Abstract
Species survival depends on the faithful replication of genetic information, which is continually monitored and maintained by DNA repair pathways thatcorrect replication errors and the thousands of lesions that arise daily from the inherent chemical lability of DNA and the effects of genotoxic agents. Nonetheless,neutrally evolving DNA (not under purifying selection) accumulates base substitutions with time (the neutral mutation rate). Thus, repair processes are not 100% efficient. The neutral mutation rate varies both between and within chromosomes. For example it is 10 – 50 fold higher at CpGsthan at non-CpG positions. Interestingly, the neutral mutation rate at non-CpG sites is positively correlated with CpG content. Althoughthe basis of this correlation was not immediately apparent,some bioinformatic results were consistent with the induction of non-CpGmutations byDNA repairat flanking CpG sites. Recent studies with a model system showed that in vivo repair of preformed lesions (mismatches, abasic sites, single stranded nicks) can in factinduce mutations in flanking DNA. Mismatch repair (MMR) is an essential component for repair-induced mutations, which can occur as distant as 5 kb from the introduced lesions. Most, but not all, mutations involved the C of TpCpN (G of NpGpA) which is the target sequence of the C-preferringsingle-stranded DNA specific APOBEC deaminases. APOBEC-mediated mutations are not limited to our model system: Recent studies by others showed that some tumors harbor mutations with the same signature, as can intermediates in RNA-guided endonuclease-mediated genome editing. APOBEC deaminases participate in normal physiological functions such as generating mutations that inactivate viruses or endogenous retrotransposons, or that enhance immunoglobulin diversity in B cells. The recruitment of normally physiological errorprone processes during DNA repairwould have important implications for disease, aging and evolution. This perspective briefly reviews both the bioinformatic and biochemical literature relevant to repair-induced mutagenesis and discussesfuture directions required to understand the mechanistic basis of this process.
Keywords: DNA repair, mutagenesis, APOBEC deaminase, base excision repair, mismatch repair, cancer, CRISPR/Cas
1. Introduction
Species survival depends on maintaining the integrity of genetic information and to this end all organisms contain enzymatic pathways that sense and repair the numerous lesions that can afflict DNA[1-9]. However, genetic diversity (mutational differences within a population) is essentialforadaptive (and evolutionary) responses of a species to environmental exigencies. Mutations due to base substitutions are the most common source of genetic diversity and divergence (mutational differences between species) [10]. The accumulation rate of base mutations in a given population (species) depends on natural selection and chance (genetic drift),which are influenced by demographic features such as population size and migration. However, the rate at which they arise in a genomeis determined biochemically; i.e., a consequenceof the frequency with which DNA replication errors and damage occur, and the efficiency and fidelity of their repair. Although it had been commonly assumed that base substitutions were singular events, recent results indicate that 1-2% can occur as multiples, usually doublets, during normal development [11, 12].
2. Bioinformatic data suggesting error-prone repair
2.1 Variation in the neutral mutation rate
The accumulation of base mutations that are not subject to natural selection is consideredthe neutral mutation rate [13, 14], and numerous studies showed that in a given species this rate can vary both between and within chromosomes froma megabase scale tothat of neighboring bases.For example, the neutral mutation rate on the Y and X chromosomes differ from each other and from autosomes, being highest on the Y and lowest on the X. The ratio of these mutation rates was originally modeled as a function of the relative number of replications undergone by the Y (continuous throughout life) and the X (ceasing at egg maturity in the embryo) chromosomes[15-17]. This model implies thatuncorrected DNA replication errors are the major driverof the neutral mutation rate. However, this conclusion was not supported by subsequent estimates of the neutral mutation rates on the X and Y chromosomes[18-20],and others suggested that differences betweenthe mutational processes or DNA repair efficiency in male and female gametes could account for Y/X differences[21-23].
Intra-autosomal differences in theneutral mutation rate,whichare clearly independent of chromosomal replication, have been correlated withnumerous factors includingcompositionalfeatures of the genome: e.g., base type, sequence context and GC content. These are consistent with the inherent chemical properties of DNA. For example, C is the most mutable base in mammals [10], likely a function of its inherent susceptibility to spontaneous hydrolytic deamination to U[24]. Its mutation rate is considerably increased when followed by a G due to the preferential methylation of C in this context (CpG). This modification substantially increases the C-deamination rate to generate a T/G mismatch [25-27]. Consequently, CpG mutations comprise a third of all single nucleotide polymorphism (SNPs), are an important cause of genetic diseases and cancer and include almost half of the monogenic causes of X-linked diseases [28, 29].And finally, thatC deamination only occurs on single-stranded DNA [30]likely accounts for the negative correlation between the neutral mutation rate of CpGsandGC content [31, 32].
Also, the neutral mutation rate has been correlated with structural and functional features of the genome such as telomeric or centromeric location, nucleosome occupancy,replication timing, recombination, and transcription[33-39]. A recent comprehensive analysis of 40,000 spontaneous and presumed neutral mutations in yeast [40] revealed variation in mutation rates generally consistent with those of the earlier studies. Some of these correlations are explicable by the physical status of DNA that result from the above processes. For example, the negative correlation of mutation rate with nucleosome occupancy would be consistent with the putative protection of DNA when packaged in nucleosomes [39]. Furthermore,the positive correlation of mutation rates with transcription, recombination or replication could be explained by the increased susceptibility to damage of the transient single-strandedDNA present during these processes[41-45]. In fact,the single strandedness that occurs duing replication could explain male driven mutations. In particular, a recent study of 78 Icelandic parent–offspring triosstudies showed an increase in male derived mutations with the age of the father[46]. Thatthe mutation rate at CpGs was18 times that of any other site strongly implies that the major source of male driven mutations is DNA damage rather than the fidelity of DNA replication (also see ref. [23]).
