Abstract
High-throughput functional proteomic technologies provide a way to quantify the expression of proteins of interest. Statistical inference centers on identifying the activation state of proteins and their patterns of molecular interaction formalized as dependence structure. Inference on dependence structure is particularly important when proteins are selected because they are part of a common molecular pathway. In that case, inference on dependence structure reveals properties of the underlying pathway. We propose a probability model that represents molecular interactions at the level of hidden binary latent variables that can be interpreted as indicators for active versus inactive states of the proteins. The proposed approach exploits available expert knowledge about the target pathway to define an informative prior on the hidden conditional dependence structure. An important feature of this prior is that it provides an instrument to explicitly anchor the model space to a set of interactions of interest, favoring a local search approach to model determination. We apply our model to reverse-phase protein array data from a study on acute myeloid leukemia. Our inference identifies relevant subpathways in relation to the unfolding of the biological process under study.
Keywords: AML, Graphical models, Mixture models, POE, RJ-MCMC, RPPA
1. INTRODUCTION
In this article, we consider the statistical analysis of high-throughput protein expression data. We focus on the identification of patterns of protein interactions, capitalizing on both a molecular interrogation protocol called reverse-phase protein array (RPPA) technology and prior biological pathway knowledge.
RPPAs (Tibes et al. 2006) represent a high-throughput proteomic technology that provides a quantification of the expression for specifically targeted proteins selected from molecular pathways. Unlike traditional microarrays in which thousands of gene probes are immobilized on glass or fabric slides and are hybridized against samples from individuals, in RPPA, one immobilizes individual samples on the slides and hybridizes them against a single antibody that recognizes only one protein. This is why the technology is named reverse phase. Figure 1 presents an image of RPPA, in which 40 individual patient samples containing whole cellular protein repertoires are printed in 40 batches. Each batch can be recognized as a square of four-by-four dots. For quantification purposes, each sample is diluted serially in eight steps so that the resulting strength of the dilutions are full strength, 1/2, 1/4, 1/8, 1/16, 1/32, 1/64, and 1/128 of the full strength. Duplicates are produced for each dilution, resulting in a total of 16 spots for each sample, shown as the four-by-four square on the slide. Each slide is probed with an antibody that represents one specific protein. The antibody can be detected by amplified fluorescent, colorimetric, or chemiluminescent assays. After each slide is hybridized against the antibody, signal intensities for all the samples on the slide are measured by scanning the slide using a specially designed scanner. Quantification of each sample is based on the intensities of the 16 spots. In our application, we base our analysis on results obtained from the SuperCurve software (http://bioinformatics.mdanderson.org/Software/OOMPA). Currently, researchers are able to deposit over a thousand patient samples on one RPPA, making the technology particularly attractive for pathway analysis. In general, the availability of protein-specific antibodies remains a limitation of RPPA, but this issue lies outside the scope of this article.
Figure 1.
(a) A typical reverse-phase protein array with 40 samples shown as the 40 batches on the slide. Each batch represents one individual sample with 16 spots, which are the results of duplicates of eight-step dilutions. (b) Normalized RPPA intensities for 51 proteins and 531 AML patients.
Statistical research in the estimation of dependence patterns has mainly centered on Bayesian networks (Jansen et al. 2003; Friedman 2004; Pittman et al. 2004; Sebastiani and Ramoni 2005). Other recent attempts on network models include the work on finding differentially expressed gene sets (often belonging to a genetic pathway) by Efron and Tibshirani (2006) and Newton et al. (2007). Recent extensions to this framework (Shojaie and Michailidis 2009, 2010) show how the introduction of explicit prior information about gene–gene interactions improves power in the differential analysis of gene sets. The innovative work by these authors still stops short of modeling specific interaction among the genes. In fact, in the setting of estimating gene–gene interactions, recent contributions have highlighted how well-established computational techniques often perform poorly in very-high-dimensional settings (Dobra et al. 2004; Scott and Carvalho 2008). In this regard, some progress has been reported when the analysis is carried out using convenient probabilistic schemes, such as Gaussian Markov random fields (MRFs) (Wong, Carter, and Khon 2003; Jones et al. 2005; Scott and Carvalho 2008), or functional transformation schemes (Telesca et al. 2009).
In the case of RPPA data, the curse of dimensionality is somewhat mitigated by the fact that the technology looks at a limited set (typically < 200) of proteins for expression and activation states, which represents a fraction of the estimated 4 million proteins and activation states thought to occur in the human proteome. Furthermore, in typical protocols, one is not dealing with the n ≪ p paradigm, as large sample sizes can easily be analyzed. On the other hand, when investigating dependence, some issues that are inherently associated with high-throughput molecular interrogation persist: (1) the sampling distribution of proteomic abundance is usually non-Gaussian, rendering the convenient Gaussian graphical models framework inadequate as a model for protein interactions; and (2) RPPA experiments often target specific biological pathways. It is therefore important that the prior specification exploit available prior information on such targeted pathways.
