Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that function in gene regulatory processes in plants and animals by targeting sites within messenger RNA. In insects, miRNAs have been shown to regulate a variety of physiological processes throughout insect development, including molting, metamorphosis, oogenesis, embryogenesis, behavior and host-pathogen interactions. The roles of miRNAs in the model organism, Drosophila melanogaster, have been studied extensively due to the conserved nature of miRNA function among highly divergent species. However, seeking to understand miRNA function in non-drosophilid insect species has become a growing trend in insect science. Here, we highlight the recent discoveries regarding miRNA function in insect physiology and development.
Introduction
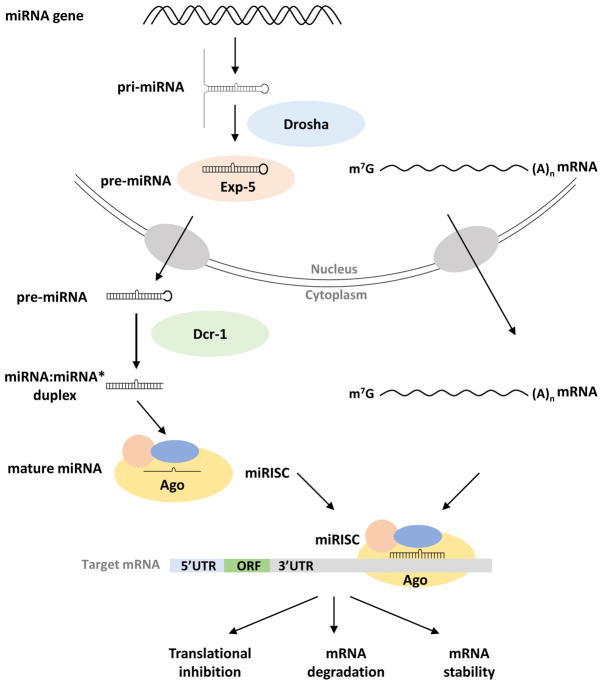
MicroRNAs (miRNAs) are endogenous non-coding RNAs that post-transcriptionally regulate transcript levels and translational status of messenger RNA (mRNA). These small molecules are essential regulators of various physiological processes in plants and animals. In insects, canonical miRNA loci are transcribed similarly to mRNAs by RNA polymerase II, resulting in the formation of the primary miRNA (pri-miRNA) transcript [1]. Pri-miRNAs undergo a series of processing and sorting events before mature miRNAs can act on target mRNAs (Figure 1). First, the pri-miRNA is processed in the nucleus by the RNase III enzyme Drosha, liberating an approximately 70-nucleotide (nt) stem-loop structure known as the precursor miRNA (pre-miRNA) [2, 3]. The pre-miRNA is exported to the cytoplasm by Exportin-5 (Exp-5) where its terminal loop structure is cleaved by another RNase III endonuclease, Dicer-1 (Dcr-1), producing a 22-nt miRNA duplex [4–6]. Duplex strands are sorted and loaded into their respective Argonaute (Ago) proteins and complete their action on target sequences through the RNA-induced silencing complex (RISC). The RISC typically targets the 3 untranslated region (UTR) of target transcripts, promoting translational repression and mRNA degradation [7]*. More recently, several reports have revealed that mRNA-miRNA interactions may lead to the stabilization of mRNA [8].
Figure 1.
Schematic diagram of microRNA (miRNA) biogenesis in insects. miRNA transcripts, termed primary miRNAs (pri-miRNA), fold into hairpin structures and are processed in the nucleus by the Drosha to form a ~70-nt precursor miRNA (pre-miRNA). The pre-miRNA is exported to the cytoplasm by Exportin-5 (Exp-5) where it is processed by Dicer-1 (Dcr-1) to form the miRNA:miRNA* duplex. The duplex strands are then sorted and loaded into the RNA induced silencing complex (RISC) with its respective Argonaute protein (Ago). The miRNA-RISC (miRISC) acts on the 3′ untranslated region of messenger RNAs (mRNA) by binding to complementary seed match sites, resulting in translational inhibition and subsequent mRNA degradation or mRNA stability.
Conserved and lineage-specific miRNAs have been identified in several insect species. Conserved miRNAs can elicit preserved functions among organisms, as well as non-conserved actions. Additionally, lineage- and species-specific miRNAs may contribute to developmental novelties within particular insects. Due to the conserved function of miRNAs, the fruit fly Drosophila melanogaster has attracted much attention for understanding miRNA biogenesis and mode of action, as well as the functional role of miRNAs in animals. miRNAs have been shown to function in various biological processes, including animal development and disease. Because of their important role in animal physiology, studying the function of miRNAs in other insects of great medical and agricultural importance has become a growing trend in insect science. Here, we report on recent work undertaken to understand the functional roles of miRNAs in insect species.
