Abstract
Pancreatic cancer remains one of the most lethal of malignancies and a major health burden. We performed whole-genome sequencing and copy number variation (CNV) analysis of 100 pancreatic ductal adenocarcinomas (PDACs). Chromosomal rearrangements leading to gene disruption were prevalent, affecting genes known to be important in pancreatic cancer (TP53, SMAD4, CDKN2A, ARID1A and ROBO2) and new candidate drivers of pancreatic carcinogenesis (KDM6A and PREX2). Patterns of structural variation (variation in chromosomal structure) classified PDACs into 4 subtypes with potential clinical utility: the subtypes were termed stable, locally rearranged, scattered and unstable. A significant proportion harboured focal amplifications, many of which contained druggable oncogenes (ERBB2, MET, FGFR1, CDK6, PIK3R3 and PIK3CA), but at low individual patient prevalence. Genomic instability co-segregated with inactivation of DNA maintenance genes (BRCA1, BRCA2 or PALB2) and a mutational signature of DNA damage repair deficiency. Of 8 patients who received platinum therapy, 4 of 5 individuals with these measures of defective DNA maintenance responded.
Pancreatic cancer (PC) has a median survival of 6 months and a 5-year survival that remains less than 5% despite 50 years of research and therapeutic development1. It is the fourth commonest cause of cancer death in Western societies and is projected to be the second leading cause within a decade. As a consequence, there is an urgent need to better select patients for current therapies and develop novel therapeutic strategies.
Recent exome and CNV analyses of pancreatic ductal adenocarcinoma have revealed a complex mutational landscape2,3.Activating mutations of KRAS are near ubiquitous and inactivation of TP53, SMAD4 and CDKN2A occur at rates of >50%. The prevalence of recurrently mutated genes then drops to ~ 10% for a handful of genes involved in chromatin modification, DNA damage repair and other mechanisms known to be important in carcinogenesis; however, a long tail of infrequently mutated genes dominates, resulting in significant intertumoural heterogeneity. Faced with this diversity, it is not surprising that therapeutic development using an unselected approach to patient recruitment for clinical trials has been challenging2–4.
Somatic structural rearrangement of chromosomes represents a common class of mutation that is capable of causing gene disruption (such as deletion or rearrangement), gene activation (for example, copy number gain or amplification) and the formation of novel oncogenic gene products (gene fusions). Many of these events actively drive carcinogenesis5,6 and in some instances present therapeutic targets. Early karyotyping7 and more recent genomic sequencing of small numbers of primary tumours (n = 3) and metastases (n = 10) suggests that PDAC genomes contain widespread and complex patterns of chromosomal rearrangement8,9.
Here we performed deep whole-genome sequencing of 100 PDACs and show that structural variation (variation in chromosomal structure) is an important mechanism of DNA damage in pancreatic carcinogenesis. We classify PDAC into four subtypes based on structural variation profiles and implicate molecular mechanisms underlying some of these events. Finally, as proof of concept, we use a combination of structural variation, mutational signatures and gene mutations to define putative biomarkers of therapeutic responsiveness for platinum-based chemotherapy, which are current therapeutic options for PDAC10–14, and for therapeutics that target similar molecular mechanisms such as PARP inhibitors15 that are currently being tested in clinical trials.
Genomic landscape of pancreatic cancer
Patients were recruited and consent obtained for genomic sequencing through participating institutions of the Australian Pancreatic Cancer Genome Initiative (APGI; htpp://www.pancreaticcancer.net.au) as part of the International Cancer Genome Consortium (ICGC; http://www.icgc.org)16 (Supplementary Table 1). Array-based CNV was analysed using GAP17 and tumour cellularity estimated with qPure18. Whole-genome sequencing was performed on 100 primary PDACs with an epithelial cellularity of ≥ 40% (n = 75), and complemented by cell lines derived from APGI participants (n = 25) to an average depth of 65×, and compared to the germline (average depth 38×) (Supplementary Table 2). Mutations were detected using qSNP19 and GATK and indels called with Pindel and GATK.
Point mutations and structural variation in PDAC
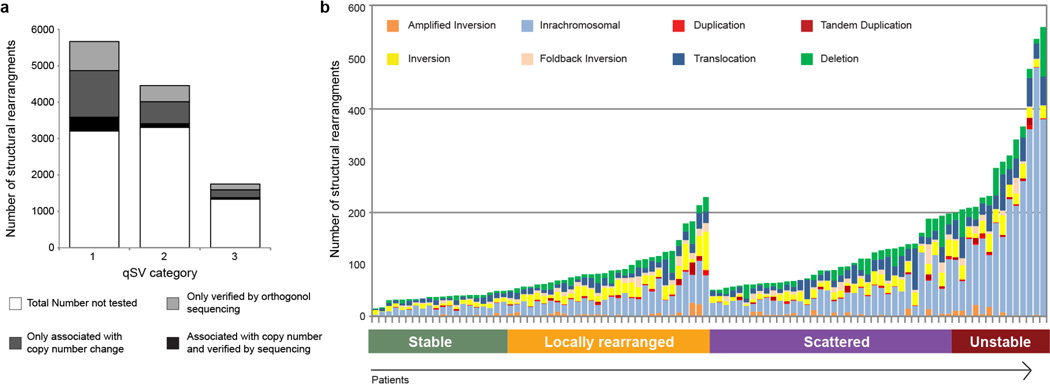
A total of 857,971 somatic point mutations and small insertions and deletions were detected in the cohort: 7,888 were non-silent mutations in 5,424 genes (Supplementary Tables 3 and 4). Orthogonal validation of >3,000 exonic mutations estimated the accuracy of mutation calls at >95% (Methods). Consistent with previous estimates20, the average mutational burden across the cohort was 2.64 per Mb (range 0.65–28.2 per Mb). Somatic structural variants were identified with the qSV package, which uses multiple lines of evidence to define events (discordant pairs, soft clipping and split reads). Events verified using an orthogonal sequencing method were also included (Methods and Extended Data Fig. 1a). Where possible, these events were cross-referenced with CNV data (Methods). In total, 11,868 somatic structural variants were detected at an average of 119 per individual (range 15–558) (Supplementary Table 5 and Extended Data Fig. 1b). The majority of structural variants were intra-chromosomal (10,114) and were classified into 7 types: intra-chromosomal rearrangements (5,860), deletions (1,393), duplications (128), tandem duplications (179), inversions (1,629), fold-back inversions (579) and amplified inversions (346); inter-chromosomal translocations were less prevalent (1,754) (Supplementary Table 6). A total of 6,908 rearrangements directly disrupted gene sequences and 1,220 genes contained a breakpoint in 2 or more patients (Supplementary Table 7). Recurrent gene fusions were not detected: 1,236 structural variants led to the joining of two gene loci, however, only 183 of these events were fused in an orientation and frame that was capable of expressing a product, and none of these predicted fusion events occurred in more than one sample.
Genes affected by mutation and structural variation
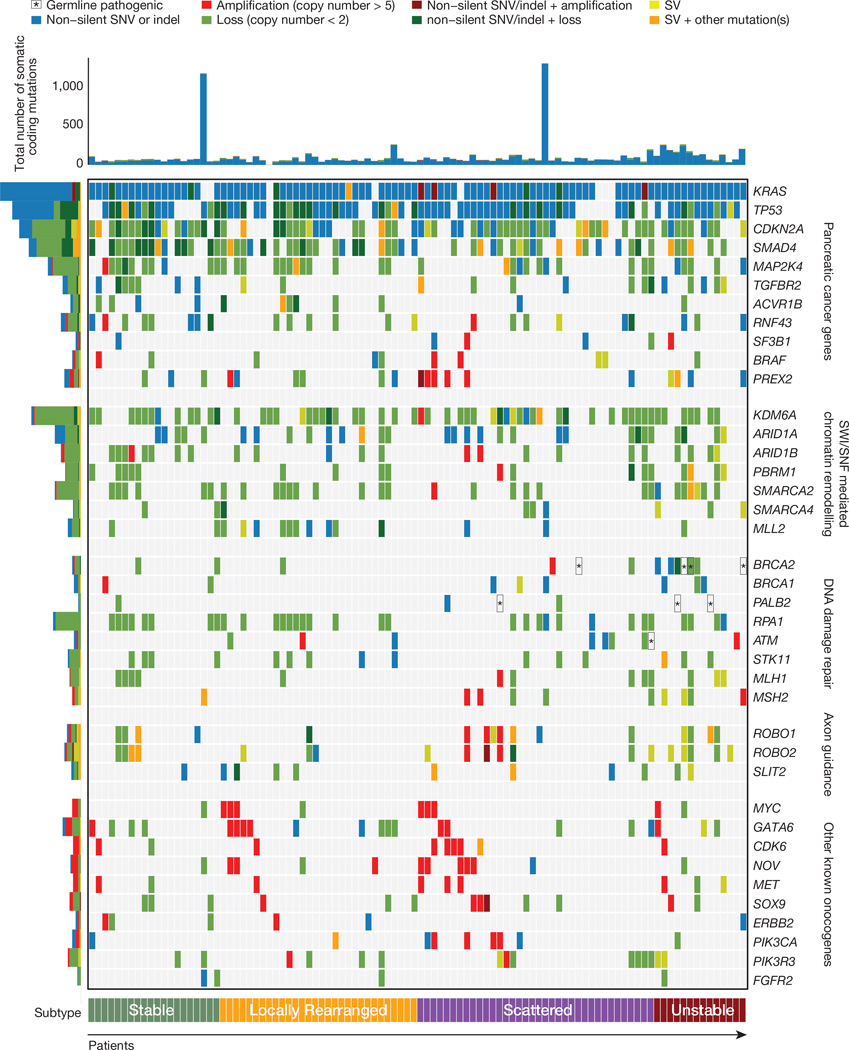
Commonly mutated genes that characterize PDAC (KRAS, TP53, SMAD4 and CDKN2A)2,3 were reaffirmed as significant using MutSig21 analysis (Supplementary Table 8). Combining structural variation events with deleterious point mutations increased the prevalence of inactivation events for TP53 to 74% (3 structural variants and 71mutations), 31%for SMAD4 (9 structural variants and 22 mutations) and 35%for CDKN2A (11 structural variants and 24 mutations). Two additional genes not previously described in human PDAC (KDM6A and PREX2) had recurrent pathogenic mutations and structural variants at a rate of 10% or more. KDM6A is a SWI/SNF interacting partner that was identified in a pancreatic sleeping-beauty transposon mutagenesis screen22, and is mutated in RCC and medulloblastoma. In our cohort, KDM6A was inactivated in 18% of patients, (4 frame shifts, 1 in-frame deletion and 2 missense mutations, 5 structural variants and 8 homozygous deletions). In most cases (n = 15), both alleles of KDM6A were affected. The RAC1 guanine nucleotide exchange factor PREX2, mutated in melanoma23 was inactivated in 10% of PDAC patients (1 frame shift, 1 splice site and 5 missense mutations, 2 structural variants and1 homozygous deletion). In addition, the tumour suppressor gene RNF43, originally identified in cystic tumours of the pancreas, was inactivated in 10% of PDAC patients (4 frameshift and 4 nonsense mutations, 2 structural variants). Two of these PDACs had an associated intraductal papillary mucinous neoplasm (IPMNs). Recent studies have suggested that loss of functional RNF43 may confer sensitivity to WNT inhibitors24. Figure 1 shows the prevalence of aberrations in key driver genes and pathways in PDAC; implicating structural variation as an important mutational mechanism in pancreatic carcinogenesis.
Figure 1. Mutations in key genes and pathways in pancreatic cancer.
The upper panel shows non-silent single nucleotide variants and small insertions or deletions. The central matrix shows: non-silent mutations (blue), copy number changes (amplification (>5 copies) represented in red and loss represented in green) and genes affected by structural variants (SV, yellow). Pathogenic germline variants are highlighted with asterisk (*) symbols. The histogram on the left shows the number of each alteration in each gene.
