholar]
- 5.Milbradt J, Auerochs S, Marschall M. Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J Gen Virol. 2007;88:2642–2650. doi: 10.1099/vir.0.82924-0. [DOI] [PubMed] [Google Scholar]
- 6.Johnson RA, Huong SM, Huang ES. Activation of the mitogen-activated protein kinase p38 by human cytomegalovirus infection through two distinct pathways: a novel mechanism for activation of p38. J Virol. 2000;74:1158–1167. doi: 10.1128/jvi.74.3.1158-1167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson RA, Ma XL, Yurochko AD, Huang ES. The role of MKK1/2 kinase activity in human cytomegalovirus infection. J Gen Virol. 2001;82:493–497. doi: 10.1099/0022-1317-82-3-493. [DOI] [PubMed] [Google Scholar]
- 8.Johnson RA, Wang X, Ma XL, Huong SM, Huang ES. Human cytomegalovirus up-regulates the phosphatidylinositol 3-kinase (PI3-K) pathway: inhibition of PI3-K activity inhibits viral replication and virus-induced signaling. J Virol. 2001;75:6022–6032. doi: 10.1128/JVI.75.13.6022-6032.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hertel L, Chou S, Mocarski ES. Viral and cell cycle-regulated kinases in cytomegalovirus-induced pseudomitosis and replication. PLoS Pathog. 2007;3:e6. doi: 10.1371/journal.ppat.0030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Stinski MF. Role of regulatory elements and the MAPK/ERK or p38 MAPK pathways for activation of human cytomegalovirus gene expression. J Virol. 2002;76:4873–4885. doi: 10.1128/JVI.76.10.4873-4885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terry LJ, Vastag L, Rabinowitz JD, Shenk T. Human kinome profiling identifies a requirement for AMP-activated protein kinase during human cytomegalovirus infection. Proc Natl Acad Sci USA. 2012;109:3071–3076. doi: 10.1073/pnas.1200494109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moorman NJ, Shenk T. Rapamycin-resistant mTORC1 kinase activity is required for herpesvirus replication. J Virol. 2010;84:5260–5269. doi: 10.1128/JVI.02733-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou S. Cytomegalovirus UL97 mutations in the era of ganciclovir and maribavir. Rev Med Virol. 2008;18:233–246. doi: 10.1002/rmv.574. [DOI] [PubMed] [Google Scholar]
- 14.Krosky PM, Baek MC, Coen DM. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J Virol. 2003;77:905–914. doi: 10.1128/JVI.77.2.905-914.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamirally S, Kamil JP, Ndassa-Colday YM, Lin AJ, et al. Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 2009;5:e1000275. doi: 10.1371/journal.ppat.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hume AJ, Finkel JS, Kamil JP, Coen DM, et al. Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science. 2008;320:797–799. doi: 10.1126/science.1152095. [DOI] [PubMed] [Google Scholar]
- 17.Prichard MN, Sztul E, Daily SL, Perry AL, et al. Human cytomegalovirus UL97 kinase activity is required for the hyperphosphorylation of retinoblastoma protein and inhibits the formation of nuclear aggresomes. J Virol. 2008;82:5054–5067. doi: 10.1128/JVI.02174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaguchi Y, Matsumura T, Roizman B, Hirai K. Cellular elongation factor 1delta is modified in cells infected with representative alpha-, beta-, or gammaherpesviruses. J Virol. 1999;73:4456–4460. doi: 10.1128/jvi.73.5.4456-4460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baek MC, Krosky PM, Pearson A, Coen DM. Phosphorylation of the RNA polymerase II carboxyl-terminal domain in human cytomegalovirus-infected cells and in vitro by the viral UL97 protein kinase. Virology. 2004;324:184–193. doi: 10.1016/j.virol.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 20.van Zeijl M, Fairhurst J, Baum EZ, Sun L, Jones TR. The human cytomegalovirus UL97 protein is phosphorylated and a component of virions. Virology. 1997;231:72–80. doi: 10.1006/viro.1997.8523. [DOI] [PubMed] [Google Scholar]
- 21.Kamil JP, Coen DM. Human cytomegalovirus protein kinase UL97 forms a complex with the tegument phosphoprotein pp65. J Virol. 2007;81:10659–10668. doi: 10.1128/JVI.00497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prichard MN, Gao N, Jairath S, Mulamba G, et al. A recombinant human cytomegalovirus with a large deletion in UL97 has a severe replication deficiency. J Virol. 1999;73:5663–5670. doi: 10.1128/jvi.73.7.5663-5670.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu D, Silva MC, Shenk T. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci U S A. 2003;100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baek MC, Krosky PM, He Z, Coen DM. Specific phosphorylation of exogenous protein and peptide substrates by the human cytomegalovirus UL97 protein kinase. Importance of the P+5 position. J Biol Chem. 2002;277:29593–29599. doi: 10.1074/jbc.M202312200. [DOI] [PubMed] [Google Scholar]
