Abstract
Melanoma is a malignant tumor of melanocytes with high capability of invasion and rapid metastasis to other organs. Malignant melanoma is the most common metastatic malignancy found in Gastrointestinal Tract (GI). In this work, the 1H NMR-based metabolomics approach is used to investigate the metabolite profile differences of stomach tissue extracts of metastatic B16-F10 melanoma and control groups in C57BL/6J mouse and to search for specific metabolite biomarker candidates. Principal Component Analysis (PCA), an unsupervised multivariate data analysis method, is used to detect possible outliers, while Orthogonal Projection to Latent Structure (OPLS), a supervised multivariate data analysis method, is employed to evaluate important metabolites responsible for discriminating the control and the melanoma groups. Both PCA and OPLS results reveal that the melanoma group can be well separated from its control group. Among the 50 identified metabolites, it is found that the concentrations of 19 metabolites are significantly changed with the levels of O-phosphocholine and hypoxanthine down-regulated while the levels of isoleucine, leucine, valine, isobutyrate, threonine, cadaverine, alanine, glutamate, glutamine, methionine, citrate, asparagine, tryptophan, glycine, serine, uracil, and formate up-regulated in the melanoma group. These significantly changed metabolites are associated with multiple biological pathways and may be potential biomarkers for metastatic melanoma in stomach.
Keywords: 1H NMR Metabolomics, B16-F10 melanoma, Stomach, Metabolomics, Multivariate analysis, OPLS, PCA
Introduction
Malignant melanoma is one of the most common cancers in the United States and many other western countries, especially in white populations [1-3]. Melanoma is also the most dangerous form of skin cancer due to its high capability of invasion and rapid metastasis to other organs, accounting for 90% of skin cancer mortality [2,4]. Every one person out of 75 in the United States suffers from this aggressive disease [5]. Malignant melanoma is the most common metastatic malignancy found in the Gastrointestinal Tract (GI), and GI metastasis of melanoma can cause a variety of non-specific symptoms, including abdominal pain, fatigue, dysphagia, constipation, tenesmus, small bowel obstruction, and weight loss [6,7]. GI metastatic melanoma is present in 60% of patients who die of disseminated melanoma. Operative intervention is discouraging, as the five-year survival rates post operation are less than 10% [6-9]. NMR-based metabolomics, i.e. the combined application of NMR metabolic profiling and multivariate data analysis is an efficient, quantitative and robust method capable not only of identifying metabolite biomarkers for early detection of various diseases, but also providing insight into the pathologies of these diseases through the identification of disturbed metabolic pathways [10-13].
In this study, we evaluated the metabolic changes in stomach after metastasis of primary melanoma cells implanted at the flank location on the legs of a B16-F10 melanoma mouse model. We choose stomach as it is one of the two most common sites (stomach and small intestine) involved in GI tract metastasis of melanoma [8]. The GI tract is a complex ecosystem containing approximately 100 trillion of microbial organisms, the varying metabolic activities of which not only determine absorption, digestion, metabolism, and excretion of dietary nutrients but also shape region-specifically the surrounding and distant host cell biochemical processes [14]. Herein, 1H NMR metabolomics is utilized for metabolic profiling of both hydrophilic and hydrophobic extracts from excised stomachs of the control and the melanoma mice. Multivariate data analysis (both PCA and OPLS) are carried out for pattern recognition and for identifying a set of metabolites whose concentrations are significantly changed as a result of the melanoma metastasis in stomach. Based on the findings, the potential metabolic pathways/networks that are affected by the disease are discussed. The present study is a complementary work of our previous investigation of metastatic melanoma, as well as a further application of the data processing method proposed in our previous publication [13].
Materials and Methods
Animal experiments and sample preparation
A total of 12 six-week-old C57BL/6J male mice were purchased from Jackson Labs (Maine, USA) and were housed at the Pacific Northwest National Laboratory (PNNL) animal facility. After acclimation at the facility for one week, at the age of week-7 the mice were randomly divided into two groups, an experimental group (n=7) that received subcutaneous injection with suspended 105 B16-F10 tumor cells at four flank locations on each leg, and a control group (n=5) that were injected with the same amount of PBS at same places. The individually housed animals were fed a standard diet and maintained in a temperature-controlled room with an ambient temperature of 22-25°C and 45% humidity on 12 h light, 12 h dark cycle. All mice, each of which was weighted weekly, were allowed free access to water and food before being sacrificed by CO2 asphyxiation at the age of fourteen weeks. Twelve hours prior to the sacrificing, foods except water were not supplied in order to empty the stomach. Two mice from the experimental group died during the feeding process resulting in five mice per group. Stomach tissues were dissected and snap-frozen at −80°C before NMR spectroscopy and morphology analysis. The presence of malignant melanoma in stomach can be observed by histological images (Figure S1 in Supporting Information), which is consistent with prior literature reports [15,16]. All animal work was approved by the Institutional Animal Care and Use Committee (IACUC) at Pacific Northwest National Laboratory.
Polar and lipophilic metabolites were extracted using a modified Folch method by following published protocol [17], which was considered to generate the highest yields in mild condition [18,19]. It has been generally accepted that about 95% of tissue lipids can be extracted during the first step [20]. The detailed recipes for NMR sample preparation can be found in our previous publications [13,21]. The extraction steps are briefly summarized below.
Step 1: After randomization of the samples, each pre-weighted intact frozen tissue was homogenized in ice bath using a Tissue Tearor (Model 985-370, BioSpec Products, Inc.) after adding 0.25 ml MeOH and 0.053 ml H2O for each sample containing 30 to 60 mg of tissue, followed by vortexing the mixture and then adding 0.125 ml chloroform, vortexing again. This process took 7 minutes and kept exactly the same for each sample to minimize the possibility of introducing difference in sample degradation that may occur during this very first but critical extraction step, where enzymes are partially active during the earlier times of the procedure. Step 2: 0.125 ml chloroform and 0.125 ml water was added to the sample and then vortexed again, followed by transferring different layers into glass vials separately with syringes after the mixture being left on ice for 15 minutes and centrifuged at 8000 rpm for 2 minutes at 4°C. Finally, the solvents were removed by employing a lyophilizer (MeOH/H2O layer, hydrophilic metabolites) or by evaporating under dry nitrogen gas (CHCl3 layer, hydrophobic metabolites). The extracts were then stored at −80°C before performing NMR measurements.
1H NMR spectroscopy
The hydrophilic metabolites were reconstituted in 600 μl of D2O containing 0.5 mM 3-(trimethylsilyl)-2,2’,3,3’-tetradeuteropropionate (TSP-d4) as chemical shift reference and internal concentration standard. About 550 μl of the prepared sample was loaded into a standard 5 mm NMR tube (Wilmad, Buena, NJ). 0.2% sodium azide (w/v) was added into the mixture to prevent biodegradation. All 1H NMR spectra were recorded on a Varian 800 MHz NMR spectrometer equipped with a Z axis-gradient 5 mm HCN probe at 20°C. One-dimension 1H NMR spectra were acquired from each sample using the standard Varian PRESAT pulse sequence with a single pulse excitation and 1.5 s low power pre-saturation at the water peak position to suppress the residual H2O signal. A total of 2048 transients were collected into 32 k complex data points. The acquisition time and recycle delay (RD) of a single scan were 2 s and 1.5 s, respectively, resulting in a total pulse sequence duration of about 3.5 s. The total experimental time for acquiring each spectrum was about 2 h.