2.2 Correlation between neutral mutation rate and CpG content
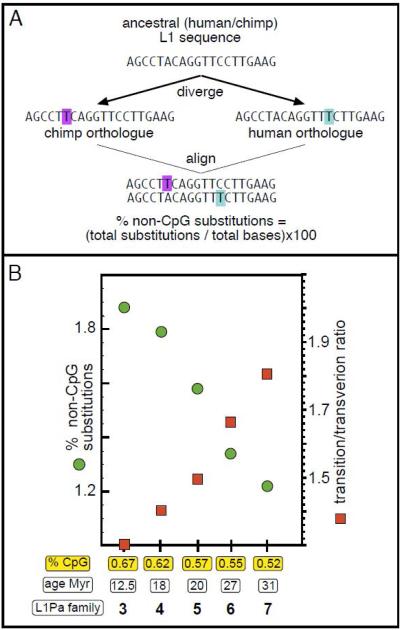
In contrast to the foregoing, the basis of the positive correlationbetween CpG content and the neutral mutation rate of non-CpG flanking DNA was not readily apparent.In fact, it was originally assumed to represent the joint manifestation of high CpG content and some other unidentified cause of mutations [33, 36, 37].However, comparisons of the frequency of base substitutions (i.e., divergence) averaged overmany thousands of pairs of neutrally evolving orthologous autosomal sequences in chimpanzee and human that differed only in CpG content strongly suggested that CpGsdirectly affectthe non-CpG mutation rate[20, 47]. These orthologue pairs were inserted into the common ancestor of chimpanzees and humans at different timesby different families of the L1 retrotransposon that had been active and gone extinct prior to the divergence of human and chimpanzee from their common ancestor[33, 48].Thus, while very similar in sequence, they differ in their content of CpG, which had been converted with the passage of time to TpG/CpA[10, 49, 50]. Because theorthologue pairs of all the members of any given L1 family were randomly inserted in all autosomes,their mean divergencereflects the average effects of allchromosomal contexts on the neutral mutation rate. Thus,CpG content is theonlyvariable common to all members of a given L1 family that could affectits mean divergence, i.e., mutation rate(Figure 1A).
Figure 1.
Bioinformatic data suggesting a mutagenic effect of DNA repair
(A) Alignment of theoretical human and chimpanzee L1 orthologues with base substitutionshighlighted. (B) Relationship of the percent non-CpG base substitutions and transition/transversion ratio for different L1Pa family members. The CpG content (%) and age of the indicated L1Pa family is shown. Compiled from data in Figure 2A and 2B in [47].
Figure 1 shows the straightforward methodology for determining the relationship between CpG content and mutation rate. Pairs of orthologous L1 inserts representing different ancestral L1 familieswere retrieved from the chimpanzee and human genome data bases.They were aligned, the CpG sites (or those sites that had been derived from CpG sites) were masked, and the non-CpG substitutions between the orthologous pairs were counted (top panel). The % non-CpG substitutions (the number of mutations that accumulated since chimpanzees and humans split from their common ancestor) are plotted as a function of the CpG content of their respective orthologue pairs (bottom panel), which was the same for either the chimp or human orthologue of a given family [20, 47]. The base substitution rate decreased as a function of the decrease in CpG content (Figure 1). Theratio of transition and transversion mutations was also a function of CpG content. As transitions and transversions are generated by different mechanisms, the mutational environment of the L1 orthologues is also a function of CpG content. We could not identify any other genomic variable (e.g., transcriptional orientation, recombination rate, etc.) except CpG content that was correlated with either the substitution rate or transition / transversion ratio of the L1 orthologues. Thus, thesefindings strongly implicate a direct effect of CpG (methyl-CpG) on these mutational processes.
One explanation that could account forsuch a direct effectisrelated to the fact thatmethyl-CpG sites are mutational hotspots and thus foci of intense DNA repair. For that matter, but to a lesser extent, so are un-methylated Cs. This is due to spontaneous hydrolytic deamination of methyl-C to T (or C to U)[10, 23, 27, 51]. The ensuing T/G or U/G mispairs are substrates for base excision repair (BER), which is usually error free(see section 3.1) [2, 4, 9, 52, 53]. However,in some instances BER-processed U/G mispairs serve as the entry point for mismatch repair (MMR) to carry out an essentialstep in the physiological error prone process of somatic hypermutation (SHM).This non-canonical MMR (canonical MMR repairs mismatches that arise during DNA replication) was thoughtto be limited to immunoglobulin producing (lymphoid B) cells where it enhances immunoglobulin diversity[54-56]. However, others recently showed that non-canonical MMR can also generate somatic mutations in non-lymphoidcells [57, 58].
To determine whether repair of T/G or U/G mismatches could induce mutations inflanking DNA, we insertedthese mismatchesin a shuttle vector that is capable of replication in both mammalian and bacterial cells[59]. After transferinto various mammalian cell lines we determined the outcome and mutageniceffect of their repair [60]. In the next section we give a brief review of BER and MMR and outline the major elements of the experimental system to examine this issue.
3. Experimental demonstration that T/G and U/G repair renders flanking normal DNA susceptible to mutation
3.1 Brief summary of BER
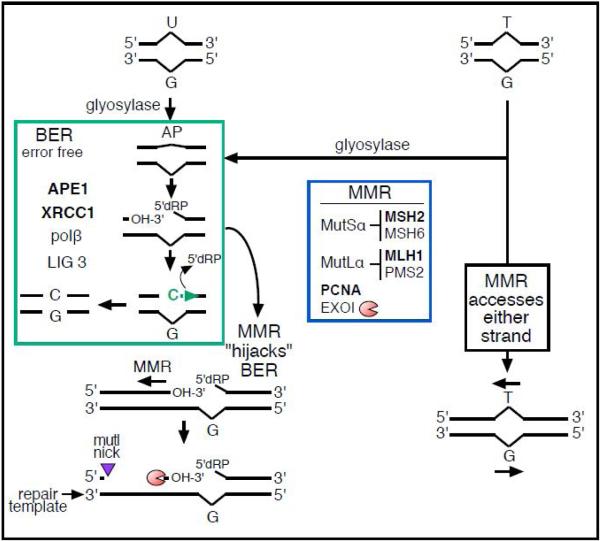
Figure 2 shows the major components of BERand presents simplified versions of how it processesT/G orU/G mismatches.Although cells contain numerous glycosylases that continually scan DNA for miss-paired or abnormal bases, the major ones that sense and excise mismatched T and U are thymine DNA glycosylase (TDG) and uracil-N glycosylase (UNG) respectively (glycosylase, in Figure 2),reviewed in [4]. UNG is highly specific for U-mispairs, howeverTDG is more promiscuous. For example, TDGcan also excise a T opposite a damaged A (e.g., hypoxanthine) or an abasic site [61] and a number of bases mismatched with G including U. Furthermore, the enzymology of TDG is more complex than that of UNG. TDG interacts with its substrate with higher affinitythan UNG, is subject to SUMOylation[4]and interacts with a number of other non-repair proteins including transcription factors [2]. In the end however,the ensuing abasic (apurinic/apyrimidinic – AP) site is passed on with varying efficiency,at least in vitro (UNG very efficiently, TDG very slowly),for further processing by the highly specific and precise apurinic/apyrimidinic endonuclease 1 (APE1)[62].
Figure 2.