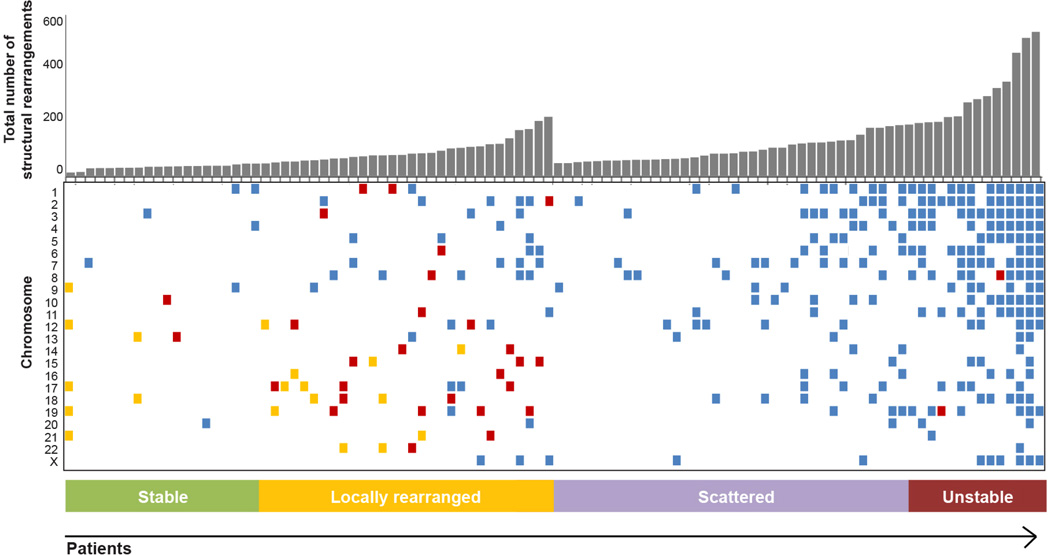
Subtyping using structural rearrangements
The distribution of events was used to classify tumours into the following four subtypes (Fig. 2 and Extended Data Fig. 2 and Methods).
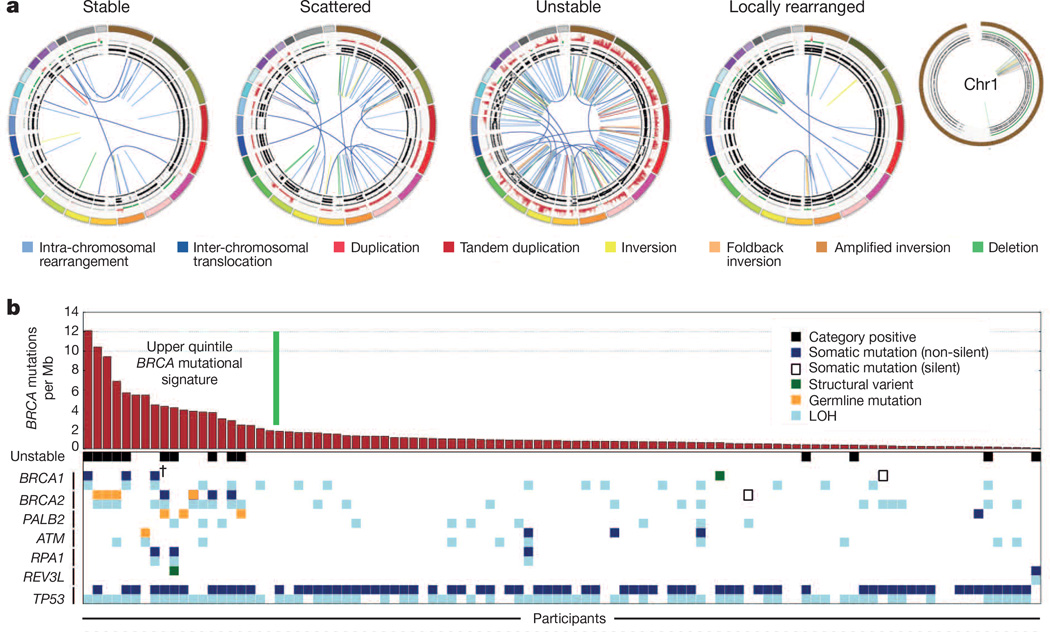
Figure 2. Subtypes of pancreatic cancer.
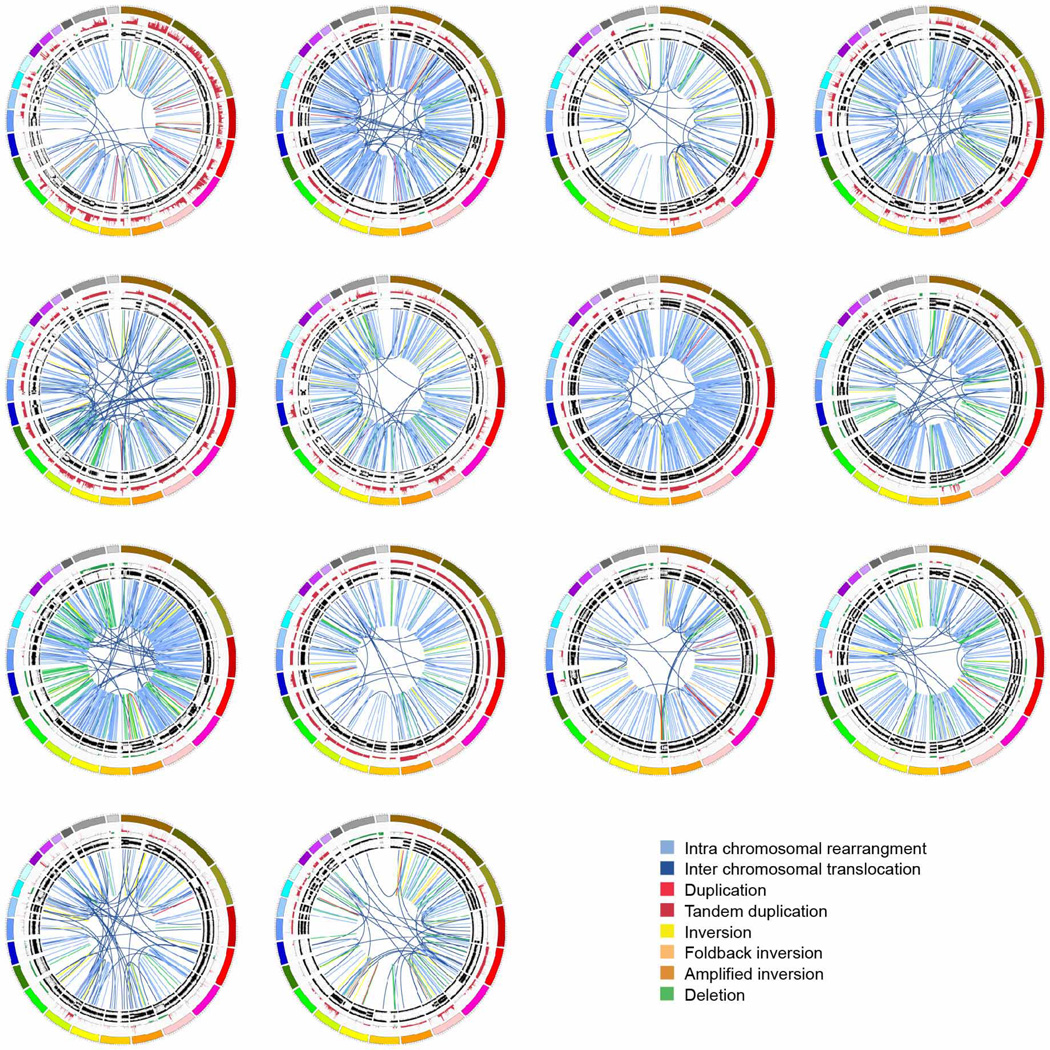
a, Subgroups of PDAC based on the frequency and distribution of structural rearrangements. Representative tumours of each group are shown. The coloured outer rings are chromosomes, the next ring depicts copy number (red represents gain and green represents loss), the next is the B allele frequency (proportion of the B allele to the total quantity of both alleles). The inner lines depict chromosome structural rearrangements. b, The contribution of the BRCA mutational signature within each tumour ranked by prevalence (red bars). Unstable tumours are associated with a high BRCA mutation signature and deleterious mutations in BRCA pathway genes. The dagger (†) symbol indicates predicted only as possibly damaging by Polyphen2.
Stable subtype
Subtype 1 was classified as ‘stable’ (20% of all samples). These tumour genomes contained ≤ 50 structural variation events and often exhibited widespread aneuploidy suggesting defects in cell cycle/mitosis (Extended Data Fig. 3). Point mutation rates for KRAS and SMAD4 were similar to the rest of the cohort, and the prevalence of TP53 mutations was only slightly less (61% versus a mean of 70%across all samples). In addition, telomere length was no different in comparison to other subgroups.
Locally rearranged subtype
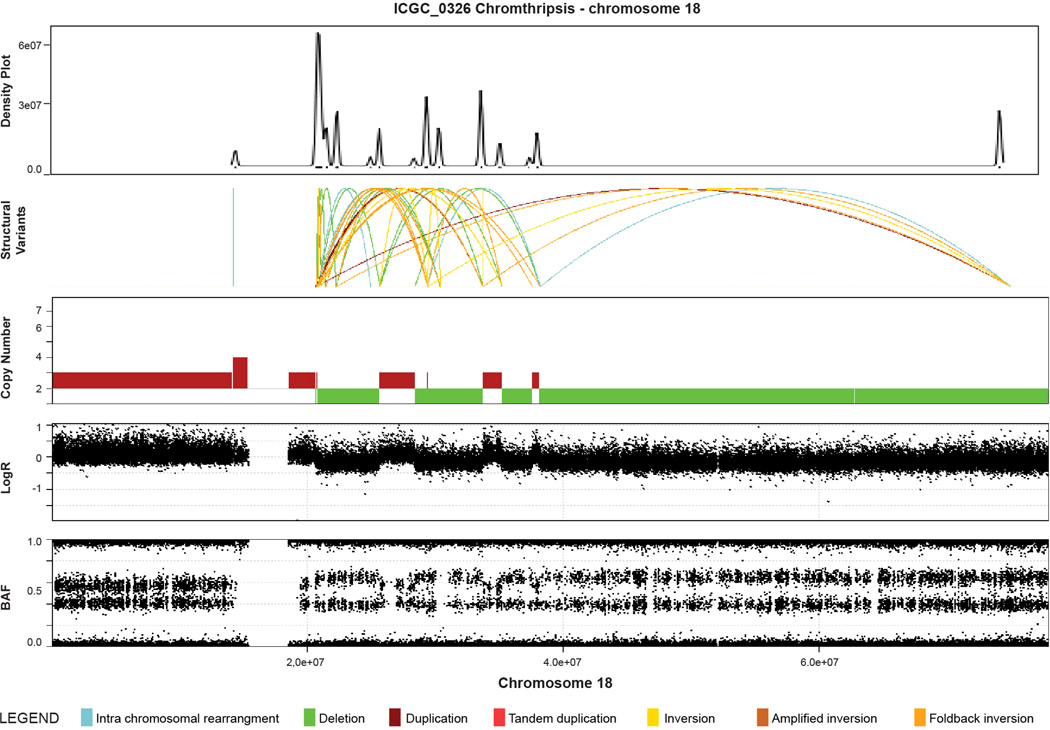
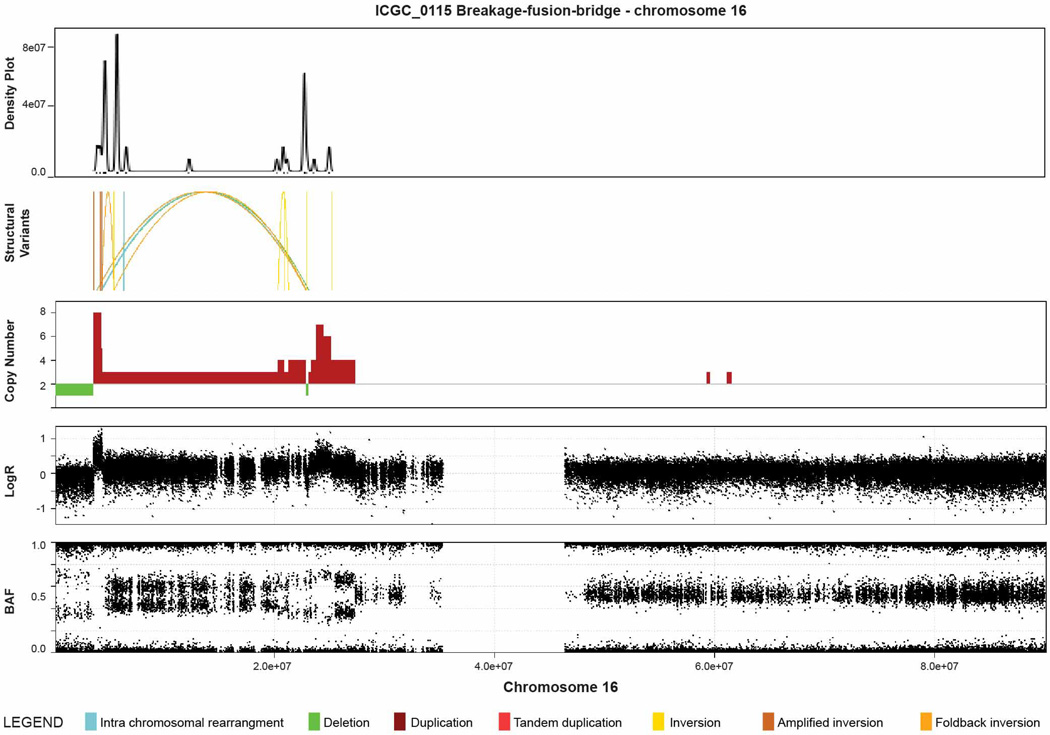
Subtype 2 was classified as ‘locally rearranged’ (30% of all samples). This subtype exhibited a significant focal event on one or two chromosomes. The group could be further divided into those with focal regions of gain/amplification and those that contained complex genomic rearrangements (Extended Data Fig. 4). Approximately one-third of locally rearranged genomes contained regions of copy number gain that harboured known oncogenes (Supplementary Table 9). These included common focal amplifications in KRAS, SOX9 and GATA6 and often included therapeutic targets such as ERBB2, MET, CDK6, PIK3CA and PIK3R3, but at low individual prevalence (1–2% of patients) (Supplementary Table 9). The remaining local rearrangements involved complex genomic events such as breakage–fusion–bridge (BFB, n = 9) or chromothripsis5,25 (n = 15), which resulted in a ring chromosome in at least one case (ICGC_0059) (Extended Data Figs 5 and 6). Chromothripsis is linked to TP53 mutations in medullobastoma and acute myeloid leukaemia and here, 10/13 chromothriptic tumours had a TP53 mutation, 5 of which were bi-allelic (Fig. 1). Five of these chromothriptic events occurred after chromosomal duplication suggesting that they are less likely to be driving carcinogenesis (Methods).
