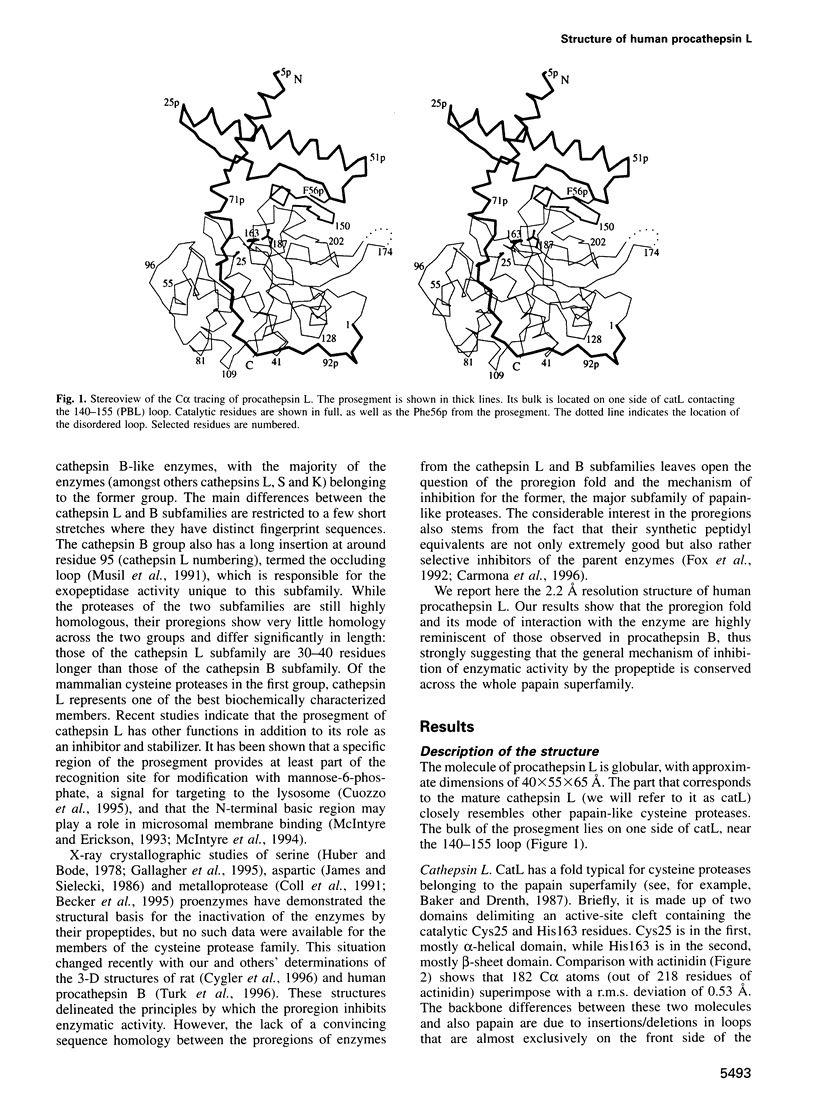
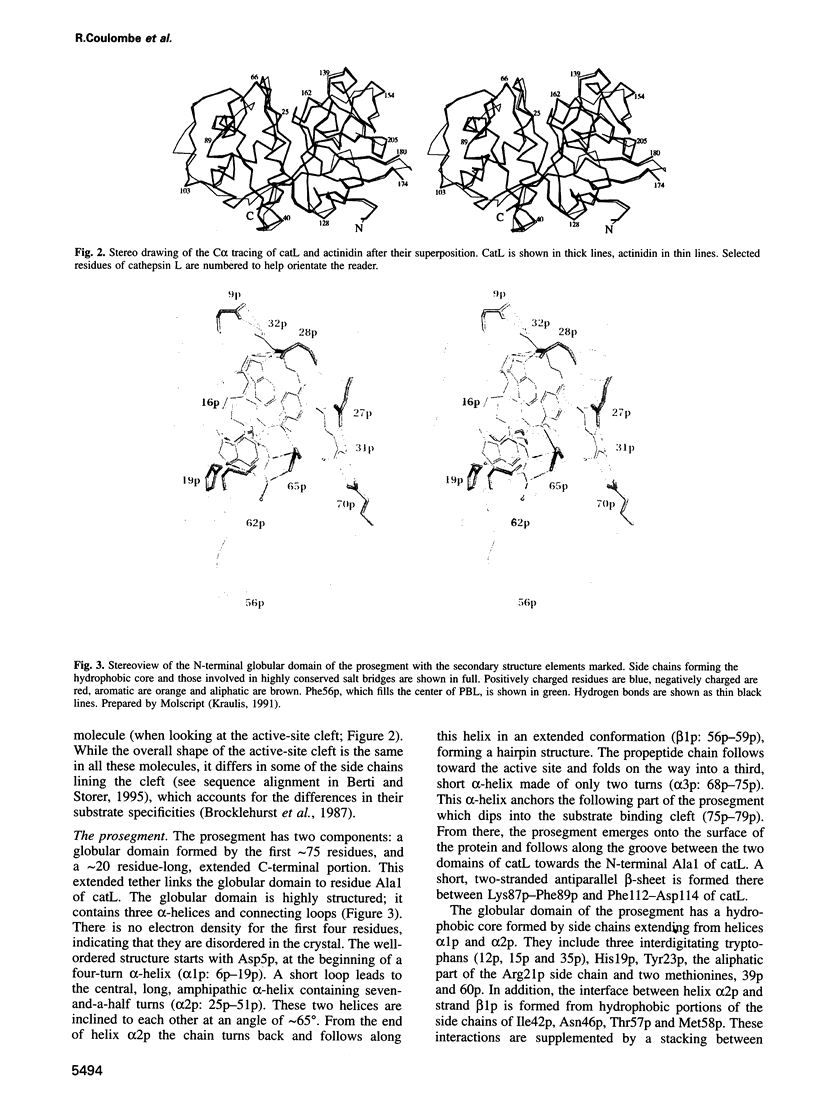
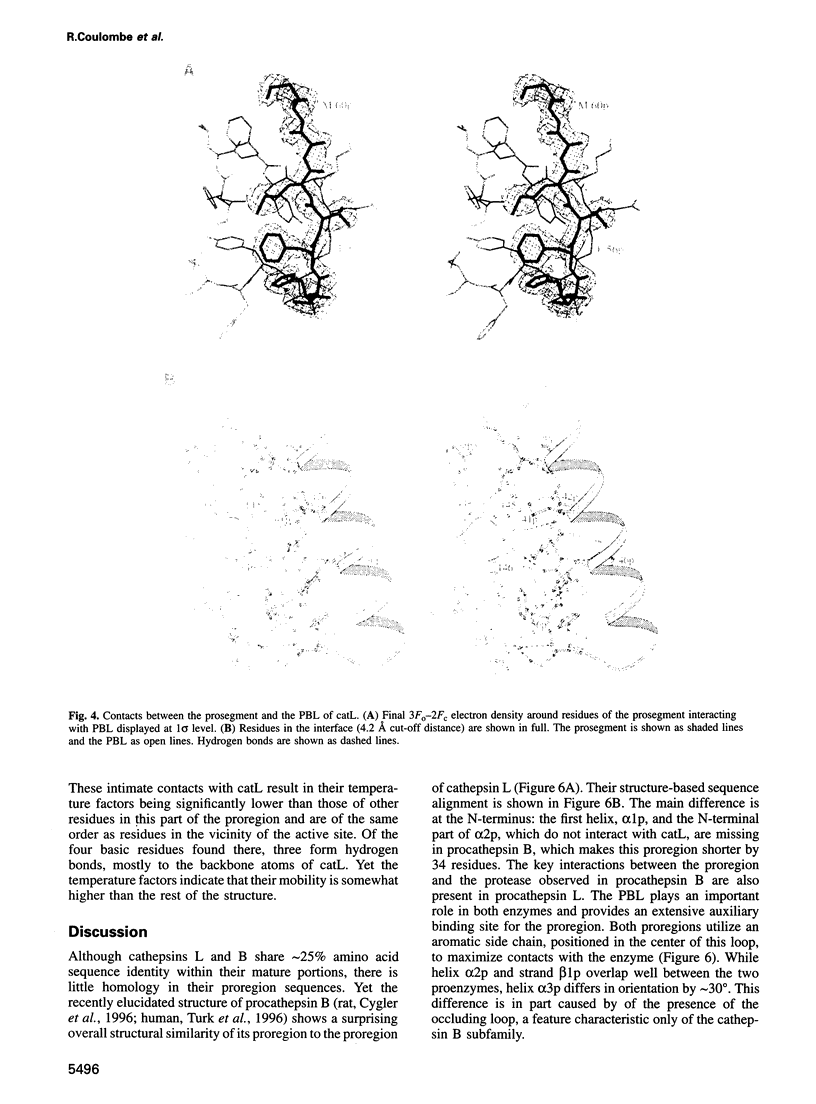
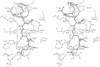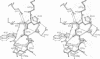Abstract
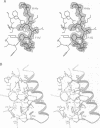
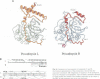
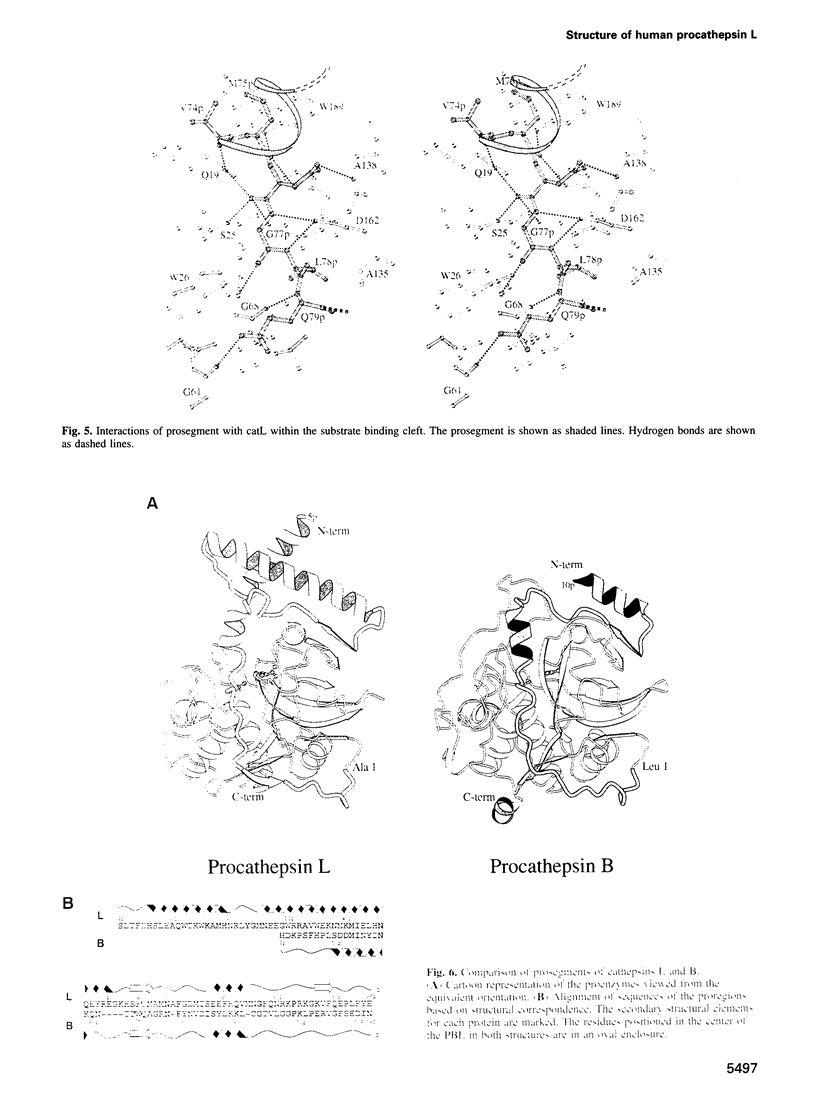
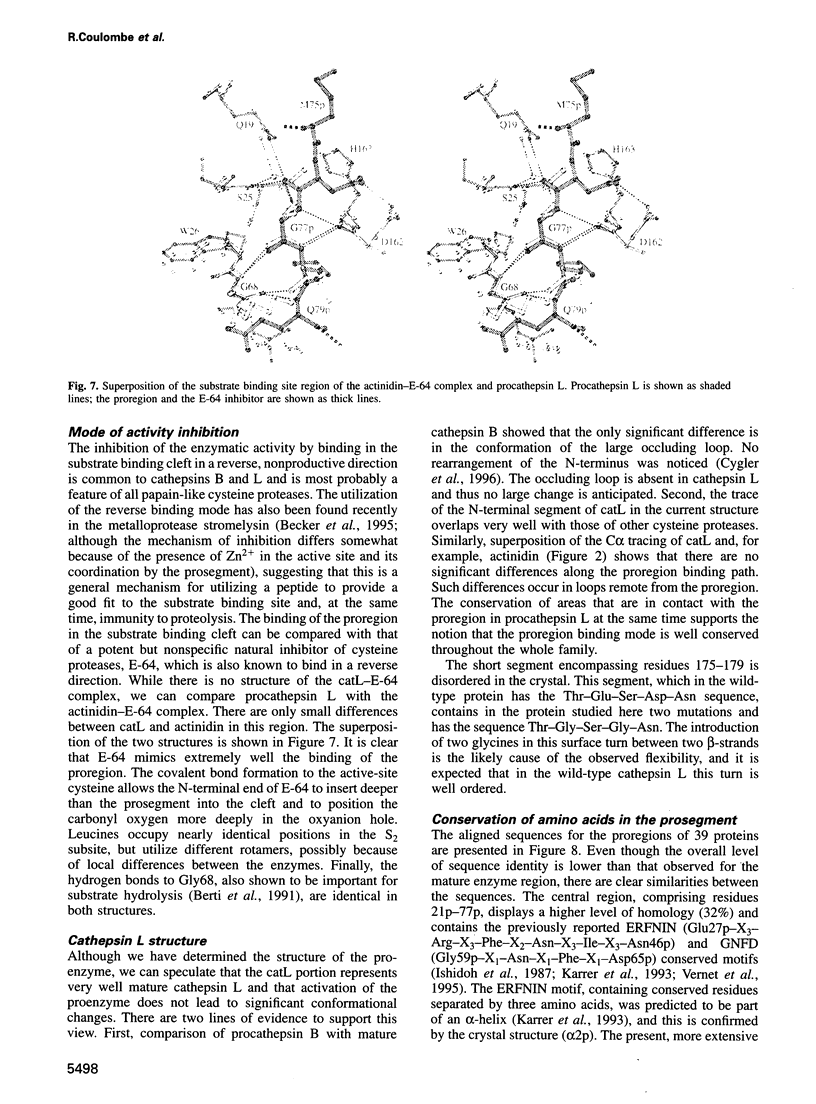
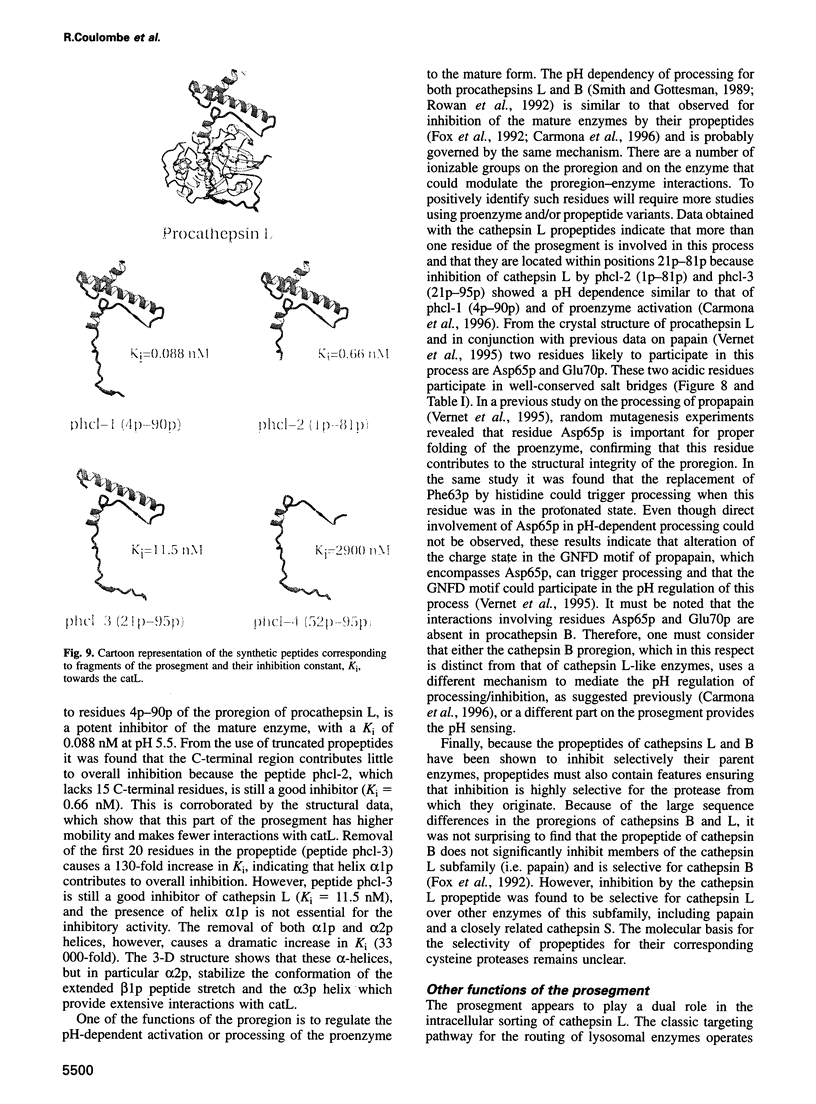
Cathepsin L is a member of the papain superfamily of cysteine proteases and, like many other proteases, it is synthesized as an inactive proenzyme. Its prosegment shows little homology to that of procathepsin B, whose structure, the first for a cysteine protease proenzyme, has been determined recently. We report here the 3-D structure of a mutant of human procathepsin L determined at 2.2 A resolution, describe the mode of binding employed by the prosegment and discuss the molecular basis for other possible roles of the prosegment. The N-terminal part of the prosegment is globular and contains three alpha-helices with a small hydrophobic core built around aromatic side chains. This domain packs against a loop on the enzyme's surface, with the aromatic side chain from the prosegment being located in the center of this loop and providing a large contact area. The C-terminal portion of the prosegment assumes an extended conformation and follows along the substrate binding cleft toward the N-terminus of the mature enzyme. The direction of the prosegment in the substrate binding cleft is opposite