Similarly, the hydrophobic metabolites were reconstituted in 600 μl of deuterated chloroform (CDCl3) containing 0.03% (v/v) tetramethylsilane (TMS) shortly before the NMR experiments. About 550 μl of the prepared sample was loaded into a standard 5 mm NMR tube. 1D NMR spectra were acquired at 20 °C using a single pulse sequence with 1024 transients for each spectrum. The acquisition time was 1.9 s and the recycle delay was 2 s, resulting in a total experimental time of about 1 h.
NMR data processing and multivariate data analysis
All free induction decays were multiplied by an exponential function with 0.5 Hz Lorentz line broadening prior to Fourier transformation. And, all 1H NMR spectra were manually phased and baseline corrected using the Processor module of Chenomx (NMR suite 7.6, Professional) and referenced to the chemical shift of TSP-d4 at 0 ppm. For peak assignment purposes, targeted profiling of hydrophilic extracts using spectral deconvolution method was employed to get the absolute metabolite concentrations with the help of the Profiler module of Chenomx (NMR suite 7.6, Professional) which contains a database of more than 300 common metabolites associated with mammals and bacteria [22]. The detailed description of spectral deconvolution method to obtain the absolute concentration of each metabolite in the NMR tube can be found in our previous publication [13]. The metabolite concentrations were then normalized to per milligram of tissue before extraction followed by multivariate data analysis in SIMCA (Version 13.0.3 64-bit, Umetrics, Umea, Sweden), i.e. Principal Component Analysis (PCA) and Orthogonal Projection to Latent Structure (OPLS). For hydrophobic extracts, spectral binning was used for data reduction before statistical data analysis. The spectral regions at δ 0.6 ~ 6.0 were segmented into discrete bins with equal width of 0.003 ppm using the Profiler module of Chenomx (NMR suite 7.6, Professional). The integral areas of all bins were then normalized to per milligram of tissue before extraction followed by multivariate data analysis in SIMCA.
PCA was performed first using the mean-centered and unit-variance scaled data, i.e. metabolites concentrations for hydrophilic extracts and spectral binning data for hydrophobic extracts, to obtain an overview and detect possible outliers. Subsequently, OPLS was conducted using the autoscaled data as X-matrix (with each row representing a sample, each column representing a variable) and class information as Y-matrix to find significant variables, i.e. metabolites, responsible for the discrimination of two different classes [23,24]. y = 0 was assigned to the control group and y = 1 to the tumor group before building an OPLS model, so that positive loadings mean up-regulated while negative loadings mean down-regulated [25]. Both PCA and OPLS models were constructed using the non-linear iterative partial least squares (NIPALS) algorithm and model complexity (number of components) was determined by a 7-fold cross-validation method. Model quality was evaluated from the parameters R2, revealing the interpretability of the model, and Q2, indicating the predictability of the model. Finally, the model significance was further assessed by the CV-ANOVA test at the level of p<0.05 [26]. OPLS loadings plot with color-coded correlation coefficients, was employed to evaluate the variable importance for class separation with warm colored (e.g. red) metabolites being more significant than cold colored (e.g. blue) ones [25]. A cutoff value of |r| > 0.811 for correlation coefficients is chosen to select metabolites responsible for between group variation based on the discrimination significance (p<0.05) [27-29].
Results
NMR spectra of stomach tissue extracts
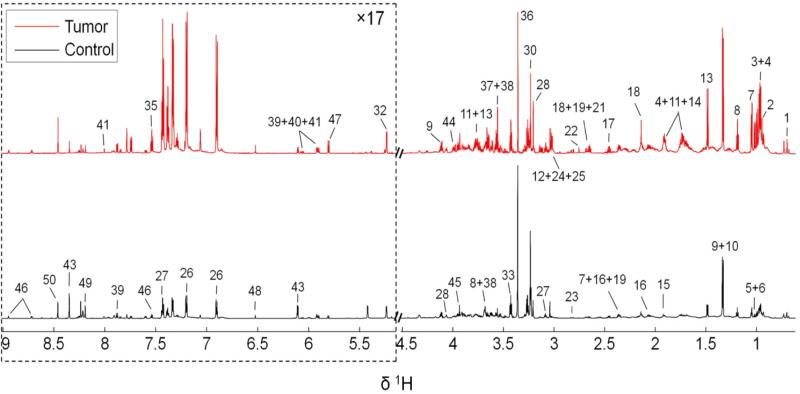
Examples of typical 1H NMR spectra of hydrophilic extracts obtained from a control mouse and a melanoma mouse were shown in Figure 1. The peak intensities were normalized to per unit weight of tissue before extraction, so that the concentrations of a given metabolite in the tumor group and the control group can be directly compared visually according to the peak intensities in the corresponding spectrum. Peak assignments were listed in Table 1. A total of 50 metabolites were identified with good confidence based on spectral deconvolution using Chenomx. Chemical shift identities of metabolites were assigned according to both the scientific literature [13,28,30-33], and the Chenomx metabolite library. A wide range of amino acids, carbohydrates, glycolysis and tricarboxylic acid cycle (TCA cycle) intermediates were detected. Other observed metabolites included choline metabolites, organic bases, purine and pyrimidine derivatives. Visual inspection of the 1H NMR spectra revealed apparent metabolic alterations induced by melanoma, i.e. the intensities of nearly all peaks were enhanced by melanoma in the upper trace of Figure 1 when compared with the lower trace from the control group, with only one exception that hypoxanthine (peak 49) was decreased. To identify significantly changed metabolites, thus taking within-group variations into consideration, both PCA and OPLS modeling were performed on the entire spectra set (i.e. five from tumor group and another five from control group).
Figure: 1.
800 MHz liquid state 1H NMR metabolite spectra of the hydrophilic extracts of stomachs excised from the control (black) and the melanoma mice (red). The peak intensities were normalized to per unit weight of tissue before extraction. The dotted regions were vertically expanded 17 times as denoted in the figure. A total of 50 metabolites were identified with metabolite numbers, i.e. the metabolite keys shown in Table 1
Table 1.