Scattered subtype
Subtype 3 was classified as ‘scattered’ (36% of all samples). Tumours in this class exhibited a moderate range of non-random chromosomal damage and less than 200 structural variation events (Extended Data Fig. 7).
Unstable subtype
Subtype 4 was classified as ‘unstable’ (14%of all samples). The tumours exhibited a large number of structural variation events (>200; maximum of 558) (Extended Data Fig. 8). This scale of genomic instability suggested defects in DNA maintenance26, which potentially defines sensitivity to DNA-damaging agents (Fig. 3a; Methods).
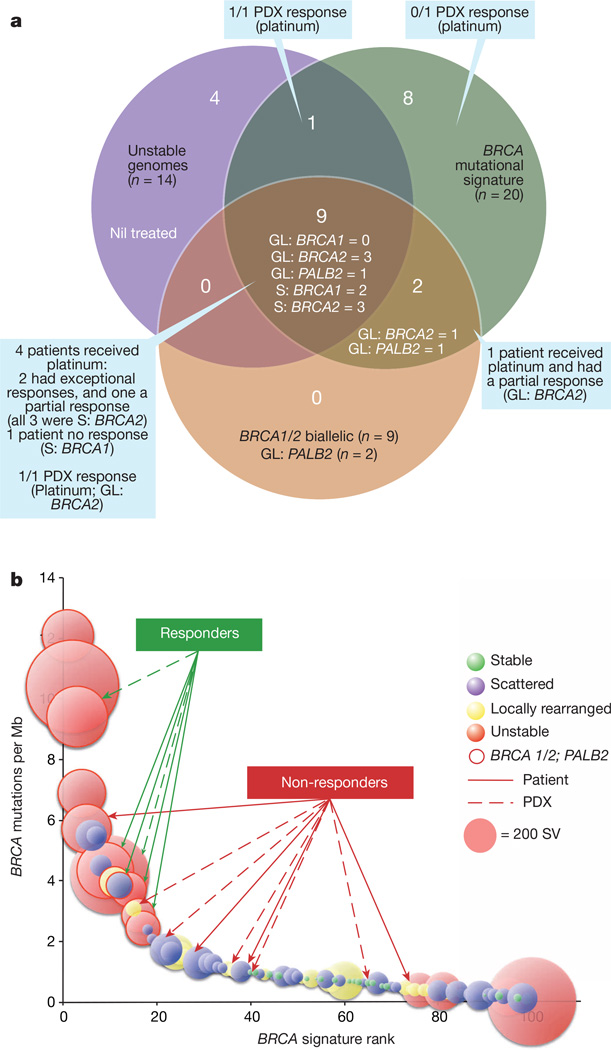
Figure 3. Putative biomarkers of platinum and PARP inhibitor responsiveness.
a, A Venn diagram showing the overlap of surrogate measures of defects in DNA maintenance (unstable genomes and BRCA mutational signature), with mutations in BRCA pathway genes. Of a total of 24 patients (24%), 10 have both unstable genomes and the BRCA mutational signature. The majority of patients with mutations in BRCA pathway genes (9) are within this intersect, however 2 have the mutational signature, but are classified either as scattered (n = 1) or locally rearranged (n = 1). GL, germline; S, somatic. b, Individual tumours are ranked based on their BRCA mutational signature burden, with the diameter of each circle representing the number of structural variants in each. Those encircled by a solid line have mutations in BRCA pathway genes. Responders and non-responders to platinum-based therapy are indicated with solid lines for patients and broken lines for patient-derived xenografts (PDX).
Genomic markers of defective DNA maintenance
We mapped the relationship between the unstable subtype, mutations in BRCA pathway genes and a recently described mutational signature associated with deleterious mutations in BRCA1 or BRCA2 in breast, ovarian and pancreatic cancer20. The majority of unstable tumours (10 of 14) fell within the top quintile of the BRCA signature when ranked by prevalence per Mb (Fig. 2b). In addition, the top quintile of the BRCA signature was associated with deleterious mutations of BRCA1 (n = 2), BRCA2 (n = 7), and PALB2 (n = 2) (Fig. 2b) (Supplementary Table 10). Four of the BRCA2 mutations were germline in origin (3 frameshift and 1 nonsense), and in each case, the wild-type allele was inactivated in the tumour. A further 2 patients had somatic mutations in BRCA1 (both with splice site mutations), and another 3 had somatic BRCA2 mutations (1 indel and 2 splice site mutations). All deleterious BRCA1 and BRCA2 mutations had inactivation of the second allele. Three patients had pathogenic germline PALB2 mutations that were associated with the BRCA mutational signature. One of these was a TGTT deletion, which is known to occur in pancreatic cancer27 (this tumour also had a somatic BRCA2 mutation), and the mutations of PALB2 in both the other 2 cases are associated with an inherited predisposition to breast cancer28. Germline PALB2 mutation carriers did not have evidence of somatic loss of the second allele; however, heterozygous germline mutation of PALB2 appears sufficient to cause DNA replication and damage response defects29. In contrast, tumours containing a somatic heterozygous silent mutation of BRCA2, a heterozygous intronic structural variation and 2 unclassified heterozygous missense mutations in BRCA1 (predicted to be benign or only possibly damaging by Polyphen2) were not associated with a high-ranking BRCA mutational signature (<1 BRCA signature mutation per Mb) or an unstable genome (Supplementary Table 10). Overlapping deleterious mutations in BRCA1, BRCA2 and PALB2 with unstable genomes and the BRCA mutational signature showed that mutations in these genes were associated with the top quintile of the BRCA mutational signature, and the majority (9 of 11) also exhibited unstable genomes (Fig. 3a).
Defective DNA repair without BRCA pathway mutations
Mutations in BRCA pathway genes accounted for approximately half of patients with a high BRCA mutational signature and/or an unstable genome (Fig. 3a). Hyper-methylation is known to play a role in silencing BRCA1, BRCA2 and PALB2 in some breast and ovarian cancers; however, high-density methylome array profiling of this cohort30 allowed us to exclude this as a contributing mechanism. Single instances of biallelic, inactivating, somatic mutation was observed for two genes known to induce genomic instability and chemosensitivity when inactivated: RPA1 (ref. 31) (splice site and loss of heterozygosity (LOH)), and the DNA polymerase zeta catalytic unit/REV3L32 (nonsense and LOH). We also detected mutations in other genes involved in DNA maintenance such as ATM, FANCM, XRCC4 and XRCC6 in tumours with an unstable genome or the BRCA mutational signature; however, they are yet to be causally linked to these genomic events or sensitivity to DNA-damaging agents.
Putative genotypes of platinum responsiveness
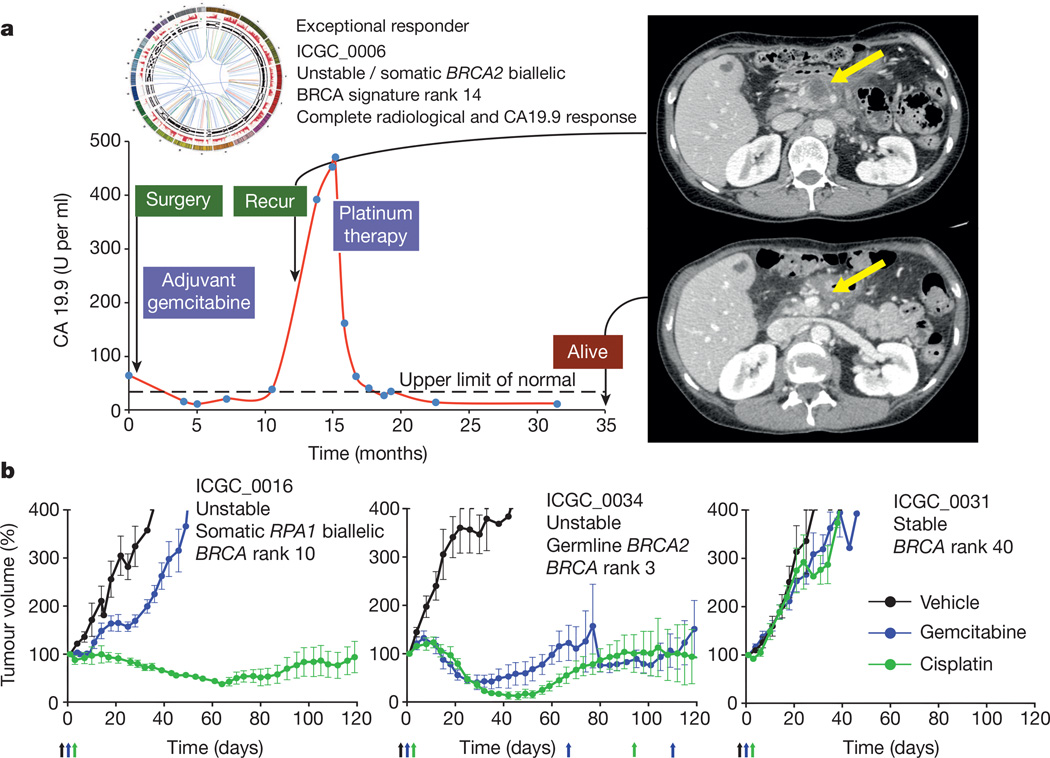
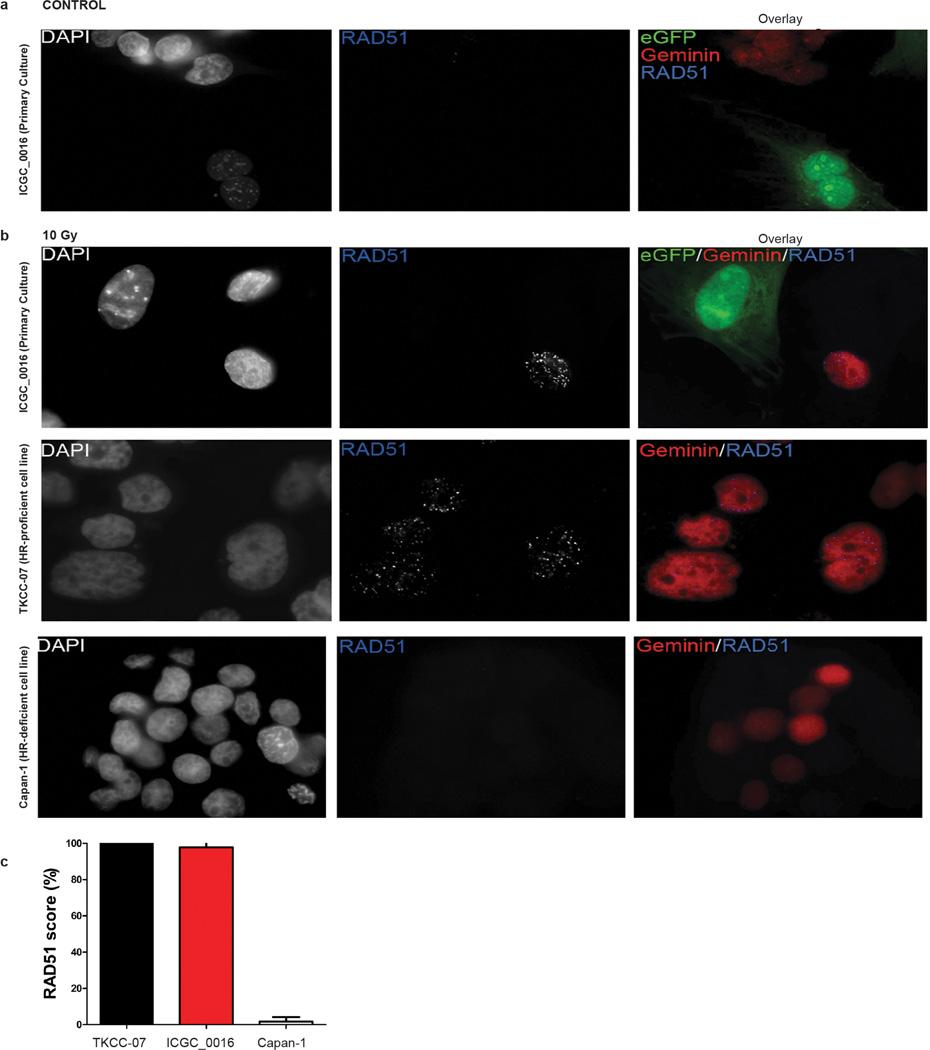
As the APGI was a prospective observational cohort study with extensive clinical follow-up, it was possible to track therapeutic responsiveness of participants that received chemotherapy when their disease recurred. At the time of analysis, 53 patients had documented recurrences and 25 received a variety of chemotherapeutic agents (Supplementary Table 11). This analysis was complemented through therapeutic testing of patient-derived xenografts (PDXs) generated from APGI participants. Overall, 8 patients received a platinum-based therapy and 7 PDXs were treated with gemcitabine and cisplatin (Fig. 3b). Of 5 patients with unstable genomes and/or a high BRCA mutational signature burden (designated as ‘on-genotype’) 2 had exceptional responses (defined as complete radiological resolution of disease and normalization of CA19.9 levels33), and 2 had robust partial responses based on RECIST1.1 criteria34 (Fig. 4a), while 3 patients who did not have any of these characteristics (‘off-genotype’) did not respond. These observations were supported by PDX studies where 2 of 3 on-genotype PDXs responded to cisplatin (one BRCA2 mutant responded and one carrying bi-allelic inactivation of RPA1, which notably retained RAD51 foci (Extended Data Fig. 9) also responded. Another, with a mutational signature but not an unstable genome, and without a mutation in a BRCA pathway gene, did not respond. This compares to no responses in the 4 PDXs in the off-genotype group (Figs 3 and 4b). Combining patient and PDX response data, on-genotype tumours were associated with response to platinum-based therapy (P = 0.0070, Fisher’s exact test, Fig. 3b) (Supplementary Table 11).