Growth and Development
Insect models have revealed that numerous conserved miRNAs target signaling pathways important for animal development. Several studies have shown that dme-miR-8 regulates multiple peptide hormones in the fat body, contributing to cell growth [9–11]. dme-miR-14 was shown to target Hedgehog (Hg), a core component of the highly conserved Hg signaling pathway important for various developmental processes [12]. This miRNA also functions in the degradation of the larval salivary gland during pupal development by regulating autophagy in salivary gland cells through the modulation of inositol 1,4,5-trisphosphate kinase 2 (ip3k2) [13]*. Furthermore, the Hippo signaling pathway regulates pro-apoptotic gene expression during tissue growth through the dme-miR-2a cluster of miRNAs in Drosophila [14]. Additionally, dme-bantam controls growth and proliferation in the posterior signaling center, a stem cell-like niche, by functioning upstream of the insulin signaling pathway [15]. dme-miR-305 also interacts with the insulin signaling pathway, as well as the Notch signaling pathway, in Drosophila intestinal stem cells to control self-renewal and differentiation [16]*. Yet another miRNA, dme-miR-7, controls cell growth and cell cycle progression during Drosophila wing development by targeting the cell cycle regulator dacapo and the Notch signaling pathway [17].
The insect steroid hormone, 20-hydroxyecdysone (20E), coordinates multiple developmental events in insects. Several insect miRNAs have been recently shown to act in insect development and metamorphosis by regulating genes in the ecdysone cascade. In Bombyx mori, bmo-miR-281 participates in silkworm developmental regulation in the Malpighian tubules by suppression of silkworm EcR-B in an isoform-specific manner [18]. Unlike EcR in Drosophila, the silkworm 3′ UTR of EcR-A differs substantially from the EcR-B 3′ UTR, indicating that the conserved miR-281 functions as a non-conserved regulator of the EcR-B isoform in silkworm. In the first report of the utilization of the transgenic miRNA sponge (miR-SP) technology combined with the binary Gal4-UAS system in silkworm, bmo-let-7 miR-SP was shown to trigger deleterious effects during the larval–larval and larval–pupal transitions [19]**. This work indicated the importance of bmo-let-7 in silkworm molting and metamorphosis, during which bmo-let-7 may regulate molting by targeting FTZ–F1 and E74, key regulatory genes of the ecdysone signaling cascade.
Neuronal development has been shown to be modulated by miRNAs in insect species. Drosophila dme-miR-8 is required for neuromuscular junction (NMJ) expansion during the late larval phase by targeting the actin-regulatory protein Enabled [20]. Furthermore, dme-miR-8 plays a role in early NMJ development by functioning upstream of the synaptic cell adhesion molecules, Fasciclin III (FasIII) and Neuroglian (Nrg). dme-miR-8 also modulates atrophin levels to prevent neurodegradation, during which dme-miR-8 mutant flies exhibit significant apoptosis in larval brains and developmental defects in adults [21]. Consistent with these results, in the cockroach Blattella germanica, bge-miR-8-3p and bge-miR-8-5p targets atrophin, and misregulation of atrophin levels results in impaired motor coordination [22].
In the brown plant lopper, Nilaparvata lugens, nlu-miR-8-5p and nlu-miR-2a-3p targets the chitin biosynthesis pathway components, membrane-bound trehalase (Tre-2) and phosphoacetylglucosamine mutase (PAGM), respectively [23]. Overexpression of nlu-miR-8-5p and nlu-miR-2a-3p resulted in reduced survival among individuals and decreased chitin content. Furthermore, nlu-miR-8-5p and nlu-miR-2a-3p levels were shown to be negatively regulated by 20E through the Broad Complex (BR-C). In Spodoptera exigua, commonly known as the beet army worm, sex-miR-4924 modulates larval development and molting, possibly through the regulation of the chitinase 1 [24]. Moreover, silencing of Dcr-1 in the migratory locust, Locusta migratoria, reduced miRNA content and disrupted two types of molt—nymph-nymph and nymph-adult [25].