Assignments and Concentration of Metabolites from hydrophilic tissue extracts of stomach
| Key | Metabolites | δ 1H (ppm) and multiplicity* | Concentration (μM/mg) Mean ± SD | Estimates of absolute concentrations in tissue (mM) Mean ± SD | ||
|---|---|---|---|---|---|---|
| Tumor | Control | Tumor | Control | |||
| 1 | Cholate | 0.68(s),0.93(s), 0.96(d),1.00(m), 1.16(m),1.30(m), 1.42(m),1.51(m), 1.60(m),1.70(m), 1.80(m),1.89(m), 2.00(m),2.10(m), 2.23(m),3.51(m), 3.91(m), 4.08(m) | 2.94 ± 2.09 | 1.15 ± 0.72 | 1.86 ± 1.32 | 0.73 ± 0.45 |
| 2 | Isoleucine | 0.94(t), 0.99(d), 1.27(m), 1.45(m), 1.99(m), 3.67(d) | 9.82 ± 2.94 | 3.30 ± 2.73 | 6.20 ± 1.86 | 2.08 ± 1.72 |
| 3 | Alloisoleucine | 0.96(d), 0.97(t), 1.37(m), 1.42(m), 2.02(m), 3.70(d) | 7.69 ± 2.93 | 2.78 ± 2.55 | 4.86 ± 1.85 | 1.76 ± 1.61 |
| 4 | Leucine | 0.96(d), 0.97(d), 1.69(m), 1.70(m), 1.74(m), 3.74(dd) | 8.97 ± 2.38 | 3.04 ± 1.59 | 5.67 ± 1.50 | 1.92 ± 1.00 |
| 5 | 2-Aminobutyrate | 0.99(t), 1.90(m), 3.73(dd) | 5.86 ± 1.96 | 1.94 ± 1.16 | 3.70 ± 1.24 | 1.23 ± 0.73 |
| 6 | Valine | 0.97(d), 1.02(d), 2.28(m), 3.61(d) | 8.39 ± 2.47 | 3.00 ± 2.58 | 5.30 ± 1.56 | 1.89 ± 1.63 |
| 7 | Isobutyrate | 1.05(d), 2.37(m) | 5.88 ± 1.30 | 2.36 ± 1.66 | 3.71 ± 0.82 | 1.49 ± 1.05 |
| 8 | Ethanol | 1.19(t), 3.66(q) | 13.11 ± 3.53 | 6.11 ± 1.33 | 8.28 ± 2.23 | 3.86 ± 0.084 |
| 9 | Lactate | 1.33(d), 4.11(q) | 28.34 ± 6.45 | 22.17 ± 3.92 | 17.90 ± 4.07 | 14.00 ± 2.48 |
| 10 | Threonine | 1.33(d), 3.58(d), 4.26(m) | 6.06 ± 1.90 | 1.63 ± 1.13 | 3.83 ± 1.20 | 1.03 ± 0.71 |
| 11 | Lysine | 1.44(m), 1.51(m), 1.73(m), 1.89(m), 1.92(m), 3.03(t), 3.77(t) | 10.06 ± 3.92 | 3.08 ± 2.52 | 6.35 ± 2.48 | 1.95 ± 1.59 |
| 12 | Cadaverine | 1.46(m), 1.70(m), 3.03(m) | 1.11 ± 0.18 | 0.43 ± 0.29 | 0.70 ± 0.11 | 0.27 ± 0.18 |
| 13 | Alanine | 1.48(d), 3.78(q) | 21.38 ± 4.49 | 9.39 ± 4.91 | 13.50 ± 2.84 | 5.93 ± 3.10 |
| 14 | Arginine | 1.66(m), 1.73(m), 1.91(m), 1.92(m), 3.25(t), 3.78(t) | 10.90 ± 4.93 | 3.55 ± 2.08 | 6.88 ± 3.11 | 2.24 ± 1.31 |
| 15 | Acetate | 1.92(s) | 0.82 ± 0.26 | 1.14 ± 0.69 | 0.52 ± 0.16 | 0.72 ± 0.44 |
| 16 | Glutamate | 2.06(m), 2.14(m), 2.34(m), 2.37(m), 3.76(dd) | 10.08 ± 2.03 | 6.66 ± 1.51 | 6.37 ± 1.28 | 4.21 ± 0.95 |
| 17 | Glutamine | 2.13(m), 2.15(m), 2.44(m), 2.47(m), 3.76(t), 6.85(s) | 9.70 ± 2.12 | 5.04 ± 1.62 | 6.13 ± 1.34 | 3.18 ± 1.02 |
| 18 | Methionine | 2.13(m), 2.14(s), 2.21(m), 2.65(t), 3.86(dd) | 4.25 ± 1.16 | 1.66 ± 1.01 | 2.68 ± 0.73 | 1.05 ± 0.64 |
| 19 | Malate | 2.38(dd), 2.67(dd), 4.30(m) | 2.09 ± 0.66 | 1.23 ± 0.32 | 1.32 ± 0.42 | 0.78 ± 0.20 |
| 20 | Citrate | 2.53(d), 2.67(d) | 0.31 ± 0.05 | 0.19 ± 0.07 | 0.20 ± 0.03 | 0.12 ± 0.04 |
| 23 | Asparagine | 2.85(dd), 2.95(dd), 4.00(dd) | 2.83 ± 0.44 | 0.61 ± 0.45 | 1.79 ± 0.28 | 0.39 ± 0.28 |
| 24 | Creatine phosphate | 3.04(s), 3.93(s) | 3.02 ± 0.97 | 2.06 ± 0.48 | 1.91 ± 0.61 | 1.30 ± 0.30 |
| 25 | Creatine | 3.04(s), 3.93(s) | 2.37 ± 0.46 | 1.61 ± 0.39 | 1.50 ± 0.29 | 1.02 ± 0.25 |
| 26 | Tyrosine | 3.06(dd), 3.21(dd), 3.95(dd), 6.90(m), 7.20(m) | 4.74 ± 1.60 | 1.93 ± 1.24 | 2.99 ± 1.01 | 1.22 ± 0.78 |
| 27 | Phenylalanine | 3.13(dd), 3.29(dd), 4.00(dd), 7.33(m), 7.38(m), 7.43(m) | 6.90 ± 2.29 | 2.49 ± 1.80 | 4.36 ± 1.45 | 1.57 ± 1.14 |
| 28 | Choline | 3.21(s), 3.52(m), 4.07(m) | 4.85 ± 0.83 | 2.79 ± 1.61 | 3.06 ± 0.52 | 1.76 ± 1.02 |
| 29 | O-Phosphocholine | 3.22(s), 3.58(m), 4.17(m) | 0.23 ± 0.06 | 2.07 ± 0.94 | 0.15 ± 0.04 | 1.31 ± 0.59 |
| 30 | sn-Glycero-3-phosphocholine | 3.23(s), 3.62(dd), 3.67(m), 3.68(m), 3.88(m), 3.92(m), 3.96(m), 4.33(m) | 7.02 ± 1.73 | 8.95 ± 1.98 | 4.43 ± 1.09 | 5.65 ± 1.25 |
| 31 | Trimethylamine N-oxide | 3.27(s) | 0.27 ± 0.10 | 0.35 ± 0.10 | 0.17 ± 0.06 | 0.22 ± 0.06 |
| 32 | Glucose | 3.23(m), 3.40(m), 3.48(m), 3.54(dd), 3.71(dd), 3.73(dd), 3.76(m), 3.82(m), 3.84(m), 3.90(dd), 4.65(d), 5.23(d) | 4.19 ± 3.30 | 2.96 ± 1.30 | 2.65 ± 2.08 | 1.87 ± 0.82 |
| 33 | Taurine | 3.26(t), 3.43(t) | 21.33 ± 2.96 | 16.32 ± 2.92 | 13.47 ± 1.87 | 10.31 ± 1.84 |
| 34 | myo-Inositol | 3.29(t), 3.53(dd), 3.62(t), 4.06(m) | 3.41 ± 0.35 | 3.10 ± 0.56 | 2.15 ± 0.22 | 1.96 ± 0.35 |