Figure 4. Responses to platinum therapy.
a, ICGC_0006: one of two patients who had an exceptional response to platinum-based therapy. Treatment of the recurrence with FOLFOX resulted in an exceptional response with recanalization of the portal vein which was previously obliterated by tumour and resolution of the mass with complete normalization of CA19.9 levels. b, Platinum responsiveness in patient-derived xenografts. Curves represent relative tumour volume (0.5 × length × width2, y axis) over time (days, x axis). Arrows indicate drug treatment. Responses remain stable for >210 days. Error bars indicate standard error of the mean.
Discussion
This study provides the most comprehensive description, to date, of the genomic events that characterize pancreatic cancer and demonstrates that structural variation is a prominent mechanism of genomic damage in this disease. It reinforces the importance of KRAS, TP53, SMAD4, CDKN2A and ARID1A gene mutations, in addition to numerous genes mutated at low prevalence. Recurrent mutations identified in KDM6A further highlights the role of chromatin modification and a broader role for aberrant WNT signalling is implicated through the relatively frequent inactivation of suppressor genes such as ROBO1, ROBO2, SLIT2 and RNF43.
Structural variant analysis classifies PDAC into four subtypes with potential clinical relevance. A significant proportion of tumours contain amplifications and copy-number gains of known oncogenes, but most occur at low individual prevalence, suggesting significant diversity of mechanisms involved in PDAC progression. Several of these constitute known therapeutic targets with available inhibitors (ERBB2, MET, FGFR1). Others include: GATA6, which is known to be amplified in PDAC and correlates with poor survival in other cancer types35; PIK3CA, which is amplified in ovarian36 and lung squamous cell carcinomas37; PIK3R3 amplified in ovarian cancer; and CDK6, amplified in oesophageal cancer. These may present opportunities for therapeutic intervention, either alone or in combination with other agents.
Multiple studies of platinum-based therapies in PDAC have shown borderline signals, and some meta-analyses show a benefit11,12, suggesting that individual studies were underpowered, and that these signals could be driven by subgroups of responders. More recently, addition of oxaliplatin has shown efficacy in second line therapy14, and FOLFIRINOX, a platinum-containing combination therapy is emerging as a treatment option for advanced PDAC. Most patients do not receive this therapy due to its toxicity, or it is substantially modified38. There are, however, significant responses in subgroups that are not well-defined39,40, and improved survival reported in patients with germline BRCA1 and BRCA2 mutations who receive platinum-based therapies41. Defining biomarkers of platinum responsiveness would significantly alter current treatment approaches to PDAC and improve overall outcomes. Current patient recruitment strategies for clinical trials of PARP inhibitors, thought to target similar mechanisms, are mostly based on germline deleterious mutations of BRCA1 and BRCA2. If we take into account mutations in BRCA pathway components, both germline and somatic, as well as putative surrogate measures of deficiencies in DNA maintenance, that is, unstable genomes and the BRCA mutational signature, germline mutations in BRCA1 and BRCA2 only account for as few as 4 of a potential 24 (17%), and only4%of all patients. Genomic instability and BRCA mutational signature status based on whole-genome sequencing also provide independent evidence of putative deficiencies in DNA damage repair. It remains to be seen whether these surrogate measures are predictive of therapeutic response in the absence of BRCA or PALB2 mutations. However, the presence of mutations in non-BRCA pathway genes that are associated with both genomic instability and chemosensitivity in 2/14 unstable tumours suggests that diagnostic whole-genome sequencing to detect surrogate measures of defects in DNA maintenance may ultimately be a better method of identifying potential responders to platinum and PARP inhibitor therapy.
The proof of concept data presented here suggest that mutations in BRCA pathway component genes and surrogate measures of defects in DNA maintenance (genomic instability and the BRCA mutational signature) have potential implications for therapeutic selection for pancreatic cancer. These data define a putative biomarker hypothesis that needs testing in a clinical trial, as these results are from a small number of patients selected based on high tumour cellularity; patients often received combination therapies, and the primary tumour was sequenced rather than the recurrence. As only selected gene sets can be tested in the clinic at this time, surrogate measures of molecular mechanisms identified using whole-genome sequencing can be used to inform individual gene selection for clinical use. As diagnostic genomic approaches continue to evolve and become more affordable, whole-genome sequencing may provide new opportunities in the clinic. However, there are significant hurdles still to overcome. These include the technical challenge of whole-genome sequencing using small diagnostic samples that are preserved in fixatives such as formalin, analytical demands and the return of results within a clinically relevant timeframe. Major initiatives are emerging that aim to address these challenges (such as Genomics England and the Scottish Genomes Partnership) to ultimately advance and assess these approaches for their potential to improve human health for many diseases including cancer.
METHODS
Human research ethical approvals
Australian Pancreatic Cancer Genome Initiative: Sydney South West Area Health Service Human Research Ethics Committee, western zone (protocol number 2006/54); Sydney Local Health District Human Research Ethics Committee (X11-0220); Northern Sydney Central Coast Health Harbour Human Research Ethics Committee (0612-251M); Royal Adelaide Hospital Human Research Ethics Committee (091107a); Metro South Human Research Ethics Committee (09/QPAH/220); South Metropolitan Area Health Service Human Research Ethics Committee (09/324); Southern Adelaide Health Service/Flinders University Human Research Ethics Committee (167/10); Sydney West Area Health Service Human Research Ethics Committee (Westmead campus) (HREC2002/3/4.19); The University of Queensland Medical Research Ethics Committee (2009000745); Greenslopes Private Hospital Ethics Committee (09/34);North Shore Private Hospital Ethics Committee. Johns Hopkins Medical Institutions: Johns Hopkins Medicine Institutional Review Board (NA00026689). ARC-NET, University of Verona: approval number 1885 from the Integrated University Hospital Trust (AOUI) Ethics Committee (Comitato Etico Azienda Ospedaliera Universitaria Integrata) approved in their meeting of 17 November 2010 and documented by the ethics committee 52070/CE on 22 November 2010 and formalized by the Health Director of the AOUI on the order of the General Manager with protocol 52438 on 23 November 2010. Ethikkommission an der Technischen Universität Dresden (Approval numbers EK30412207 and EK357112012).
Animal experiment approvals
Mouse experiments were carried out in compliance with Australian laws on animal welfare. Mouse protocols were approved by the Garvan Institute/St Vincent’s Hospital Animal Ethics Committee (ARA 09/19, 11/23 and 12/21 protocols). Female NOD/SCID/interleukin 2 receptor [IL2R] gamma (null) (NSG) mice and athymic Balb-c-nude mice were housed with a 12 h light, 12 h dark cycle, receiving food ad libitum.
Sample acquisition
Samples used were prospectively acquired and restricted to primary operable, non-pretreated pancreatic ductal adenocarcinoma. After ethical approval was granted, individual patients were recruited preoperatively and consented using an ICGC approved process. Immediately following surgical extirpation, a specialist pathologist analysed specimens macroscopically and samples of the tumour, normal pancreas and duodenal mucosa were snap frozen in liquid nitrogen (for full protocol see APGI website: http://www.pancreaticcancer.net.au/). The remaining resected specimen underwent routine histopathologic processing and examination. Once the diagnosis of pancreatic ductal adenocarcinoma was made, representative sections were reviewed independently by at least one other pathologist with specific expertise in pancreatic diseases (authors: A.G., D.M., R.H.H. and A.C.), and only those where there was no doubt as to the histopathological diagnosis were entered into the study. Co-existent intraductal papillary mucinous neoplasms in the residual specimen were not excluded provided the bulk of the tumour was invasive carcinoma, and the invasive carcinoma samples were used for sequencing. All samples were stored at −80 °C. Duodenal mucosa or circulating lymphocytes were used for generation of germline DNA. A representative sample of duodenal mucosa was excised and processed in formalin to confirm non-neoplastic histology before processing. All participant information and biospecimens were logged and tracked using a purpose-built data and biospecimen information management system (Cansto Pancreas). Median survival was estimated using the Kaplan–Meier method and the difference was tested using the log-rank test. P values of less than 0.05 were considered statistically significant. Statistical analysis was performed using StatView 5.0 Software (Abacus Systems, Berkeley, CA, USA). Disease-specific survival was used as the primary endpoint.
Sample extraction
Samples were retrieved, and either had full face sectioning performed in OCT or the ends excised and processed in formalin to verify the presence of carcinoma in the sample to be sequenced and to estimate the percentage of malignant epithelial nuclei in the sample relative to stromal nuclei. Macrodissection was performed if required to excise areas of non-malignant tissue. Nucleic acids were then extracted using the Qiagen Allprep Kit in accordance with the manufacturer’s instructions with purification of DNA and RNA from the same sample. DNA was quantified using Qubit HS DNA Assay (Invitrogen). Throughout the process, all samples were tracked using unique identifiers.
Patient material
One hundred matched normal and tumour derived samples were obtained from patients with PDAC. DNA was extracted from the samples using the QiagenAllprep DNA/RNA mini kit method. Tumour cellularity was determined from SNP array data using qpure18. Clinical and sample data are summarized in (Supplementary Table 2). Patients were recruited and consent obtained for genomic sequencing through the Australian Pancreatic Cancer Genome Initiative (APGI) as part of the International Cancer Genome Consortium (ICGC)16.
