Abstract
The auto-phosphorylation of murine receptor-interacting protein 3 (Rip3) on Thr 231 and Ser 232 in the necrosome is required to trigger necroptosis. However, how Rip3 phosphorylation is regulated is still largely unknown. Here we identified protein phosphatase 1B (Ppm1b) as a Rip3 phosphatase and found that Ppm1b restricts necroptosis in two settings: spontaneous necroptosis caused by Rip3 auto-phosphorylation in resting cells, and tumour necrosis factor-α (TNF)-induced necroptosis in cultured cells. We revealed that Ppm1b selectively suppresses necroptosis through the dephosphorylation of Rip3, which then prevents the recruitment of mixed lineage kinase domain-like protein (Mlkl) to the necrosome. We further showed that Ppm1b deficiency (Ppm1bd/d) in mice enhanced TNF-induced death in a Rip3-dependent manner, and the role of Ppm1b in inhibiting necroptosis was evidenced by elevated Rip3 phosphorylation and tissue damage in the caecum of TNF-treated Ppm1bd/d mice. These data indicate that Ppm1b negatively regulates necroptosis through dephosphorylating Rip3 in vitro and in vivo.
Necroptosis is a kind of programmed necrosis that relies on the receptor-interacting serine/threonine kinase 1 (Rip1) and Rip3 (ref. 1), although Rip1 is dispensable in some conditions2–4. It has been found to be involved in many physiological and pathological processes, such as host defence against viruses, inflammation, tissue injury, eye diseases and cancers5,6. Although necroptosis can be triggered by various stimuli, our knowledge on the necroptotic pathway is mainly obtained through the study of that induced by TNF. Under TNF stimulation, a multiple protein complex (termed the necrosome) forms to initiate necroptosis1. Rip1 and Rip3 are the core components in the necrosome and interact through their RIP homotypic interaction motif7–11 (RHIM). Rip3 in the necrosome recruits and phosphorylates the pseudokinase Mlkl (refs 12–14). The phosphorylated Mlkl translocates to the cytomembrane to execute necroptosis15–18. The phosphorylation of Rip3 (Thr 231 and Ser 232 in mouse Rip3; Ser 227 in human Rip3) is essential for the recruitment of Mlkl to the necrosome13,19. Here we identified Ppm1b as a Rip3 phosphatase and showed that Ppm1b suppressed necroptosis both in culture cells and in vivo by dephosphorylating Rip3.
RESULTS
Identification of Ppm1b as a Rip3 phosphatase
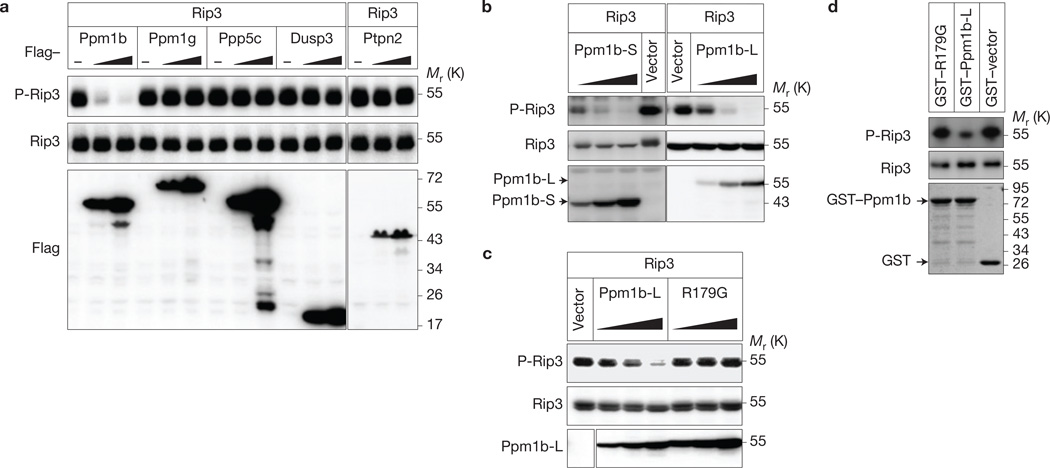
As a result of the requirement of Rip3 auto-phosphorylation in necroptosis13,19, we searched for Rip3 phosphatase(s) that regulate Rip3 phosphorylation. By mass spectrometry analysis of murine Rip3 immunocomplexes, we identified five phosphatases, Ppm1b, protein phosphatase 1G (Ppm1g), serine/threonine-protein phosphatase 5 (Ppp5c), dual-specificity phosphatase 3 (Dusp3) and tyrosine-protein phosphatase non-receptor type 2 (Ptpn2; Supplementary Table 1). The three known proteins in the Rip3 complex including Rip1, FAS-associated death domain protein (Fadd) and Mlkl were found in the immunocomplexes, so these phosphatases could be Rip3-interacting proteins. As Rip3 underwent auto-phosphorylation when overexpressed in 293T cells19, we coexpressed each of these phosphatases with Rip3 in 293T cells to evaluate their phosphatase activity towards Rip3 and found that Ppm1b but not the other phosphatases reduced the phosphorylation of Rip3 (Fig. 1a).
Figure 1.
Identification of Ppm1b as a Rip3 phosphatase. (a) Ppm1b dephosphorylates Rip3 in 293T cells. Rip3 was coexpressed with or without increasing amounts of phosphatases in 293T cells. The cells were lysed and immunoblotted with the indicated antibodies 36 h post transfection. The vertical line represents a splice mark. The samples were obtained and processed in the same experiment, and the gels/blots were processed in parallel. (b) Both Ppm1b-L and Ppm1b-S dephosphorylate Rip3 in a dose-dependent manner. Rip3 was coexpressed with or without increasing amounts of Ppm1b-S and Ppm1b-L in 293T cells. The cells were lysed and immunoblotted with the indicated antibodies 36 h post transfection. (c) Ppm1b phosphatase activity is required for dephosphorylating Rip3. Rip3 was coexpressed with or without increasing amounts of Ppm1b-L and its phosphatase-deficient mutant R179G in 293T cells. The cells were analysed as in b. The vertical line represents a splice mark. The spliced images were from the same blot. (d) Ppm1b dephosphorylates Rip3 in vitro. Flag–Rip3 (purified from 293T cells) was incubated with recombinant GST–Ppm1b-L, GST–R179G and GST–vector in vitro. The phosphorylated Rip3 and total Rip3 were detected by immunoblotting with the indicated antibodies. Recombinant Ppm1b and R179G were shown by Coomassie blue staining. Data shown are representative of two or more independent experiments. Uncropped images of blots are shown in Supplementary Fig. 7.
There are seven splicing variants of murine Ppm1b encoding four proteins with different carboxy-terminal sequences. Splicing variants encoding isoform 1 (relative molecular mass 55,000 (Mr55K), termed Ppm1b-L) and isoform 2 (Mr43K, termed Ppm1b-S) are expressed ubiquitously20–22. The Ppm1b complementary DNA used in the above experiments encodes Ppm1b-L. Ppm1b-S is also capable of lowering Rip3 phosphorylation in a dose-dependent manner (Fig. 1b). The phosphatase dead mutant of Ppm1b (R179G, Arg 179 to glycine mutation23) failed to reduce the phosphorylation of Rip3, confirming that Ppm1b is a phosphatase of Rip3 (Fig. 1c). The data obtained by using bacterially expressed recombinant GST–Ppm1b-L and GST–R179G mutant demonstrated that Ppm1b dephosphorylates Rip3 directly (Fig. 1d).
Ppm1b is likely to associate with Rip3 in resting cells because we can detect its interaction with Rip3 in L929 cells (Supplementary Fig. 1a) and when the two proteins were coexpressed in 293T cells (Supplementary Fig. 1b). The interaction is specific as Ppm1b interacts with Rip3 but not the other proteins involved in the necroptosis, such as Fadd, caspase-8, Mlkl or Rip1 (Supplementary Fig. 1c). Ppm1b contains an amino-terminal phosphatase domain (PD) and a C-terminal domain (CD; Supplementary Fig. 1d). Whereas the CD was suggested to determine the substrate specificity of Ppm1b (ref. 23), we found that PD is responsible for the interaction with Rip3 (Supplementary Fig. 1e). We also found that the kinase domain (KD) of Rip3 is responsible for its interaction with Ppm1b (Supplementary Fig. 1f).
Ppm1b protects cells from Rip3-dependent spontaneous necroptosis
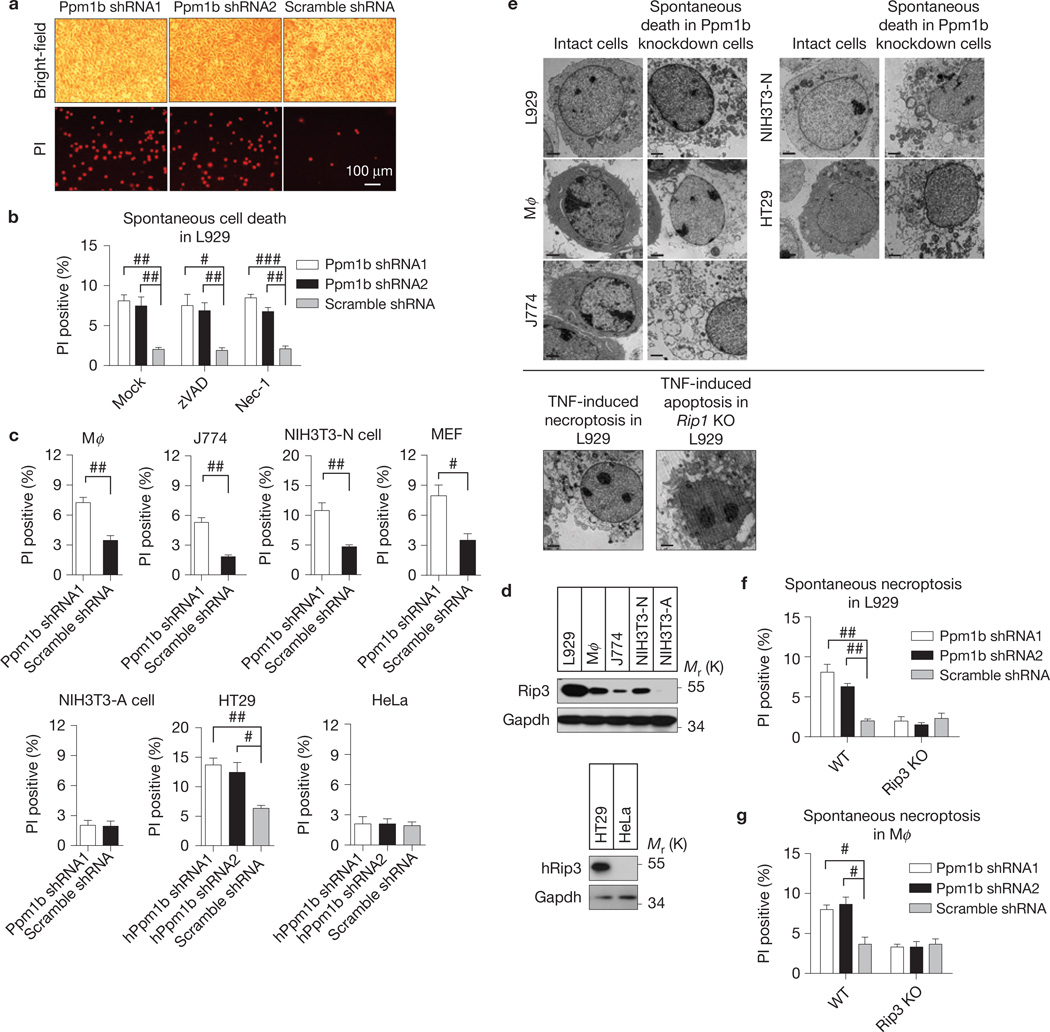
Both Ppm1b-L and Ppm1b-S are expressed in L929, a cell line commonly used in studying TNF-induced necroptosis (Supplementary Fig. 2a). Knockdown of Ppm1b by short hairpin RNAs (shRNAs; targeting both L and S isoforms, Supplementary Fig. 2a) increased spontaneous cell death (Fig. 2a). Different L929 sublines vary in their sensitivity to zVAD-induced cell death24. To avoid zVAD-induced cell death, we used a L929 line that resists zVAD-induced death to determine whether the spontaneous cell death is caspase-dependent. The spontaneous cell death is not dependent on caspase activity or Rip1 kinase activity, as neither general caspase inhibitor zVAD nor Rip1 kinase inhibitor necrostatin-1 (Nec-1; ref. 25) blocked the cell death (Fig. 2b). Ppm1b-L and Ppm1b-S seemed to have an additive effects because the spontaneous cell death became obvious only in Ppm1b-L and Ppm1b-S double-knockdown cells (Supplementary Fig. 2b). Unless specifically indicated, Ppm1b knockdown in the rest of our studies refers to knockdown of both Ppm1b-L and Ppm1b-S.