- 25.Krosky PM, Baek MC, Jahng WJ, Barrera I, et al. The human cytomegalovirus UL44 protein is a substrate for the UL97 protein kinase. J Virol. 2003;77:7720–7727. doi: 10.1128/JVI.77.14.7720-7727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Z, He YS, Kim Y, Chu L, et al. The human cytomegalovirus UL97 protein is a protein kinase that autophosphorylates on serines and threonines. Journal of virology. 1997;71:405–411. doi: 10.1128/jvi.71.1.405-411.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marschall M, Freitag M, Suchy P, Romaker D, et al. The protein kinase pUL97 of human cytomegalovirus interacts with and phosphorylates the DNA polymerase processivity factor pUL44. Virology. 2003;311:60–71. doi: 10.1016/s0042-6822(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 28.Thomas M, Rechter S, Milbradt J, Auerochs S, et al. Cytomegaloviral protein kinase pUL97 interacts with the nuclear mRNA export factor pUL69 to modulate its intranuclear localization and activity. J Gen Virol. 2009;90:567–578. doi: 10.1099/vir.0.005827-0. [DOI] [PubMed] [Google Scholar]
- 29.Li R, Zhu J, Xie Z, Liao G, et al. Conserved herpesvirus kinases target the DNA damage response pathway and TIP60 histone acetyltransferase to promote virus replication. Cell Host Microbe. 2011;10:390–400. doi: 10.1016/j.chom.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 31.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 32.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 33.Yu LR, Veenstra T. Phosphopeptide enrichment using offline titanium dioxide columns for phosphoproteomics. Methods Mol Biol. 2013;1002:93–103. doi: 10.1007/978-1-62703-360-2_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 35.Taus T, Kocher T, Pichler P, Paschke C, et al. Universal and confident phosphorylation site localization using phosphoRS. J Proteome Res. 2011;10:5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
- 36.R Core Team. R Foundation for Statistical Computing. 2014 [Google Scholar]
- 37.Dutang MLD-MaRPaJ-BDaC. 2014 [Google Scholar]
- 38.Mikis Stasinopoulos BRVV. gamlss: Generalised Additive Models for Location Scale and Shape. 2014. Calliope Akantziliotou, and Marco Enea. [Google Scholar]
- 39.Wu R, Dephoure N, Haas W, Huttlin EL, et al. Correct interpretation of comprehensive phosphorylation dynamics requires normalization by protein expression changes. Mol Cell Proteomics. 2011;10:M111 009654. doi: 10.1074/mcp.M111.009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Breitwieser FP, Colinge J. IsobarPTM: A software tool for the quantitative analysis of post-translationally modified proteins. Journal of Proteomics. 2013;90:77–84. doi: 10.1016/j.jprot.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou MF, Schwartz D. Biological sequence motif discovery using motif-x. In: Baxevanis Andreas D, et al., editors. Current protocols in bioinformatics. Unit 13. Chapter 13. 2011. pp. 15–24. [DOI] [PubMed] [Google Scholar]
- 42.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 43.Wang T, Kettenbach AN, Gerber SA, Bailey-Kellogg C. MMFPh: a maximal motif finder for phosphoproteomics datasets. Bioinformatics. 2012;28:1562–1570. doi: 10.1093/bioinformatics/bts195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shannon P, Markiel A, Ozier O, Baliga NS, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bindea G, Mlecnik B, Hackl H, Charoentong P, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michel D, Pavic I, Zimmermann A, Haupt E, et al. The UL97 gene product of human cytomegalovirus is an early-late protein with a nuclear localization but is not a nucleoside kinase. Journal of virology. 1996;70:6340–6346. doi: 10.1128/jvi.70.9.6340-6346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma M, Bender BJ, Kamil JP, Lye MF, et al. Human Cytomegalovirus UL97 Phosphorylates the Viral Nuclear Egress Complex. Journal of virology. 2014 doi: 10.1128/JVI.02426-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma M, Coen DM. Comparison of effects of inhibitors of viral and cellular protein kinases on human cytomegalovirus disruption of nuclear lamina and nuclear egress. Journal of virology. 