Patient-derived cell line(PDCL) generation
The PDX-derived primary cell lines, named The Kinghorn Cancer Centre (TKCC) lines, were generated in the laboratory. All cell lines were profiled by short tandem repeat (STR) DNA profiling as unique (http://www.cellbankaustralia.com). Briefly, patient-derived tumours established in immunocompromised mice were mechanically and enzymatically dissociated using collagenase (Stem Cell Technologies, USA) and plated onto flasks coated with 0.2mg ml−1 rat tail collagen (BD Biosciences, USA). Subsequently, epithelial cultures were enriched and purified using a FACS Aria III Cell sorter (BD Biosciences, USA), using a biotinylated anti-mouse MHCI antibody (1:200 dilution; eBiosciences, USA) coupled with Streptavidin AlexaFluor 647 secondary step (1:1,000; Invitrogen, USA) and anti-mouse CD140a-PE antibody (1:300; BD Biosciences, USA) to remove mouse stroma. Dead cells were removed using propidium iodide (Sigma-Aldrich, Australia). Following establishment, all patient-derived (TKCC) cell lines were profiled by short tandem repeat (STR) DNA profiling as unique (http://www.cellbankaustralia.com).
Sequencing
DNA (1 µg) was diluted to 52.5 µl in DNase-/RNase-free molecular biology grade water before fragmentation to approximately 300 bp using the Covaris S2 sonicator with the following settings Duty Cycle 10%, intensity 5, cycles per burst 200, time 50 s or 45 s for PCR-Free libraries. Following fragmentation libraries for sequencing were prepared using the standard Illumina library preparation technique of end-repair, adenylate 3′ ends, indexed adaptor ligation, size selection and finally PCR enrichment for adaptor ligated library molecules following the manufacturer’s recommendations (Part no. 15026486 Rev. C July 2012). A subset of libraries was generated omitting the final PCR enrichment step to generate PCR-Free libraries as per the manufacturer’s recommendations (Part no. 15036187 Rev. A Jan 2013). For standard libraries commercially available TruSeq DNALT Sample Prep Kit v2 (Catalogue no. FC-121-2001) were used for all steps with the following exceptions. Size selections of the Adaptor Ligated fragments were completed using two rounds of SPRI bead purifications (AxyPrepMag PCR Clean-upCatalog no. MAG-PCR-CL-250) using a final bead to DNA volume ratio of 0.60:1 followed by 0.70:1, selecting for molecules with an average size of 500 bp. Size-selected libraries were then amplified for a total of 8 cycles of PCR to enrich for DNA fragments both compatible with sequencing and containing the ligated indexed adaptor. For PCR-Free libraries commercially available TruSeq PCR-Free DNA LT Sample Preparation Kit (Catalog no.FC-121-3001 and FC-121-3002) was used following the 350 bp library LT protocol for all steps with no modifications. The final whole-genome libraries were qualified (amplified and PCR-Free libraries) and quantified (amplified libraries only) via the Agilent BioAnalsyser 2100 (Catalog ID:G2940CA) instrument using the DNA High Sensitivity kit (Catalog ID:5067-4626). Quantification of PCR-Free libraries was performed using the KAPA Library Quantification Kits For Illumina sequencing platforms (Kit code KK4824) in combination with Life Technologies Viia 7 real time PCR instrument.
Whole genome libraries were prepared for cluster generation by cBot (catalogue no. SY-301-2002) and sequencing as per the manufacturer’s guidelines. Individual libraries were clustered on a single lane of a HiSeq v3 flowcell using the TruSeq PE Cluster Kit v3-cBot-HS kit (Catalogue no. PE-401-3001). Illumina supplied control library PhiX (10pM) was spiked into each lane at a concentration of 0.3% to provide real time analysis metrics. Final library concentrations of 8 pM (amplified) and 14 pM (PCR-free) were used for cluster generation. Clustered flow cells were sequenced on the Illumina HiSeq 2000 instrument (HiSeq control software v1.5/Real Time Analysis 1.13) using TruSeq SBS Kit v3-HS (200 cycles, Catalog no. FC-401-3001). Paired reads each of 101 bp were generated for all libraries and in total approximately 220-million paired reads were generated per lane, in line with the manufacturer’s specification. Real time analysis of the control library PhiX showed cluster density, error rates, quality scores, mapping rates and phasing rates were also in line with published specifications.
Sequence alignment and data management
Sequence data was mapped to a genome based on the Genome Reference Consortium (http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/) GRCh37 assembly using BWA42.Multiple BAM files from the same sequence library were merged and within library duplicates were marked. Resulting final BAMs were used as input into variant calling. All BAM files have been deposited in the EGA (Accession number: EGAS 00001000154).
Copy number analysis
Matched tumour and normal patient DNA was assayed using Illumina SNP BeadChips as per manufacturer’s instructions (Illumina, San Diego CA) (HumanOmni1-Quad or HumanOmni2.5–8 BeadChips). SNP arrays were scanned and data was processed using the Genotyping module (v1.8.4) in Genomestudio v2010.3 (Illumina, San Diego CA) to calculate B-allele frequencies (BAF) and logR values. GenoCN43 and GAP17 were used to call somatic regions of copy number change – gain, loss or copy neutral LOH. Recurrent regions of copy number change were determined and genes within these regions were extracted using ENSEMBL v70 annotations.
Identification of structural variations
Somatic structural variants were identified using the qSV tool (manuscript in preparation). qSV uses independent lines of evidence to call structural variants including discordant reads, soft clipping and split read. Breakpoints are also identified using both de novo assembly of abnormally mapping reads and split contig alignment to enhance break point resolution. Depending on the level of evidence qSV bins calls into different categories and calls were considered high confidence if: (i) they were category 1 and therefore contain multiple lines of evidence (discordant pairs, soft clipping on both sides and split reads); (ii) they were category 2 and therefore there was 2 lines of evidence: discordant pairs (both breakpoints) and soft clipping; or discordant pairs (both breakpoints) and split read; or soft clipping (double sided) and split read; (iii) they were category 3 with 10 or more supporting events (discordant read pairs or soft clipping at both ends). Only high confidence calls were used in further downstream analysis. Copy number variation was estimated using SNP arrays and the GAP tool17. Depending on the read pair types supporting an aberration or the associated of copy number events each structural variant was classified as: deletion, duplication, tandem duplication, foldback inversion, amplified inversion, inversion, intrachromosomal or translocation. Essentially, the type of rearrangement is initially inferred from the orientation information of discordant read pairs, soft clipping clusters and assembled contigs which span the breakpoints. This allows identification of 4 groups of events: duplications/intra-chromosomal rearrangements, deletions/intra-chromosomal rearrangements, inversions and inter-chromosomal translocations. Boundaries of segments of copy number that occur in close proximity to each breakpoint were then used to aid further classification of the events. Structural variants with breakpoints that flanked a copy number segment of loss were annotated as deletions. Duplications and inversions associated with increases in copy number enabled the characterization of tandem duplications and amplified or foldback inversions. Events within the same chromosome which linked the ends of copy number segments of similar copy number levels were often identified and were called intra-chromosomal rearrangements.
Events were then annotated if they were within 100 kb of a centromere or telomere and genes which were affected by breakpoints were annotated using ENSEMBL v70. Structural variants and copy number data were visualized using circos44.
The landscape of structural rearrangements in pancreatic ductal adenocarcinoma
In total 11,868 structural variants were detected within the 100 PDAC cohort with an average of 119 events per patient (range 15–558). Each event was classified into one of 8 categories: deletion, duplication, tandem duplication, foldback inversion, amplified inversion, inversion, intra chromosomal and translocation. Within the cohort there was inter patient heterogeneity in terms of total number of events (range of events per patient 15–558) and proportion of event type (Extended Data Fig. 1).
Classification of subtypes based on the pattern or structural rearrangements
Each tumour was classified into one of four subtypes based on the volume of events, the predominance of specific types of structural rearrangement events and the distribution of events across the genome in each patient. In addition to counting structural variation events, two analyses were carried out to detect localized events. Non-random chromosomal clustering of structural variants was detected using an approach originally described by Korbel and Campbell25. Significant clustering of structural variation events was determined by a goodness-of-fit test against the expected exponential distribution of (with a significance threshold of <0.0001). Highly focal events were detected using an adaptation of a method45 where chromosomes with a high structural variant mutation rate per Mb exceeded 5 times the length of the interquartile range from the 75th percentile of the chromosome counts for each patient. The rules used to determine these subtypes are as follows:
Stable
These tumours contain few structural rearrangements (<50) which are located randomly through the genome.
Locally rearranged
The intra-chromosomal rearrangements in these tumours are not randomly positioned through the genome, instead they are clustered on one or few chromosomes. To correct for the different chromosome lengths, the number of events per Mb was calculated for each chromosome within each tumour. Tumours were considered locally rearranged if they harboured at least 50 somatic events within the genome and contained a locally rearranged chromosome. Chromosomes were considered locally rearranged if the number of intrachromosomal events exceeded 5 times the length of the interquartile range from the 75th percentile of the chromosome counts per Mb for that patient. The events in the locally rearranged tumours are broadly comprised of either: (1) focal amplifications—the majority of events are gain (tandem duplication, duplication, foldback inversion or amplified inversion) or (2) complex rearrangements—the events are part of a complex event such as chromothripsis or breakage–fusion–bridge.
Scattered
These tumours contain 50–200 structural rearrangements which are scattered throughout the genome.
Unstable
These tumours are massively rearranged as they contain >200 structural rearrangements which are generally scattered throughout the genome.
Classification of complex localized events
Evidence of clustering of breakpoints was estimated as proposed by Korbel and Campbell25. Chromosomes with clustering of structural variants were reviewed for evidence of chromothripsis (oscillation of copy number, random joins and retention of heterozygosity) and breakage–fusion–bridge (BFB for loss of telomeric region with neighbouring highly amplified region with inversions).
Verification of structural variations
We used two methods of verification for structural variants: (1) an in silico approach, which considers events with multiple lines of evidence (qSV category 1: discordant pairs, soft clipping on both sides and split read evidence) as verified, as well as events which were associated with a copy number change (gain or loss) and (2) orthogonal sequencing methods including SOLiD long mate pair and capillary sequencing.
Long mate pair sequencing and verification of structural rearrangements
Long mate-paired libraries were made according to Applied Biosystems Mate-Paired Library Preparation 5500 Series SOLiD systems protocol using 5 µg of DNA which was sheared using the CovarisS220 System. Long mate pair libraries were sequenced using the SOliD v4 (Applied Biosystems). Sequence data was mapped to a genome based on the Genome Reference Consortium (http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/). GRCh37 assembly using bioscope v1.2.1 (Applied Biosystems). Each sample was sequenced to an average non-redundant physical coverage of 180× (64–333) in the tumour and 187× (52–503) in the control sample. Structural rearrangements were determined by analysing clusters of discordant read pairs using the qSV tool. Events identified by Hiseq sequencing were considered verified if the right and left breakpoint of these events were within 500 bases of the right and left breakpoint of an event identified by SOLiD sequencing.
PCR and capillary sequencing for verification of structural rearrangements
For PCR and capillary sequencing PCR primers were designed with primer BLAST (NCBI) to span the predicted breakpoint, primers were designed with primer BLAST (NCBI). PCR was carried out in the tumour and matched normal genomic DNA using, respectively, a 25 or 50 µl reaction volume composed of 22 or 44 µl of Platinum Taq DNA polymerase (Invitrogen, Carlsbad, Ca), 2 or 4 µl of 10 µM primer (Integrated DNA Technology) and 1 or 2 µl of genomic DNA as template (1 ng µl−1). The following parameters was used for the PCR. Initial denaturation at 94 °C for 2 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s and extension at 68 °C for 1 min; followed by final extension at 68 °C for 15 min. PCR products were visualized by gel electrophoresis and classified into one of four categories: (1) validated—strong and specific PCR band of the expected size was observed only in the tumour and not in the normal sample, this indicates a somatic rearrangement; (2) germline—clear PCR band of the expected size both in the tumour and normal; (3) not validated—PCR yields smears or multiple bands, this potentially indicates non-specific primer pair; (4) not tested—no PCR band was observed in tumour and normal.
Verification of structural variations—results
In total 7,105 events were verified in silico. Of these 5,666 events contained multiple lines of evidence (qSV category 1), 2,904 events were associated with a copy number change (events classified as deletion, duplication, tandem duplication, amplified inversion and foldback inversion) and 1,871 contained multiple lines of evidence and were associated with a copy number change.
We also verified structural variant events using long mate pair resequencing (SOLiD paired 50 bp) or sequencing of a different sample from the same patient of 33 tumours. Using this approach 1,924 events were confirmed and the verification status of structural variant events was recorded in Supplementary Table 5 in the “validation_status_id” column where 0 = untested and 1 = verified. In total 7,228 of the 11,868 events identified (61%) were verified (Supplementary Table 5 and Extended Data Fig. 1) the remaining events remain untested.