to that of substrates. The previously described role of the prosegment in the interactions with membranes is supported by the structure of its N-terminal domain. The fold of the prosegment and the mechanism by which it inhibits the enzymatic activity of procathepsin L is similar to that observed in procathepsin B despite differences in length and sequence, suggesting that this mode of inhibition is common to all enzymes from the papain superfamily.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker E. N. Structure of actinidin: details of the polypeptide chain conformation and active site from an electron density map at 2-8 A resolution. J Mol Biol. 1977 Sep 25;115(3):263–277. doi: 10.1016/0022-2836(77)90154-1. [DOI] [PubMed] [Google Scholar]
- Becker J. W., Marcy A. I., Rokosz L. L., Axel M. G., Burbaum J. J., Fitzgerald P. M., Cameron P. M., Esser C. K., Hagmann W. K., Hermes J. D. Stromelysin-1: three-dimensional structure of the inhibited catalytic domain and of the C-truncated proenzyme. Protein Sci. 1995 Oct;4(10):1966–1976. doi: 10.1002/pro.5560041002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti P. J., Faerman C. H., Storer A. C. Cooperativity of papain-substrate interaction energies in the S2 to S2' subsites. Biochemistry. 1991 Feb 5;30(5):1394–1402. doi: 10.1021/bi00219a033. [DOI] [PubMed] [Google Scholar]
- Berti P. J., Storer A. C. Alignment/phylogeny of the papain superfamily of cysteine proteases. J Mol Biol. 1995 Feb 17;246(2):273–283. doi: 10.1006/jmbi.1994.0083. [DOI] [PubMed] [Google Scholar]
