Abstract
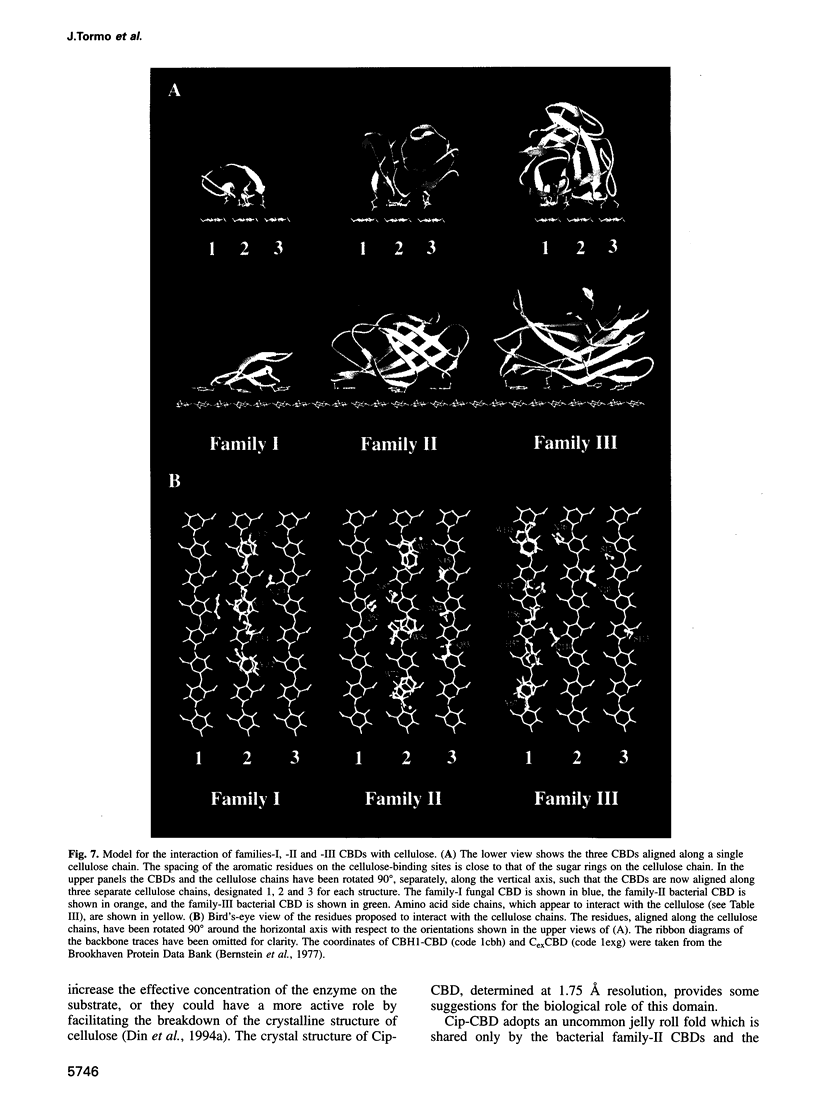
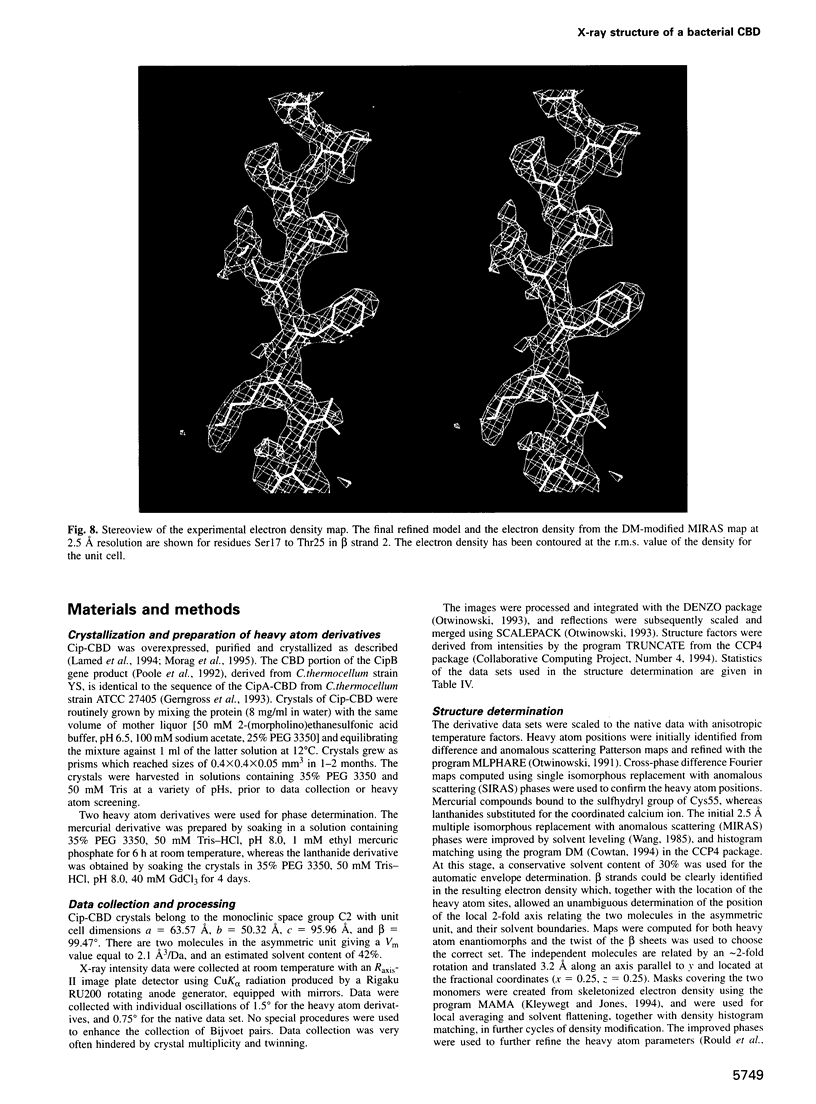
The crystal structure of a family-III cellulose-binding domain (CBD) from the cellulosomal scaffoldin subunit of Clostridium thermocellum has been determined at 1.75 A resolution. The protein forms a nine-stranded beta sandwich with a jelly roll topology and binds a calcium ion. conserved, surface-exposed residues map into two defined surfaces located on opposite sides of the molecule. One of these faces is dominated by a planar linear strip of aromatic and polar residues which are proposed to interact with crystalline cellulose. The other conserved residues are contained in a shallow groove, the function of which is currently unknown, and which has not been observed previously in other families of CBDs. On the basis of modeling studies combined with comparisons of recently determined NMR structures for other CBDs, a general model for the binding of CBDs to cellulose is presented. Although the proposed binding of the CBD to cellulose is essentially a surface interaction, specific types and combinations of amino acids appear to interact selectively with glucose moieties positioned on three adjacent chains of the cellulose surface. The major interaction is characterized by the planar strip of aromatic residues, which align along one of the chains. In addition, polar amino acid residues are proposed to anchor the CBD molecule to two other adjacent chains of crystalline cellulose.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayer E. A., Morag E., Lamed R. The cellulosome--a treasure-trove for biotechnology. Trends Biotechnol. 1994 Sep;12(9):379–386. doi: 10.1016/0167-7799(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Béguin P., Aubert J. P. The biological degradation of cellulose. FEMS Microbiol Rev. 1994 Jan;13(1):25–58. doi: 10.1111/j.1574-6976.1994.tb00033.x. [DOI] [PubMed] [Google Scholar]
- Choe S., Bennett M. J., Fujii G., Curmi P. M., Kantardjieff K. A., Collier R. J., Eisenberg D. The crystal structure of diphtheria toxin. Nature. 1992 May 21;357(6375):216–222. doi: 10.1038/357216a0. [DOI] [PubMed] [Google Scholar]