Identification of substitutions and small insertion/deletions
Substitutions are called using 2 variant callers: qSNP19 an in-house heuristics-driven somatic/germline caller; and GATK46 which is a Bayesian caller. The two callers were chosen because they use very different calling strategies and while each maybe subject to artefacts (as are all variant callers), they will be subject to different artefacts. Each compared variant falls into one of three categories: seen only by qSNP, seen only by GATK, and seen by both qSNP and GATK. Mutations identified by both callers or those that were unique to a caller and verified by an orthogonal sequencing approach were considered high confidence and used in all subsequent analyses (Supplementary Table 3). Small indels (<200 bp)were identified using Pindel47; each indel was visually inspected in the Integrative Genome Browser (IGV)48. Once somatic mutations were called, their effects on any alternative transcripts were annotated using a local install of the Ensembl database (v70) and the Ensembl Perl API.
Verification of substitutions and small insertion/deletions
In total 3,304 of the 10,335 events identified were verified (Supplementary Tables 3 and 12) the remaining events remain untested. Substitutions and indels were verified using orthogonal sequence data which included data produced on different sequencing platforms (Hiseq or SOLiD exome or long mate pair SOLiD sequencing) or data from related nucleotide samples (RNA-seq). For example, if orthogonal tumour sequence data was available (DNA from a cell line, RNA from the primary sample etc.) and a somatic variant was also observed in the second tumour sample then that would add support for the variant. It should be noted that tumour samples can only be used to support an existing somatic variant and the absence of a called variant in a second tumour sample does not discredit the original call. Conversely, a second normal sample will only discredit somatic variants and the absence of the called variant in the second normal does not support the original call. This approach is designed to be conservative. In order to be considered for verification, an additional BAM should have a minimum of 10 reads at the variant position, and at least two reads must show the variant. If multiple additional BAMs are available, each BAM votes independently and the concordance of the votes is used to classify the verification of the variant. Each variant examined by qVerify is assigned to one of four categories:
Verified—one or more additional tumour BAMs showed evidence of the variant and no additional normal BAMs showed the variant.
False Positive—one or more additional normal BAMs showed evidence of the variant indication that it is likely to be a germline variant.
Mixed—across multiple additional BAMs, there was conflicting evidence – one or more additional tumour BAMs showed the variant as did one or more additional normal BAMs. This could also be evidence of a germline variant incorrectly called somatic.
Untested—there were no additional BAMs or there were additional BAMs but none passed the minimum coverage threshold or there were additional BAMs that did not show the variant and so did not provide evidence for or against it.
Telomere length analysis
Reads containing the telomeric repeat (TTAGGG) × 3 or (CCCTAA) × 3 were counted and normalized to the average genomic coverage (the average base coverage of each genome). The normalized telomere count was obtained separately for each tumour and its matching normal. A ratio was calculated by tumour normalized counts/normal normalized counts.
Determination of the BRCA signature
High confidence somatic mutations that were called by both qSNP and GATK across the genome were used to determine the proportion of the BRCA signature in each sample using a published computational framework20,49. In this way, the 96 substitution classification (as determined by substitution class and sequence context) was determined for each sample and compared to the validated BRCA signature20 and the proportion of the BRCA signature in a given sample was ascertained.
Patient derived xenograft (PDX) mouse model generation
Six female eight-week-old NOD/SCID/interleukin 2 receptor [IL2R] gamma(null) (NOG) mice and athymic Balb-c-nude mice were used for the establishment of the patient derived xenograft (PDX) model. All mice were bred at the Australian Bioresources (ABR) under research protocols approved by the Garvan Animal Ethics Committee (09/19, 11/23, 11/09).
The PDXs were generated according to methodology published elsewhere with modifications50. Briefly, surgical non-diagnostic specimens of patients operated at APGI clinical sites were implanted subcutaneously (s.c.) into three NOG and three Balb-c-nude mice for each patient, with two small pieces per mouse (left and right flank; engraftment stage). Once established, tumours were grown to a size of 1,500mm3, at which point they were harvested, divided, and re-transplanted into further mice to bank sufficient tissues for experimentation (first passage and second passage). After expansion, passaged tumours were excised and propagated to cohorts of 40 female Balb-c-nude mice or greater at an average of 8 weeks old, which constituted the treatment cohort (third passage). Utilization of the NOG mouse model, which is characterized by high immune deficiency in this study has enabled establishment of a significant cohort of PDXs (80) xenografts, with a high rate of successful engraftment and propagation (76%, data not shown).
In vivo therapeutic testing
Tumour-bearing mice with a palpable tumour (volume (V) = 150mm3; V = 0.5 × length × width2) were treated with various agents at maximum tolerable dose (MTD) or vehicle treatment based on previously established schedules50,51, where gemcitabine (140 mg per kg) was administered intraperitoneally on day 1 and day 4 for 4weeks and cisplatin (6mg per kg) intravenously on day 1 and day 14. The investigators were not blinded to the group allocation. To avoid accumulating toxicity of repeated injections, an additional treatment was given after the recovery time of two weeks only when no tumour regression was observed, otherwise treatment was continued once the tumour relapsed to its original size (100%). Measurement of chemotherapy response was based on published methodology51, where primary xenografts were treated with the specified monotherapy and their growth characteristics mapped from the time resistance developed (characterized by progressive tumour growth in the presence of drug), until euthanasia. Mice were euthanized and tissues collected for further analyses when tumour size reached 400% (600–700mm3).
RAD51 foci formation assay
Antibodies used included RAD51 (Clone 14B4, GeneTex), γH2AX (phospho-histone H2AX Ser129 clone 20E3, Cell signaling), and geminin (10802-1-AP, ProteinTech Group, Chicago, IL). Primary culture of PDX from patient ICGC_0016 was established by plating and growing cells from an enzymatically digested xenograft on a collagen matrix for approximately 1 week before irradiation and immunofluorescence staining. For this experiment, xenograft was established in a NSG-eGFP mouse. This mouse model allowed us to efficiently visualize eGFP positive mouse stromal cells and eGFP negative tumour cells under the microscope. Briefly, the eGFP expressing NSG mouse was generated in our laboratory by crossing previously established heterozygous eGFPNOD.CB17-Prkdcscid mice52 with the the NOD/SCID/interleukin 2 receptor (IL2R) gamma (null) (NOG) strain in our laboratory. eGFP expressing offspring was backcrossed five times onto the parental line to ensure homozygosity for IL2Rgamma deletion and confirmed by genotyping (Transnetyx).
Cell lines of interest were grown on coverslips overnight and irradiated with 10 Gy or left untreated. Subsequently coverslips were fixed with 4% paraformaldehyde (in PBS) 6 h post-irradiation and stained with RAD51, γH2AX and geminin antibodies as previously described53.DAPI was used as a nuclear stain. RAD51 focus assay scoring was performed as previously established53.
Extended Data
Extended Data Figure 1. Summary of structural rearrangements.
a, Histogram showing the number of events verified in silico or by orthogonal sequencing methods (Methods). In total 7,228 of the 11,868 events identified (61%) were verified, the others remain untested. These included 5,666 events which contained multiple lines of evidence (qSV category 1: discordant pairs, soft clipping on both sides and split read evidence, Methods) thus were considered verified. Of these events 2,463 events were also verified by orthogonal sequencing methods (SOLiD long mate pair or PCR amplicon sequencing) or the event was associated with a copy number change which was determined using SNP arrays. The remaining 1,562 events were verified using orthogonal sequencing methods or the event was associated with a copy number change (qSV category 2 and 3, Methods). b, Histogram showing the number of structural rearrangements in each pancreatic cancer. 100 PDACs were sequenced using HiSeq paired-end whole-genome sequencing. Structural rearrangements were identified and classified into 8 categories (deletions, duplications, tandem duplications, foldback inversions, amplified inversions, inversions, intra-chromosomal and inter-chromosomal translocations, Methods). The number and type of event for each patient is shown. PDAC shows a high degree of heterogeneity in both the number and types of events per patient. The structural rearrangements were used to classify the tumours into four categories (stable, locally rearranged, scattered and unstable, Methods).
Extended Data Figure 2. Distribution of structural variant breakpoints within each patient.
The 100 patients are plotted along the x axis. The upper plot shows the number of structural rearrangements (y axis) in each tumour. The lower plot shows which chromosomes (y axis) harbour clusters of breakpoints. The distribution of breakpoints (events per Mb) within each chromosome for each sample was evaluated using two methods to identify clusters of rearrangements or chromosomes which contain a large number of events. Method 1: chromosomes with a significant cluster of events were determined by a goodness-of-fit test against the expected exponential distribution (with a significance threshold of <0.0001). Chromosomes which pass these criteria are coloured blue. Method 2: chromosomes were identified which contain significantly more events per Mb than other chromosomes for that patient. Chromosomes were deemed to harbour a high number of events if they had a mutation rate per Mb which exceeds 1.5 times the length of the interquartile range from the 75th percentile of the chromosome counts for each patient. Chromosomes which pass these criteria are coloured orange. Chromosomes which pass both tests they are coloured red. These criteria show that the unstable tumours which contain many events often have significant clusters of events. In contrast locally rearranged tumours are associated with both clusters of events and a high number of events within that chromosome when compared to other chromosomes.
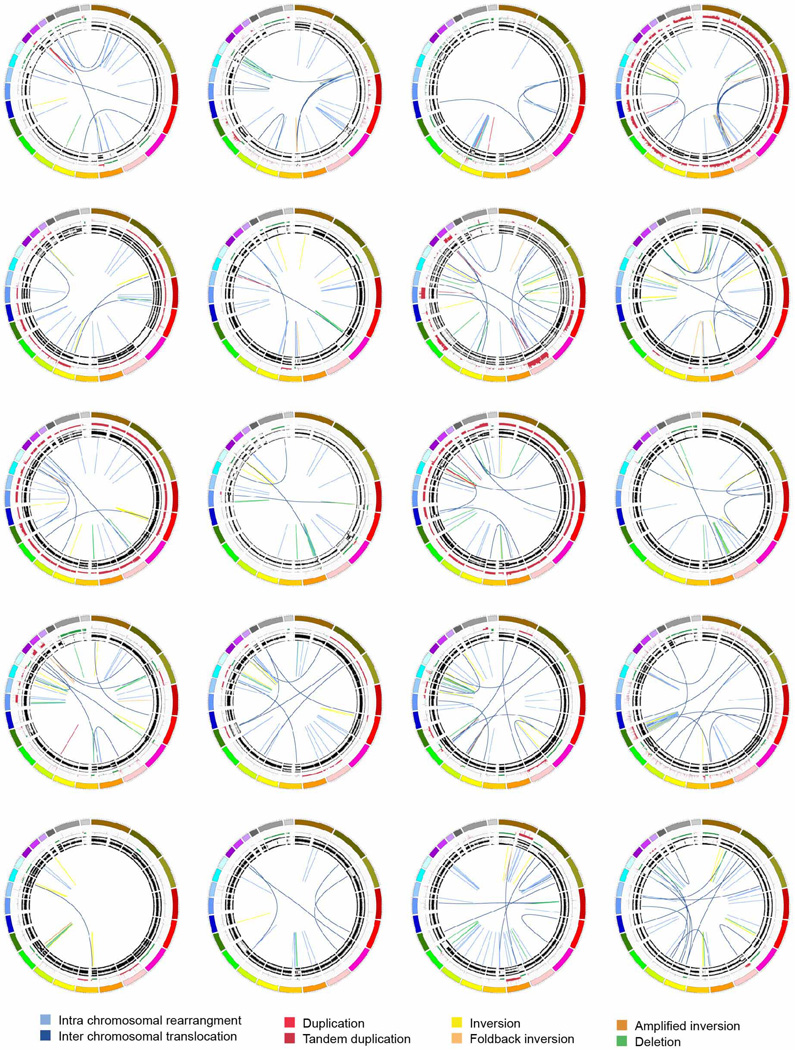
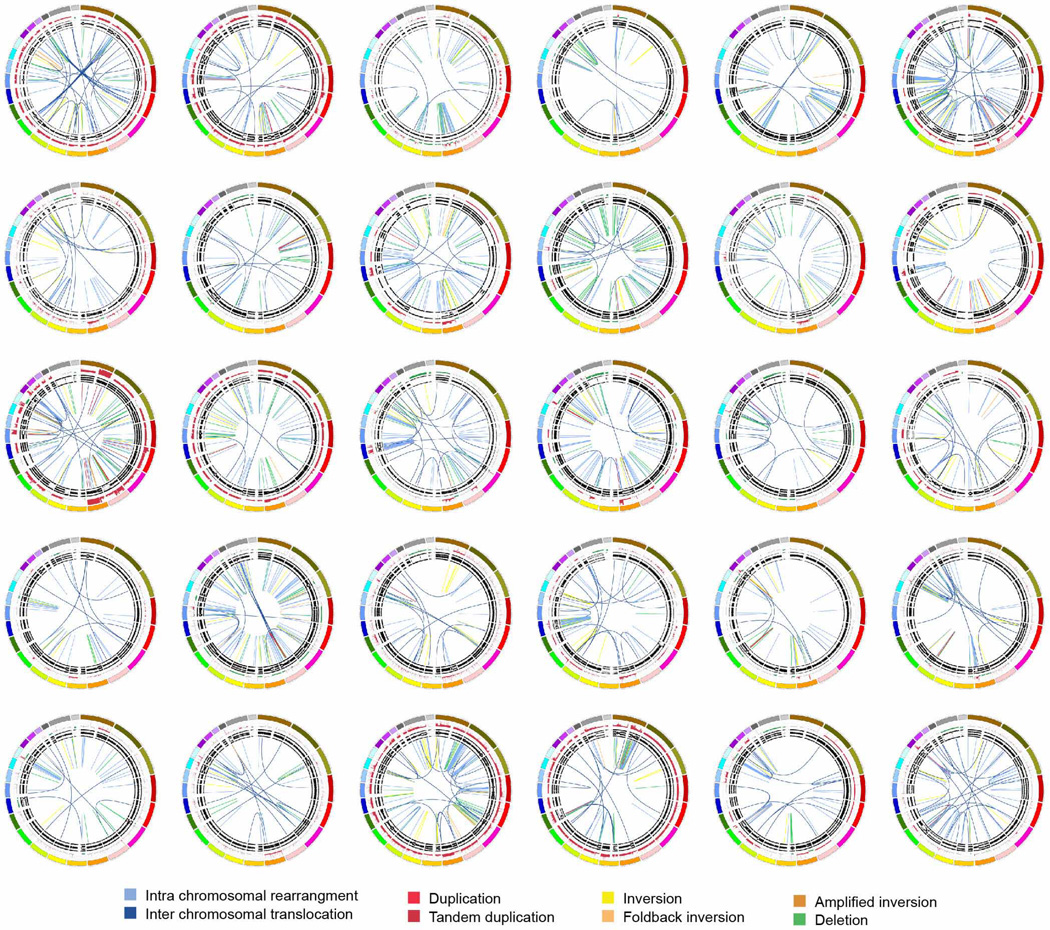
Extended Data Figure 3. The stable subtype in pancreatic ductal adenocarcinoma.