- Carmona E., Dufour E., Plouffe C., Takebe S., Mason P., Mort J. S., Ménard R. Potency and selectivity of the cathepsin L propeptide as an inhibitor of cysteine proteases. Biochemistry. 1996 Jun 25;35(25):8149–8157. doi: 10.1021/bi952736s. [DOI] [PubMed] [Google Scholar]
- Coll M., Guasch A., Avilés F. X., Huber R. Three-dimensional structure of porcine procarboxypeptidase B: a structural basis of its inactivity. EMBO J. 1991 Jan;10(1):1–9. doi: 10.1002/j.1460-2075.1991.tb07914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe R., Li Y., Takebe S., Ménard R., Mason P., Mort J. S., Cygler M. Crystallization and preliminary X-ray diffraction studies of human procathepsin L. Proteins. 1996 Jul;25(3):398–400. doi: 10.1002/(SICI)1097-0134(199607)25:3<398::AID-PROT11>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Cuozzo J. W., Tao K., Wu Q. L., Young W., Sahagian G. G. Lysine-based structure in the proregion of procathepsin L is the recognition site for mannose phosphorylation. J Biol Chem. 1995 Jun 30;270(26):15611–15619. doi: 10.1074/jbc.270.26.15611. [DOI] [PubMed] [Google Scholar]
- Cygler M., Sivaraman J., Grochulski P., Coulombe R., Storer A. C., Mort J. S. Structure of rat procathepsin B: model for inhibition of cysteine protease activity by the proregion. Structure. 1996 Apr 15;4(4):405–416. doi: 10.1016/s0969-2126(96)00046-9. [DOI] [PubMed] [Google Scholar]
- Drenth J., Jansonius J. N., Koekoek R., Swen H. M., Wolthers B. G. Structure of papain. Nature. 1968 Jun 8;218(5145):929–932. doi: 10.1038/218929a0. [DOI] [PubMed] [Google Scholar]