Figure 2.
Ppm1b protects cells from Rip3-dependent spontaneous necroptosis. (a) Ppm1b was knocked down by lentivirus-delivered shRNAs targeting both the Ppm1b-L and Ppm1b-S isoforms in L929 cells. Forty-eight hours after infection, the cells were stained with propidium iodide (PI) and analysed under a microscope. Dead cells were PI positive. Scale bar, 100 µm. shScramble: non-target control shRNA. The protein expression levels are shown in Supplementary Fig. 2a. (b) Ppm1b was knocked down by lentivirus-delivered shRNA in L929 cells. Six hours after infection, these cells were treated with DMSO, zVAD (20 µM) and Nec-1 (30 µM). Spontaneous cell death was analysed by PI staining under a microscope 48 h later. n = 3,000 cells pooled from three independent experiments. (c) Ppm1b was knocked down in different mouse and human cell lines. Spontaneous cell death was counted by PI staining under a microscope 48 h (72 h for Mφ and J774) later. Mφ: mouse peritoneal macrophage. The protein expression levels are shown in Supplementary Fig. 2c. n = 3,000 cells pooled from three independent experiments. (d) The Rip3 expression level correlates with the cell death caused by Ppm1b knockdown in the cells analysed in c. (e) The morphology of Ppm1b knockdown-induced spontaneous dead cells was analysed by transmission electron microscopy. Scale bar, 2 µm. TNF-induced necroptosis in L929 cells and TNF-induced apoptosis in Rip1 KO L929 cells were included as a control. (f,g) Ppm1b was knocked down in WT, Rip3 KO L929 cells (f) and WT, Rip3 KO Mφ (g). Then, spontaneous cell death was analysed. n = 3,000 cells pooled from three independent experiments. The protein expression levels are shown in Supplementary Fig. 2d,e. For b,c,f,g, results shown are mean ± s.e.m.; #P <0.05, ##P <0.01, ###P <0.001. For a,d,e data shown are representative of two or more independent experiments. Statistics source data for this figure can be found in Supplementary Table 2. Uncropped images of blots are shown in Supplementary Fig. 7.
We next examined the effect of Ppm1b knockdown on cell viability in a panel of different types of cell. Knockdown of Ppm1b in primary peritoneal macrophages (Mφ), macrophage cell line J774, NIH3T3-N (ref. 8), mouse embryonic fibroblast (MEF) and HT29 cells (Supplementary Fig. 2c) increased spontaneous cell death (Fig. 2c), but had no effect on cell death in NIH3T3-A (ref. 8) and HeLa cells (Fig. 2c and Supplementary Fig. 2c). As NIH3T3-A and HeLa cells lack Rip3 expression, whereas all the others express Rip3 (ref. 8; Fig. 2d), the Ppm1b knockdown-mediated increase of spontaneous cell death seems to be dependent on the presence of Rip3. Meanwhile, using transmission electron microscopy, we found necrotic morphology in Ppm1b knockdown-induced death of L929, Mφ, J774, NIH3T3-N and HT29 cells (Fig. 2e). Furthermore, Ppm1b knockdown (Supplementary Fig. 2d,e) increased spontaneous cell death in WT but not Rip3 KO L929 and Mφ cells (Fig. 2f,g), confirming that Ppm1b knockdown-mediated cell death is Rip3-dependent.
Ppm1b prevents Rip3 auto-activation in resting cells
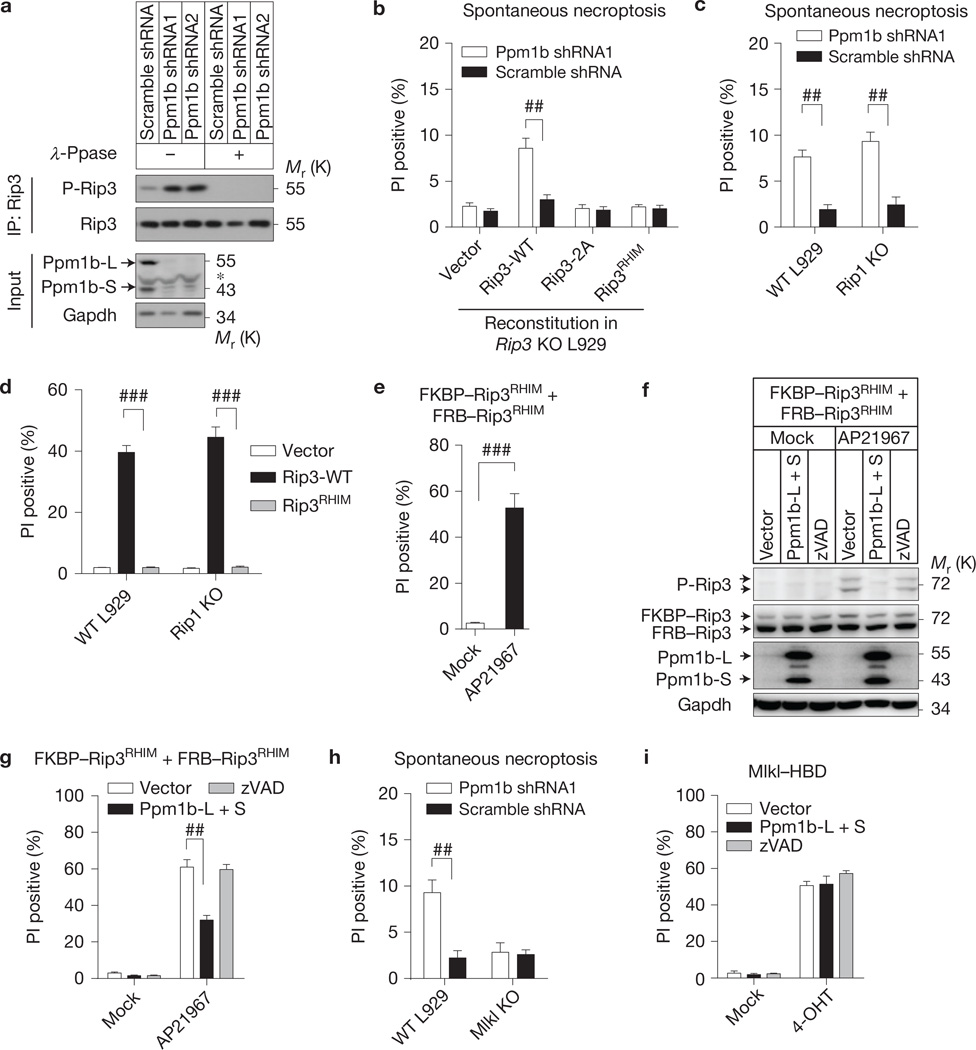
We addressed whether knockdown of Ppm1b would increase the basal phosphorylation level of Rip3. Rip3 was immunoprecipitated from WT and Ppm1b knockdown L929 cells, and its phosphorylation level was measured by immunoblotting with anti-phospho-Rip3 antibody. Rip3 phosphorylation was indeed increased in Ppm1b knockdown cells (Fig. 3a). The specificity of the phospho-Rip3 antibody was confirmed by immunoblotting with the samples treated with λ-phosphatase (λ-Ppase). Reintroducing Rip3 WT but not the Rip3 phosphorylation-deficient mutant (Thr 231 and Ser 232 mutated to alanine, termed Rip3-2A) into Rip3 KO L929 cells restored the sensitivity of L929 cells to Ppm1b depletion-induced necroptosis (Fig. 3b and Supplementary Fig. 3a), indicating that the upregulation of Rip3 phosphorylation corresponds to spontaneous necroptosis.
Figure 3.