2014;88:10982–10985. doi: 10.1128/JVI.01391-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharma M, Kamil JP, Coughlin M, Reim NI, Coen DM. Human cytomegalovirus UL50 and UL53 recruit viral protein kinase UL97, not protein kinase C, for disruption of nuclear lamina and nuclear egress in infected cells. Journal of virology. 2014;88:249–262. doi: 10.1128/JVI.02358-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamirally S, Kamil JP, Ndassa-Colday YM, Lin AJ, et al. Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 2009;5:e1000275. doi: 10.1371/journal.ppat.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krosky PM, Baek MC, Jahng WJ, Barrera I, et al. The human cytomegalovirus UL44 protein is a substrate for the UL97 protein kinase. J Virol. 2003;77:7720–7727. doi: 10.1128/JVI.77.14.7720-7727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma M, Bender BJ, Kamil JP, Lye MF, et al. Human cytomegalovirus UL97 phosphorylates the viral nuclear egress complex. J Virol. 2015;89:523–534. doi: 10.1128/JVI.02426-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas M, Rechter S, Milbradt J, Auerochs S, et al. Cytomegaloviral protein kinase pUL97 interacts with the nuclear mRNA export factor pUL69 to modulate its intranuclear localization and activity. The Journal of general virology. 2009;90:567–578. doi: 10.1099/vir.0.005827-0. [DOI] [PubMed] [Google Scholar]
- 55.Baek MC, Krosky PM, Pearson A, Coen DM. Phosphorylation of the RNA polymerase II carboxyl-terminal domain in human cytomegalovirus-infected cells and in vitro by the viral UL97 protein kinase. Virology. 2004;324:184–193. doi: 10.1016/j.virol.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 56.Bigley TM, Reitsma JM, Mirza SP, Terhune SS. Human cytomegalovirus pUL97 regulates the viral major immediate early promoter by phosphorylation-mediated disruption of histone deacetylase 1 binding. Journal of virology. 2013;87:7393–7408. doi: 10.1128/JVI.02825-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baek MC, Krosky PM, He Z, Coen DM. Specific phosphorylation of exogenous protein and peptide substrates by the human cytomegalovirus UL97 protein kinase. Importance of the P+5 position. The Journal of biological chemistry. 2002;277:29593–29599. doi: 10.1074/jbc.M202312200. [DOI] [PubMed] [Google Scholar]
- 58.Wang T, Kettenbach AN, Gerber SA, Bailey-Kellogg C. MMFPh: a maximal motif finder for phosphoproteomics datasets. Bioinformatics. 2012;28:1562–1570. doi: 10.1093/bioinformatics/bts195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malumbres M. Cyclin-dependent kinases. Genome Biol. 2014;15:122. doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szklarczyk D, Franceschini A, Wyder S, Forslund K, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prokocimer M, Davidovich M, Nissim-Rafinia M, Wiesel-Motiuk N, et al. Nuclear lamins: key regulators of nuclear structure and activities. J Cell Mol Med. 2009;13:1059–1085. doi: 10.1111/j.1582-4934.2008.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Katahira J. mRNA export and the TREX complex. Biochimica et biophysica acta. 2012;1819:507–513. doi: 10.1016/j.bbagrm.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 64.Silva LA, Strang BL, Lin EW, Kamil JP, Coen DM. Sites and roles of phosphorylation of the human cytomegalovirus DNA polymerase subunit UL44. Virology. 2011;417:268–280. doi: 10.1016/j.virol.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Michel D, Kramer S, Hohn S, Schaarschmidt P, et al. Amino acids of conserved kinase motifs of cytomegalovirus protein UL97 are essential for autophosphorylation. Journal of virology. 1999;73:8898–8901. doi: 10.1128/jvi.73.10.8898-8901.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baek MC, Krosky PM, Coen DM. Relationship between autophosphorylation and phosphorylation of exogenous substrates by the human cytomegalovirus UL97 protein kinase. Journal of virology. 2002;76:11943–11952. doi: 10.1128/JVI.76.23.11943-11952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 68.Atherton-Fessler S, Hannig G, Piwnica-Worms H. Reversible tyrosine phosphorylation and cell cycle control. Semin Cell Biol. 1993;4:433–442. doi: 10.1006/scel.1993.1051. [DOI] [PubMed] [Google Scholar]