- Fox T., de Miguel E., Mort J. S., Storer A. C. Potent slow-binding inhibition of cathepsin B by its propeptide. Biochemistry. 1992 Dec 22;31(50):12571–12576. doi: 10.1021/bi00165a005. [DOI] [PubMed] [Google Scholar]
- Gallagher T., Gilliland G., Wang L., Bryan P. The prosegment-subtilisin BPN' complex: crystal structure of a specific 'foldase'. Structure. 1995 Sep 15;3(9):907–914. doi: 10.1016/S0969-2126(01)00225-8. [DOI] [PubMed] [Google Scholar]
- George D. G., Barker W. C., Hunt L. T. Mutation data matrix and its uses. Methods Enzymol. 1990;183:333–351. doi: 10.1016/0076-6879(90)83022-2. [DOI] [PubMed] [Google Scholar]
- Gour-Salin B. J., Lachance P., Magny M. C., Plouffe C., Ménard R., Storer A. C. E64 [trans-epoxysuccinyl-L-leucylamido-(4-guanidino)butane] analogues as inhibitors of cysteine proteinases: investigation of S2 subsite interactions. Biochem J. 1994 Apr 15;299(Pt 2):389–392. doi: 10.1042/bj2990389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann U., Pal G. P., Hilgenfeld R., Saenger W. Crystal and molecular structure of the sulfhydryl protease calotropin DI at 3.2 A resolution. J Mol Biol. 1982 Nov 15;161(4):591–606. doi: 10.1016/0022-2836(82)90410-7. [DOI] [PubMed] [Google Scholar]
- Ishidoh K., Kominami E. Procathepsin L degrades extracellular matrix proteins in the presence of glycosaminoglycans in vitro. Biochem Biophys Res Commun. 1995 Dec 14;217(2):624–631. doi: 10.1006/bbrc.1995.2820. [DOI] [PubMed] [Google Scholar]