| 35 | Tryptophan | 3.29(dd), 3.47(dd), 4.06(dd), 7.21(m), 7.29(m), 7.32(s), 7.54(d), 7.74(d) | 1.39 ± 0.44 | 0.44 ± 0.34 | 0.88 ± 0.28 | 0.28 ± 0.21 |
| 36 | Methanol | 3.36(s) | ||||
| 37 | Glycine | 3.56(s) | 8.68 ± 1.71 | 3.87 ± 1.83 | 5.48 ± 1.08 | 2.44 ± 1.16 |
| 38 | Glycerol | 3.56(dd), 3.65(dd), 3.78(m) | 9.50 ± 4.14 | 3.16 ± 1.72 | 6.00 ± 2.61 | 2.00 ± 1.09 |
| 39 | Uridine | 3.80(dd), 3.92(dd), 4.13(m), 4.23(t), 4.36(t), 5.90(d), 5.92(d), 7.88(d) | 0.48 ± 0.16 | 0.32 ± 0.10 | 0.30 ± 0.10 | 0.20 ± 0.06 |
| 40 | Cytidine | 3.80(dd), 3.92(dd), 4.12(m), 4.20(t), 4.30(m), 5.91(d), 6.06(d), 7.85(d) | 0.19 ± 0.06 | 0.10 ± 0.04 | 0.12 ± 0.04 | 0.06 ± 0.03 |
| 41 | Guanosine | 3.81(dd), 3.88(dd), 4.22(m), 4.41(dd), 4.75(t), 5.92(d), 8.00(s) | 0.19 ± 0.11 | 0.10 ± 0.04 | 0.12 ± 0.07 | 0.06 ± 0.03 |
| 42 | Adenosine | 3.83(dd), 3.91(dd), 4.28(m), 4.42(dd), 4.79(dd), 6.07(d), 8.23(s), 8.35(s) | 0.11 ± 0.03 | 0.05 ± 0.02 | 0.07 ± 0.02 | 0.03 ± 0.01 |
| 43 | Inosine | 3.84(dd), 3.92(dd), 4.27(m), 4.44(m), 4.76(t), 6.10(d), 8.23(s), 8.35(s) | 0.50 ± 0.12 | 0.99 ± 0.42 | 0.32 ± 0.08 | 0.63 ± 0.27 |
| 44 | Serine | 3.84(dd), 3.95(dd), 3.99(dd) | 8.40 ± 1.11 | 3.22 ± 2.13 | 5.31 ± 0.70 | 2.03 ± 1.35 |
| 45 | Glycolate | 3.93(s) | 1.36 ± 0.35 | 0.66 ± 0.40 | 0.86 ± 0.22 | 0.42 ± 0.25 |
| 46 | Nicotinurate | 3.98(m), 7.60(dd), 8.25(m), 8.71(dd), 8.94(m) | 0.50 ± 0.03 | 0.42 ± 0.08 | 0.32 ± 0.02 | 0.27 ± 0.05 |
| 47 | Uracil | 5.80(d), 7.53(d) | 1.21 ± 0.17 | 0.50 ± 0.23 | 0.76 ± 0.11 | 0.32 ± 0.15 |
| 48 | Fumarate | 6.52(s) | 0.13 ± 0.03 | 0.09 ± 0.02 | 0.08 ± 0.02 | 0.06 ± 0.01 |
| 49 | Hypoxanthine | 8.19(s), 8.21(s) | 0.44 ± 0.10 | 0.95 ± 0.12 | 0.28 ± 0.06 | 0.60 ± 0.08 |
| 50 | Formate | 8.46(s) | 2.87 ± 0.59 | 1.63 ± 0.46 | 1.81 ± 0.37 | 1.03 ± 0.29 |
s: singlet; d: doublet; t: triplet; q: quartet; m: multiplet; dd: doublet of doublet.
Melanoma induced metabolic changes in stomach tissue
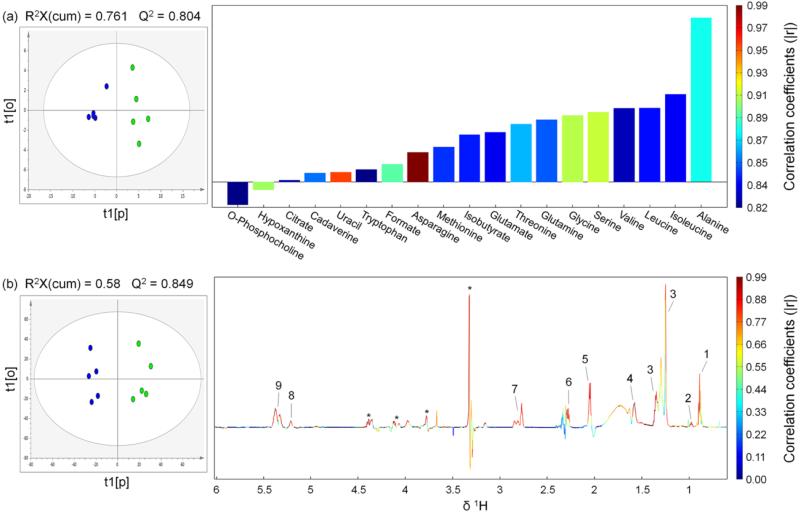
Unsupervised (or exploratory) data analysis, i.e. PCA in this case, was conducted firstly to get an overview and detect possible outliers. PCA scores plots (Figure S2 in Supporting Information) showed clear classification of the control and tumor cell treated mice without any outlier. Therefore all 10 samples were kept for further OPLS modeling. In order to maximize the correlation between the X-matrix (i.e. metabolites concentrations or spectral binning data for hydrophilic and hydrophobic extracts, respectively) and Y-matrix (class information, i.e. the control versus melanoma groups) as well as the variation in X-matrix, OPLS was performed to assess variable importance and determine discriminatory variables responsible for separation of different groups [24]. Values of the resulting model parameters, i.e. R2 and Q2, showed good qualities of the generated OPLS models (Figure 2). CV- ANOVA results further confirmed the model validity (p values of 0.0506 and 0.028 for models (a) and (b), respectively). The key variables showing significant differences between the control group and the melanoma group were extracted from the correlation coefficients-coded OPLS loadings plots (Figure 2). Based on the results of OPLS modeling, a group of metabolites were identified to be responsible for the discrimination of the tumor group from the control group with their correlation coefficients tabulated in Table 2. It's known from Tables 1 and 2 that, compared with the control group, in the tumor group the concentrations of O-phosphocholine and hypoxanthine were decreased (with negative loadings in Figure 2 (a)), while the concentrations of alanine, isoleucine, leucine, valine, serine, glycine, glutamine, threonine, glutamate, isobutyrate, methionine, asparagine, formate, tryptophan, uracil, cadaverine and citrate were increased (with positive loadings in Figure 2 (a)). Figure 2 (b) showed the scores and loadings plot of the OPLS model constructed using the NMR spectral data of hydrophobic contents. Lipid levels were elevated by metastatic melanoma in the stomach, with discriminatory variables showing positive loadings in Figure 2 (b). We consider all these metabolites to be potential biomarker candidates for metastatic melanoma in the stomach.