- Din N., Damude H. G., Gilkes N. R., Miller R. C., Jr, Warren R. A., Kilburn D. G. C1-Cx revisited: intramolecular synergism in a cellulase. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11383–11387. doi: 10.1073/pnas.91.24.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din N., Forsythe I. J., Burtnick L. D., Gilkes N. R., Miller R. C., Jr, Warren R. A., Kilburn D. G. The cellulose-binding domain of endoglucanase A (CenA) from Cellulomonas fimi: evidence for the involvement of tryptophan residues in binding. Mol Microbiol. 1994 Feb;11(4):747–755. doi: 10.1111/j.1365-2958.1994.tb00352.x. [DOI] [PubMed] [Google Scholar]
- Gerngross U. T., Romaniec M. P., Kobayashi T., Huskisson N. S., Demain A. L. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol Microbiol. 1993 Apr;8(2):325–334. doi: 10.1111/j.1365-2958.1993.tb01576.x. [DOI] [PubMed] [Google Scholar]
- Gerwig G. J., Kamerling J. P., Vliegenthart J. F., Morag E., Lamed R., Bayer E. A. The nature of the carbohydrate-peptide linkage region in glycoproteins from the cellulosomes of Clostridium thermocellum and Bacteroides cellulosolvens. J Biol Chem. 1993 Dec 25;268(36):26956–26960. [PubMed] [Google Scholar]
- Gilkes N. R., Henrissat B., Kilburn D. G., Miller R. C., Jr, Warren R. A. Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991 Jun;55(2):303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes N. R., Kilburn D. G., Miller R. C., Jr, Warren R. A., Sugiyama J., Chanzy H., Henrissat B. Visualization of the adsorption of a bacterial endo-beta-1,4-glucanase and its isolated cellulose-binding domain to crystalline cellulose. Int J Biol Macromol. 1993 Dec;15(6):347–351. doi: 10.1016/0141-8130(93)90052-n. [DOI] [PubMed] [Google Scholar]
- Gilkes N. R., Warren R. A., Miller R. C., Jr, Kilburn D. G. Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J Biol Chem. 1988 Jul 25;263(21):10401–10407. [PubMed] [Google Scholar]
- Goldstein M. A., Doi R. H. Mutation analysis of the cellulose-binding domain of the Clostridium cellulovorans cellulose-binding protein A. J Bacteriol. 1994 Dec;176(23):7328–7334. doi: 10.1128/jb.176.23.7328-7334.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffrén A. M., Teeri T. T., Teleman O. Molecular dynamics simulation of fungal cellulose-binding domains: differences in molecular rigidity but a preserved cellulose binding surface. Protein Eng. 1995 May;8(5):443–450. doi: 10.1093/protein/8.5.443. [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993 Sep 5;233(1):123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Klyosov A. A. Trends in biochemistry and enzymology of cellulose degradation. Biochemistry. 1990 Nov 27;29(47):10577–10585. doi: 10.1021/bi00499a001. [DOI] [PubMed] [Google Scholar]