- Ishidoh K., Towatari T., Imajoh S., Kawasaki H., Kominami E., Katunuma N., Suzuki K. Molecular cloning and sequencing of cDNA for rat cathepsin L. FEBS Lett. 1987 Oct 19;223(1):69–73. doi: 10.1016/0014-5793(87)80511-2. [DOI] [PubMed] [Google Scholar]
- James M. N., Sielecki A. R. Molecular structure of an aspartic proteinase zymogen, porcine pepsinogen, at 1.8 A resolution. Nature. 1986 Jan 2;319(6048):33–38. doi: 10.1038/319033a0. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kakegawa H., Nikawa T., Tagami K., Kamioka H., Sumitani K., Kawata T., Drobnic-Kosorok M., Lenarcic B., Turk V., Katunuma N. Participation of cathepsin L on bone resorption. FEBS Lett. 1993 Apr 26;321(2-3):247–250. doi: 10.1016/0014-5793(93)80118-e. [DOI] [PubMed] [Google Scholar]
- Karrer K. M., Peiffer S. L., DiTomas M. E. Two distinct gene subfamilies within the family of cysteine protease genes. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):3063–3067. doi: 10.1073/pnas.90.7.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschke H., Wiederanders B., Brömme D., Rinne A. Cathepsin S from bovine spleen. Purification, distribution, intracellular localization and action on proteins. Biochem J. 1989 Dec 1;264(2):467–473. doi: 10.1042/bj2640467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamzin V. S., Wilson K. S. Automated refinement of protein models. Acta Crystallogr D Biol Crystallogr. 