Figure: 2.
OPLS scores (left) and coefficients-coded loadings plot (right) of the model discriminating the control (blue dots) and the tumor (green dots) groups. (a) Using the hydrophilic metabolites concentrations obtained by spectral deconvolution (only metabolites with correlation coefficients |r| > 0.811 were plotted, correlation coefficients of all metabolites used for OPLS analysis were listed in Table S1 in Supporting Information). (b) Using the data derived from binning results of 1H NMR spectra from hydrophobic extracts. CV-ANOVA results gave p values of 0.0506 and 0.028 for models (a) and (b), respectively. A total of 2 components were extracted for both model (a) and model (b). R2, i.e. fraction of Y variation modeled by X, was 0.93 and 0.961 for model (a) and model (b), respectively. Lipids peak assignments in model (b): 1, CH3CH2CH2C=C; 2, CH3CH2; 3, (CH2)n, CH3CH2(CH2)n, CH2CH2CH2CO, CH2; 4, CH2CH2CO; 5, CH2C=C; 6, CH2CO; 7, C=CHCH2CH=C; 8, -CH=CHCH2CH=CH- (Unsaturated lipid), =CHCH2CH2; 9, =CHCH2CH2 (Unsaturated lipid). “Asterisk” indicates unassigned lipid contents
Table 2.
Melanoma induced metabolic changes in hydrophilic tissue extracts of stomach
| Key | Metabolites | Correlation coefficients |
|---|---|---|
| 2 | Isoleucine | 0.835 |
| 4 | Leucine | 0.839 |
| 6 | Valine | 0.825 |
| 7 | Isobutyrate | 0.841 |
| 10 | Threonine | 0.868 |
| 12 | Cadaverine | 0.856 |
| 13 | Alanine | 0.882 |
| 16 | Glutamate | 0.835 |
| 17 | Glutamine | 0.852 |
| 18 | Methionine | 0.846 |
| 20 | Citrate | 0.823 |
| 23 | Asparagine | 0.986 |
| 29 | O-Phosphocholine | −0.817 |
| 35 | Tryptophan | 0.817 |
| 37 | Glycine | 0.910 |
| 44 | Serine | 0.913 |
| 47 | Uracil | 0.954 |
| 49 | Hypoxanthine | −0.907 |
| 50 | Formate | 0.894 |
Discussion
Melanoma is a malignant tumor of melanocytes with high capability of rapid metastasis to other organs, and is the most common metastatic malignancy found in gastrointestinal tract with currently no effective treatment [6]. The ultimate goal of the present study is to understand the metabolic perturbations caused by metastatic melanoma in stomach and to identify the affected metabolic pathway networks for novel therapeutic targets for treating this fatal disease.
Notably, 12 of the 19 identified discriminatory metabolites responsible for the separation of the melanoma group from the control group are amino acids including alanine, isoleucine, leucine, valine, serine, glycine, glutamine, threonine, glutamate, methionine, asparagine, and tryptophan. Among these metabolites, isoleucine, leucine, valine, threonine, methionine, and tryptophan are essential amino acids that can only be obtained from diet, while alanine, serine, glycine, glutamine, glutamate, asparagine are non-essential amino acids. Valine, leucine and isoleucine are branched-chain amino acids (BCAAs), and the degradation of these three amino acids leads to the production of acetyl-CoA which is then oxidized via the citric acid cycle (TCA cycle). Substantial amounts of valine, leucine and isoleucine will be generated by protein breakdown under fasting conditions [34]. Elevation of these three amino acids in tumor bearing mice observed in this study may be attributed to enhanced protein breakdown induced by metastatic melanoma in stomach. Increased protein degradation is consistent with the increased cadaverine levels in melanoma mice; cadaverine is observed in only small quantities in normal, healthy animals and is primarily produced by protein hydrolysis during putrefaction of animal tissue [35]. Further evidence for enhanced proteolysis in melanoma mice is the observation of increased levels of isobutyrate that originates from bacterial fermentation of amino acids [36]. These observations reflect disturbed microbiota metabolism induced by metastatic melanoma.
Glutamate is a multifunctional precursor involved in a variety of metabolic pathways as depicted in the Small Molecule Pathway Database (SMPDB) (http://www.smpdb.ca/). It interconverts with many other metabolites via the actions of different enzymes. For example, glutamate can be generated from alanine or aspartate in combination with 2-Oxoglutarate catalyzed by transaminase, the resulting byproducts (pyruvate and oxaloacetate) of which are key components in glycolysis, gluconeogenesis and the TCA cycle. Glutamate can also be metabolized to glutamine through the action of enzyme glutamine synthetase, which connects the pyruvate metabolism and the pyrimidine metabolism, as well as the purine metabolism [34]. The increased level of glutamate in melanoma group suggests that the above mentioned pathways may have been profoundly disturbed. This is confirmed by observed elevation of glutamine, alanine, citrate, asparagine, and decreased hypoxanthine in the tumor bearing mice which are intermediates of the pyrimidine metabolism, the pyruvate metabolism, the TCA cycle, the glutamate metabolism, and the purine metabolism, respectively. Glutamate signaling in cancer has stimulated extensive research interest recently and has shown great importance in human melanoma development [37-40]. Increased glutamate concentration has also been reported by previous studies as a common phenomenon in tumors [41]. Furthermore, Magnus Karlsson and coworkers have reported that increased glutamate pools in tumors leads to increased leucine formation, a consistent result reflected in this study [42]. The blockade of glutamate receptors has been proposed as a promising novel therapy for treating melanoma [43]. Catalyzed by carbamoyl phosphate synthetase, glutamine is converted to carbamoyl phosphate and then used for pyrimidine metabolism in which uracil, a pyrimidine derivative, is observed evidently up-regulated in tumor bearing mice in the present study. Through a series of reactions, glutamine can also be metabolized to Guanosine Monophosphate (GMP) during purine nucleotide biosynthesis. Successive dephosphorylation and deamination of GMP will lead to the production of xanthine, an important purine that can alternatively be produced from another notable purine, i.e. hypoxanthine, via the action of xanthine dehydrogenase/oxidase [34]. Significantly increased levels of glutamine and uracil as well as decreased hypoxanthine reflect deviations of the nucleotide pools and perturbed pyrimidine/purine metabolism caused by metastatic melanoma in the stomach, which has been observed in many cancers and has proved to be of central importance in malignant transformation and tumor cell differentiation [44].