- Kraulis J., Clore G. M., Nilges M., Jones T. A., Pettersson G., Knowles J., Gronenborn A. M. Determination of the three-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei. A study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. Biochemistry. 1989 Sep 5;28(18):7241–7257. doi: 10.1021/bi00444a016. [DOI] [PubMed] [Google Scholar]
- Lamed R., Tormo J., Chirino A. J., Morag E., Bayer E. A. Crystallization and preliminary X-ray analysis of the major cellulose-binding domain of the cellulosome from Clostridium thermocellum. J Mol Biol. 1994 Nov 25;244(2):236–237. doi: 10.1006/jmbi.1994.1721. [DOI] [PubMed] [Google Scholar]
- Linder M., Mattinen M. L., Kontteli M., Lindeberg G., Ståhlberg J., Drakenberg T., Reinikainen T., Pettersson G., Annila A. Identification of functionally important amino acids in the cellulose-binding domain of Trichoderma reesei cellobiohydrolase I. Protein Sci. 1995 Jun;4(6):1056–1064. doi: 10.1002/pro.5560040604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhalen C. A., Strynadka N. C., James M. N. Calcium-binding sites in proteins: a structural perspective. Adv Protein Chem. 1991;42:77–144. doi: 10.1016/s0065-3233(08)60535-5. [DOI] [PubMed] [Google Scholar]
- Morag E., Lapidot A., Govorko D., Lamed R., Wilchek M., Bayer E. A., Shoham Y. Expression, purification, and characterization of the cellulose-binding domain of the scaffoldin subunit from the cellulosome of Clostridium thermocellum. Appl Environ Microbiol. 1995 May;61(5):1980–1986. doi: 10.1128/aem.61.5.1980-1986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno J. D., Bayer R. The limits of the ledger in public health promotion. Hastings Cent Rep. 1985 Dec;15(6):37–41. [PubMed] [Google Scholar]
- Pagès S., Belaich A., Tardif C., Reverbel-Leroy C., Gaudin C., Belaich J. P. Interaction between the endoglucanase CelA and the scaffolding protein CipC of the Clostridium cellulolyticum cellulosome. J Bacteriol. 1996 Apr;178(8):2279–2286. doi: 10.1128/jb.178.8.2279-2286.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole D. M., Hazlewood G. P., Huskisson N. S., Virden R., Gilbert H. J. The role of conserved tryptophan residues in the interaction of a bacterial cellulose binding domain with its ligand. FEMS Microbiol Lett. 1993 Jan 1;106(1):77–83. doi: 10.1111/j.1574-6968.1993.tb05938.x. [DOI] [PubMed] [Google Scholar]