BER and MMR
Glycosylases generate the substrate for the subsequent steps of the BER pathway (green box). Cleavage of the DNA 5’ of the ensuing abasic site (AP) by APE1generates a 3’-OH and a 5’dRP. The concerted removal of 5’dRP and insertion of dCMP by pol, followed by LIG3 are facilitated by XRCC1.The blue box encloses components of the MMR pathway. Non-canonical MMR (MutSαand MutLα/PCNA) can hijack U/G-BER intermediates and MutLαintroduces nicks 5’ of the lesion, after which EXO1 would generate gapped DNA 5’ of the lesion.The right side of the figure shows direct access of a T/G mismatch by MMR and either the G-strand or T-strand can serve as the repair template for T/G repair. Adapted from Figure 10, [60].
This step and subsequent ones in the BER pathway are coordinated and channeled by a series of hand-offs involving protein complexes, which protect the cell from the potentially toxic and mutagenic effects of the abasic site and its downstream products [52, 63-66]. The left side of Figure 2 illustrates single nucleotideBER, which processes the 3’-OH and 5’-deoxyribose phosphate (5’dRP) that were generated by APE1 cleavage. Polymerase (pol) catalyzes a concerted removal of 5’dRP and insertion of dCMP. BER is completedby ligation with ligase III (LIG3),and all these reactions are facilitated by the scaffolding protein, X-ray cross complementing factor1 (XRCCI), the foregoing references and reviewed in [9, 53].
3.2 Brief summary of canonical MMR
Canonical MMRis a high fidelity process that functions at replication forks (not illustrated in Figure 2)to remove miss-incorporated bases from nascent DNA strands[3, 5, 8]. Essential components include the heterodimer of MSH2 and MSH6 (MutS), which recognizes mismatches, and the heterodimer of MLH1 and PMS2 (MutLα), which accesses the mismatch-containing strand. This process[54-56] requires the multipurpose replication clamp, proliferating cell nuclear antigen (PCNA),reviewed in [67, 68]. PCNA also activates a latent endonuclease in MutLα[69, 70] that provides access for the EXO1 nuclease,which excises the mismatch-containing nascent strand in the 5’ to 3’ direction to expose the repair template for re-copying by a high fidelity DNA polymerase, such as pol δ.T/G mispairs could arise during replication and thus be a substrate for MMR, and studies in vitro showed that components of MMR can directly access T/G mismatches in a PCNA-dependent reaction [69, 70]. Thus T/G can be processed by both BER and MMR (Figure 2). On the other hand, most U-containing “mispairs” would likely arise via incorporation of dUMP opposite A, andare rapidly and efficiently removed by UNG2 and BER at the replication fork[71]. Therefore, U/G mispairs (that would arise from deamination of C)would not seem to be much of an issue for canonical MMR.
3.3 Brief summary of non-canonical MMR
MMR can generate substrates for a physiologic error-prone process in lymphoid (B) cells that contributes to the somatic hypermutation (SHM) that enhances immunoglobulin diversity. This process has been called non-canonical MMR, and in this instance MMR accesses BER processed U/G mismatches that had been generated by the activation induced cytidine deaminase (AID, a member of the AID/apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC) family of cytidine deaminases, which are specific for single stranded nucleic acids[72, 73]. In B cells deamination of C to U occurs on the transient single stranded regions that are generated during transcription. As Figure 2 shows MMR can hijack BER intermediates generated from U/Gmismatches and generates a substrate that could be subject to mutagenic processes[55]. In one instance, the strand exposed by EXO1 is copied by the error-prone DNA pol that is recruited by mono-ubiquitinated PCNA.Features of non-canonical MMR have been reproduced in vitro, including by extracts of non-lymphoid cells on substrates that contained U/G mispairs and nicks at abasic sites[57, 74]. Additionally,these studies implicated MMR in the mutagenesis exhibited by these non-lymphoid cells in vivo;more mutationswere produced in MMR-proficient than MMR-deficient cells that had been stressed with the alkylating agent, N-methyl-N-nitro-N-nitrosoguanidine (MNNG).
3.4 The mutagenic effect of repairing T/G or U/G mismatches
Non-replicative T/G or U/G mismatches would be substrates for BER, which as outlined above is a coordinated process that protects its intermediatesfrom exposure to DNA damage sensors or other repair processes that could introduce mutations in the DNA that flanks the mismatch. For exampleBER could become mutagenic if the protection process failed and allowedthe single strand break (SSB)that APE produced 5’ of the mismatch to became vulnerable to MMR as discussed in the foregoing paragraph and illustrated in Figures 2 and 3. There could be any number of causes for such failure, including an imbalance in the various metabolic or enzymatic components that either underlie the orderly progression of BER or compete for its DNA intermediates[52, 75]. But whatever the cause,MMR could expose the bottomnon-lesion containing strand(the repairtemplate) to at least two (not necessarily exclusive) processes that could introduce mutations in the normal flanking DNA: (1)Error-prone copying of the repair template by an error prone polymerase that was inappropriately recruited during gaprepair [76, 77]. (2)Damage of the repair template by chemical (oxidation, hydrolytic deamination) or enzymatic processes (APOBEC-mediated deamination) that would degrade its genetic information. Figure 3 also shows the generation of single stranded regions in T/G containing vectors that have been accessed directly by MMR.Direct access of MMR on T/G mispairs was demonstrated in vitro[57, 70]. We do not know the mechanistic details of this process, but as we discuss below, mutagenesis associated with T/G repair, while dependent on MMR, can occur independently of BER.
Figure 3.
Possible fates of repair template exposed by MMR
DNA repair pathways remove the naturally occurring mismatches such as T/G and U/G and generate a single stranded region (see text and Figure 2). The single stranded region, which serves as the template for DNA repair, can be potentially accessed by an error-prone polymerase or by APOBECs that deaminates TpC to TpU to generate an altered repair template. This template can also be potentially copied by an error-prone polymerase (or a high fidelity polymerase, shown in the figure) that could then undergo the illustrated processes to generate the possible mutational outcomes at the U position, the APOBEC product. The same mutagenic processes can take place on the repair template exposed by MMR that directly access a T/G mismatch. Adapted from Figure 10, [60].