Alanine is most commonly produced from pyruvate via alanine transaminase. Elevation of alanine in melanoma mice may be due to the perturbed expression of alanine transaminase, the level of which is used to evaluate tissue damage and adverse events in preclinical and clinical researches [45,46]. Enhanced production and excretion of alanine has been reported previously as a metabolic signature of melanoma [13,47]. In addition to inter-conversion with alanine, pyruvate also plays a central role in key pathways of energy metabolism. It can either be converted by the pyruvate dehydrogenase complex to produce acetyl-CoA for fatty acid synthesis, or carboxylated by pyruvate carboxylase in gluconeogenesis to produce oxaloacetate which participates the TCA cycle [34]. Alternatively, through the transformation to serine catalyzed by serine dehydratase/threonine deaminase, pyruvate is associated with the glycine and serine metabolism in which the concentrations of methionine, serine, and glycine are significantly up-regulated in this study due to metastasis of melanoma to gastrointestinal tract. Recent developments of genetic and functional research have identified that bioactivities of glycine, serine, sarcosine, methionine, and threonine, as well as their related metabolic pathways play essential roles in rapid cell proliferation, transformation, malignancy, and function as drivers of cancer pathogenesis and metastasis [48,49]. Serine is an essential metabolite for biosynthetic reactions including synthesis of proteins, nucleotides, and phosphatidyl-serine, as well as sphingosine that are required for cell proliferation. Functional genomics studies have revealed significant overexpression of phosphoglycerate dehydrogenase (PHGDH) in breast cancer, melanoma, and several other cancer types, thereby causing flux to the serine biosynthetic pathway in cancer cells and therefore resulting in elevated levels of serine [50]. As a nonessential amino acid, alanine can also be produced as a byproduct of tryptophan metabolism as depicted in Figure 3. Two other metabolites involved in tryptophan metabolism, i.e. tryptophan and formate, are also significantly increased in melanoma mice, further confirming alteration of tryptophan metabolism induced by metastatic melanoma in stomach. Tryptophan plays essential roles in many physiological processes, and biological metabolism of tryptophan include protein synthesis and degradation through three different routes, i.e. the kynurenine pathway, the serotonin pathway and the tryptamine pathway, with the kynurenine pathway being the predominant degradation route (responsible for about 95% of tryptophan metabolization) when tryptophan is not used for protein synthesis [51]. Increased concentration of tryptophan in melanoma mice may be caused by enhanced protein breakdown, echoing with up-regulated levels of valine, leucine, isoleucine, isobutyrate and cadaverine as afore discussed. Degradation of tryptophan through the kynurenine pathway is catalyzed by tryptophan 2,3-dioxygenase (TDO) and indoleamine 2,3-dioxygenase (IDO) in the initial rate-limiting step of this biochemical process [51]. Previous studies have reported up-regulation of IDO in metastatic malignant melanoma cells, and its strong association with increased number of regulatory T cells transformed from active T lymphocytes, which facilitate tumors to evade immune targeting [52]. Observed accumulation of tryptophan metabolic compounds, i.e. formate and alanine in the present study, may be partly attributed to the up-regulation of IDO in the melanoma bearing mice.
Figure: 3.
Schematic diagram of potentially disturbed metabolic pathway networks associated with the significantly altered metabolites during melanoma development based on the findings from this work and the diverse metabolic fates depicted in the Small Molecule Pathway Database (SMPDB) (http://www.smpdb.ca/) and literatures. Solid line indicates one-step process, while dash line indicates multi-steps process. Metabolites colored green are not detected. Metabolic pathways under the numbered shadow squares are parts of metabolism networks of particular metabolites: 1 purine metabolism, 2 glycine and serine metabolism, 3 pyrimidine metabolism, 4 pyruvate metabolism, 5 glutamate metabolism, 6 tryptophan metabolism, 7 threonine degradation. Other significantly affected metabolites pathways are valine, leucine, isoleucine degradation, lipids metabolism, methionine metabolism and TCA cycle
The down-regulated level of O-phosphocholine and up-regulated level of lipids contents imply enhanced lipids metabolism in melanoma mouse stomach. Morvan and coworkers have previously reported strong phospholipids metabolism perturbations in melanoma tumors as a mechanism participating in tumor cell redifferentiation and/or survival [53,54]. Tumor metastasis can be activated during cell migration, invasion and angiogenesis by modulating phosphorylation and acetylation of proteins, a process that sphingolipids are involved by mediating the expression of specific genes, such as MMP-2, uPA, ZNF580, MMP-9 [55]. Elevated levels of sphingolipids metabolites, including sphingosine, ceramide, dihydroceramide, sphinganine, sphingosine-1-phophate, as well as sphinganine-1-phosphate, are typical characteristics in tumor tissues of a diversity of cancers, including human endometrial cancer, malignant breast tumor and melanoma, when compared with those in normal tissues [56]. Activation of sphingolipid synthesis is proved to be closely related to tumor formation and growth in mice with melanoma, and suppression of sphingolipids metabolism has been proposed as a potential target for melanoma therapy [56]. The observed elevation of lipids, amino acids, and nucleic acids in the present study may also partly be attributed to promoted tumor cell growth and proliferation of metastatic melanoma in stomach, since these metabolites are needed for construction of new cellular components and the maintenance of various cellular activities [48].
Collectively, the potentially affected metabolic pathway networks associated with metastatic melanoma in stomach are illustrated in Figure 3, based on the results from our metabolomics study of this disease and the metabolite fates described in the Small Molecule Pathway Database (http://www.smpdb.ca/) and the literatures as previously discussed.
Conclusions
We have shown that the combined application of 1H NMR metabolic profiling and multivariate statistical analysis (i.e. PCA and OPLS) is a powerful tool for investigating metastatic melanoma in mouse stomach. Both PCA and OPLS show that the tumor group is well separated from the control group based on either the metabolites found from the hydrophilic extracts or metabolites from the hydrophobic extracts. A total of 50 metabolites with concentration in stomach ranged from as low as 30 μM to as high as 17.9 mM are identified in the hydrophilic tissue extracts [21]. Severe metabolic perturbations have been observed in stomach of melanoma mice. The potentially disturbed metabolic pathways revealed by our metabolomics study include pyrimidine metabolism, purine metabolism, pyruvate metabolism, tryptophan metabolism, glycine/serine metabolism, glutamate metabolism, threonine degradation, methionine metabolism, valine/leucine/isoleucine degradation, lipids metabolism, and TCA cycle. Additionally, the microbiota metabolism has also been evidently altered resulting in enhanced protein breakdown. Therefore, our 1H NMR metabolomics study clearly show that multiple biological pathways are potentially affected by metastatic melanoma in the stomach. Identification of these altered metabolic pathway networks provides important information for improved understanding of the molecular mechanisms associated with melanoma metastasis in stomach and suggests potential therapeutic targets for treating this fatal disease.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of Envionmental Health Sciences of the National Institute of Health (NIH) under Award Number R01ES022176. All of the NMR experiments were performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the DOE's Office of Biological and Environmental Research, and located at Pacific Northwest National Laboratory (PNNL). PNNL is a multiprogram national laboratory operated for the DOE by Battelle Memorial Institute under Contract DE-AC06-76RLO 1830. And finally we thank Dr. Xihai Wang for his contributions in pathology and animal work.