In addition to conserved miRNAs, recent investigations of lineage-specific miRNAs have identified their important role in insect development. In Ae. albopictus larvae, depletion of the mosquito- and larva-specific miRNA, aael-miR-2942, results in a reduced eclosion rate indicating an important role in regulating eclosion events in mosquito larvae [26]. Sequencing of small RNA libraries from developmental stages of two lepidopterans, Cameraria ohridella (Horse chestnut Leafminer) and Pararge aegeria (Speckled Wood butterfly), identified many conserved and lineage-specific miRNAs [27]*. The lepidopteran-specific miRNA, miR-2768, was found to arise from an intron of the invected (inv) gene and targets the coding region of cubitus interruptus (ci), possibly playing a role in segmentation of the embryo or during postembryonic patterning of the wing primordia.
Behavior
Recent studies have revealed an important role of miRNAs in regulating insect behavioral plasticity, performance and memory. More recently, in Lo. migratoria, lmi-miR-133 was shown to mediate phenotypic plasticity and behavior changes between social and solitary phases by targeting key players in the dopamine synthesis pathway, henna and pale [28]*. Furthermore, in Drosophila, dme-miR-124 knockout flies display severely reduced locomotion, flight and fertility during the adult stage, indicating that this miRNA is required for normal adult behavior [29]. In the honeybee, Apis mellifera, depletion of ame-miR-932 impairs long-term memory formation, indicating a regulatory role in learning and memory. Ame-miR-932 was shown to interact with Act5C, which potentially affect learning and memory and activity-dependent neuronal plasticity [30]*.
Sex determination
The action miRNAs in regulating sex determination has been explored in several insect species. miRNAs are important for establishing sexually dimorphic traits and controlling sexual behavior. Repertoires of male- and female-biased, and ovary- and testis-enriched miRNAs reveal the overall complexity of miRNA regulation on sex determination and gametogenesis [31]**. Depletion of dme-let-7 results in abnormal somatic cell behavior, resulting in a delay of early ovarian germline differentiation and large aggregates of somatic cells in the testes. dme-let-7 plays an important role in male and female sexual identity during the late-larval to late-pupal stages, and ecdysone signaling continues to regulate sexual identity throughout adulthood via dme-let-7 [31]. In addition, dme-miR-124 controls male sexual differentiation by targeting the sex-specific splicing factor transformer in the sex-determination pathway [32]*. miR-124 mutant male flies produce aberrant pheromone levels, resulting in reduced mating success and receptivity by female flies, and increased male–male courtship. Furthermore, miRNA expression is altered in multiple tissues post-mating following receipt of sex peptide (SP) from male flies [33]. Female flies lacking miRNAs (dme-miR-279, dme-miR-317, dme-miR-278, dme-miR-184) display altered sexual receptivity responses to SP.
The female honeybee has been of particular interest due to its ability to exist as two different phenotypes (castes)—the worker and the queen bee—while sharing the same genotypic background. miRNAs have been shown to play a role in cast determination in these bees (Ap. mellifera ligustica); miRNAs are seemingly more abundant in worker jelly than royal jelly [34]. Addition of miRNAs, most notably ame-miR-184, to the food of queen larvae impacts the morphology of the adult bee to that of the worker phenotype and results in dynamic changes in mRNA expression profiles.
Oogenesis and Embryogenesis
A number of miRNAs have been shown to function in oogenesis and embryogenesis. In Drosophila, dme-miR-989 is active in somatic follicle cells and plays a role in border cell migration toward the oocyte during oogenesis [35]. Bithorax-Complex (BX-C) HOX cluster miRNAs—dme-miR-iab-4 and dme-miR-iab-8—regulate neural patterning and reproduction in Drosophila by restricting HOX gene targets [36]*. Furthermore, female dme-miR-124 knockout flies display reduced fertility, possibly due to behavioral defects brought about by abnormal NMJ formation [29]. Mutant flies with a deletion of the miR-iab-4/8 hairpin are sterile and lay no eggs due to defective passage of eggs through the reproductive tract from the ovary. Lack of dme-miR-282 in Drosophila results in a reduction of viability and egg-laying due to elevated apoptotic activity in the ovary originating from the soma as a result of neurological defects [37].
In the mosquito Aedes albopictus, aal-miR-286b is highly expressed in the embryo, and depletion results in a delayed hatching rate, clearly suggesting its important role in embryogenesis [26]. In the silk worm, it was shown that bmo-miR-1a-3p may function to down-regulate bmVMP23, a vitelline membrane protein that may contribute to the integrity and structure of the egg, via near-perfect binding to target sites within its 3′ UTR [38]. In Locusta migratoria, depletion of Argonaute-1 adult females causes a pronounced decrease in the expression of vitellogenin (Vg), an important yolk protein precursor (YPP), greatly impacting oocyte maturation and ovarian development [39].