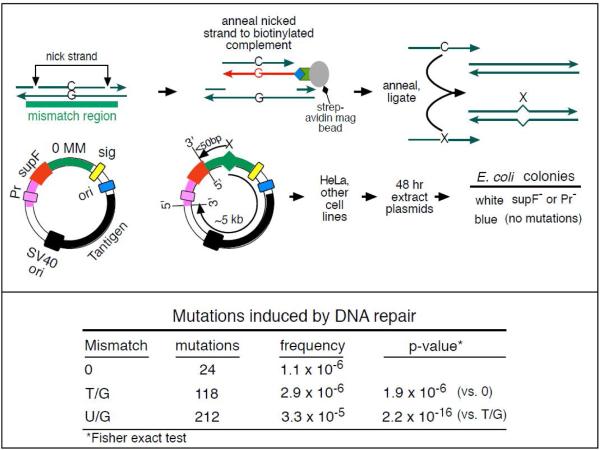
The top panel of Figure 4 shows the method of introducingpreformed lesions into a shuttle vector [78] modified by the presence ofa mismatch (MM) region that contains a pair of single strand cutting restriction enzymes specific for the top or bottom strands [60]. The method involves removing the nicked strand and replacing it with either an exact copy (0 mismatch)or lesion-containing strand where X could be a T, U, or another lesion such as an abasic site using the method described in [79]. After passage in theHeLa-JMcell line[60]the plasmids were rescued and screened by blue / white selection for mutations in the supF gene or its promoter (collectively, the reporter region). We sequenced the reporter region of all the white clones and several hundred blue clones. None of the latter contained mutations.
Figure 4.
Experimental system to detect the mutagenic effect of DNA repair. Mismatch region with nicking sites (vertical arrows) is digested with a single strand restriction enzyme on the top (or bottom) strand, and the nicked strand is removed by hybridization to a 5’ biotin (blue diamond) labeled complementary DNA (red). The hybrid is then tethered to a streptavidin (green polygon)-coated magnetic bead (gray oval). The purified gapped episome is reconstituted by ligation to its perfect complement (C containing strand) or an oligonucleotide that contains lesions(X containing strand) to generate vectors with a top (or bottom) strand lesion or its corresponding 0 MM control. These vectors are transferred to mammalians cells, harvested after 48 hours and subjected to blue/white screening. The Table at the bottom part of Figure gives the frequency of mutations induced by T/G and U/G repair. Adapted from Figure 1B, [60].
The table in Figure 4 shows that repair of T/G or U/G produced a statistically significant increase in the mutation frequency (number of mutations / number of nucleotides screened)in the reporter region over the 0 mismatch control. An abasic site or single-strandedbreak (SSB) induced as much mutagenesis as the U/G mismatch. The mutational effect can be propagated over the entire 5 kb of the plasmid, undiminished in intensity for T/G but showing a strong 3’ polarity for U/G (oran abasic site or SSB) – i.e., as the distance between the U and the cis 3’ end of the reporter region increase, the frequency of mutations in the reporter region decrease. Thus, repair of a top strand U/G (cis 3’ distance, ~ 50 bp) was far more mutagenic to the reporter region than a bottom strand G/U where this distance is ~5 kb. However, top and bottom strand T/Gs were equally mutagenic. SiRNA knockdowns of BER and MMR components (indicated in bold font in Figure 2) showed that both pathways were involved in repair-induced mutagenesis of U/G but that about half of the mutations induced by T/G repair were generated by just MMR alone (right side of Figures 2 and 3).
Most, but not all, of the mutations involved the C of TpCpN, read out as the G of NpGpA on the opposite strand. This mutational signature corresponds to the target of theTpC-preferring APOBEC C-deaminases (i.e., APOBEC3B, 3F, and 3C – abbreviated respectively, A3B, A3F, A3C). That A3B played a major role in generating these mutations was corroborated by a decrease in repair-induced mutagenesis by siRNA knockdown of the relevant APOBEC enzymes, its rescue by exogenous siRNA-resistant A3B, and by ChIP analysis,which showed the mismatch-dependent presence of A3B on the shuttle vector in amounts consistent with the extent of the mutagenic effect; i.e., roughly evenly distributed over the entire plasmid for T/G but with a strong 3’ polarity for U/G.
The bottom part of Figure 3 illustrates possiblemutagenic processes subsequent to the APOBEC catalyzed C deamination ofa repair template TpC that had been exposed by MMR. Thus, APOBEC acts downstream of the BER and MMR pathways which indicates that the intermediates generated by these pathways are vulnerable to APOBEC activity [60]. This scenario is opposite to the situation in antibody producing cells wherein error-prone non-canonical MMR is recruited to the sites of BER activity on AID-generated U residues in immunoglobulin genes.
Repair-induced mutagenesis was not limitedto HeLa-JM cells.Furthermore,the presence of APOBEC deaminases was not sufficient for U/G induced mutagenesis. Cell lines, including a second HeLa cell line, that contained ample A3B, A3C and A3F exhibited greatly reduced U/G induced mutagenesis.However, such cells retained at least 50% of the T/G induced mutagenesis, presumptive evidence that the BER-independent T/G mutagenic pathway was operative in all cells tested. Thus it seems that breakdown in the tight handoff between BER intermediates was a major factor in the mutagenic effect of U/G repair. That cells differ in their response to BER-dependent repair-induced mutagenesis indicates that the mutagenic effect of U/G (and abasic site and SSB) repairis not a function of the shuttle vector assay, but reflects the integrity of the BER pathway in various cells. That 35 – 55% of the APOBEC mutations induced by either T/G or U/G repair occur in multiplesis consistent with the processive behavior of the AID/ABOBEC family of deaminases on its substrates [80], reviewed in [55]. Although some unclustered mutations also exhibited a TpCpN mutational signature, some did not, indicating that non-APOBEC mediated mutational processes also accompanied the mutagenesis induced by repair, particularly of T/G.
4. APOBEC mediated mutagenesis in tumors and as a result of genome editing
4.1 Mutator phenotype of tumorscan include APOBEC-mediated mutations
An enhanced mutation rate in tumors, termed a mutator phenotype, hasbeen a longrecognizedbut not inevitable feature of variouscancers[reviewed in 81, 82, 83]. These mutations can include rearrangements, insertions / deletions (indels), and base substitutions, the later of whichexhibit a variety of mutational signatures [83-86]. The mutational mechanism(s) that account for these base substitution mutations are largely unknown, though some reflect the known DNA damaging effects of the predisposing carcinogenic agent. However, numerous recent studies showed that anywhere from a few to more than ninety percent of the base substitutionsin some tumors bear amutational signaturethat corresponds to ABOBEC3 C-deaminase target sites, i.e.,C mutations inTpCpN (or its complement). Thesemutations can accompany the progression of,and are thought topossibly initiate some cancers [85, 87-91]. Furthermore, consistent with the processivity of these deaminases, these mutations can appear as long strand coordinated clusters.