Ppm1b prevents Rip3 auto-activation and it mediated spontaneous necroptosis. (a) Endogenous Rip3 was immunoprecipitated with anti-Rip3 antibody in Ppm1b knockdown and control cells. The immunoprecipitates were treated with or without λ-Ppase and subsequently immunoblotted with the indicated antibodies. (b) Rip3 KO L929 cells reconstituted with Rip3-WT, Rip3-2A and Rip3RHIM mutants were infected with Ppm1b or control shRNA. Spontaneous necroptosis was analysed. Protein expression levels are shown in Supplementary Fig. 3a. (c) Ppm1b knockdown-induced spontaneous cell death in WT and Rip1 KO L929 cells was analysed. Protein expression levels are shown in Supplementary Fig. 3b. (d) WT Rip3, Rip3RHIM and empty vector were introduced into WT and Rip1 KO L929 by lentiviral vectors. Necroptosis was analysed 24 h later. Protein expression levels are shown in Supplementary Fig. 3c. (e) FKBP–Rip3RHIM and FRB–Rip3RHIM mutants were introduced into Rip1–Rip3 DKO L929 and forced to interact with each other by treatment of dimerizer AP21967; subsequent cell death was analysed (see Methods). (f,g) Cells expressing FKBP–Rip3RHIM and FRB–Rip3RHIM were infected with lentiviral vector encoding Ppm1b (co-infection of Ppm1b-L and Ppm1b-S) or with empty vector, or treated with zVAD (20 µM). Twenty-four hours later, cells were treated with or without AP21967 (250 nM) for 2 h. (f) cells were then lysed and immunoblotted; (g) cell death was analysed. (h) Ppm1b knockdown-induced spontaneous cell death in WT and Mlkl KO L929 cells was analysed. Protein expression levels are shown in Supplementary Fig. 3d. (i) Mlkl KO L929 cells expressing Mlkl–HBD were analysed as in g, except that 4-OHT (1 µM) rather than AP21967 was used to induce Mlkl oligomerization-mediated cell death. For b–e,g–i, results shown are mean ± s.e.m.; n = 3,000 cells pooled from three independent experiments; ##P <0.01, ###P <0.001. For a,f, data shown are representative of two or more independent experiments. The asterisk denotes a nonspecific band. Statistics source data for this figure can be found in Supplementary Table 2. Uncropped images of blots are shown in Supplementary Fig. 7.
Reintroducing the Rip3 RHIM domain mutant (QIG449–451AAA mutation in RHIM, termed Rip3RHIM) to Rip3 KO L929 cells cannot rescue the spontaneous necroptosis (Fig. 3b and Supplementary Fig. 3a), suggesting that the RHIM-mediated Rip3 interaction is essential for the spontaneous necroptosis. Rip3 also interacts with Rip1 through its RHIM (ref. 26), but Ppm1b knockdown increased spontaneous death in Rip1 KO L929 cells to a level similar to that observed in WT L929 cells (Fig. 3c and Supplementary Fig. 3b), indicating that Ppm1b depletion-induced spontaneous necroptosis is not Rip1-dependent. This is consistent with the data of Nec-1 treatment (Fig. 2b). As RHIM is essential for Rip3 overexpression-induced Rip1-independent cell death process (Fig. 3d and Supplementary Fig. 3c), we examined whether RHIM is directly required for targeting Rip3 by Ppm1b. We expressed FK506 binding protein (FKBP)-fused Rip3RHIM (FKBP–Rip3RHIM) and FKBP–rapamycin binding domain (FRB)-fused Rip3RHIM (FRB–Rip3RHIM) in Rip1–Rip3 double knockout cells by lentiviral vectors27 and created RHIM-independent Rip3 homo-interaction by the addition of AP21967 to induce interaction of FKBP and FRB (Fig. 3e). This RHIM-independent Rip3–Rip3 interaction drove the auto-phosphorylation of Rip3 (Fig. 3f), and Ppm1b still attenuated the phosphorylation level (Fig. 3f) and blocked the cell death (Fig. 3g).
Mlkl is a downstream effector of Rip3 in the necroptosis pathway12–15,17,18,28 and we confirmed that Ppm1b knockdown did not increase spontaneous cell death in Mlkl KO cells (Fig. 3h and Supplementary Fig. 3d). Consistently, the cell death caused by the hormone binding domain of oestrogen receptor (HBD)-mediated oligomerization of the Mlkl–HBD fusion protein17 cannot be affected by Ppm1b (Fig. 3i). We also excluded a role of TNF autocrine29 in Ppm1b depletion-induced spontaneous necroptosis by using TNFR1 KO L929 (ref. 30; Supplementary Fig. 3e). Collectively, our data suggest that the spontaneous cell death is initiated by Rip3 RHIM-mediated auto-activation and its downstream mechanismismost likely the same as TNF-induced necroptosis.
Ppm1b negatively regulates TNF-induced necroptosis
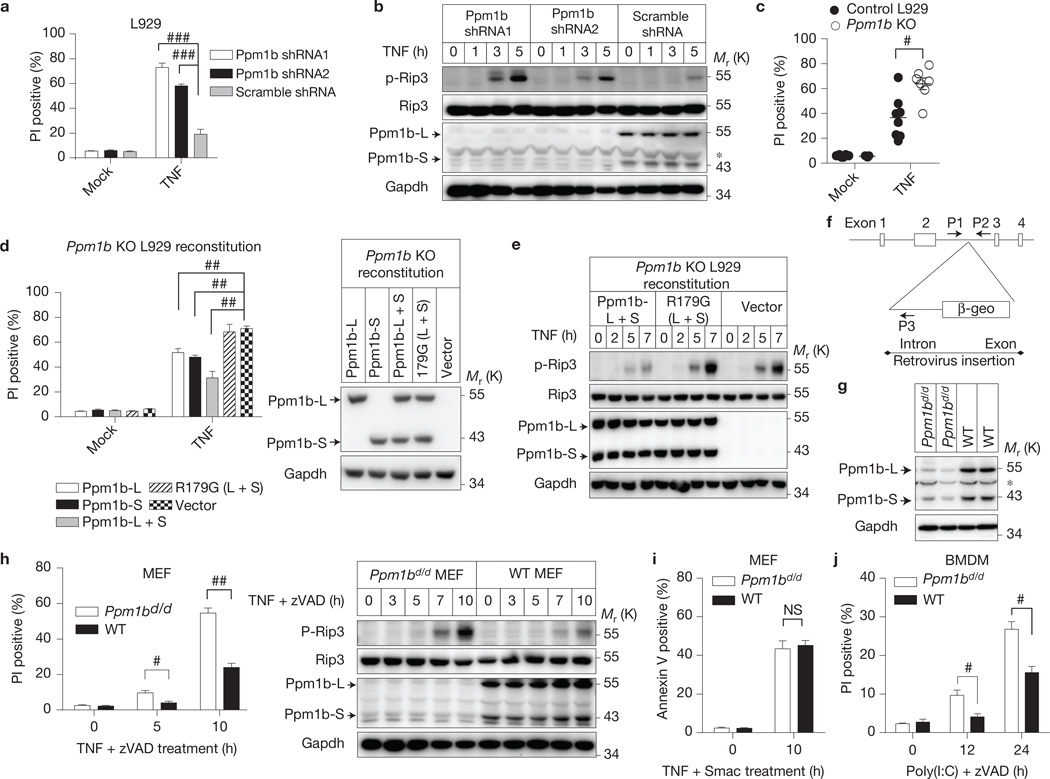
As Rip3 phosphorylation is required for TNF-induced necroptosis13,19, we next investigated whether Ppm1b affects TNF-induced necroptosis. The dead cells caused by spontaneous death in culture were removed before the analyses (see Methods). Compared with the control, knockdown of Ppm1b enhanced TNF-induced necroptosis in L929 cells (Fig. 4a and Supplementary Fig. 4a,b). This enhanced necroptosis correlates with the increase of Rip3 phosphorylation in L929 cells (Fig. 4b). In contrast, knockdown of Ppm1b in NIH3T3-A cells did not affect TNF-induced apoptosis (Supplementary Fig. 4c). Both Ppm1b-L and Ppm1b-S are important in this process as knockdown of either of them exhibited a less enhancing effect than double knockdown (Supplementary Fig. 4d). We knocked out the Ppm1b gene in L929 cells and selected a number of Ppm1b knockout lines. As controls, we also isolated several Ppm1b-expressing colonies during the process of knocking out Ppm1b. Perhaps owing to the growth selection in culture, none of the Ppm1b KO lines showed a higher level of spontaneous cell death than control L929 cells. However, Ppm1b knockout cells were more sensitive to TNF-induced cell death (Fig. 4c). Reconstitution of either Ppm1b-L or Ppm1b-S expression in Ppm1b KO cells attenuated TNF-induced cell death, and re-expression of both of them led to a severe inhibitory effect on TNF-induced cell death, but re-expression of R179G mutants showed no difference from the control (Fig. 4d). It seemed that these two Ppm1b isoforms exhibited no functional difference in inhibiting necroptosis and the inhibitory effect was decided by their overall expression level. We observed that the infection of L929 cells with 10 moi virus encoding Ppm1b-L (L) + 10 moi virus encoding Ppm1b-S (S) (20 moi in total) had a similar effect to the infection with 20 moi L or 20 moi S alone (Supplementary Fig. 4e); and the infection of L929 cells with 20 moi L + 20 moi S had a stronger effect than that with 20 moi L or S alone (Supplementary Fig. 4e). The level of TNF-induced Rip3 phosphorylation in Ppm1b-reconstituted L929 cells was lower than in control cells (Fig. 4e), but the level in Ppm1b R179G mutant-expressing cells was not (Fig. 4e), supporting the role of Ppm1b in dephosphorylating Rip3. Importantly, Ppm1b is specific for TNF-induced phospho-Rip3 dephosphorylation in L929 cells, as Ppm1b knockdown had no effect on TNF-induced p65 and p38 phosphorylation (Supplementary Fig. 4f). Together, these data demonstrate that Ppm1b dephosphorylates Rip3 and thus negatively regulates TNF-induced necroptosis in L929 cells.
Figure 4.
Ppm1b negatively regulates TNF-induced necroptosis. (a,b) L929 cells were infected with Ppm1b or control shRNA. Forty-eight hours later, spontaneous dead cells were removed by re-plating the cells. (a) These cells were then treated with or without TNF (10 ng ml−1) for 5 h. Cell death was analysed by flow cytometer; n = 3 independent experiments. (b) These cells were treated with TNF for the indicated time and then analysed by immunoblotting. (c) TNF-induced necroptosis was analysed in Ppm1b KO (n = 7 clones) and WT (n = 8 clones) L929 cells. n is representative of a single experiment where two independent experiments were performed to assess reproducibility. (d) Ppm1b KO L929 cells were reconstituted with Ppm1b-L, Ppm1b-S, both isoforms or the R179G mutants. Left, TNF-induced necroptosis was analysed. n = 3 independent experiments. R179G (L + S): Ppm1b-L R179G and Ppm1b-S R179G mutants. Right, cell lysates were immunoblotted. (e) Ppm1b KO L929 cells reconstituted with Ppm1b or its mutants were treated with TNF for the indicated time. Then cell lysates were immunoblotted. (f) Ppm1d/d mice with a gene-trap vector inserted in the second intron of Ppm1b genomic locus. (g) Cell lysates of primary Ppm1bd/d and WT MEF cells were immunoblotted. (h) The immortalized Ppm1bd/dand WT MEF cells were treated with TNF (30 ng ml−1) plus zVAD (20 µM) for the indicated time. Left, cell death was analysed. n = 3 independent experiments. Right, cell lysates were immunoblotted. (i) Ppm1bd/d and WT MEF cells were treated with TNF (30 ng ml−1) plus Smac mimetic (Smac; 100 nM) for the indicated time. Then apoptosis was measured by Annexin V staining. n = 3 independent experiments. (j) BMDMs from Ppm1bd/d and WT mice were treated with poly(I:C) (100 µg ml−1) plus zVAD (20 µM) for the indicated time, and then cell death was analysed. n = 3 independent experiments. For a,c,d,h–j, results shown are mean ± s.e.m.; NS: no significant difference, #P < 0.05, ##P < 0.01, ###P < 0.001. For b,e,g,h, data shown are representative of two independent experiments. The asterisk denotes a nonspecific band. Statistics source data can be found in Supplementary Table 2. Uncropped images of blots are shown in Supplementary Fig. 7.