Several miRNAs have been shown to function in anautogenous mosquito species to regulate events related to blood feeding and reproduction. One recent study has shown that the lineage-specific miRNA, miR-1174, displays predominant expression in the posterior midgut in two blood-feeding mosquito species—Aedes aegypti and Anopheles gambiae [40]*. Depletion of miR-1174 in Ae. aegypti and An. gambiae demonstrated that miR-1174 is critical for sugar absorption, fluid excretion, blood intake and digestion, and, as a result, egg development. Aae-miR-1174 was shown to target the highly conserved Serine hydroxymethyltransferase (SHMT), indicating that lineage-specific miRNAs have the ability to modulate conserved functions to influence lineage-specific events. In Ae. aegypti, aae-miR-8 is induced in the fat body post blood meal and functions in the Wingless (Wg) signaling pathway by targeting Secreted Wg-interacting molecule (swim) to regulate reproductive processes [41]*. In the first application of miR-SP combined with the Gal4-UAS system in Ae. aegypti, fat body-specific depletions of aae-miR-8 resulted in defects related to egg development and deposition, possibly due to defects in secretion of YPPs. Furthermore, in Ae. albopictus, the mosquito-specific miR-1891 plays a role in blood meal-associated events [26]. Depletion of aal-miR-1890 results in reduced fecundity in adult insects, indicating a role in blood digestion or egg development.
It has been demonstrated that miRNAs participate in maternal effects caused by cytoplasmic inheritance. More recently, miRNAs were shown to be maternally deposited in Drosophila unfertilized oocytes [42]*. The conserved miR-34 is maternally inherited in Drosophila and zebrafish (Danio rerio), and knockdown of maternal dre-miR-34 in unfertilized zebrafish oocytes leads to neuronal growth abnormalities. The majority of maternally inherited miRNAs are highly adenylated at their 3 ends during early development in Drosophila, facilitating down-regulation during maternal-to-zygotic transition [43]**.
Host-Pathogen Interactions and Immunity
Several miRNAs have been demonstrated to function in processes related to insect host-pathogen interactions and immunity. A recent report in Drosophila indicated that null miR-8 mutant flies display abnormal expression of Drosomycin and increased lethality [44]. dme-miR-8 miRNA was shown to play a role in maintaining immune homeostasis by targeting multiple effectors in the Toll immune pathway, including Toll and Dorsal, and a member of the insulin signaling pathway, U-shaped [45]*. In the Cabbage Moth, Plutella xylostella, pxy-miR-8 regulates anti-microbial peptide production and innate immune homeostasis by targeting Serpin 27, a player in the Toll immune pathway [46]. The understanding of host-pathogen interactions in mosquitoes has received substantial attention because of their medical importance as disease vectors [47]. In Ae. aegypti, a recent study reported that aae-miR-375 is induced by a blood meal and targets cactus and REL1, two immune-related genes [48].
Introduction of the endosymbiotic bacterium Wolbachia pipientis into the mosquito is considered a promising biocontrol strategy for mosquito-borne disease due to its ability to interfere with viral replication. Wolbachia manipulates Ae. aegypti miRNAs after infection to promote its stability. In Wolbachia-infected mosquito cells, Ago-1 translocation to the nucleus is blocked because of an increase in aae-miR-981, which regulates importin β-4, an important gene in Ago-1 distribution [49]*. The absence of Ago-1 in the nucleus may affect trafficking of miRNAs, which, in turn, facilitates stability of Wolbachia. Furthermore, Wolbachia-infected Ae. aegypti display exclusive expression of the mosquito-specific aae-miR-2940 [50–52]. Three genes—metalloprotease m41 FtsH [50]*, DNA methyltransferase (AaDnmt2) [51] and arginine methyltransferase 3 (AaArgM3) [52]—have recently been identified as targets of aae-miR-2940. While Wolbachia-infected mosquitoes display repressed AaDnmt2 via aae-miR-2940, dengue virus (DENV) induces AaDnmt2 expression [51]. Metalloprotease m41 FtsH mRNA is stabilized by aae-miR-2940, and its depletion reduces the metalloprotease m41 FtsH levels and restricts West Nile virus (WNV) replication [50, 53]. Overexpression of AaDnmt2 inhibits Wolbachia replication and promotes Dengue virus (DENV) replication. Although silencing of AaArgM3 has been shown to have no effect on DENV replication, mosquito cells display reduction in Wolbachia replication [52]. It has been demonstrated that metalloprotease m41 FtsH is required for efficient WNV replication.