Given the strong preference for single-stranded DNA by these deaminases [reviewed in 73], an important issue is the source of the single stranded DNAAPOBEC3 substrate. With regard to strand coordinated clustered mutations, experiments in yeast showed that the transitory single strand regions that arise at replication forks or during double strand break repair could accumulate such mutations upon chronic alkylation of DNA [92]. And mutations with an APOBEC signature were observed in yeast at deliberately introduced double strand breaks in the presence of ectopically overexpressed AID/APOBEC deaminases [93]. We know of no experiments that address the source of the APOBEC substrates in mammalian cells. However, our findings using a model system in the absence of genotoxic agents indicates that such substrates can be generated during repair of U/G (or its downstream BER intermediates) and T/G (both via hijacking BER and direct access of MMR).Thousands of BER substrates and hundreds of T/G mismatches arise daily even in the absence genotoxic stress due to the inherent susceptibility of DNA to hydrolytic deamination or oxidation [27, 94].Additionally, mismatches betweenG and the oxidation derivatives of methyl-C that are generated during its physiological demethylation[95, 96]would also add to the DNA repair burden.A mutagenic effect of repairing these mismatches that occur during physiological cycling of epigenetic marks could possibly contribute to the high mutation rate of regulatory sequences involved in both disease [97] and evolutionary novelty [98].
4.2 Genome editing by RNA-guided DNA endonucleases can be susceptible to APOBEC mediated mutagenesis
The RNA-guided DNA endonuclease system (e.g., CRISPR-Cas9) is a highly specific genome-engineering tool, whereby a guide RNA (gRNA) directs the Cas9 endonuclease to the intended genomic site by hybridizingto target DNA [99, 100]. Endogenous DNA repair pathways repair the nicked target DNA that was introduced by the Cas9 endonucleaseto accomplish the desired genome editing[101, 102]. However, the CRISPR-Cas9 system can also generate unwanted off-target effects at a fairly high rate. This is because large eukaryotic genomes can containseveral potential binding sitesfor a particulargRNAgiven its propensity to tolerate some mismatches at its target site[103, 104]. A recent improvement of CRISPR-Cas9 system by Tsai et alshowed that fusing the FokI endonuclease to an endonuclease defective Cas9 (dCas9) can greatly decrease the chance of off-target effectsdue to the more stringent target site requirement of the dimeric FokI endonuclease[105]. However, this methodalso generateda previously unidentified side effect, base substitutions at the target site.Mostinvolved the C of TpC dinucleotides,the mutational signature of APOBEC cytidine deaminase-mediated mutations. As these experiments were carried out in human osteosarcoma (U2OS) and embryonic kidney (HEK293) cells, these results indicate that the SSB generated in this procedure in a near normal chromatin environment and cellular milieu is vulnerable toAPOBEC-mediated mutagenesis.
As DNA nicking is essential forgenome editing and APOBECSs are commonly expressed in most cells and tissues [106, 107], the possibility of APOBEC-mediated mutationsis an important concern for the highly precise Cas9-directed genome editing that is necessary for therapeutic gene correction [108-110].As most APOBEC3-mediated mutations occur on the C of TpCpN, these mutations could be avoided by targeting the relevant endonucleases to the sites that lack TpC dinucleotides. Thus, in those cases that permit some flexibility in the selection of endonuclease target sites, this strategy could eliminate such potentially deleterious base substitutions.However, this procedure would not necessarily eliminate non-APOBEC3 mediated or other mechanisms of repair-induced mutagenesis. While not compromising the use of the CRISPR-Cas9genome editing system for experimental studies, the threat of repair-induced mutations at sites of genetic remodeling could render it unsuitable for gene therapy.
5. Concluding remarks
In mammals, 60 – 90 % of CpGs are methylated [111, 112], which further enhances the already high rate of spontaneous deamination of C (to U), but in this case of methyl-C to T. Despite repair mechanisms biased to remove the T of a T/G mismatch [113, 114] and the error-prone polymerase,pol ι, that preferentially inserts a G opposite a T, which could eventually ensure replicative restoration of the C/G pair [115], restoration of TpG to C/G is only about 90% efficient [116]. Consequently, neutrally evolving CpGs are converted with time to TpG / CpA. However, essential CpGs that are under strong purifying selection are retained in the population as highly mutable sites[117] and thus, foci of chronic DNA repair.
We devised a model system to determine whether repair of preformed T/G (or U/G) mismatches could induce mutations in the normal DNA that flanked these introduced lesions[60]. The motivation for this study was to test the idea that the positive correlation between CpG content and the neutral mutation rate of non-CpG DNA was due to DNA repair processes that would be recruited to T/G (or U/G)mismatches and induce mutations in normal flanking DNA. We found the introduced lesions were invariably repaired and induced statistically significant numbers of mutations in flanking DNA, some of whichas distant as 5 kb from the introduced lesion. Most, but not all, of the mutations bore the mutational spectrum corresponding to the target sites of single-strand specific, C-preferring APOBEC deaminases. Recently, APOBEC-mediated mutations have received considerable attention in cancer studies as they can account for anywhere from a few to ninety percent of the mutational load (i.e., mutator phenotype) that many tumors acquire. The mechanism that renders the tumor genome susceptible to the C-preferring APOBEC deaminase is not known, but our model system suggests that DNA repair could be involved.
Additionally, the recapitulation by our model system of some of the salient features of APOBEC mediated mutagenesis in tumors suggest that it can serve as a tractable and reliable proxy for investigating the effect of repair on the mutational fate of normal flanking DNA in cells unstressed by genotoxic agents. The mutagenic effect of T/G repair appears to require only MMR to generate a single stranded repair template that then becomes vulnerable to mutagenic processes (APOBEC and others). Thus, it might now be experimentally feasible to isolate MMR-intermediates that have been parasitized by APOBECto determine what factors allowed APOBEC access. In contrast, the mutagenesis induced by U/G repair requires BER that is dysfunctional to the extent that it is vulnerable to MMR, which is not usually the case in non-lymphoid cells. That this vulnerability differs between various cell lines indicates that the appropriate comparisons between these cells could reveal the basis of the BER dysfunction. Such an analysis could also have relevance to tumor biology as not all tumor cells exhibit APOBEC-mediated mutations.