As Ppm1b KO mouse is pre-implantation lethal31, we generated a Ppm1b gene-trap mouse line (Ppm1b+/d) using embryonic stem cells containing a retroviral gene trap in the second intron of the Ppm1b locus (Fig. 4f). The insertion resulted in the expression of a Ppm1b-β-galactosidase-neomycin (β-geo) fusion protein containing 1–282 amino acids of Ppm1b, which lacks a functional phosphatase domain (1–300 amino acids; Supplementary Fig. 4g) and thus is a loss-of-function mutant. The Ppm1bd/d mice are viable and born with a normal Mendelian distribution. The normal development of Ppm1bd/d mice is due to the expression of a small amount of wild-type Ppm1b because we detected Ppm1b-L and Ppm1b-S protein in MEF cells isolated from Ppm1bd/d mice (Fig. 4g). The expression of Ppm1b most probably resulted from the splicing out of the intron in which the retroviral vector inserts. Similar to what was observed in L929 cells, Ppm1bd/d MEF cells are more sensitive to TNF + zVAD-induced Rip3 phosphorylation and necroptosis (Fig. 4h). It was reported that kinase dead Rip3 promotes Rip1-dependent apoptosis in mice and MEF cells32.We questioned whether Ppm1b regulates Rip1-dependent apoptosis and found that TNF plus Smac mimetic (Smac)-induced apoptosis33,34 in Ppm1bd/d and WT MEF cells were comparable (Fig. 4i). It should be mentioned that the dephosphorylation of Rip3 should not affect the kinase activity of Rip3 because mutation of the phosphorylation sites (Thr 231 and Ser 232 to alanine) did not affect the kinase activity of Rip3 (ref. 19). We also found that Ppm1b knockdown enhanced poly(I:C)+zVAD-induced necroptosis of bone marrow-derived macrophages (BMDMs; Fig. 4j).
The regulation of necroptosis by Ppm1b is independent of the NF-κB pathway
Ppm1b can dephosphorylate inhibitor of nuclear factor κ-B kinase subunit β (IKKβ) and negatively regulate NF-κB activation in HeLa cells35 (Supplementary Fig. 5a). However, the knockdown of Ppm1b in L929 cells (Supplementary Fig. 4f) and HT29 cells (Supplementary Fig. 5b) did not affect NF-κB p65 phosphorylation. TNF-induced interleukin 6 (IL-6) production was also not influenced by Ppm1b knockdown in L929 cells (Supplementary Fig. 5c). Thus, Ppm1b regulates NF-κB in some but not all types of cell. Using the IKKβ inhibitors TPCA-1 (ref. 36) and IMD 0354 (ref. 37), and an IKKβ knockout L929 line, we showed that the role of Ppm1b in necroptosis has no linkage to IKKβ (Supplementary Fig. 5d,e,f). TGF-β activated kinase 1 (TAK1) was also reported to be dephosphorylated by Ppm1b (ref. 38). However, Ppm1b knockdown still enhanced TNF-induced necroptosis in the presence of the TAK1 inhibitor 5Z-7-oxozeaenol (5z-7; ref. 39; Supplementary Fig. 5g). NF-κB activation is known to play a pro-survival role in TNF-stimulated cells, which was also the result we saw here in that the IKKβ and TAK1 inhibition or depletion increased cell death in TNF-treated cells (Supplementary Fig. 5e,g). However, our data demonstrate that the regulation of necroptosis by Ppm1b is independent of the NF-κB pathway.
As Ppm1b inhibits IKKβ in HeLa but not in L929 and HT-29 cells, and Rip3 is expressed in L929 and HT-29 but not in HeLa (Fig. 2d), one would ask whether the presence of Rip3 makes Ppm1b unable to target IKKβ. We used HeLa-hRip3 (HeLa cells stably expressing human Rip3) to address this question. Similar to that in control HeLa cells (Supplementary Fig. 5a), the knockdown of Ppm1b enhanced the NF-κB p65 phosphorylation in HeLa-hRip3 cells (Supplementary Fig. 5h). Besides, spontaneous and TNF (TNF plus Smac mimetic plus zVAD (TSZ; ref. 13))-induced necroptosis in HeLa-hRip3 cells was also enhanced by Ppm1b knockdown (Supplementary Fig. 5i). The effects of IKKβ inhibitors were the same as what was observed in L929 cells (Supplementary Fig. 5i). These data again indicate that the effects of Ppm1b on Rip3 and IKKβ are independent of each other. Different from L929 and HT-29 cells, the two functions of Ppm1b in HeLa-hRip3 cells coexist. As Ppm1b depletion promotes necroptosis, the Ppm1b-Rip3 pathway seems to be more dominant than the Ppm1b- IKKβ pathway in HeLa-hRip3 cells. We also measured the serum IL-6 levels and found that they were comparable in TNF-treated Ppm1bd/d and WT mice (Supplementary Fig. 5j), which suggested that the Ppm1b-Rip3 pathway might be the predominant one in vivo.
Ppm1b suppresses Mlkl recruitment to the necrosome on TNF stimulation
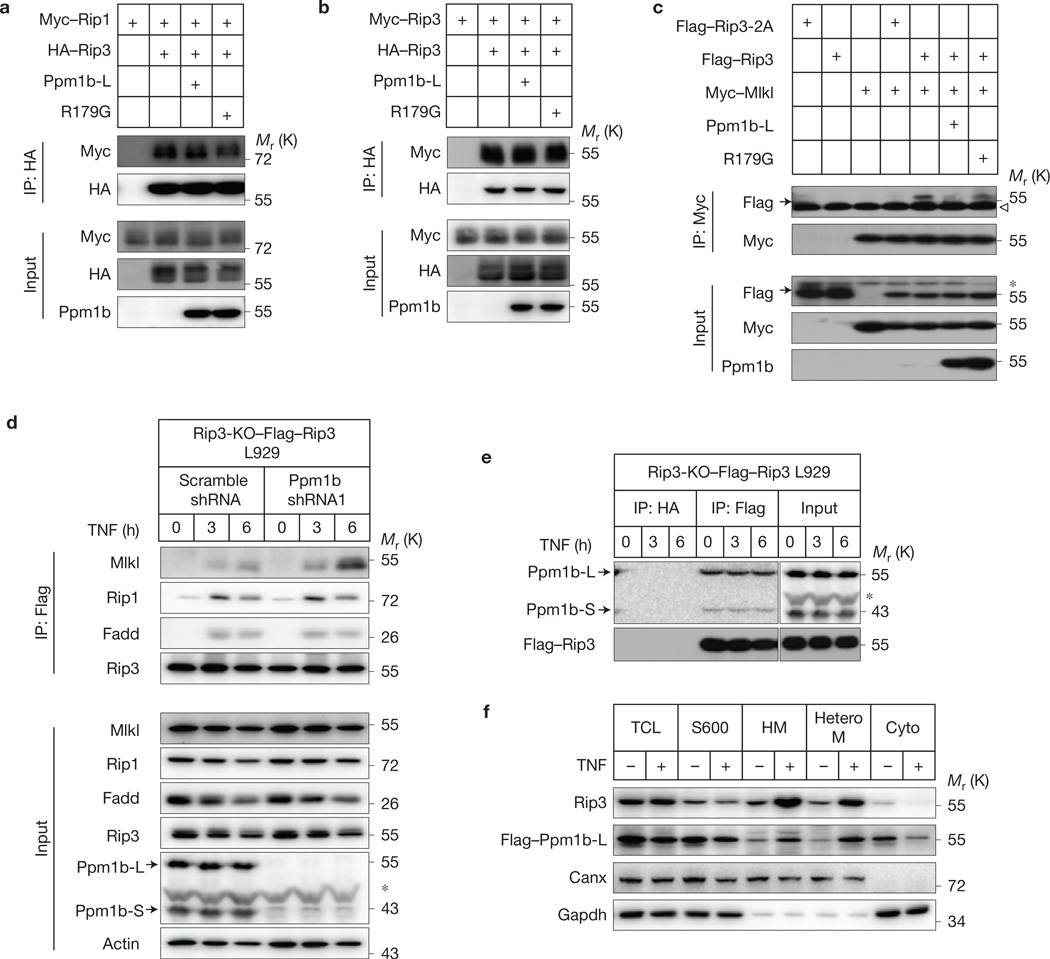
As Rip3 phosphorylation is essential for the Rip3–Mlkl interaction but not the Rip1–Rip3 interaction13,19,40, we tested whether Ppm1b affects the interaction of Rip1–Rip3, Rip3–Rip3 and Rip3–Mlkl by co-immunoprecipitation in 293T cells and found that Ppm1b-L attenuated the interaction between Rip3 and Mlkl, but not the interaction between Rip1 and Rip3 or Rip3 and Rip3 (Fig. 5a – c). To analyse the recruitment of Mlkl to the necrosome in TNF-stimulated cells, we used Rip3 KO L929 cells reconstituted with Flag–Rip3 (Rip3-KO– Flag–Rip3) because Rip3-KO–Flag–Rip3 cells behave similarly to WT L929 cells in necroptosis and the Flag tag facilitates immunoprecipitation with the advantage of specific anti-Flag monoclonal antibody. Whereas knockdown of Ppm1b did not affect the assembly of the necrosome, as we were able to detect similar levels of Rip1 and Fadd proteins in the immunoprecipitates of Flag–Rip3 in control and Ppm1b knockdown cells after TNF stimulation (Fig. 5d), the recruitment of Mlkl to Flag–Rip3-containing necrosome was significantly increased in Ppm1b knockdown compared with control cell. It is unclear how Ppm1b is regulated, as we observed consistent association of Ppm1b with Rip3 before and after TNF stimulation in L929 cells (Fig. 5e) and translocation of both Ppm1b and Rip3 to a heterotypic membrane fraction on TNF treatment41 (Fig. 5f). Nonetheless, these data demonstrate that Ppm1b-mediated dephosphorylation of Rip3 impairs Rip3-mediated recruitment of Mlkl to the necrosome.
Figure 5.