Further elucidating miRNA function in viral immunity, Ae. albopictus aal-miR-252 was shown to be an inhibitor of DENV Envelope protein, a protein required for cell attachment and viral entry [54]. This interaction between the host miRNA, aal-miR-252, and the viral genome implies that aal-miR-252 may act as an antiviral regulator in Ae. albopictus. In contrast, in the same species, an abundant midgut-specific miRNA, aal-miR-281, promotes DENV replication [55]. Inhibition of this mRNA in female mosquitoes led to a significant reduction in DENV-2 abundance. This miRNA was also shown to potentially target the DENV-2 genomic 5′ UTR SLA region, indicating an interaction between a host miRNA and the DENV genome, with DENV exploiting host miRNAs for the benefit of its replication. Furthermore, miRNAs were detected in Ae. aegypti and Ae. albopictus saliva, 31 of which were previously unidentified and were designated as novel [56]. The effect of CHIKV infection in miRNA expression in the saliva resulted in the up-regulation of 59 and 30 known miRNAs in Ae. aegypti and Ae. albopictus, respectively.
In addition to miRNA regulation of anti-viral immunity, miRNAs have been shown to play some roles in malaria infection. Recent research has identified that aga-miR-305 is involved in function of anti-Plasmodium defense in An. gambiae and regulation of mosquito gut microbiota [57]. Inhibition of aga-miR-305 results in increased resistance to P. falciparum infection and suppression of the midgut microbiota; aga-miR-305 mimics increased susceptibility to P. falciparum infection and results in expansion of midgut microbiota. Furthermore, several immune effectors are computationally predicted as candidate targets of aga-miR-305.
Conclusions
These recent studies signify the importance of miRNAs in regulating insect physiology, including molting, metamorphosis, neuronal development, behavior, oogenesis, embryogenesis, and host-pathogen interactions. With the advent of improved bioinformatic and genetic tools required for the study of miRNAs in non-model organisms, a greater understanding of the roles of miRNAs in these insects is beginning to emerge. While miRNAs have been studied intensely in model organisms with well-established classical genetics and transgenic tools, until recently little work had been done to investigate their functional role in non-drosophilid insects. Limitation of genetic tools remains a severe hindrance in understanding the role of miRNAs in non-model organisms. However, the recent application of Gal4-UAS systems with the miR-SP transgenic method has begun to shed new light on understanding miRNA function in non-drosophilid insects [19, 41]. Additionally, investigations of the CRISPR-Cas9 system offers promising results that will allow genome engineering previously not feasible for non-model organisms [58–62].
Highlights.
miRNAs are non-coding RNAs that post-transcriptionally regulate gene expression
Conserved and lineage-specific miRNAs regulate insect physiology and development
Studying miRNA function is a growing trend in non-drosophilid insects
Advances in genetics and transgenic tools influence miRNA functional analysis
Acknowledgments
This work was supported by National Institutes of Health Grant R01 AI113729 (to A.S.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Keira J. Lucas, Email: kneum001@ucr.edu.
Bo Zhao, Email: bzhao002@ucr.edu.
Shiping Liu, Email: shiping.liu@ucr.edu.
Alexander S. Raikhel, Email: alexander.raikhel@ucr.edu.
References
- 1.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 3.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 4.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 5.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15(20):2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293(5538):2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukaya T, Iwakawa HO, Tomari Y. MicroRNAs block assembly of eIF4F translation initiation complex in Drosophila. Mol Cell. 2014;56(1):67–78. doi: 10.1016/j.molcel.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA. 2012;3(3):311–330. doi: 10.1002/wrna.121. [DOI] [PubMed] [Google Scholar]
- 9.Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, Chung J, Kim VN. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell. 2009;139(6):1096–1108. doi: 10.1016/j.cell.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 10.Jin H, Kim VN, Hyun S. Conserved microRNA miR-8 controls body size in response to steroid signaling in Drosophila. Genes Dev. 2012;26(13):1427–1432. doi: 10.1101/gad.192872.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee GJ, Jun JW, Hyun S. MicroRNA miR-8 regulates multiple growth factor hormones produced from Drosophila fat cells. Insect Mol Biol. 2015;24(3):311–318. doi: 10.1111/imb.12156. [DOI] [PubMed] [Google Scholar]