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and by ShanghaiTech University start-up funds to JC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- [1].Cannavo E, Gerrits B, Marra G, Schlapbach R, Jiricny J. Characterization of the interactome of the human MutL homologues MLH1, PMS1, and PMS2. J Biol Chem. 2007;282:2976–2986. doi: 10.1074/jbc.M609989200. [DOI] [PubMed] [Google Scholar]
- [2].Cortázar D, Kunz C, Saito Y, Steinacher R, Schär P. The enigmatic thymine DNA glycosylase. DNA Repair. 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- [3].Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jacobs AL, Schar P. DNA glycosylases: in DNA repair and beyond. Chromosoma. 2012;121:1–20. doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jiricny J. Postreplicative mismatch repair. Cold Spring Harb Perspect Biol. 2013;5:a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kunz C, Saito Y, Schar P. DNA Repair in mammalian cells: Mismatched repair: variations on a theme. Cell Mol Life Sci. 2009;66:1021–1038. doi: 10.1007/s00018-009-8739-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liu P, Kenney JM, Stiller JW, Greenleaf AL. Genetic Organization, Length Conservation, and Evolution of RNA Polymerase II Carboxyl-Terminal Domain. Mol. Biol. Evol. 2010;27:2628–2641. doi: 10.1093/molbev/msq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Robertson AB. Base excision repair: The long and short of it. Cellular and molecular life sciences. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hwang DG, Green P. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc Natl Acad Sci U S A. 2004;101:13994–13401. doi: 10.1073/pnas.0404142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Michaelson JJ, Shi YJ, Gujral M, Zheng HC, Malhotra D, Jin X, Jian MH, Liu GM, Greer D, Bhandari A, Wu WT, Corominas R, Peoples A, Koren A, Gore A, Kang SL, Lin GN, Estabillo J, Gadomski T, Singh B, Zhang K, Akshoomoff N, Corsello C, McCarroll S, Iakoucheva LM, Li YR, Wang J, Sebat J. Whole-Genome Sequencing in Autism Identifies Hot Spots for De Novo Germline Mutation. Cell. 2012;151:1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Schrider DR, Hourmozdi JN, Hahn MW. Pervasive Multinucleotide Mutational Events in Eukaryotes. Current Biology. 2011;21:1051–1054. doi: 10.1016/j.cub.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ochman H. Neutral mutations and neutral substitutions in bacterial genomes. Mol. Biol. Evol. 2003;20:2091–2096. doi: 10.1093/molbev/msg229. [DOI] [PubMed] [Google Scholar]
- [15].Miyata T, Hayashida H, Kuma K, Mitsuyasu K, Yasunaga T. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harb. Symp. Quant. Biol. 1987;52:863–867. doi: 10.1101/sqb.1987.052.01.094. [DOI] [PubMed] [Google Scholar]
- [16].Li WH, Yi S, Makova K. Male-driven evolution. Curr. Opin. Genet. Dev. 2002;12:650–656. doi: 10.1016/s0959-437x(02)00354-4. [DOI] [PubMed] [Google Scholar]
- [17].Makova KD, Li W-H. Strong male-driven evolution of DNA sequences in humans and apes. Nature. 2002;416:624–626. doi: 10.1038/416624a. [DOI] [PubMed] [Google Scholar]
- [18].Bohossian HB, Skaletsky H, Page DC. Unexpected similar rates of nucleotide substitution found in male and female hominids. Nature. 2000;406:622–625. doi: 10.1038/35020557. [DOI] [PubMed] [Google Scholar]
- [19].Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. Genetic evidence for complex speciation of humans and chimpanzees. Nature. 2006;441:1103–1108. doi: 10.1038/nature04789. [DOI] [PubMed] [Google Scholar]
- [20].Walser JC, Ponger L, Furano AV. CpG dinucleotides and the mutation rate of non-CpG DNA. Genome Res. 2008;18:1403–1414. doi: 10.1101/gr.076455.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huttley GA, Wakefield MJ, Easteal S. Rates of genome evolution and branching order from whole genome analysis. Molecular Biology and Evolution. 2007;24:1722–1730. doi: 10.1093/molbev/msm094. [DOI] [PubMed] [Google Scholar]
- [22].Huttley GA, Jakobsen IB, Wilson SR, Easteal S. How important is DNA replication for mutagenesis? Mol Biol Evol. 2000;17:929–937. doi: 10.1093/oxfordjournals.molbev.a026373. [DOI] [PubMed] [Google Scholar]
- [23].Arnheim N, Calabrese P. Understanding what determines the frequency and pattern of human germline mutations. Nat Rev Genet. 2009;10:478–488. doi: 10.1038/nrg2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- [25].Shen JC, Rideout WM, 3rd, Jones PA. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 1994;22:972–976. doi: 10.1093/nar/22.6.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- [28].Antonarakis SE. eLS. John Wiley & Sons, Ltd; 2001. CpG Dinucleotides and Human Disorders. [Google Scholar]
- [29].Cooper DN, Youssoufian H. The CpG dinucleotide and human genetic disease. Human genetics. 1988;78:151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- [30].Frederico LA, Kunkel TA, Shaw BR. Cytosine deamination in mismatched base pairs. Biochemistry. 1993;32:6523–6530. doi: 10.1021/bi00077a005. [DOI] [PubMed] [Google Scholar]
- [31].Zhao Z, Jiang C. Methylation-dependent transition rates are dependent on local sequence lengths and genomic regions. Molecular Biology and Evolution. 2007;24:23–25. doi: 10.1093/molbev/msl156. [DOI] [PubMed] [Google Scholar]
- [32].Fryxell KJ, Moon WJ. CpG mutation rates in the human genome are highly dependent on local GC content. Molecular Biology and Evolution. 2005;22:650–658. doi: 10.1093/molbev/msi043. [DOI] [PubMed] [Google Scholar]
- [33].Chimpanzee-Sequencing-Analysis-Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- [34].Gaffney DJ, Keightley PD. The scale of mutational variation in the murid genome. Genome Res. 2005;15:1086–1094. doi: 10.1101/gr.3895005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hardison RC, Roskin KM, Yang S, Diekhans M, Kent WJ, Weber R, Elnitski L, Li J, O'Connor M, Kolbe D, Schwartz S, Furey TS, Whelan S, Goldman N, Smit A, Miller W, Chiaromonte F, Haussler D. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 2003;13:13–26. doi: 10.1101/gr.844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hellmann I, Prufer K, Ji H, Zody MC, Paabo S, Ptak SE. Why do human diversity levels vary at a megabase scale? Genome Res. 2005;15:1222–1231. doi: 10.1101/gr.3461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tyekucheva S, Makova KD, Karro JE, Hardison RC, Miller W, Chiaromonte F. Human-macaque comparisons illuminate variation in neutral substitution rates. Genome Biol. 2008;9:R76. doi: 10.1186/gb-2008-9-4-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Furano AV, Walser JC. Encyclopedia of Life Sciences (ELS) John Wiley & Sons, Ltd; Chichester: 2009. Mutation rate of non-CpG DNA. [Google Scholar]