Ppm1b suppresses Mlkl recruitment to the necrosome under TNF stimulation. (a–c) 293T cells were transfected with the plasmids as indicated. Cell lysates were immunoprecipitated with anti-HA antibody (IP: HA) or anti-Myc antibody (IP: Myc). The lysates (Input) and immunoprecipitates were immunoblotted. (d) Ppm1b was knocked down in Rip3-KO–Flag–Rip3 L929 cells, and then the cells were treated with TNF (10 ng ml−1) for the indicated time. The necrosome was analysed by immunoprecipitation with anti-Flag antibody (IP: Flag). The cell lysates (Input) and immunocomplexes were analysed by immunoblotting with the indicated antibodies. (e) Rip3-KO–Flag– Rip3 L929 cells were treated with TNF for the indicated time. The cell lysates were immunoprecipitated with anti-HA (IP: HA) and anti-Flag (IP: Flag) antibodies and analysed by immunoblotting with the indicated antibodies. The vertical line represents a splice mark. The samples were obtained and processed in the same experiment, and the gels/blots were processed in parallel, except that the exposure time of Ppm1b for the immunoprecipitates is ten times that for the input. (f) L929 cells expressing Flag–Ppm1b-L were treated with or without TNF for 6 h. Then cells were subjected to subcellular fractionation (see Methods). Each fraction was subjected to immunoblotting with the indicated antibodies. TCL: total cell lysate; S600: supernatant fraction after centrifugation at 600g; HM: heavy membrane fraction; HeteroM: heterotypic membrane fraction. Canx: ER marker. Data shown are representative of two or more independent experiments. The arrowhead denotes the heavy chain and the asterisk denotes a nonspecific band. Uncropped images of blots are shown in Supplementary Fig. 7.
Ppm1a does not regulate necroptosis
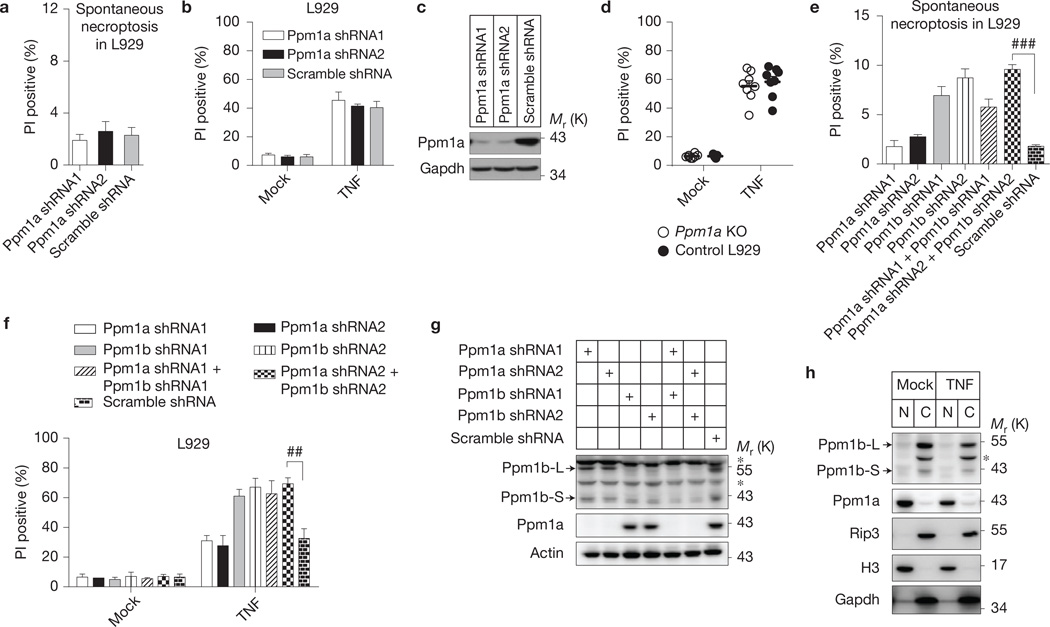
Protein phosphatase 1A (Ppm1a) is the closest homologue of Ppm1b. However, knockdownofPpm1a neither caused spontaneous cell death nor changed the sensitivity of L929 cells to TNF-induced cell death (Fig. 6a – c). Knockout of Ppm1a by CRIPSR-Cas9 technology also did not sensitize L929 cells to TNF killing (Fig. 6d). Double knockdown of Ppm1a and Ppm1b did not exhibit an additive effect on spontaneous necroptosis or TNF-induced cell death compared with Ppm1b single knockdown (Fig. 6e – g). The lack of effect on necroptosis could be due to the nuclear localization of Ppm1a (ref. 42), because Ppm1b and Rip3 were found in the cytosol fraction (Fig. 6h).
Figure 6.
Ppm1a does not negatively regulate necroptosis. (a–c) Ppm1a was knocked down in L929 cells. (a) Forty-eight hours later, spontaneous cell death was measured; n = 3,000 cells pooled from three independent experiments. (b) The cells were treated with or without TNF (10 ng ml−1) for 6 h and cell death was analysed by flow cytometer; n = 3 independent experiments. (c) The cell lysates were immunoblotted with the indicated antibodies. (d) Ppm1a KO L929 cells were generated by CRISPR/Cas9 technology. Ppm1a KO (n = 8 clones) and WT (n = 8 clones) L929 were treated with or without TNF. Then cell death was analysed. n is representative of a single experiment where two independent experiments were performed to assess reproducibility. (e–g) Ppm1a does not compensate for Ppm1b in restricting necroptosis. Ppm1a and Ppm1b were knocked down respectively or together. (e) Forty-eight hours later, spontaneous cell death was recorded; (f) the cells were treated with or without TNF and then cell death was analysed; (g) the cell lysates were immunoblotted with the indicated antibodies. n = 3 independent experiments. (h) Different subcellular localization of Ppm1a and Ppm1b. L929 cells were treated with or without TNF for 6 h. Then nucleus and cytosol fractions of the cells were isolated by subcellular fractionation. These fractions were analysed by immunoblotting with the indicated antibodies. N: nucleus fraction; C: cytosol fraction; H3: Histone H3, a nucleus marker; Gapdh: a cytosol marker. For a,b,d–f, results shown are mean ± s.e.m.; ##P < 0.01; ###P <0.001. For c,g,h, data shown are representative of two or more independent experiments. The asterisk denotes a nonspecific band. Statistics source data can be found in Supplementary Table 2. Uncropped images of blots are shown in Supplementary Fig. 7.
Ppm1b protects mice from TNF-induced caecum damage through dephosphorylating Rip3
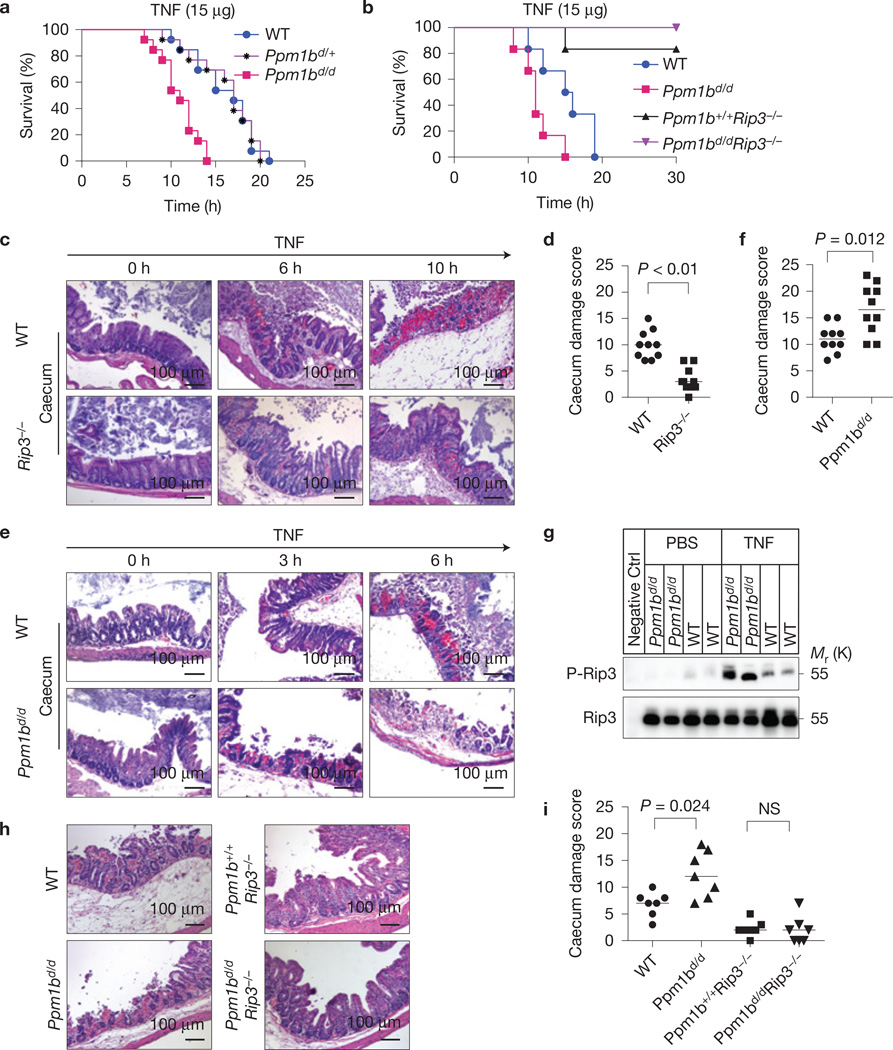
TNF-induced necroptosis is essential for TNF-induced systemic inflammatory response syndrome (SIRS) as Rip3 KO mice are resistant to TNF-induced death32,43,44. Littermates of WT and Ppm1bd/d mice were injected intravenously with mouse TNF, with Ppm1bd/d mice being more sensitive to TNF-induced death than WT mice (Fig. 7a). An underlying mechanism of this in vivo result is likely to be that Ppm1b suppresses TNF-induced necroptosis in this mouse model. However, Ppm1b has been reported to negatively the regulate the NF-κB pathway35, which might also affect TNF-induced SIRS. We examined Ppm1bd/d Rip3−/− mice and found that the effect of Ppm1b deficiency on TNF-induced mouse death was completely prevented by Rip3 knockout (Fig. 7b). Thus, the dephosphorylation of Rip3 should be the in vivo function of Ppm1b in TNF-treated mice.
Figure 7.