- 12.Kim K, Vinayagam A, Perrimon N. A rapid genome-wide microRNA screen identifies miR-14 as a modulator of Hedgehog signaling. Cell Rep. 2014;7(6):2066–2077. doi: 10.1016/j.celrep.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson C, Ambros V, Baehrecke EH. miR-14 regulates autophagy during developmental cell death by targeting ip3-kinase 2. Mol Cell. 2014;56(3):376–388. doi: 10.1016/j.molcel.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W, Cohen SM. The Hippo pathway acts via p53 and microRNAs to control proliferation and proapoptotic gene expression during tissue growth. Biol Open. 2013;2(8):822–828. doi: 10.1242/bio.20134317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam V, Tokusumi T, Tokusumi Y, Schulz RA. bantam miRNA is important for Drosophila blood cell homeostasis and a regulator of proliferation in the hematopoietic progenitor niche. Biochem Biophys Res Commun. 2014;453(3):467–472. doi: 10.1016/j.bbrc.2014.09.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foronda D, Weng R, Verma P, Chen YW, Cohen SM. Coordination of insulin and Notch pathway activities by microRNA miR-305 mediates adaptive homeostasis in the intestinal stem cells of the Drosophila gut. Genes Dev. 2014;28(21):2421–2431. doi: 10.1101/gad.241588.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aparicio R, Simoes Da Silva CJ, Busturia A. MicroRNA miR-7 contributes to the control of Drosophila wing growth. Dev Dyn. 2015;244(1):21–30. doi: 10.1002/dvdy.24214. [DOI] [PubMed] [Google Scholar]
- 18.Jiang J, Ge X, Li Z, Wang Y, Song Q, Stanley DW, Tan A, Huang Y. MicroRNA-281 regulates the expression of ecdysone receptor (EcR) isoform B in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2013;43(8):692–700. doi: 10.1016/j.ibmb.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Ling L, Ge X, Li Z, Zeng B, Xu J, Aslam AF, Song Q, Shang P, Huang Y, Tan A. MicroRNA Let-7 regulates molting and metamorphosis in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2014;53:13–21. doi: 10.1016/j.ibmb.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 20.Loya CM, McNeill EM, Bao H, Zhang B, Van Vactor D. miR-8 controls synapse structure by repression of the actin regulator enabled. Development. 2014;141(9):1864–1874. doi: 10.1242/dev.105791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131(1):136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 22.Rubio M, Montañez R, Perez L, Milan M, Belles X. Regulation of atrophin by both strands of the mir-8 precursor. Insect Biochem Mol Biol. 2013;43(11):1009–1014. doi: 10.1016/j.ibmb.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Chen J, Liang Z, Liang Y, Pang R, Zhang W. Conserved microRNAs miR-8-5p and miR-2a-3p modulate chitin biosynthesis in response to 20-hydroxyecdysone signaling in the brown planthopper, Nilaparvata lugens. Insect Biochem Mol Biol. 2013;43(9):839–848. doi: 10.1016/j.ibmb.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Zhang YL, Huang QX, Yin GH, Lee S, Jia RZ, Liu ZX, Yu NT, Pennerman KK, Chen X, Guo AP. Identification of microRNAs by small RNA deep sequencing for synthetic microRNA mimics to control Spodoptera exigua. Gene. 2015;557(2):215–221. doi: 10.1016/j.gene.2014.12.038. [DOI] [PubMed] [Google Scholar]
- 25.Wang YL, Yang ML, Jiang F, Zhang JZ, Kang L. MicroRNA-dependent development revealed by RNA interference-mediated gene silencing of LmDicer1 in the migratory locust. Insect Sci. 2013;20(1):53–60. doi: 10.1111/j.1744-7917.2012.01542.x. [DOI] [PubMed] [Google Scholar]
- 26.Puthiyakunnon S, Yao Y, Li Y, Gu J, Peng H, Chen X. Functional characterization of three MicroRNAs of the Asian tiger mosquito, Aedes albopictus. Parasit Vectors. 2013;6(1):230. doi: 10.1186/1756-3305-6-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quah S, Hui JH, Holland PW. A Burst of miRNA Innovation in the Early Evolution of Butterflies and Moths. Mol Biol Evol. 2015;32(5):1161–1174. doi: 10.1093/molbev/msv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang M, Wei Y, Jiang F, Wang Y, Guo X, He J, Kang L. MicroRNA-133 inhibits behavioral aggregation by controlling dopamine synthesis in locusts. PLoS Genet. 2014;10(2):e1004206. doi: 10.1371/journal.pgen.1004206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, Feng T, Wan Q, Kong Y, Yuan L. miR-124 controls Drosophila behavior and is required for neural development. Int J Dev Neurosci. 2014;38:105–12. doi: 10.1016/j.ijdevneu.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Cristino AS, Barchuk AR, Freitas FC, Narayanan RK, Biergans SD, Zhao Z, Simoes ZL, Reinhard J, Claudianos C. Neuroligin-associated microRNA-932 targets actin and regulates memory in the honeybee. Nat Commun. 2014;5(5529) doi: 10.1038/ncomms6529. [DOI] [PubMed] [Google Scholar]