- [39].Ananda G, Chiaromonte F, Makova KD. A genome-wide view of mutation rate co-variation using multivariate analyses. Genome Biology. 2011;12:R27. doi: 10.1186/gb-2011-12-3-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lujan S, Clausen A, Clark A, MacAlpine H, MacAlpine D, Malc E, Mieczkowski P, Burkholder A, Fargo D, Gordenin D, Kunkel T. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome research. 2014;24:1751–1764. doi: 10.1101/gr.178335.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Green P, Ewing B, Miller W, Thomas PJ, Green ED. Transcription-associated mutational asymmetry in mammalian evolution. Nat. Genet. 2003;33:514–517. doi: 10.1038/ng1103. [DOI] [PubMed] [Google Scholar]
- [42].Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- [43].Huvet M, Nicolay S, Touchon M, Audit B, d'Aubenton-Carafa Y, Arneodo A, Thermes C. Human gene organization driven by the coordination of replication and transcription. Genome Res. 2007;17:1278–1285. doi: 10.1101/gr.6533407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mugal CF, von Grunberg HH, Peifer M. Transcription-induced mutational strand bias and its effect on substitution rates in human genes. Mol. Biol. Evol. 2009;26:131–142. doi: 10.1093/molbev/msn245. [DOI] [PubMed] [Google Scholar]
- [45].Polak P, Arndt PF. Transcription induces strand-specific mutations at the 5' end of human genes. Genome Res. 2008;18:1216–1223. doi: 10.1101/gr.076570.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, Jonasdottir A, Wong WSW, Sigurdsson G, Walters GB, Steinberg S, Helgason H, Thorleifsson G, Gudbjartsson DF, Helgason A, Magnusson OT, Thorsteinsdottir U, Stefansson K. Rate of de novo mutations and the importance of father/'s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Walser JC, Furano AV. The mutational spectrum of non-CpG DNA varies with CpG content. Genome Res. 2010;20:875–882. doi: 10.1101/gr.103283.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].IHGS-Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- [49].Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978;274:775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- [50].Sved J, Bird A. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc. Natl. Acad. Sci. U. S. A. 1990;87:4692–4696. doi: 10.1073/pnas.87.12.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cooper DN, Youssoufian H. The CpG dinucleotide and human genetic disease. Hum. Genet. 1988;78:151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- [52].Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer. 2012;12:104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell research. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Martomo SA, Gearhart PJ. Somatic hypermutation: subverted DNA repair. Curr Opin Immunol. 2006;18:243–248. doi: 10.1016/j.coi.2006.03.007. [DOI] [PubMed] [Google Scholar]
- [55].Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, Scharff MD. The biochemistry of somatic hypermutation. Ann. Rev. Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- [56].Teng G, Papavasiliou FN. Immunoglobulin Somatic Hypermutation. Ann. Rev. Genetics. 2007;41:107–120. doi: 10.1146/annurev.genet.41.110306.130340. [DOI] [PubMed] [Google Scholar]
- [57].Peña-Diaz J, Bregenhorn S, Ghodgaonkar M, Follonier C, Artola-Borán M, Castor D, Lopes M, Sartori A, Jiricny J. Noncanonical Mismatch Repair as a Source of Genomic Instability in Human Cells. Molecular Cell. 2012;47:669–680. doi: 10.1016/j.molcel.2012.07.006. [DOI] [PubMed] [Google Scholar]
- [58].Peña-Diaz J, Jiricny J. Mammalian mismatch repair: error-free or error-prone? Trends Biochem Sci. 2012;37:206–214. doi: 10.1016/j.tibs.2012.03.001. [DOI] [PubMed] [Google Scholar]
- [59].Seidman MM, Dixon K, Razzaque A, Zagursky RJ, Berman ML. A shuttle vector plasmid for studying carcinogen-induced point mutations in mammalian cells. Gene. 1985;38:233–237. doi: 10.1016/0378-1119(85)90222-7. [DOI] [PubMed] [Google Scholar]
- [60].Chen J, Miller BF, Furano AV. Repair of naturally occurring mismatches can induce mutations in flanking DNA. Elife. 2014;3:e02001. doi: 10.7554/eLife.02001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Talhaoui I, Couve S, Gros L, Ishchenko AA, Matkarimov B, Saparbaev MK. Aberrant repair initiated by mismatch-specific thymine-DNA glycosylases provides a mechanism for the mutational bias observed in CpG islands. Nucleic Acids Research. 2014;42:6300–6313. doi: 10.1093/nar/gku246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tsutakawa SE, Lafrance-Vanasse J, Tainer JA. The cutting edges in DNA repair, licensing, and fidelity: DNA and RNA repair nucleases sculpt DNA to measure twice, cut once. DNA Repair (Amst) 2014;19:95–107. doi: 10.1016/j.dnarep.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Simonelli V, Narciso L, Dogliotti E, Fortini P. Base excision repair intermediates are mutagenic in mammalian cells. Nucleic Acids Res. 2005;33:4404–4411. doi: 10.1093/nar/gki749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Prasad R, Shock DD, Beard WA, Wilson SH. Substrate channeling in mammalian base excision repair pathways: passing the baton. J Biol Chem. 2010;285:40479–40488. doi: 10.1074/jbc.M110.155267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Prasad R, Williams JG, Hou EW, Wilson SH. Pol beta associated complex and base excision repair factors in mouse fibroblasts. Nucleic Acids Research. 2012;40:11571–11582. doi: 10.1093/nar/gks898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kuznetsova A, Kuznetsov N, Ishchenko A, Saparbaev M, Fedorova O. Pre-steady-state fluorescence analysis of damaged DNA transfer from human DNA glycosylases to AP endonuclease APE1. Biochim Biophys Acta. 2014;1840:3042–3051. doi: 10.1016/j.bbagen.2014.07.016. [DOI] [PubMed] [Google Scholar]
- [67].Lee K.-y., Myung K. PCNA Modifications for Regulation of Post-Replication Repair Pathways. Mol. Cells. 2008;26:5–11. [PMC free article] [PubMed] [Google Scholar]
- [68].Moldovan G-L, Pfander B, Jentsch S. PCNA, the Maestro of the Replication Fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- [69].Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- [70].Pluciennik A, Dzantiev L, Iyer RR, Constantin N, Kadyrov FA, Modrich P. PCNA function in the activation and strand direction of MutLalpha endonuclease in mismatch repair. Proc Natl Acad Sci U S A. 2010;107:16066–16071. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, Aas PA, Steinsbekk K, Bakke O, Krokan HE. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Conticello S. The AID/APOBEC family of nucleic acid mutators. Genome Biology. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Bransteitter R, Prochnow C, Chen XS. The current structural and functional understanding of APOBEC deaminases. Cell Mol Life Sci. 2009;66:3137–3147. doi: 10.1007/s00018-009-0070-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Schanz S, Castor D, Fischer F, Jiricny J. Interference of mismatch and base excision repair during the processing of adjacent U/G mispairs may play a key role in somatic hypermutation. Proc Natl Acad Sci U S A. 2009;106:5593–5598. doi: 10.1073/pnas.0901726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Prasad R, Beard WA, Batra VK, Liu Y, Shock DD, Wilson SH. A review of recent experiments on step-to-step "hand-off" of the DNA intermediates in mammalian base excision repair pathways. Molekuliarnaia biologiia. 2011;45:586–600. [PMC free article] [PubMed] [Google Scholar]