Footnotes
Citation: Wang X, Hu M, Liu M, Hu JZ (2014) Metastatic Melanoma Induced Metabolic Changes in C57BL/6J Mouse Stomach Measured by 1H NMR Spectroscopy. Metabolomics 4: 135. doi: 10.4172/2153-0769.1000135
References
- 1.La Torre M, Frezza B, Fransvea P, Ziparo V, Balducci G, et al. Gastrointestinal metastases of cutaneous melanoma. Am Surg. 2014;80:E45–46. [PubMed] [Google Scholar]
- 2.Maire C, Vercambre-Darras S, Devos P, D'Herbomez M, Dubucquoi S, et al. Metastatic melanoma: spontaneous occurrence of auto antibodies is a good prognosis factor in a prospective cohort. J Eur Acad Dermatol and Venereol. 2013;27:92–96. doi: 10.1111/j.1468-3083.2011.04364.x. [DOI] [PubMed] [Google Scholar]
- 3.Bourne RM, Stanwell P, Stretch JR, Scolyer RA, Thompson JF, et al. In vivo and ex vivo proton MR spectroscopy of primary and secondary melanoma. Eur J Radiol. 2005;53:506–513. doi: 10.1016/j.ejrad.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Bailet O, Fenouille N, Abbe P, Robert G, Rocchi S, et al. Spleen tyrosine kinase functions as a tumor suppressor in melanoma cells by inducing senescence-like growth arrest. Cancer Res. 2009;69:2748–2756. doi: 10.1158/0008-5472.CAN-08-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orlov NV, Weeraratna AT, Hewitt SM, Coletta CE, Delaney JD, et al. Automatic detection of melanoma progression by histological analysis of secondary sites. Cytometry A. 2012;81:364–373. doi: 10.1002/cyto.a.22044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang KV, Sanderson SO, Nowakowski GS, Arora AS. Metastatic malignant melanoma of the gastrointestinal tract. Mayo Clin Proc. 2006;81:511–516. doi: 10.4065/81.4.511. [DOI] [PubMed] [Google Scholar]
- 7.Syngal S, Kaptain S, O'Neill MJ, Isselbacher KJ, Sober AJ, et al. A 50-year-old woman with severe diarrhea during radiation treatment for resected metastatic melanoma Cutaneous melanoma metastasized to the gastrointestinal tract, with protein-losing enteropathy. Adrenal cortical adenoma. N Engl J Med. 1999;340:789–796. doi: 10.1056/NEJM199903113401008. [DOI] [PubMed] [Google Scholar]
- 8.Akaraviputh T, Arunakul S, Lohsiriwat V, Iramaneerat C, Trakarnsanga A. Surgery for gastrointestinal malignant melanoma: Experience from surgical training center. World J Gastroenterol. 2010;16:745–748. doi: 10.3748/wjg.v16.i6.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotteas EA, Adamopoulos A, Drogitis PD, Zalonis A, Giannopoulos KV, et al. Gastrointestinal bleeding as initial presentation of melanoma of unknown primary origin: report of a case and review of the literature. In Vivo. 2009;23:487–489. [PubMed] [Google Scholar]
- 10.Graham SF, Holscher C, McClean P, Elliott CT, Green BD. 1H NMR metabolomics investigation of an Alzheimer's disease (AD) mouse model pinpoints important biochemical disturbances in brain and plasma. Metabolomics. 2013;9:974–983. [Google Scholar]
- 11.Tian N, Wang JS, Wang PR, Song XF, Yang MH, et al. NMR-based metabonomic study of Chinese medicine Gegen Qinlian Decoction as an effective treatment for type 2 diabetes in rats. Metabolomics. 2013;9:1228–1242. [Google Scholar]
- 12.Huang RZ, Gao HC, Ma LH, Wang X, Jia JM, et al. Dynamic 1H NMR-based extracellular metabonomic analysis of oligodendroglia cells infected with herpes simplex virus type 1. Metabolomics. 2014;10:33–41. [Google Scholar]
- 13.Wang X, Hu M, Feng J, Liu M, Hu JZ. 1H NMR Metabolomics Study of Metastatic Melanoma in C57BL/6J Mouse Spleen. Metabolomics. 2014;10:1129–1144. doi: 10.1007/s11306-014-0652-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin FP, Collino S, Rezzi S, Kochhar S. Metabolomic applications to decipher gut microbial metabolic influence in health and disease. Front Physiol. 2012;3:113. doi: 10.3389/fphys.2012.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gheorgheosu D, Dehelean C, Cristea M, Muntean D. Devolopment of the B16 Murine Melanoma Model. Annals of RSCB. 2011:148–152. [Google Scholar]
- 16.Roomi MW, Kalinovsky T, Roomi NW, Ivanov V, Rath M, et al. Suppression of growth and hepatic metastasis of murine B16FO melanoma cells by a novel nutrient mixture. Oncol Rep. 2008;20:809–817. [PubMed] [Google Scholar]
- 17.Beckonert O, Keun HC, Ebbels TM D, Bundy JG, Holmes E, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 18.Washburn KW. A Modification of the Folch Method of Lipid Extraction for Poultry. Poultry Science. 1989;68:1425–1427. [Google Scholar]
- 19.Iverson SJ, Lang SL, Cooper MH. Comparison of the Bligh and Dyer and Folch methods for total lipid determination in a broad range of marine tissue. Lipids. 2001;36:1283–1287. doi: 10.1007/s11745-001-0843-0. [DOI] [PubMed] [Google Scholar]
- 20.Akoh CC, Min DB. Food Lipids: Chemistry, Nutrition, and Biotechnology. Third Edtn CRC Press; 2008. [Google Scholar]
- 21.Feng J, Isern NG, Burton SD, Hu JZ. Studies of Secondary Melanoma on C57BL/6J Mouse Liver Using 1H NMR Metabolomics. Metabolites. 2013;3:1011–1035. doi: 10.3390/metabo3041011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal Chem. 2006;78:4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 23.Cloarec O, Dumas ME, Trygg J, Craig A, Barton RH, et al. Evaluation of the orthogonal projection on latent structure model limitations caused by chemical shift variability and improved visualization of biomarker changes in 1H NMR spectroscopic metabonomic studies. Anal Chem. 2005;77:517–526. doi: 10.1021/ac048803i. [DOI] [PubMed] [Google Scholar]
- 24.Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS). Journal of Chemometrics. 2002;16:119–128. [Google Scholar]
- 25.Wiklund S, Johansson E, Sjöström L, Mellerowicz EJ, Edlund U, et al. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem. 2008;80:115–122. doi: 10.1021/ac0713510. [DOI] [PubMed] [Google Scholar]
- 26.Eriksson L, Trygg J, Wold S. CV-ANOVA for significance testing of PLS and OPLS (R) models. J Chemometrics. 2008;22:594–600. [Google Scholar]
- 27.An Y, Xu W, Li H, Lei H, Zhang L, et al. High-fat diet induces dynamic metabolic alterations in multiple biological matrices of rats. J Proteome Res. 2013;12:3755–3768. doi: 10.1021/pr400398b. [DOI] [PubMed] [Google Scholar]