The 20 stable tumours are shown using circos. The coloured outer ring represents the chromosomes, the next ring depicts copy number (red represents gain and green represents loss), the next is the B allele frequency. The inner lines represent chromosome structural rearrangements detected by whole genome paired sequencing and the legend indicates the type of rearrangement. Stable tumours contained less than 50 structural rearrangements in each tumour.
Extended Data Figure 4. The locally rearranged subtype in pancreatic ductal adenocarcinoma.
The 30 locally rearranged tumours are shown using circos. The coloured outer rings represent the chromosomes, the next ring depicts copy number (red represents gain and green represents loss), the next is the B allele frequency. The inner lines represent chromosome structural rearrangements detected by whole-genome paired sequencing and the legend indicates the type of rearrangement. In the locally rearranged subtype over 25% of the structural rearrangements are clustered on one of few chromosomes.
Extended Data Figure 5. Example of evidence for chromothripsis in a pancreatic ductal adenocarcinoma (ICGC_0109).
Upper plot is a density plot showing a concentration of break-points on chromosome 5. Next panel shows the structural rearrangements which are coloured as presented in the legend. The lower panels show copy number, logR ratio and B allele frequency derived from SNP arrays. This chromosome showed a complex localization of events similar to chromothripsis. Copy number profile and structural rearrangements suggest a shattering of chromosome 5 with a high concentration of structural rearrangements, switches in copy number state and retention of heterozygosity, which are characteristics of a chromothriptic event.
Extended Data Figure 6. Example of evidence for breakage-fusion-bridge (BFB) in a pancreatic ductal adenocarcinoma (ICGC_0042).
Upper plot is a density plot showing a concentration of break-points on chromosome 5. Next panel shows the structural rearrangements which are coloured as presented in the legend. The lower panels show copy number, logR ratio and B allele frequency derived from SNP arrays. This chromosome showed a complex localization of events similar to BFB. Copy number profile suggests loss of telomeric q arm and a high concentration of structural rearrangements suggesting a series of BFB cycles, with multiple inversions mapped to the amplified regions.
Extended Data Figure 7. The scattered subtype in pancreatic ductal adenocarcinoma.
The 36 tumours classified as scattered are shown using circos. The coloured outer rings represent the chromosomes, the next ring depicts copy number (red represents gain and green represents loss), the next shows the B allele frequency. The inner lines represent chromosome structural rearrangements detected by whole genome paired end sequencing. The legend indicates the type of rearrangement. The scattered tumours contained 50–200 structural rearrangements in each tumour.
Extended Data Figure 8. The unstable subtype in pancreatic ductal adenocarcinoma.
The 14 unstable tumours are shown using circos. The coloured outer rings are chromosomes, the next ring depicts copy number (red represents gain and green represents loss), the next is the B allele frequency. The inner lines represent chromosome structural rearrangements detected by whole genome paired sequencing and the legend indicates the type of rearrangement. The unstable tumours contained a large degree of genomic instability and harboured over 200 structural rearrangements in each tumour which were predominantly intra-chromosomal rearrangements evenly distributed through the genome.
Extended Data Figure 9. RAD51 foci formation in a primary culture of genomically unstable PDAC.
a, RAD51 and geminin fluorescence in untreated cells derived from an unstable pancreatic tumour with a somatic mutation in the RPA1 gene (ICGC_0016). Primary culture of ICGC_0016 consists of eGFP+ mouse stromal and eGFP− tumour cells. b, Upper panel: irradiated unstable pancreatic cancer cells (ICGC_0016), middle panel: HR competent (TKCC-07) and lower panel: HR-deficient (Capan-1) pancreatic tumour cells. Cells were irradiated in vitro with 10Gy, and 6 h post-irradiation examined by immunofluorescence microscopy. eGFP negative tumour cells from ICGC_0016 readily form RAD51 foci following induction of DNA damage. TKCC-07 is a pancreas cancer cell line generated from a homologous recombination (HR) pathway competent patient-derived xenograft and served as a positive control for staining and RAD51 foci formation after DNA damage. Capan-1 cells which are HR-deficient do not form RAD51 foci. c, RAD51 score (percentage of geminin positive cells that have RAD51 foci) in examined pancreatic tumour cells.
Supplementary Material
Acknowledgements
We would like to thank C. Axford, M.-A. Brancato, S. Rowe, M. Thomas, S. Simpson and G. Hammond for central coordination of the Australian Pancreatic Cancer Genome Initiative, data management and quality control; M. Martyn-Smith, L. Braatvedt, H. Tang, V. Papangelis and M. Beilin for biospecimen acquisition; and D. Gwynne for support at the Queensland Centre for Medical Genomics. We also thank M. Hodgins, M. Debeljak and D. Trusty for technical assistance at Johns Hopkins University. N. Sperandio and D. Filippini for technical assistance at Verona University. We acknowledge the following funding support: National Health and Medical Research Council of Australia (NHMRC; 631701, 535903, 427601); Queensland Government (NIRAP); University of Queensland; Institute for Molecular Bioscience; Cancer Research UK (C596/A18076, C29717/A17263); Australian Government: Department of Innovation, Industry, Science and Research (DIISR); Australian Cancer Research Foundation (ACRF); Cancer Council NSW: (SRP06-01, SRP11-01. ICGC); Cancer Institute NSW: (10/ECF/2-26; 06/ECF/1-24; 09/CDF/2-40; 07/CDF/1-03; 10/CRF/1-01, 08/RSA/1-15, 07/CDF/1-28, 10/CDF/2-26,10/FRL/2-03, 06/RSA/1-05, 09/RIG/1-02, 10/TPG/1-04, 11/REG/1-10, 11/CDF/3-26); Garvan Institute of Medical Research; Avner Nahmani Pancreatic Cancer Research Foundation; University of Glasgow; Cancer Research UK; Howat Foundation; R.T. Hall Trust; Petre Foundation; Philip Hemstritch Foundation; Gastroenterological Society of Australia (GESA); American Association for Cancer Research (AACR) Landon Foundation – INNOVATOR Award; Royal Australasian College of Surgeons (RACS); Royal Australasian College of Physicians (RACP); Royal College of Pathologists of Australasia (RCPA); Italian Ministry of Research (Cancer Genome Project FIRB RBAP10AHJB); Associazione Italiana Ricerca Cancro (12182); Fondazione Italiana Malattie Pancreas – Ministero Salute (CUP_J33G13000210001); Wilhelm Sander Stiftung 2009.039.2; National Institutes of Health grant P50 CA62924.
Appendix
Australian Pancreatic Cancer Genome Initiative (Participants are arranged by institution.)
Garvan Institute of Medical Research Andrew V. Biankin1,2,22, Amber L. Johns1, Amanda Mawson1, David K. Chang1,2,22, Christopher J. Scarlett1, Mary-Anne L. Brancato1, Sarah J. Rowe1, Skye H. Simpson1, Mona Martyn-Smith1, Michelle T. Thomas1, Lorraine A. Chantrill1, Venessa T. Chin1, Angela Chou1, Mark J. Cowley1, Jeremy L. Humphris1, Marc D. Jones1,2, R. Scott Mead1, Adnan M. Nagrial1, Marina Pajic1, Jessica Pettit1, Mark Pinese1, Ilse Rooman1, Jianmin Wu1, Jiang Tao1, Renee DiPietro1, Clare Watson1, Angela Steinmann1,Hong Ching Lee1, Rachel Wong1, Andreia V. Pinho1, Marc Giry-Laterriere1, Roger J. Daly1, Elizabeth A. Musgrove1, Robert L. Sutherland1‡; Queensland Centre for Medical Genomics / Institute for Molecular Biosciences Sean M. Grimmond3, Nicola Waddell3, Karin S. Kassahn3, David K. Miller3, Peter J. Wilson3, Ann-Marie Patch3, Sarah Song3, Ivon Harliwong3, Senel Idrisoglu3, Craig Nourse3, Ehsan Nourbakhsh3, Suzanne Manning3, Shivangi Wani3, Milena Gongora3, Matthew Anderson3, Oliver Holmes3, Conrad Leonard3, Darrin Taylor3, Scott Wood3, Christina Xu3, Katia Nones3, J. Lynn Fink3, Angelika Christ3, Tim Bruxner3, Nicole Cloonan3, Felicity Newell3, John V. Pearson3, Peter Bailey3, Michael Quinn3, Shivashankar Nagaraj3, Stephen Kazakoff3, Nick Waddell3, Keerthana Krisnan3, Kelly Quek3, David Wood3, Muhammad Z. H. Fadlullah3; Royal North Shore Hospital Jaswinder S. Samra4, Anthony J. Gill4, Nick Pavlakis4, Alex Guminski4, Christopher Toon4; Bankstown Hospital Ray Asghari5, Neil D. Merrett5, Darren Pavey5, Amitabha Das5; Liverpool Hospital Peter H. Cosman6, Kasim Ismail6, Chelsie O’Connnor6; Westmead Hospital Vincent W. Lam7 Duncan McLeod7, Henry C. Pleass7, Arthur Richardson7, Virginia James7; Royal Prince Alfred Hospital James G. Kench8, Caroline L. Cooper8, David Joseph8, Charbel Sandroussi8, Michael Crawford8, James Gallagher8; Fremantle Hospital Michael Texler9, Cindy Forest9, Andrew Laycock9, Krishna P. Epari9, Mo Ballal9, David R. Fletcher9, Sanjay Mukhedkar9; Sir Charles Gairdner Hospital Nigel A. Spry10, Bastiaan DeBoer10, MingChai10; St John of God Healthcare Nikolajs Zeps11, Maria Beilin11, Kynan Feeney11; Royal Adelaide Hospital Nan Q. Nguyen12, Andrew R. Ruszkiewicz12, Chris Worthley12, Chuan P. Tan12, Tamara Debrencini12; Flinders Medical Centre John Chen13, Mark E. Brooke-Smith13, Virginia Papangelis13; Greenslopes Private Hospital Henry Tang14, Andrew P. Barbour14; Envoi Pathology Andrew D. Clouston15, Patrick Martin15; Princess Alexandria Hospital Thomas J. O’Rourke16,Amy Chiang16, Jonathan W. Fawcett16, Kellee Slater16, Shinn Yeung16, Michael Hatzifotis16, Peter Hodgkinson16; Austin Hospital Christopher Christophi17, Mehrdad Nikfarjam17, Angela Mountain17; Victorian Cancer Biobank18; Johns Hopkins Medical Institutes James R. Eshleman19, Ralph H. Hruban19, Anirban Maitra19, Christine A. Iacobuzio-Donahue19, Richard D. Schulick19, Christopher L. Wolfgang19, Richard A Morgan19, Mary Hodgin19; ARC-Net Centre for Applied Research on Cancer Aldo Scarpa20, Rita T. Lawlor20, Stefania Beghelli20, Vincenzo Corbo20, Maria Scardoni20, Claudio Bassi20; University of California, San Francisco Margaret A. Tempero21; University of Glasgow Andrew V. Biankin1,2,22, Sean M. Grimmond2,3, David K. Chang1,2,22, Elizabeth A. Musgrove2, Marc D. Jones1,2, Craig Nourse2,3, Nigel B. Jamieson2,22, Janet S, Graham2,22; Greater Glasgow & Clyde National Health Service Andrew V. Biankin1,2,22, David K. Chang1,2,22, Nigel B. Jamieson2,22, & Janet S. Graham2,22, Karen Oien2,22 & Jane Hair22
1The Kinghorn Cancer Centre, Garvan Institute of Medical Research, 370 Victoria Street, Darlinghurst, Sydney, New South Wales 2010, Australia. 2Wolfson Wohl Cancer Research Centre, Institute of Cancer Sciences, University of Glasgow, Garscube Estate, Switchback Road, Bearsden, Glasgow, Scotland G61 1BD, UK. 3Queensland Centre for Medical Genomics, Institute for Molecular Bioscience, University of Queensland, St Lucia, Queensland 4072, Australia. 4Royal North Shore Hospital, Westbourne Street, St Leonards, New South Wales 2065, Australia. 5Bankstown Hospital, Eldridge Road, Bankstown, New South Wales 2200, Australia. 6Liverpool Hospital, Elizabeth Street, Liverpool, New South Wales 2170,Australia. 7Westmead Hospital, Hawkesbury and Darcy Roads, Westmead, New South Wales 2145, Australia. 8Royal Prince Alfred Hospital, Missenden Road, Camperdown, New South Wales 2050, Australia. 9Fremantle Hospital, Alma Street, Fremantle, Western Australia 6959, Australia. 10Sir Charles Gairdner Hospital, Hospital Avenue, Nedlands, Western Australia 6009, Australia. 11St John of God Healthcare, 12 Salvado Road, Subiaco, Western Australia 6008, Australia. 12Royal Adelaide Hospital, North Terrace, Adelaide, South Australia 5000, Australia. 13Flinders Medical Centre, Flinders Drive, Bedford Park, South Australia 5042, Australia. 14Greenslopes Private Hospital, Newdegate Street, Greenslopes, Queensland 4120, Australia. 15Envoi Pathology, 1/49 Butterfield Street, Herston, Queensland 4006, Australia. 16Princess AlexandriaHospital,237 Ipswich Road, Woolloongabba, Queensland 4102,Australia. 17AustinHospital,145 Studley Road, Heidelberg, Victoria3084,Australia. 18Victorian Cancer Biobank, 1 Rathdown Street, Carlton, Victoria 3053, Australia. 19Johns Hopkins Medical Institute, 600 North Wolfe Street, Baltimore, Maryland 21287, USA. 20ARC-NET Center for Applied Research on Cancer, University of Verona, Via dell’Artigliere, 19 37129 Verona, Province of Verona, Italy. 21University of California, San Francisco, 500 Parnassus Avenue, San Francisco, California 94122, USA. 22Greater Glasgow and Clyde National Health Service,1053GreatWestern Road, Glasgow G12 0YN, UK.