- [76].Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- [77].Rattray AJ, Strathern JN. Error-prone DNA polymerases: when making a mistake is the only way to get ahead. Annu. Rev. Genet. 2003;37:31–66. doi: 10.1146/annurev.genet.37.042203.132748. [DOI] [PubMed] [Google Scholar]
- [78].Parris CN, Seidman MM. A signature element distinguishes sibling and independent mutations in a shuttle vector plasmid. Gene. 1992;117:1–5. doi: 10.1016/0378-1119(92)90482-5. [DOI] [PubMed] [Google Scholar]
- [79].Hou EW, Prasad R, Asagoshi K, Masaoka A, Wilson SH. Comparative assessment of plasmid and oligonucleotide DNA substrates in measurement of in vitro base excision repair activity. Nucl. Acids Res. 2007;35:1–10. doi: 10.1093/nar/gkm639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3' --> 5' on single-stranded DNA. Nat Struct Mol Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- [81].Venkatesan RN, Bielas JH, Loeb LA. Generation of mutator mutants during carcinogenesis. DNA Repair. 2006;5:294–302. doi: 10.1016/j.dnarep.2005.10.012. [DOI] [PubMed] [Google Scholar]
- [82].Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci U S A. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Roberts SA, Gordenin DA. Clustered and genome-wide transient mutagenesis in human cancers: Hypermutation without permanent mutators or loss of fitness. Bioessays. 2014 doi: 10.1002/bies.201300140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer. 2014;14:786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nature Genet. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, Menzies A, Martin S, Leung K, Chen L, Leroy C, Ramakrishna M, Rance R, Lau KW, Mudie LJ, Varela I, McBride DJ, Bignell GR, Cooke SL, Shlien A, Gamble J, Whitmore I, Maddison M, Tarpey PS, Davies HR, Papaemmanuil E, Stephens PJ, McLaren S, Butler AP, Teague JW, Jonsson G, Garber JE, Silver D, Miron P, Fatima A, Boyault S, Langerod A, Tutt A, Martens JW, Aparicio SA, Borg A, Salomon AV, Thomas G, Borresen-Dale AL, Richardson AL, Neuberger MS, Futreal PA, Campbell PJ, Stratton MR. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Leonard B, Hart SN, Burns MB, Carpenter MA, Temiz NA, Rathore A, Isaksson Vogel R, Nikas JB, Law EK, Brown WL, Li Y, Zhang Y, Maurer MJ, Oberg AL, Cunningham JM, Shridhar V, Bell DA, April C, Bently D, Bibikova M, Cheetham RK, Fan J-B, Grocock R, Humphray S, Kingsbury Z, Peden J, Chien J, Swisher EM, Hartmann LC, Kalli KR, Goode EL, Sicotte H, Kaufmann SH, Harris RS. APOBEC3B upregulation and genomic mutation patterns in serous ovarian carcinoma. Cancer Research. 2013;73 doi: 10.1158/0008-5472.CAN-13-1753. doi:10.1158/0008-5472.can-1113-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, Harris S, Shah RR, Resnick MA, Getz G, Gordenin DA. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nature Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Henderson S, Chakravarthy A, Su X, Boshoff C, Fenton TR. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep. 2014;7:1833–1841. doi: 10.1016/j.celrep.2014.05.012. [DOI] [PubMed] [Google Scholar]
- [92].Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Molecular Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Atamna H, Cheung I, Ames BN. A method for detecting abasic sites in living cells: Age-dependent changes in base excision repair. Proc Natl Acad Sci U S A. 2000;97:686–691. doi: 10.1073/pnas.97.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Bhutani N, Burns DM, Blau HM. DNA Demethylation Dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, Hansen RS, Neph S, Sabo PJ, Heimfeld S, Raubitschek A, Ziegler S, Cotsapas C, Sotoodehnia N, Glass I, Sunyaev SR, Kaul R, Stamatoyannopoulos JA. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2012;13:59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- [99].Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].van der Oost J. Molecular biology. New tool for genome surgery. Science. 2013;339:768–770. doi: 10.1126/science.1234726. [DOI] [PubMed] [Google Scholar]
- [102].Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, Jaenisch R, Zhang F, Sharp PA. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- [105].Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014 doi: 10.1038/nbt.2908. DOI: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 2010;38:4274–4284. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Lackey L, Law EK, Brown WL, Harris RS. Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle. 2013;12:762–772. doi: 10.4161/cc.23713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, Yan Z, Li D, Li J. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013;13:659–662. doi: 10.1016/j.stem.2013.10.016. [DOI] [PubMed] [Google Scholar]
- [109].Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK, Nieuwenhuis EE, Beekman JM, Clevers H. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- [111].Tucker KL. Methylated cytosine and the brain: a new base for neuroscience. Neuron. 2001;30:649–652. doi: 10.1016/s0896-6273(01)00325-7. [DOI] [PubMed] [Google Scholar]
- [112].Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Wiebauer K, Jiricny J. In vitro correction of G.T mispairs to G.C pairs in nuclear extracts from human cells. Nature. 1989;339:234–236. doi: 10.1038/339234a0. [DOI] [PubMed] [Google Scholar]
- [114].Brown TC, Jiricny J. A specific mismatch repair event protects mammalian cells from loss of 5-methylcytosine. Cell. 1987;50:945–950. doi: 10.1016/0092-8674(87)90521-6. [DOI] [PubMed] [Google Scholar]
- [115].Vaisman A, Woodgate R. Unique misinsertion specificity of poliota may decrease the mutagenic potential of deaminated cytosines. EMBO J. 2001;20:6520–6529. doi: 10.1093/emboj/20.22.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Walsh CP, Xu GL. Cytosine methylation and DNA repair. Curr. Top. Microbiol. Immunol. 2006;301:283–315. doi: 10.1007/3-540-31390-7_11. [DOI] [PubMed] [Google Scholar]
- [117].Ying H, Huttley G. Exploiting CpG Hypermutability to Identify Phenotypically Significant Variation Within Human Protein-Coding Genes. Genome Biology and Evolution. 2011;3:938–949. doi: 10.1093/gbe/evr021. [DOI] [PMC free article] [PubMed] [Google Scholar]