- 69.Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol Biol Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiebusch L, Uecker R, Hagemeier C. Human cytomegalovirus prevents replication licensing by inhibiting MCM loading onto chromatin. EMBO Rep. 2003;4:42–46. doi: 10.1038/sj.embor.embor707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Biswas N, Sanchez V, Spector DH. Human cytomegalovirus infection leads to accumulation of geminin and inhibition of the licensing of cellular DNA replication. Journal of virology. 2003;77:2369–2376. doi: 10.1128/JVI.77.4.2369-2376.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qian Z, Leung-Pineda V, Xuan B, Piwnica-Worms H, Yu D. Human cytomegalovirus protein pUL117 targets the mini-chromosome maintenance complex and suppresses cellular DNA synthesis. PLoS pathogens. 2010;6:e1000814. doi: 10.1371/journal.ppat.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnson DC, Baines JD. Herpesviruses remodel host membranes for virus egress. Nature reviews Microbiology. 2011;9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 74.Mettenleiter TC. Herpesvirus assembly and egress. J Virol. 2002;76:1537–1547. doi: 10.1128/JVI.76.4.1537-1547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reim NI, Kamil JP, Wang D, Lin A, et al. Inactivation of retinoblastoma protein does not overcome the requirement for human cytomegalovirus UL97 in lamina disruption and nuclear egress. J Virol. 2013;87:5019–5027. doi: 10.1128/JVI.00007-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wild P, Senn C, Manera CL, Sutter E, et al. Exploring the nuclear envelope of herpes simplex virus 1-infected cells by high-resolution microscopy. J Virol. 2009;83:408–419. doi: 10.1128/JVI.01568-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hofemeister H, O’Hare P. Nuclear pore composition and gating in herpes simplex virus-infected cells. J Virol. 2008;82:8392–8399. doi: 10.1128/JVI.00951-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Malik P, Tabarraei A, Kehlenbach RH, Korfali N, et al. Herpes simplex virus ICP27 protein directly interacts with the nuclear pore complex through Nup62, inhibiting host nucleocytoplasmic transport pathways. The Journal of biological chemistry. 2012;287:12277–12292. doi: 10.1074/jbc.M111.331777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Toth Z, Lischka P, Stamminger T. RNA-binding of the human cytomegalovirus transactivator protein UL69, mediated by arginine-rich motifs, is not required for nuclear export of unspliced RNA. Nucleic Acids Res. 2006;34:1237–1249. doi: 10.1093/nar/gkl007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lischka P, Toth Z, Thomas M, Mueller R, Stamminger T. The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-Box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol Cell Biol. 2006;26:1631–1643. doi: 10.1128/MCB.26.5.1631-1643.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Masuda S, Das R, Cheng H, Hurt E, et al. Recruitment of the human TREX complex to mRNA during splicing. Genes & development. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sandri-Goldin RM. The many roles of the highly interactive HSV protein ICP27, a key regulator of infection. Future microbiology. 2011;6:1261–1277. doi: 10.2217/fmb.11.119. [DOI] [PubMed] [Google Scholar]
- 83.Dufu K, Livingstone MJ, Seebacher J, Gygi SP, et al. ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes & development. 2010;24:2043–2053. doi: 10.1101/gad.1898610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hautbergue GM, Hung ML, Walsh MJ, Snijders AP, et al. UIF, a New mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Current biology : CB. 2009;19:1918–1924. doi: 10.1016/j.cub.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pasdeloup D, Blondel D, Isidro AL, Rixon FJ. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J Virol. 2009;83:6610–6623. doi: 10.1128/JVI.02655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marschall M, Feichtinger S, Milbradt J. Regulatory roles of protein kinases in cytomegalovirus replication. Advances in Virus Research. 2011;80:69–101. doi: 10.1016/B978-0-12-385987-7.00004-X. [DOI] [PubMed] [Google Scholar]
- 87.Li R, Hayward SD. Potential of protein kinase inhibitors for treating herpesvirus-associated disease. Trends Microbiol. 2013 doi: 10.1016/j.tim.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Varnum SM, Streblow DN, Monroe ME, Smith P, et al. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. Journal of virology. 2004;78:10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Loret S, Guay G, Lippe R. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. Journal of virology. 2008;82:8605–8618. doi: 10.1128/JVI.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nature Biotechnology. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.