Ppm1b protects mice from TNF-induced SIRS through dephosphorylating Rip3. (a) Ppm1bd/d, Ppm1bd/+ and WT mice raised as littermates were injected with 15 µg TNF through the tail vein. Mouse survival is presented as a Kaplan–Meyer plot and log-rank test was performed. n = 13 mice for each group pooled from three independent experiments. Ppm1bd/d versus WT, P < 0.001. (b) WT, Ppm1bd/d, Ppm1b+/+ Rip3−/−and Ppm1bd/d Rip3−/− mice were analysed as in a. n = 6 mice of a single experiment where two independent experiments were performed to assess reproducibility. (c,d) Caecum of WT and Rip3−/− mice injected with TNF for the indicated time was sectioned and stained with H&E. The representative images are shown in c. At the 6 h time point, n = 10 mice for each group. At other time points, n = 4 mice. Original magnification: ×200; scale bar, 100 µm. The TNF-induced caecum damage at the 6 h time point was scored (see Methods) and is shown in d. n is representative of a single experiment where two independent experiments were performed to assess reproducibility. Wilcoxon signed-rank test, P <0.01. (e,f) Sections of the caecum of WT and Ppm1bd/d mice were analysed as in c,d. Wilcoxon signed-rank test, P = 0.012. (g) WT and Ppm1bd/d mice were injected with PBS or TNF (15 µg) for 6 h. Then the caecum was collected and lysed. Rip3 protein was enriched from the lysate through immunoprecipitation and subsequently subjected to immunoblotting with p-Rip3 and Rip3 antibodies. A Rip3−/− mouse was used as a negative control (negative CTRL). (h,i) WT, Ppm1bd/d Ppm1b+/+Rip3−/− and Ppm1bd/d Rip3−/− mice were injected with TNF for 6 h and subsequently analysed as in c,d, except that n = 7 mice for each group of a single experiment where two independent experiments were performed to assess reproducibility; Wilcoxon signed-rank test; NS: no significant difference. Statistics source data for this figure can be found in Supplementary Table 2. Uncropped images of blots are shown in Supplementary Fig. 7.
To better understand the role of Ppm1b in vivo, we first analysed necroptosis-mediated tissue injury in TNF-treated mice. Unlike in previous reports43,44, we did not find that Rip3 deletion affects TNF-induced injuries in liver and kidney (Supplementary Fig. 6). Among spleen, thymus, small intestine, liver, kidney, heart, lung, pancreas and caecum, caecum was the only organ whose injury was attenuated by Rip3 knockout (Fig. 7c,d and Supplementary Fig. 6). It is worth noting that caecum was described as particularly sensitive to TNF-induced injury in the report of the first TNF-SIRS model45. We then analysed the effect of Ppm1b and found that TNF-induced caecum damage was enhanced in Ppm1bd/d mice (Fig. 7e,f). Analysis of the caecum lysate showed that TNF-induced phosphorylation of Rip3 in Ppm1bd/d was higher than that in WT mice (Fig. 7g). Moreover, the enhanced caecum damage in TNF-treated Ppm1bd/d mice was completely prevented by Rip3 knockout (Fig. 7h,i). These data further support that Ppm1b functions in restricting necroptosis by dephosphorylating Rip3 in vivo.
DISCUSSION
In this study we identified a Rip3 phosphatase that negatively regulates Rip3-dependent spontaneous and TNF-induced necroptosis. Although the interaction of Ppm1b with Rip3 in the resting stage explains the role of Ppm1b in preventing spontaneous necroptosis, how Ppm1b is regulated during TNF-induced necroptosis is unclear. The expression level and the cytosolic localization of Ppm1b did not change during the course of TNF stimulation. Protein kinase A (PKA) was shown to regulate protein stability of Ppm1b (ref. 46), but activation of PKA by forskolin treatment did not change the protein abundance of Ppm1b in L929. As Ppm1b can be activated by fatty acids in vitro47 and Ppm1b translocated to the heterotypic membrane fraction on TNF treatment, whether Ppm1b activity was increased in the lipid-enriched fraction is interesting to know but technically inaccessible at this moment. Consistent association with its substrates has been observed in many phosphatases, including protein phosphatase 1D (also termed as Wip1), which interacts with ataxia-telangiectasia mutated kinase (ATM) before and after ionizing radiation and regulates the ATM-dependent signalling pathways48.
It was shown that gene deletion of Ppm1b led to early pre-implantation lethality in mice31. Although Rip3 knockout can rescue spontaneous necroptosis in Ppm1b knockdown cells, Rip3 KO cannot rescue the lethality of Ppm1b gene deletion. A Rip3-independent function of Ppm1b should be responsible for the normal development through the period of pre-implantation. The identification of Ppm1b as a Rip3 phosphatase is encouraging because it not only advances our understanding of the necroptosis pathway but also provides a tool to manipulate necroptosis in vitro and in vivo. The information presented in this report could be useful in exploring the contribution of necroptosis to various physiopathological conditions.
METHODS
Antibodies and reagents
Anti-mRip3, p-Rip3, Fadd and Mlkl antibodies were described previously19. Anti-Ppm1b antibody was raised in rabbit using GST-tagged full-length human Ppm1b-L as the antigen. Anti-human Rip3 (ab72106, dilution 1:1,000) and anti-Ppm1a (ab14824, dilution 1:1,000) antibodies were from Abcam. Anti-Flag (F7425, dilution 1:500), Myc (A7470, agarose affinity gel) and HA (A2095, agarose affinity gel) antibodies were obtained from Sigma. Anti-Myc (sc-40, dilution 1:500), HA (sc-805, dilution 1:500) and anti-β-actin (sc-47778, dilution 1:10,000) antibodies were obtained from Santa Cruz Biotechnology. Anti-Rip1 (610459, dilution 1:1,000) antibody was obtained from BD Biosciences. Anti-Gapdh (60004-1-Ig, dilution 1:2,000) and anti-Canx (10427-2-AP, dilution 1:500) antibodies were from Proteintech. Anti-p-p65 (3033s, dilution 1:1,000), p65 (8242s, dilution 1:200), p-p38 (9216S, dilution 1:1,000), p38 (9212s, dilution 1:200) and H3 (9715, 1:5,000) antibodies were obtained from Cell Signaling Technology. Mouse TNFoc was purchased from eBioscience. zVAD was obtained from Calbiochem. Propidium iodide (PI), 4-hydroxytamoxifen (4-OHT) and Protease Inhibitor Mix were from Sigma. λ-phosphatase (λ-Ppase) was from NEB. AP21967 (A/C Heterodimerizer) was obtained from ARIAD Pharmaceuticals.
Plasmids
Ppm1b-L and S (NM_001159496 and NM_011151) were amplified from mouse cDNA and cloned into pBOBI or pLV lentivirus vector with no tag or C-terminally tagged by the Exo III-assisted ligase-free cloning method. Deletion mutants and point mutations of Ppm1b and Rip3 were introduced using standard PCR or two rounds of PCR.
RNA interference
All lentiviral-shRNAs were constructed into pLV-H1-EF1α-puro vector following the manufacturer’s instruction (Biosettia). The indicated shRNA target sequences are: Ppm1b no. 1: 5′-ACAAGTGTGTGGATGGCAA-3′; Ppm1b no. 2: 5′-GGAGAGTAGAAATGGAAGA-3′; Ppm1b-L no. 1: 5′-GGCTAG AGCTGGCCTATAA-3′; Ppm1b-L no. 2: 5′-GAGGAGTGCTGAAAATATA-3′; Ppm1b-S no. 1: 5′-CTAAGGAAATACAGAGATA-3′; Ppm1b-S no. 2: 5′-TGCTGA ACATGAAGAGTAA-3′; hPpm1b no. 1: 5′-CAGTGGGAGTTATGATTTC-3′; hPp m1b no. 2: 5′-GGTGATTCACGTGCTGTTC-3′; Ppm1a no. 1: 5′-CCAAAGATGG AGAAGCATA-3′; Ppm1a no. 2: 5′-GAAGAAACATGGTGCAGAT-3′.
Lentivirus infection
For lentivirus production, 293T cells were transfected with lentiviral vectors carrying cDNAs or shRNAs of interest (or its mutants) and lentivirus-packing plasmids (PMDL/REV/VSVG) by the calcium phosphate precipitation method. Twelve hours later, cell culture medium was changed and the virus-containing medium was collected 36 h later. For infection, 1 ml virus-containing medium and 1.5 ml fresh medium containing 10 µg ml−1 Polybrene were added to cells plated in 6-well plates. The plates were centrifuged at 1,500g for 30 min and returned to the cell incubator. The infectious medium was changed 12 h later.
KO cell lines and cell culture
Rip1 KO, Rip3 KO and Mlkl KO L929 were described previously17,19. Ppm1b, Ppm1a, TNFRI and IKKβ KO cell lines were generated using the CRISPR/Cas9 technology30,49. Rip1–Rip3 DKO L929 cells were generated by targeting Rip1 in Rip3 KO L929 cells. The targeting sequences are: Ppm1b-5′-CTGGGAATGGTCTGCGTTA-3′; Ppm1a-5′-TGTAATCGAAATCCCCAA −3′; Rip1–5′-AACCGCGCTGAGTGAGTTGG-3′; TNFRI-5′-GCTTCAACGGC ACCGTGACA-3′; IKKβ-5′-CACCGTGACCGTTGACTAC-3′. Thioglycollateelicited peritoneal macrophages were obtained from eight-week-old WT or Rip3 KO mice using a method described previously50. Primary WT and Ppm1bd/d MEF cells were generated from day 14 embryos of the same litter of Ppm1b+/d heterozygous breeding. The primary MEF cells were immortalized by infection with retrovirus encoding E1A. Mouse BMDMs were differentiated in vitro from isolated bone marrow cells. Bone marrow cells collected from mouse femurs and tibias were incubated for seven days in DMEM containing 20% heat-inactivated FBS, penicillin, streptomycin and 30%L929 conditional medium28. Three pairs of Ppm1bd/d and WT mice were used to produce MEF cells and BMDM cells in this work, respectively. HT-29 cells were cultured in McCoy’s 5A culture medium (Invitrogen); L929, HEK-293T, NIH3T3-A, NIH3T3-N, HeLa, peritoneal macrophage, J774 and MEF cells were cultured in DMEM. All media were supplemented with 10% FBS (vol/vol), 2 mM L-glutamine, 100 IU penicillin and 100 mg ml−1 streptomycin at 37 °C in a humidified incubator containing 5% CO2.
Cell death assay
Cell death was analysed by measuring plasma membrane integrity to incorporate propidium iodide (PI). To measure the spontaneous cell death, PI (5 µg ml−1) was directly added in the medium and incubated for 10 min. Total cell numbers and PI-positive cells were counted under an inverted fluorescence microscope. Four fields of cells for each sample (more than 1,000 cells) were counted. The data are expressed as the percentage of PI-positive cells per total cells counted.
For TNF-induced cell death, cells were trypsinized and re-plated in 12-well plates to remove dead cells 12 h before TNF treatment. Cells were treated with TNF for the indicated time and then trypsinized, collected by centrifugation, and resuspended in PBS containing 5 µg ml−1 PI. The level of PI incorporation was quantified by flow cytometer (BD, FACSCalibur). Cell size was evaluated by forward-angle light scattering. PI-negative cells with a normal size were considered as living cells.