1993 Jan 1;49(Pt 1):129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- Mason R. W., Gal S., Gottesman M. M. The identification of the major excreted protein (MEP) from a transformed mouse fibroblast cell line as a catalytically active precursor form of cathepsin L. Biochem J. 1987 Dec 1;248(2):449–454. doi: 10.1042/bj2480449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason R. W., Massey S. D. Surface activation of pro-cathepsin L. Biochem Biophys Res Commun. 1992 Dec 30;189(3):1659–1666. doi: 10.1016/0006-291x(92)90268-p. [DOI] [PubMed] [Google Scholar]
- McGrath M. E., Eakin A. E., Engel J. C., McKerrow J. H., Craik C. S., Fletterick R. J. The crystal structure of cruzain: a therapeutic target for Chagas' disease. J Mol Biol. 1995 Mar 24;247(2):251–259. doi: 10.1006/jmbi.1994.0137. [DOI] [PubMed] [Google Scholar]
- McIntyre G. F., Erickson A. H. The lysosomal proenzyme receptor that binds procathepsin L to microsomal membranes at pH 5 is a 43-kDa integral membrane protein. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10588–10592. doi: 10.1073/pnas.90.22.10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre G. F., Godbold G. D., Erickson A. H. The pH-dependent membrane association of procathepsin L is mediated by a 9-residue sequence within the propeptide. J Biol Chem. 1994 Jan 7;269(1):567–572. [PubMed] [Google Scholar]
- Musil D., Zucic D., Turk D., Engh R. A., Mayr I., Huber R., Popovic T., Turk V., Towatari T., Katunuma N. The refined 2.15 A X-ray crystal structure of human liver cathepsin B: the structural basis for its specificity. EMBO J. 1991 Sep;10(9):2321–2330. doi: 