‡Deceased.
Footnotes
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
Author Contributions Biospecimens were collected at affiliated hospitals and processed at each biospecimen core resource centre. Data generation and analyses were performed by the Queensland Centre for Medical Genomics. Investigator contributions are as follows: A.V.B. and S.M.G. (concept and design); S.M.G., J.V.P. N.W., A.V.B. (project leaders); N.W., S.M.G., D.K.C., A.V.B. (writing team); J.V.P., S.M.G., N.W., A.L.J., P.B., S.S., K.S.K., Nk. W., P.J.W., A.M.P., F.N., B.P., E.M., O.H., J.L.F., C.L., D.T., S.W., Q.X., K.N., N.C., M.C.J.Q., M.J.A., M.Z.H.F., A.J.R., S.K., K.Q., M. Pi., H.C.L., M.J.C. and J.W. (bioinformatics); M. Pa., C.J.S., D.K.C., E.S.H., A.M.N., A.C., A.S., C.S., A.V.P., I.R., A.M.S., S.P.N., R. B. (preclinical testing); A.L.J., M.D.J., M.P., C.J.S., C.T., A.M.N., V.T.C., L.A.C., J.S.S., D.K.C., V.C., A.S., C.S., A.J.G., J.A.L., I.R., A.V.P., E.A.M. (sample processing and quality control); A.J.G., J.G.K., C.T., G.Z., A.S., D.A. R.H.H., A.M., C.A.I-D., A.S. (pathology assessment); A.L.J., L.A.C., A.J.G., A.C., R.S.M., C.B., M.F., G.T., J.S.S., J.G.K., C.T., K.E., N.Q.N., N.Z., H.W., N.B.J., J.S.G, R.G., C.P., R.G., C.L.W., R.A.M., R.T.L., M.F., G.Z., G.T., M.A.T., A.P.G.I., J.R.E., R.H.H.,A.M., C.A.I-D.,A.S. (sample collection and clinical annotation); D.M., T.J.C.B., A.N.C., I.H., S.I., S.M., C.N., E.N., S.W. (sequencing). All authors have read and approved the final manuscript.
BAM files and associated metadata in XML format have been uploaded to the European Genome-phenome Archive (EGA; http://www.ebi.ac.uk/ega) under accession number EGAS00001000154. All SNP array data is available via GEO (GSE61502). For more information about Australian Pancreatic Cancer Genome Initiative, see (http://www.pancreaticcancer.net.au/apgi/collaborators).
The authors declare no competing financial interests.
References
- 1.Vogelzang NJ, et al. Clinical cancer advances 2011: annual report on progress against cancer from the American Society of Clinical Oncology. J. Clin. Oncol. 2012;30:88–109. doi: 10.1200/JCO.2011.40.1919. [DOI] [PubMed] [Google Scholar]
- 2.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harada T, et al. Genome-wide DNA copy number analysis in pancreatic cancer using high-density single nucleotide polymorphism arrays. Oncogene. 2008;27:1951–1960. doi: 10.1038/sj.onc.1210832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephens PJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin CA, et al. Consistent chromosome abnormalities in adenocarcinoma of the pancreas. Cancer Res. 1995;55:2394–2399. [PubMed] [Google Scholar]
- 8.Molenaar JJ, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 9.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conroy T, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 11.Sultana A, et al. Meta-analyses of chemotherapy for locally advanced and metastatic pancreatic cancer. J. Clin. Oncol. 2007;25:2607–2615. doi: 10.1200/JCO.2006.09.2551. [DOI] [PubMed] [Google Scholar]
- 12.Ciliberto D, et al. Role of gemcitabine-based combination therapy in the management of advanced pancreatic cancer: a meta-analysis of randomised trials. Eur. J. Cancer. 2013;49:593–603. doi: 10.1016/j.ejca.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Heinemann V, Boeck S, Hinke A, Labianca R, Louvet C. Meta-analysis of randomized trials: evaluation of benefit from gemcitabine-based combination chemotherapy applied in advanced pancreatic cancer. BMC Cancer. 2008;8:82. doi: 10.1186/1471-2407-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oettle H, et al. Second-line oxaliplatin, folinic acid, and fluorouracil versus folinic acid and fluorouracil alone for gemcitabine-refractory pancreatic cancer: outcomes from the CONKO-003 Trial. J. Clin. Oncol. 2014;32:2423–2429. doi: 10.1200/JCO.2013.53.6995. [DOI] [PubMed] [Google Scholar]
- 15.Kaufman B, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J. Clin. Oncol. 2015;33:244–250. doi: 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International Cancer Genome Consortium et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popova T, et al. Genome alteration print (GAP): a tool to visualize and mine complex cancer genomic profiles obtained by SNP arrays. Genome Biol. 2009;10:R128. doi: 10.1186/gb-2009-10-11-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song S, et al. qpure: a tool to estimate tumor cellularity from genome-wide single-nucleotide polymorphism profiles. PLoS ONE. 2012;7:e45835. doi: 10.1371/journal.pone.0045835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kassahn KS, et al. Somatic point mutation calling in low cellularity tumors. PLoS ONE. 2013;8:e74380. doi: 10.1371/journal.pone.0074380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mann KM, et al. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc. Natl Acad. Sci. USA. 2012;109:5934–5941. doi: 10.1073/pnas.1202490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang X, et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc. Natl Acad. Sci. USA. 2013;110:12649–12654. doi: 10.1073/pnas.1307218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 26.Tutt A, et al. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr. Biol. 1999;9:1107–1110. doi: 10.1016/s0960-9822(99)80479-5. [DOI] [PubMed] [Google Scholar]
- 27.Jones S, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hellebrand H, et al. Germline mutations in the PALB2 gene are population specific and occur with low frequencies in familial breast cancer. Hum. Mutat. 2011;32:E2176–E2188. doi: 10.1002/humu.21478. [DOI] [PubMed] [Google Scholar]
- 29.Nikkilä J, et al. Heterozygous mutations in PALB2 cause DNA replication and damage response defects. Nature Commun. 2013;4:2578. doi: 10.1038/ncomms3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nones K, et al. Genome-wide DNA methylation patterns in pancreatic ductal adenocarcinoma reveal epigenetic deregulation of SLIT-ROBO, ITGA2 and MET signaling. Int. J. Cancer. 2014;135:1110–1118. doi: 10.1002/ijc.28765. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, et al. Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nature Genet. 2005;37:750–755. doi: 10.1038/ng1587. [DOI] [PubMed] [Google Scholar]
- 32.Doles J, et al. Suppression of Rev3, the catalytic subunit of Polξ, sensitizes drug-resistant lung tumors to chemotherapy. Proc. Natl Acad. Sci. USA. 2010;107:20786–20791. doi: 10.1073/pnas.1011409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang DK, Grimmond SM, Evans TRJ, Biankin AV. Mining the genomes of exceptional responders. Nature Rev. Cancer. 2014;14:291–292. doi: 10.1038/nrc3723. [DOI] [PubMed] [Google Scholar]
- 34.Eisenhauer EA, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur. J. Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 35.Deng N, et al. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut. 2012;61:673–684. doi: 10.1136/gutjnl-2011-301839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peddi PF, et al. Multi-institutional experience with FOLFIRINOX in pancreatic adenocarcinoma. JOP. 2012;13:497–501. doi: 10.6092/1590-8577/913. [DOI] [PubMed] [Google Scholar]
- 39.Villarroel MC, et al. Personalizing cancer treatment in the age of global genomic analyses: PALB2 gene mutations and the response to DNA damaging agents in pancreatic cancer. Mol. Cancer Ther. 2011;10:3–8. doi: 10.1158/1535-7163.MCT-10-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Showalter SL, et al. Identifying pancreatic cancer patients for targeted treatment: the challenges and limitations of the current selection process and vision for the future. Expert Opin. Drug Deliv. 2010;7:273–284. doi: 10.1517/17425240903544462. [DOI] [PubMed] [Google Scholar]
- 41.Golan T, et al. Overall survival and clinical characteristics of pancreatic cancer in BRCA mutation carriers. Br. J. Cancer. 2014;111:1132–1138. doi: 10.1038/bjc.2014.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 42.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun W, et al. Integrated study of copy number states and genotype calls using high-density SNP arrays. Nucleic Acids Res. 2009;37:5365–5377. doi: 10.1093/nar/gkp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nik-Zainal S, et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nature Genet. 2014;46:487–491. doi: 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rubio-Viqueira B, et al. An in vivo platform for translational drug development in pancreatic cancer. Clin. Cancer Res. 2006;12:4652–4661. doi: 10.1158/1078-0432.CCR-06-0113. [DOI] [PubMed] [Google Scholar]
- 51.Rottenberg S, et al. Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proc. Natl Acad. Sci. USA. 2007;104:12117–12122. doi: 10.1073/pnas.0702955104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niclou SP, et al. A novel eGFP-expressing immunodeficient mouse model to study tumor-host interactions. FASEB J. 2008;22:3120–3128. doi: 10.1096/fj.08-109611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Graeser M, et al. A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clin. Cancer Res. 2010;16:6159–6168. doi: 10.1158/1078-0432.CCR-10-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.