Apoptosis of MEF cells was measured by Annexin V staining according to the manufacturer’s instructions (Biotium).
Mass spectrometric analysis
Rip3 KO L929 cells expressing Flag-Rip3 or empty vector (control) were treated with or without TNF for 4 h. Protein complex was immunoprecipitated with M2 beads, and eluted with 3X FLAG peptides. Proteins from elution were precipitated by addition of TCA to 20% (vol/vol) and incubated on ice for at least 30 min, followed by centrifugation at 16,000g for 10 min at 4°C. Protein pellets were washed with ice-cold acetone and dried briefly by vacuum centrifugation and then resuspended in 8 M urea in 50 mM NH4HCO3. Cysteines were reduced (5 mM dithiothreitol, 30 min) and alkylated (15 mM iodoacetamide, 30 min in dark), and the urea concentration was reduced to 1.33 M by dilution with 50 mM NH4HCO3. Proteins were digested overnight at 37 °C by the addition of trypsin at a ratio of protein/trypsin (1:100). Formic acid was added to 1% (vol/vol), peptides were loaded onto a STAGEtip for desaltion, and peptides were eluted directly into inserts with 70% acetonitrile/1% formic acid. The tryptic peptides were analysed on an AB Sciex TripleTOF 5600 mass spectrometer interfaced to an Eksigent NanoLC Ultra 2D Plus HPLC system, essentially as previously described51. Peptides were chromatographed using a 60-min gradient from 2–35% (buffer A 0.1% (vol/vol) formic acid 2% (vol/vol) acetonitrile, buffer B 0.1% (vol/vol) formic acid, 98% (vol/vol) acetonitrile) after injection onto a 15-cm emitter (New Objective) packed with New Objective C18 3-µm. MS1 spectra were collected in the range 350-1,250 m/z for 250 ms, and the 20 most intense precursors with charge state 2–5 were selected for fragmentation; MS2 spectra were collected in the range 100-1,800 m/z for 100 ms. For each IP sample, triple injections were performed to achieve depth identification. All wiff files were searched against the mouse Uniprot proteome database (July 2013) appended with common contaminants and reversed-sequence decoys (101,970 sequence including decoys) using ProteinPilot V4.5 beta. ‘Iodoacetamide’ was selected for Cys alkylation, and ‘Trypsin’as protease, and Search Effort was set as ‘Rapid ID’. Proteins were filtered with 5% FDR. Phosphatases identified in either TNF-treated or untreated samples were selected as potential Rip3 phosphatases.
Immunoprecipitation and immunoblotting
Cells were lysed with lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4) on ice for 15 min. Cell lysates were then centrifuged at 20,000g for 30 min. The supernatant was immunoprecipitated with antibody-coupled beads at 4 °C for 3 h or overnight. After the immunoprecipitation, the beads were washed four times in lysis buffer and the immunoprecipitated proteins were subsequently eluted by SDS sample buffer. For endogenous Rip3 immunoprecipitation, anti-Rip3 antibody was coupled to beads by the GlycoLink Immobilization Kit according to the manufacturer’s instructions. The luminescent signals of immunoblotting were analysed using an ImageQuant LAS 4000 Scanner (GE Healthcare) or X-ray film.
In vitro phosphatase assay
GST-Ppm1b and GST-R179G were expressed and purified from Escherichia coli Flag-mRip3 protein was immunoprecipitated using anti-Flag beads from cell lysates of transfected 293T cells and eluted with ×3 Flag peptide. GST-Ppm1b, GST-R179G and GST were incubated with Flag-Rip3 in buffer (20 mM HEPES, 20 mM MgCl2, 0.03% β-mercaptoethanol) at 30 °C for 30 min with gentle shaking. The reactions were stopped by the addition of ×2 SDS sample buffer followed by boiling for 5 min.
For λ-phosphatase treatment, endogenous Rip3 was immunoprecipitated by Rip3 antibody-coupled beads and washed three times with × 1 phosphatase buffer (50 mM HEPES (pH 7.5), 100 mM NaCl, 2 mM dithiothreitol, 0.01% Brij 35, 1 mM MnCl2). The immunoprecipitate was then divided into two parts. One was augmented with 200 units of λ-phosphatase (NEB) but the other was mock treated. After incubation at 30 °C for 30 min with gentle shaking, the reactions were stopped by the addition of ×2 SDS sample buffer followed by boiling for 5 min.
Chemical-induced forced Rip3-Rip3 interaction and Mlkl oligomerization
To generate the FKBP-Rip3RHM and FRB-Rip3RHIM constructs the Rip3RHIM (QIG449–451AAA) mutant was fused to the C terminus of the FKBP domain (FK506 binding protein) and FRB domain (domain of mTOR with a point mutation T2098L). Plasmids used as templates containing FKBP and the mutant FRB domain were obtained from the ARGENT Regulated Heterodimerization Kit kindly provided by ARIAD (now iDimerizeTM Inducible Heterodimer System by Clontech). The interaction of FKBP and FRB fusion proteins can be induced by a rapamycin analogue AP21976. The FKBP-Rip3RHIM and FRB-Rip3RHM were introduced to Rip1-Rip3 DKO L929 cells by co-infection of lentiviral vectors. To evaluate the Ppm1b function, these cells were then co-infected with lentiviral vectors encoding Ppm1b-L and Ppm1b-S, or infected with empty vectors. As the infection efficiency was nearly 100% (evidenced by parallel infection or co-infection with lentiviral-GFP), it was unnecessary to select cells that express both proteins. Twenty-four hours after infection, AP21976 (250 nM) was added to induce Rip3-Rip3 interaction. Cell death was analysed 2 h later.
The chemical-induced Mlkl oligomerization has been described previously17. Briefly, Mlkl was fused to the N terminus of oestrogen-induced homodimerization of the hormone binding domain (HBD) with a G521Rmutation (termed Mlkl-HBD). A synthetic antioestrogen 4-hydroxytamoxifen (4-OHT) can induce dimerization of the HBD fusion protein. Mlkl-HBD was introduced to Mlkl KO L929 cells by lentiviral vector. Thirty-six hours after infection, 4-OHT (1 µM) was added to induce Mlkl oligomerization. Cell death was analysed 2 h later.
Generation of Ppm1bd/d and Ppm1bd/d Rip3−/− mice
The mouse Ppm1b-disrupted embryonic stem cell clone (AW0406) with a retrovirus vector based on the gene-trap technology employed at Sanger Institute was purchased from the Mutant Mouse Regional Resource Center (MMRRC, University of California Davis). The exact gene-disruption position was determined by 3′ RACE and genomic DNA PCR. Ppm1b+/d embryonic stem cells were injected into C57BL/6 blastocysts, and chimaeric male mice were bred to C57BL/6 wild-type female mice. The resulting Ppm1b+/d offspring were bred to C57BL/6 mice for more than ten generations. Genotyping was carried out by genomic DNA PCR with the following three primers: P1–5′-TCTCGTTGCATTTAGTGTTTACCACTGT-3′ (forward sequences in Ppm1b genome); P2–5′-GGCTTCTGAATAACCGAATTCAAGG-3′ (reverse sequences in Ppm1b genome); P3–5′-CTCCCTGGCCTCCAGACAAGTAGA-3′ (reverse sequences in retrovirus intron). P1 and P2 are for wild-type allele PCR to produce a 217-bp band, and P1 and P3 are for mutated allele PCR to produce a 310-bpband.
Rip3−/− mice were generated by the transcription activator-like effector nucleases (TALENs)-mediated gene-disruption method in a C57BL/6 background, as described previously28. Genotypes were confirmed by tail-snip PCR. Rip3−/− and Ppm1b+/d were subsequently intercrossed.
SIRS mice model
Mice were housed in a specific pathogen-free environment. All experiments were conducted in compliance with the regulations of Xiamen University. Six- to eight-week-old female mice (average weight approximately 20 g) were injected intravenously with 15 µg TNF (75 µg ml−1) diluted in endotoxin-free PBS. Mice injected with PBS alone were included as a control. Animals were under permanent observation and survival was checked every 30 min. Littermates of Rip3−/− and WT, Ppm1bd/d and WT, Ppm1bd/dRip3−/− and Ppm1b+/+ Rip3−/− mice were used in the experiments. The investigator was blinded to allocation when the mice were injected with TNF and when mice deaths were counted.
In total, 44 pairs of Rip3−/− and WT mice, 79 pairs of Ppm1bd/d and WT mice, and 26 pairs of Ppm1bd/d Rip3−/− and Ppm1b+/+ Rip3−/− mice were used in these experiments and the subsequent histology analyses.
Histology
After animals were euthanized, tissues of different organs were collected immediately, and fixed in 10% neutral buffered formalin for 24 h. The fixed tissues were dehydrated in ethanol, cleared in xylene, and embedded in paraffin blocks. Five-micrometre sections were cut and mounted on adhesion microscope slides (ZSGB-BIO), and then stained with haematoxylin and eosin (H&E) or periodic acid-Schiff(PAS) for analyses.
The degree of caecum damage was assessed by a method described previously52. Then the sum of the average scores of four fields per sample was calculated as the caecum damage score. Representative images were captured and processed using identical settings in the Leica DM2500 at Xiamen University.
The investigators were blinded to allocation when the histology experiments were performed and tissue damage was scored.
IL-6 level determination
Following the manufacturer’s instructions, the supernatants of the cell culture were collected and the IL-6 level was measured by commercial ELISA kits for mouse IL-6 (R&D). For the in vivo SIRS model, the sera were collected before (0 h) and after (2 h, 4 h) TNF injection, and were analysed for their IL-6 levels by ELISA kits.
Isolation of nucleus and cytosol
Cells were trypsinized, collected by centrifugation, and washed with pre-cold PBS three times. Then cells were resuspended in Buffer A (10 mM HEPES, pH 7.9; 1.5 mM MgCl2; 10 mM KCl; 300 mM sucrose; 0.5% NP-40) for 10 min on ice with gentle tapping. Cell lysate was then centrifuged at 1,000g for 5 min. The suspension (cytosol) was collected and centrifuged further at 1,000g for 10 min to get rid of nucleus contamination; the pellets (nucleus) were resuspended and washed with Buffer A twice.
Subcellular fractionation
L929 cells were fractionated and heavy membrane (HM), mitochondria (Mito) and heterotypic membrane (HeteroM) were isolated as described previously41. Briefly, approximately 3 × 108 cells were washed with cold PBS and collected into 50 ml tubes. Cells were homogenized by glass homogenizer (Kontes) in homogenization buffer (225 mM mannitol, 75 mM sucrose, 0.1 mM EGTA and 30 mM Tris-HCl (pH 7.4)). The lysate was centrifuged at 800g to remove nuclei. The supernatant was centrifuged at 100,000g for 10 min at 4 °C and the subsequent pellet was collected as HM. For further purification, HM was resuspended and loaded on 20% Percoll and centrifuged at 10,000g for 30 min at 4 °C. Mitochondria were recovered from the bottom of the gradient; the heterotypic membranes were collected from the upper gradient. Each fraction was quantified and indicated proteins were analysed by western blot.