- Poole D. M., Morag E., Lamed R., Bayer E. A., Hazlewood G. P., Gilbert H. J. Identification of the cellulose-binding domain of the cellulosome subunit S1 from Clostridium thermocellum YS. FEMS Microbiol Lett. 1992 Dec 1;78(2-3):181–186. doi: 10.1016/0378-1097(92)90022-g. [DOI] [PubMed] [Google Scholar]
- Quiocho F. A. Probing the atomic interactions between proteins and carbohydrates. Biochem Soc Trans. 1993 May;21(2):442–448. doi: 10.1042/bst0210442. [DOI] [PubMed] [Google Scholar]
- Reinikainen T., Teleman O., Teeri T. T. Effects of pH and high ionic strength on the adsorption and activity of native and mutated cellobiohydrolase I from Trichoderma reesei. Proteins. 1995 Aug;22(4):392–403. doi: 10.1002/prot.340220409. [DOI] [PubMed] [Google Scholar]
- Rini J. M. Lectin structure. Annu Rev Biophys Biomol Struct. 1995;24:551–577. doi: 10.1146/annurev.bb.24.060195.003003. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Steitz T. A. Improving multiple isomorphous replacement phasing by heavy-atom refinement using solvent-flattened phases. Acta Crystallogr A. 1992 Sep 1;48(Pt 5):751–756. doi: 10.1107/s0108767392003404. [DOI] [PubMed] [Google Scholar]
- Shoseyov O., Takagi M., Goldstein M. A., Doi R. H. Primary sequence analysis of Clostridium cellulovorans cellulose binding protein A. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3483–3487. doi: 10.1073/pnas.89.8.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D. I., Levine M., Muirhead H., Stammers D. K. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J Mol Biol. 1979 Oct 15;134(1):109–142. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- Tomme P., Van Tilbeurgh H., Pettersson G., Van Damme J., Vandekerckhove J., Knowles J., Teeri T., Claeyssens M. Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur J Biochem. 1988 Jan 4;170(3):575–581. doi: 10.1111/j.1432-1033.1988.tb13736.x. [DOI] [PubMed] [Google Scholar]
- Tomme P., Warren R. A., Gilkes N. R. Cellulose hydrolysis by bacteria and fungi. Adv Microb Physiol. 1995;37:1–81. doi: 10.1016/s0065-2911(08)60143-5. [DOI] [PubMed] [Google Scholar]
- Xu G. Y., Ong E., Gilkes N. R., Kilburn D. G., Muhandiram D. R., Harris-Brandts M., Carver J. P., Kay L. E., Harvey T. S. Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy. Biochemistry. 1995 May 30;34(21):6993–7009. [PubMed] [Google Scholar]