10.1002/j.1460-2075.1991.tb07771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara B. P., Hemmings A. M., Buttle D. J., Pearl L. H. Crystal structure of glycyl endopeptidase from Carica papaya: a cysteine endopeptidase of unusual substrate specificity. Biochemistry. 1995 Oct 10;34(40):13190–13195. doi: 10.1021/bi00040a034. [DOI] [PubMed] [Google Scholar]
- Rowan A. D., Mason P., Mach L., Mort J. S. Rat procathepsin B. Proteolytic processing to the mature form in vitro. J Biol Chem. 1992 Aug 5;267(22):15993–15999. [PubMed] [Google Scholar]
- Smith S. M., Gottesman M. M. Activity and deletion analysis of recombinant human cathepsin L expressed in Escherichia coli. J Biol Chem. 1989 Dec 5;264(34):20487–20495. [PubMed] [Google Scholar]
- Tao K., Stearns N. A., Dong J., Wu Q. L., Sahagian G. G. The proregion of cathepsin L is required for proper folding, stability, and ER exit. Arch Biochem Biophys. 1994 May 15;311(1):19–27. doi: 10.1006/abbi.1994.1203. [DOI] [PubMed] [Google Scholar]
- Tezuka K., Tezuka Y., Maejima A., Sato T., Nemoto K., Kamioka H., Hakeda Y., Kumegawa M. Molecular cloning of a possible cysteine proteinase predominantly expressed in osteoclasts. J Biol Chem. 1994 Jan 14;269(2):1106–1109. [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994 Nov 11;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabandt A., Aicher W. K., Gay R. E., Sukhatme V. P., Nilson-Hamilton M., Hamilton R. T., McGhee J. R., Fassbender H. G., Gay S. Expression of the collagenolytic and Ras-induced cysteine proteinase cathepsin L and proliferation-associated oncogenes in synovial cells of MRL/I mice and patients with rheumatoid arthritis. Matrix. 