- 31.Fagegaltier D, König A, Gordon A, Lai EC, Gingeras TR, Hannon GJ, Shcherbata HR. A genome-wide survey of sexually dimorphic expression of Drosophila miRNAs identifies the steroid hormone-induced miRNA let-7 as a regulator of sexual identity. Genetics. 2014;198(2):647–668. doi: 10.1534/genetics.114.169268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weng R, Chin JS, Yew JY, Bushati N, Cohen SM. miR-124 controls male reproductive success in Drosophila. Elife. 2013;2:e00640. doi: 10.7554/eLife.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fricke C, Green D, Smith D, Dalmay T, Chapman T. MicroRNAs influence reproductive responses by females to male sex peptide in Drosophila melanogaster. Genetics. 2014;198(4):1603–1619. doi: 10.1534/genetics.114.167320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo X, Su S, Skogerboe G, Dai S, Li W, Li Z, Liu F, Ni R, Guo Y, Chen S, Zhang S, Chen R. Recipe for a busy bee: microRNAs in Honey Bee caste determination. PLoS One. 2013;8(12):e81661. doi: 10.1371/journal.pone.0081661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kugler JM, Verma P, Chen YW, Weng R, Cohen SM. miR-989 is required for border cell migration in the Drosophila ovary. PLoS One. 2013;8(7):e67075. doi: 10.1371/journal.pone.0067075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garaulet DL, Castellanos MC, Bejarano F, Sanfilippo P, Tyler DM, Allan DW, Sánchez-Herrero E, Lai EC. Homeotic function of Drosophila Bithorax-complex miRNAs mediates fertility by restricting multiple Hox genes and TALE cofactors in the CNS. Dev Cell. 2014;29(6):635–648. doi: 10.1016/j.devcel.2014.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vilmos P, Bujna A, Szuperák M, Havelda Z, Várallyay É, Szabad J, Kucerova L, Somogyi K, Kristó I, Lukácsovich T, Jankovics F, Henn L, Erdélyi M. Viability, longevity, and egg production of Drosophila melanogaster are regulated by the miR-282 microRNA. Genetics. 2013;195(2):469–480. doi: 10.1534/genetics.113.153585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen A, Xia D, Qiu Z, Gao P, Tang S, Shen X, Zhu F, Zhao Q. Expression of a vitelline membrane protein, BmVMP23, is repressed by bmo-miR-1a-3p in silkworm, Bombyx mori. FEBS Lett. 2013;587(7):970–975. doi: 10.1016/j.febslet.2013.02.030. [DOI] [PubMed] [Google Scholar]
- 39.Song J, Guo W, Jiang F, Kang L, Zhou S. Argonaute 1 is indispensable for juvenile hormone mediated oogenesis in the migratory locust, Locusta migratoria. Insect Biochem Mol Biol. 2013;43(9):879–887. doi: 10.1016/j.ibmb.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Liu S, Lucas KJ, Roy S, Ha J, Raikhel AS. Mosquito-specific microRNA-1174 targets serine hydroxymethyltransferase to control key functions in the gut. Proc Natl Acad Sci U S A. 2014;111(40):14460–14465. doi: 10.1073/pnas.1416278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucas KJ, Roy S, Ha J, Gervaise AL, Kokoza VA, Raikhel AS. MicroRNA-8 targets the Wingless signaling pathway in the female mosquito fat body to regulate reproductive processes. Proc Natl Acad Sci U S A. 2015;12(5):1440–1445. doi: 10.1073/pnas.1424408112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soni K, Choudhary A, Patowary A, Singh AR, Bhatia S, Sivasubbu S, Chandrasekaran S, Pillai B. miR-34 is maternally inherited in Drosophila melanogaster and Danio rerio. Nucleic Acids Res. 2013;41(8):4470–4480. doi: 10.1093/nar/gkt139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee M, Choi Y, Kim K, Jin H, Lim J, Nguyen TA, Yang J, Jeong M, Giraldez AJ, Yang H, Patel DJ, Kim VN. Adenylation of maternally inherited microRNAs by Wispy. Mol Cell. 2014;56(5):696–707. doi: 10.1016/j.molcel.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi IK, Hyun S. Conserved microRNA miR-8 in fat body regulates innate immune homeostasis in Drosophila. Dev Comp Immunol. 2012;37(1):50–54. doi: 10.1016/j.dci.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 45.Lee GJ, Hyun S. Multiple targets of the microRNA miR-8 contribute to immune homeostasis in Drosophila. Dev Comp Immunol. 