- 28.Jiang L, Huang J, Wang Y, Tang H. Metabonomic analysis reveals the CCl4-induced systems alterations for multiple rat organs. J Proteome Res. 2012;11:3848–3859. doi: 10.1021/pr3003529. [DOI] [PubMed] [Google Scholar]
- 29.Dong FC, Zhang LL, Hao FH, Tang HR, Wang YL. Systemic Responses of Mice to Dextran Sulfate Sodium-Induced Acute Ulcerative Colitis Using H-1 NMR Spectroscopy. J Proteome Res. 2013;12:2958–2966. doi: 10.1021/pr4002383. [DOI] [PubMed] [Google Scholar]
- 30.Lindon JC, Nicholson JK, Everett JR. NMR spectroscopy of biofluids. Annual Reports on NMR Spectroscopy. 1999;38:1–88. [Google Scholar]
- 31.Martin FP, Dumas ME, Wang Y, Legido-Quigley C, Yap IK, et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol. 2007;3:112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin FPJ, Wang YL, Sprenger N, Holmes E, Lindon JC, et al. Effects of probiotic Lactobacillus Paracasei treatment on the host gut tissue metabolic profiles probed via magic-angle-spinning NMR spectroscopy. J Proteome Res. 2007;6:1471–1481. doi: 10.1021/pr060596a. [DOI] [PubMed] [Google Scholar]
- 33.Nicholson JK, Foxall PJ, Spraul M, Farrant RD, Lindon JC. 750 MHz 1H and 1H-13C NMR spectroscopy of human blood plasma. Anal Chem. 1995;67:793–811. doi: 10.1021/ac00101a004. [DOI] [PubMed] [Google Scholar]
- 34.Frolkis A, Knox C, Lim E, Jewison T, Law V, et al. SMPDB: The Small Molecule Pathway Database. Nucleic Acids Res. 2010;38:D480–487. doi: 10.1093/nar/gkp1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li M, Li D, Huang Y, Liu M, Wang H, et al. Improving the secretion of cadaverine in Corynebacterium glutamicum by cadaverine-lysine antiporter. J Ind Microbiol Biotechnol. 2014;41:701–709. doi: 10.1007/s10295-014-1409-4. [DOI] [PubMed] [Google Scholar]
- 36.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Namkoong J, Shin SS, Lee HJ, Marin YE, Wall BA, et al. Metabotropic glutamate receptor 1 and glutamate signaling in human melanoma. Cancer Res. 2007;67:2298–2305. doi: 10.1158/0008-5472.CAN-06-3665. [DOI] [PubMed] [Google Scholar]
- 38.Choi KY, Chang K, Pickel JM, Badger JD, Roch KW. Expression of the metabotropic glutamate receptor 5 (mGluR5) induces melanoma in transgenic mice. Proc Natl Acad Sci U S A. 2011;108:15219–15224. doi: 10.1073/pnas.1107304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stepulak A, Sifringer M, Rzeski W, Endesfelder S, Gratopp A, et al. NMDA antagonist inhibits the extracellular signal-reglulated kinase pathway and suppresses cancer growth. Proc Natl Acad Sci U S A. 2005;102:15605–15610. doi: 10.1073/pnas.0507679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rzeski W, Turski L, Ikonomidou C. Glutamate antagonists limit tumor growth. Proc Natl Acad Sci U S A. 2001;98:6372–6377. doi: 10.1073/pnas.091113598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor BS, Pal M, Yu JJ, Laxman B, Kalyana-Sundararri S, et al. Humoral response profiling reveals pathways to prostate cancer progression. Mol Cell Proteomics. 2008;7:600–611. doi: 10.1074/mcp.M700263-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Karlsson M1, Jensen PR, `t Zandt R, Gisselsson A, Hansson G, et al. Imaging of branched chain amino acid metabolism in tumors with hyperpolarized 13C ketoisocaproate. Int J Cancer. 2010;127:729–736. doi: 10.1002/ijc.25072. [DOI] [PubMed] [Google Scholar]
- 43.Song ZQ, He CD, Liu J, Sun CK, Lu P, et al. Blocking glutamate-mediated signalling inhibits human melanoma growth and migration. Exp Dermatol. 2012;21:926–931. doi: 10.1111/exd.12048. [DOI] [PubMed] [Google Scholar]
- 44.Hatse S, De Clercq E, Balzarini J. Role of antimetabolites of purine and pyrimidine nucleotide metabolism in tumor cell differentiation. Biochem Pharmacol. 1999;58:539–555. doi: 10.1016/s0006-2952(99)00035-0. [DOI] [PubMed] [Google Scholar]
- 45.Bernardo SG, Moskalenko M, Pan M, Shah S, Sidhu HK, et al. Elevated rates of transaminitis during ipilimumab therapy for metastatic melanoma. Melanoma Res. 2013;23:47–54. doi: 10.1097/CMR.0b013e32835c7e68. [DOI] [PubMed] [Google Scholar]
- 46.Sato A, Yoshikawa N, Kubo E, Kakuda M, Nishiuchi A, et al. Inhibitory effect of cordycepin on experimental hepatic metastasis of B16-F0 mouse melanoma cells. In Vivo. 2013;27:729–732. [PubMed] [Google Scholar]
- 47.Filipp FV, Ratnikov B, De Ingeniis J, Smith JW, Osterman AL, et al. Glutamine-fueled mitochondrial metabolism is decoupled from glycolysis in melanoma. Pigment Cell Melanoma Res. 2012;25:732–739. doi: 10.1111/pcmr.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gomes MM, Coimbra JB1, Clara RO1, Dörr FA1, Moreno AC1, et al. Biosynthesis of N,N-dimethyltryptamine (DMT) in a melanoma cell line and its metabolization by peroxidases. Biochem Pharmacol. 2014;88:393–401. doi: 10.1016/j.bcp.2014.01.035. [DOI] [PubMed] [Google Scholar]
- 52.Brody JR, Costantino CL, Berger AC, Sato T, Lisanti MP, et al. Expression of indoleamine 2,3-dioxygenase in metastatic malignant melanoma recruits regulatory T cells to avoid immune detection and affects survival. Cell Cycle. 2009;8:1930–1934. doi: 10.4161/cc.8.12.8745. [DOI] [PubMed] [Google Scholar]
- 53.Morvan D, Demidem A, Papon J, De Latour M, Madelmont JC. Melanoma tumors acquire a new phospholipid metabolism phenotype under cystemustine as revealed by high-resolution magic angle spinning proton nuclear magnetic resonance spectroscopy of intact tumor sampled. Cancer Res. 2002;62:1890–1897. [PubMed] [Google Scholar]
- 54.Morvan D, Demidem A, Papon J, Madelmont JC. Quantitative HRMAS proton total correlation spectroscopy applied to cultured melanoma cells treated by chloroethyl nitrosourea: Demonstration of phospholipid metabolism alterations. Magnetic Resonance in Medicine. 2003;49:241–248. doi: 10.1002/mrm.10368. [DOI] [PubMed] [Google Scholar]
- 55.Patwardhan GA, Liu YY. Sphingolipids and expression regulation of genes in cancer. Prog Lipid Res. 2011;50:104–114. doi: 10.1016/j.plipres.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.