Statistical evaluation
Statistical analysis was performed with Prism software (GraphPad Software). Data are expressed as means ± s.e.m. Two-tailed Student’s t-test was used to compare differences between treated groups and their paired controls. Mice survival is presented as a Kaplan-Meyer plot and compared by log-rank (Mantel-Cox) Test. To evaluate the caecum damage between the two genotypes, a Wilcoxon signed-rank test was performed. Differences in compared groups were considered statistically significantly different with P values lower than 0.05;#: P < 0.05; ##: P < 0.01; ###: P < 0.001.
No statistical method was used to predetermine sample size. The experiments were not randomized. The investigators were blinded to allocation during the SIRS mouse model experiments and histology experiments (see the SIRS mice model and Histology section above). For other experiments, the investigators were not blinded to allocation during experiments and outcome assessment.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Tamura and M. Ohnishi for Ppm1b+/− mice. This work was supported by the National Basic Research Program of China (973 Program; 2015CB553800), the National Scientific and Technological Major Project (2013ZX10002-002), the National Natural Science Foundation of China (91429301, 31420103910, 31330047, 91029304, 31221065, and 31090360), the Hi-Tech Research and Development Program of China (863 program; 2012AA02A201), the 111 Project (B12001), the Science and Technology Foundation of Xiamen (No. 3502Z20130027), the National Science Foundation of China for Fostering Talents in Basic Research (Grant No.J1310027) and the Open Research Fund of State Key Laboratory of Cellular Stress Biology, Xiamen University. This research was also partly supported by grants from MOST (2012CB966600) and NIH (R01GM63773, R01AR053591, R01CA108454). The collaborative research is also supported by the special funds for Innovation Center for Cell Biology from Zhejiang University and Xiamen University.
Footnotes
Note: Supplementary Information is available in the online version of the paper
AUTHOR CONTRIBUTIONS
W.C., J.W, L.L., Zhengmo Z., J.R., Y.L., X-H.F. and J.H. carried out the experiments. F.C, C.Y., Zhenru Z., S.S.S., X.Z., Zhirong Z., C-Q.Z., H.W, M.X. and X.L. helped to prepare cell lines, generated the ppm1b gene-trap line, provided reagents and mass spectrum analysis. J.H. contributed to the overall design of the project. WC. and J.H. interpreted the data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev. Mol. Cell. Biol. 2010;11:700–714. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- 2.Vanlangenakker N, Bertrand MJ, Bogaert P, Vandenabeele P, Vanden Berghe T. TNF-induced necroptosis in L929 cells is tightly regulated by multiple TNFR1 complex I and II members. Cell Death Dis. 2011;2:e230. doi: 10.1038/cddis.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaiser W, et al. Toll-like receptor 3-mediated necrosis via TRIF, RIP3, and MLKL. J. Biol. Chem. 2013;288:31268–31279. doi: 10.1074/jbc.M113.462341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Upton JW, Kaiser WJ, Mocarski ES. Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe. 2010;7:302–313. doi: 10.1016/j.chom.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaczmarek A, Vandenabeele P, Krysko DV. Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity. 2013;38:209–223. doi: 10.1016/j.immuni.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Galluzzi L, Kepp O, Krautwald S, Kroemer G, Linkermann A. Molecular mechanisms of regulated necrosis. Sem. Cell Dev. Biol. 2014;35:24–32. doi: 10.1016/j.semcdb.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Holler N, et al. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat. Immunol. 2000;1:489–495. doi: 10.1038/82732. [DOI] [PubMed] [Google Scholar]
- 8.Zhang DW, et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 9.He S, et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-a. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 10.Cho YS, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J, et al. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc. Natl Acad. Sci. USA. 2012;109:5322–5327. doi: 10.1073/pnas.1200012109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun L, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 14.Murphy JM, et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39:443–453. doi: 10.1016/j.immuni.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, et al. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol. Cell. 2014;54:133–146. doi: 10.1016/j.molcel.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Dondelinger Y, et al. MLKL compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Rep. 2014;7:971–981. doi: 10.1016/j.celrep.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, et al. Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res. 2014;24:105–121. doi: 10.1038/cr.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai Z, et al. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat. Cell Biol. 2014;16:55–65. doi: 10.1038/ncb2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen W, et al. Diverse sequence determinants control human and mouse receptor interacting protein 3 (RIP3) and mixed lineage kinase domain-like (MLKL) interaction in necroptotic signaling. J. Biol. Chem. 2013;288:16247–16261. doi: 10.1074/jbc.M112.435545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu C, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lattin JE, et al. Expression analysis of G protein-coupled receptors in mouse macrophages. Immunome Res. 2008;4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terasawa T, et al. Molecular cloning of a novel isotype of Mg(2+)-dependent protein phosphatase β (type 2C β) enriched in brain and heart. Arch. Biochem. Biophys. 1993;307:342–349. doi: 10.1006/abbi.1993.1598. [DOI] [PubMed] [Google Scholar]
- 23.Kusuda K, et al. Mutational analysis of the domain structure of mouse protein phosphatase 2Cβ. Biochem. J. 1998;332:243–250. doi: 10.1042/bj3320243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu T, et al. Regulator of G-protein signaling 19 (RGS19) and its partner Gα-inhibiting activity polypeptide 3 (GNAI3) are required for zVAD-induced autophagy and cell death in L929 cells. PLoS ONE. 2014;9:e94634. doi: 10.1371/journal.pone.0094634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Degterev A, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun X, Yin J, Starovasnik MA, Fairbrother WJ, Dixit VM. Identification of a novel homotypic interaction motif required for the phosphorylation of receptor-interacting protein (RIP) by RIP3. J. Biol. Chem. 2002;277:9505–9511. doi: 10.1074/jbc.M109488200. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Zheng XF, Brown EJ, Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl Acad. Sci. USA. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu J, et al. Mlkl knockout mice demonstrate the indispensable role of Mlkl in necroptosis. Cell Res. 2013;23:994–1006. doi: 10.1038/cr.2013.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu YT, et al. zVAD-induced necroptosis in L929 cells depends on autocrine production of TNFα mediated by the PKC-MAPKs-AP-1 pathway. Cell Death Differ. 2011;18:26–37. doi: 10.1038/cdd.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki M, et al. Disruption of the mouse protein Ser/Thr phosphatase 2Cβ gene leads to early pre-implantation lethality. Mech. Dev. 2007;124:489–499. doi: 10.1016/j.mod.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Newton K, et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science. 2014;343:1357–1360. doi: 10.1126/science.1249361. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Du F, Wang X. TNF-α induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 34.Bertrand MJ, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 35.Sun W, et al. PPM1A and PPM1B act as IKKβ phosphatases to terminate TNFα-induced IKKβ-NF-κB activation. Cell Signal. 2009;21:95–102. doi: 10.1016/j.cellsig.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Podolin PL, et al. Attenuation of murine collagen-induced arthritis by a novel, potent, selective small molecule inhibitor of IκB Kinase 2, TPCA-1 (2-[(aminocarbonyl)amino]-5-(4-fluorophenyl)-3-thiophenecarboxamide), occurs via reduction of proinflammatory cytokines and antigen-induced T cell Proliferation. J. Pharmacol. Exp. Ther. 2005;312:373–381. doi: 10.1124/jpet.104.074484. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka A, et al. A novel NF-κB inhibitor, IMD-0354, suppresses neoplastic proliferation of human mast cells with constitutively activated c-kit receptors. Blood. 2005;105:2324–2331. doi: 10.1182/blood-2004-08-3247. [DOI] [PubMed] [Google Scholar]
- 38.Hanada M, et al. Regulation of the TAK1 signaling pathway by protein phosphatase 2C. J. Biol. Chem. 2001;276:5753–5759. doi: 10.1074/jbc.M007773200. [DOI] [PubMed] [Google Scholar]
- 39.Ninomiya-Tsuji J, et al. A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J. Biol. Chem. 2003;278:18485–18490. doi: 10.1074/jbc.M207453200. [DOI] [PubMed] [Google Scholar]
- 40.Xie T, et al. Structural insights into RIP3-mediated necroptotic signaling. Cell Rep. 2013;5:70–78. doi: 10.1016/j.celrep.2013.08.044. [DOI] [PubMed] [Google Scholar]
- 41.Li L, et al. The Gβγ-Src signaling pathway regulates TNF-induced necroptosis via control of necrosome translocation. Cell Res. 2014;24:417–432. doi: 10.1038/cr.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin X, et al. PPM1A functions as a Smad phosphatase to terminate TGFβ signaling. Cell. 2006;125:915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linkermann A, et al. Dichotomy between RIP1- and RIP3-mediated necroptosis in tumor necrosis factor-α-induced shock. Mol. Med. 2012;18:577–586. doi: 10.2119/molmed.2011.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duprez L, et al. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity. 2011;35:908–918. doi: 10.1016/j.immuni.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 45.Tracey KJ, et al. Shock and tissue injury induced by recombinant human cachectin. Science. 1986;234:470–474. doi: 10.1126/science.3764421. [DOI] [PubMed] [Google Scholar]
- 46.Choi HK, et al. PKA negatively regulates PP2Cβ to activate NF-κB-mediated inflammatory signaling. Biochem. Biophys. Res. Commun. 2013;436:473–477. doi: 10.1016/j.bbrc.2013.05.129. [DOI] [PubMed] [Google Scholar]
- 47.Klumpp S, Selke D, Hermesmeier J. Protein phosphatase type 2C active at physiological Mg2+: stimulation by unsaturated fatty acids. FEBS Lett. 1998;437:229–232. doi: 10.1016/s0014-5793(98)01237-x. [DOI] [PubMed] [Google Scholar]
- 48.Shreeram S, et al. Wip1 phosphatase modulates ATM-dependent signaling pathways. Mol. Cell. 2006;23:757–764. doi: 10.1016/j.molcel.2006.07.010. [DOI] [PubMed] [Google Scholar]
References
- 49.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. 2008;11 doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu X, et al. Investigation of RIP3-dependent protein phosphorylation by quantitative phosphoproteomics. Mol. Cell Proteomics. 2012;11:1640–1651. doi: 10.1074/mcp.M112.019091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coburn B, Li Y, Owen D, Vallance BA, Finlay BB. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect. Immun. 2005;73:3219–3227. doi: 10.1128/IAI.73.6.3219-3227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.