2014;45(2):245–251. doi: 10.1016/j.dci.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 46.Etebari K, Asgari S. Conserved microRNA miR-8 blocks activation of the Toll pathway by upregulating Serpin 27 transcripts. RNA Biol. 2013;10(8):1356–1364. doi: 10.4161/rna.25481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lucas KJ, Myles KM, Raikhel AS. Small RNAs: a new frontier in mosquito biology. Trends Parasitol. 2013;29(6):295–303. doi: 10.1016/j.pt.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hussain M, Walker T, O’Neill SL, Asgari S. Blood meal induced microRNA regulates development and immune associated genes in the Dengue mosquito vector, Aedes aegypti. Insect Biochem Mol Biol. 2013;43(2):146–152. doi: 10.1016/j.ibmb.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 49.Hussain M, O’Neill SL, Asgari S. Wolbachia interferes with the intracellular distribution of Argonaute 1 in the dengue vector Aedes aegypti by manipulating the host microRNAs. RNA Biol. 2013;10(12):1868–1875. doi: 10.4161/rna.27392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hussain M, Frentiu FD, Moreira LA, O’Neill SL, Asgari S. Wolbachia uses host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc Natl Acad Sci U S A. 2011;108(22):9250–9255. doi: 10.1073/pnas.1105469108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang G, Hussain M, O’Neill SL, Asgari S. Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in Aedes aegypti. Proc Natl Acad Sci U S A. 2013;110 (25):10276–10281. doi: 10.1073/pnas.1303603110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang G, Hussain M, Asgari S. Regulation of arginine methyltransferase 3 by a Wolbachia-induced microRNA in Aedes aegypti and its effect on Wolbachia and dengue virus replication. Insect Biochem Mol Biol. 2014;53:81–88. doi: 10.1016/j.ibmb.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 53.Slonchak A, Hussain M, Torres S, Asgari S, Khromykh AA. Expression of mosquito microRNA Aae-miR-2940-5p is downregulated in response to West Nile virus infection to restrict viral replication. J Virol. 2014;88(15):8457–8467. doi: 10.1128/JVI.00317-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan H, Zhou Y, Liu Y, Deng Y, Chen X. miR-252 of the Asian tiger mosquito Aedes albopictus regulates dengue virus replication by suppressing the expression of the dengue virus envelope protein. J Med Virol. 2014;86(8):1428–1436. doi: 10.1002/jmv.23815. [DOI] [PubMed] [Google Scholar]
- 55.Zhou Y, Liu Y, Yan H, Li Y, Zhang H, Xu J, Puthiyakunnon S, Chen X. miR-281, an abundant midgut-specific miRNA of the vector mosquito Aedes albopictus enhances dengue virus replication. Parasit Vectors. 2014;7(1):488. doi: 10.1186/s13071-014-0488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maharaj PD, Widen SG, Huang J, Wood TG, Thangamani S. Discovery of mosquito saliva microRNAs during CHIKV infection. PLoS Negl Trop Dis. 2015;9(1):e0003386. doi: 10.1371/journal.pntd.0003386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dennison NJ, BenMarzouk-Hidalgo OJ, Dimopoulos G. MicroRNA-regulation of Anopheles gambiae immunity to Plasmodium falciparum infection and midgut microbiota. Dev Comp Immunol. 2015;49(1):170–178. doi: 10.1016/j.dci.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Basu S, Aryan A, Overcash JM, Samuel GH, Anderson MA, Dahlem TJ, Myles KM, Adelman ZN. Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti. Proc Natl Acad Sci U S A. 2015;112(13):4038–4043. doi: 10.1073/pnas.1502370112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong S, Lin J, Held NL, Clem RJ, Passarelli AL, Franz AW. Heritable CRISPR/Cas9-Mediated Genome Editing in the Yellow Fever Mosquito, Aedes aegypti. PLoS One. 2015;10(3):e0122353. doi: 10.1371/journal.pone.0122353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kistler KE, Vosshall LB, Matthews BJ. Genome Engineering with CRISPR-Cas9 in the Mosquito Aedes aegypti. Cell Rep. 2015;S2211–1247(15):00262–00264. doi: 10.1016/j.celrep.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei W, Xin H, Roy B, Dai J, Miao Y, Gao G. Heritable genome editing with CRISPR/Cas9 in the silkworm, Bombyx mori. PLoS One. 2014;9(7):e101210. doi: 10.1371/journal.pone.0101210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y, Li Z, Xu J, Zeng B, Ling L, You L, Chen Y, Huang Y, Tan A. The CRISPR/Cas system mediates efficient genome engineering in Bombyx mori. Cell Res. 2013;23(12):1414–1416. doi: 10.1038/cr.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]