1990 Dec;10(6):349–361. doi: 10.1016/s0934-8832(11)80142-3. [DOI] [PubMed] [Google Scholar]
- Turk D., Podobnik M., Kuhelj R., Dolinar M., Turk V. Crystal structures of human procathepsin B at 3.2 and 3.3 Angstroms resolution reveal an interaction motif between a papain-like cysteine protease and its propeptide. FEBS Lett. 1996 Apr 22;384(3):211–214. doi: 10.1016/0014-5793(96)00309-2. [DOI] [PubMed] [Google Scholar]
- Turk D., Podobnik M., Popovic T., Katunuma N., Bode W., Huber R., Turk V. Crystal structure of cathepsin B inhibited with CA030 at 2.0-A resolution: A basis for the design of specific epoxysuccinyl inhibitors. Biochemistry. 1995 Apr 11;34(14):4791–4797. doi: 10.1021/bi00014a037. [DOI] [PubMed] [Google Scholar]
- Vernet T., Berti P. J., de Montigny C., Musil R., Tessier D. C., Ménard R., Magny M. C., Storer A. C., Thomas D. Y. Processing of the papain precursor. The ionization state of a conserved amino acid motif within the Pro region participates in the regulation of intramolecular processing. J Biol Chem. 1995 May 5;270(18):10838–10846. doi: 10.1074/jbc.270.18.10838. [DOI] [PubMed] [Google Scholar]
- Yagel S., Warner A. H., Nellans H. N., Lala P. K., Waghorne C., Denhardt D. T. Suppression by cathepsin L inhibitors of the invasion of amnion membranes by murine cancer cells. Cancer Res. 1989 Jul 1;49(13):3553–3557. [PubMed] [Google Scholar]