We address issue (1) following the formalism introduced by Telesca et al. (2010), modeling the sampling distribution of protein abundance as a heavy-tailed mixture distribution and defining dependence at a hidden level, between binary indicators of inactive and active states. Addressing issue (2) defines a key feature of the proposed model, as our formulation introduces an informative prior that assigns high prior probability mass to the model space surrounding the target pathway. Mukherjee and Speed (2008), for example, suggested a similar approach in the context of Gaussian directed acyclic graphical (DAG) models. Our approach is also comparable to that described by Airoldi et al. (2007) in a data integration context. From a methodological perspective, we extend the work of Telesca et al. (2010) in two fundamental directions. First, we introduce a fully general dependence prior between hidden multivariate binary random quantities, and second, we propose a framework for Bayesian model determination that allows a local search for interactions, penalizing the model space as a function of a discrepancy measure between the target pathway and random departures from its graphical topology.
The rest of this article is structured as follows. We introduce the proposed analysis framework and probability model in Section 2. Statistical inference based on Markov chain Monte Carlo (MCMC) sampling from the posterior distribution of interest is discussed in Section 3. In Section 4, we illustrate our approach through the analysis of an RPPA dataset from a set of patients diagnosed with acute myeloid leukemia (AML). We conclude with a critical discussion of our contribution in Section 5.
2. DEPENDENT MIXTURE MODEL
We consider protein expression data in the form of a G × T matrix Y = [ygt], with the generic element ygt denoting the observed molecular expression for protein g in sample t, where g = 1, …, G and t = 1, … T. The sampling model for Y is defined conditionally on a latent variable egt, representing an indicator of class membership in a mixture density and is described in Section 2.1. We argue that egt is likely to represent the biologically meaningful signal of protein activation better than the noisy row intensities ygt. A key feature of the model is a graphical model
 to parameterize a dependent prior on e = [egt]. The graph
to parameterize a dependent prior on e = [egt]. The graph
 reflects the dependence structure between proteins. The strength of this dependence is indexed by an additional set of parameters β. In summary, the probability model is defined as
reflects the dependence structure between proteins. The strength of this dependence is indexed by an additional set of parameters β. In summary, the probability model is defined as
| (1) |
Underbraced numbers indicate the subsections where each sub-model is discussed.
2.1 Sampling Model: p(Y |e)
We assume that the sampling distribution of ygt has heavy tails and follows a mixture of a normal distribution and two scale mixtures of uniform distributions. Let egt denote a latent trinary indicator, egt ∈ {−1, 0, 1}, indexing three possible distribution functions. Also, let ỹgt = ygt − (αt + μg) denote a normalized measure of relative abundance, corrected for a protein-specific effect μg and a sample-specific effect αt. Using f (·) to generically denote a probability density function, we write the sampling distribution of ỹgt as
| (2) |
where , and I(A) denotes an indicator of the event A. Such normal–uniform mixtures have been previously introduced by Parmigiani et al. (2002) and Dean and Raftery (2005) for the robust analysis of high-throughput cDNA microarrays. Recently, Telesca et al. (2010) have extended the original formulation of Parmigiani et al. (2002), defining mixing proportions via a multivariate probit link.
Conditional on et = (egt, g = 1, …, G), the observed abundance measurements ygt are exchangeable in (2). Later, in Section 2.2, we will exploit the trinary indicators egt to model conditional dependence between protein abundance measurements. For each protein g, the marginal distribution of eg = (eg1, …, egT)′ is trinomial, with T possible trials, support {−1, 0, 1}, and probabilities , and .
In the foregoing formulation, denotes the variance of the baseline distribution for protein g. The mixture distribution models overdispersion relative to f0(·) through the tail parameters and . We will require that to account for heavy tails in the proteomic distribution of abundance. More precisely, marginalizing over , the sampling model (2) is heavy tailed compared with the central normal distribution.
We propose the following conditionally conjugate hierarchical normal prior for the remaining parameters in (2): , and finally, . Additionally, we will require , for likelihood identifiability.
2.2 Modeling Dependence: p(e | β,
 )
)
We model dependence between proteins using the formalism of graphical models (Lauritzen 1996). A graph
 = {V, E} is an algebraic structure composed of a set of vertices V and a set of edges E ⊆ V × V. When the set of vertices V represents a collection of random quantities, the edge structure E is often used to identify the full set of conditional independence relations between the components of V via the global Markov property associated with
= {V, E} is an algebraic structure composed of a set of vertices V and a set of edges E ⊆ V × V. When the set of vertices V represents a collection of random quantities, the edge structure E is often used to identify the full set of conditional independence relations between the components of V via the global Markov property associated with
 (Besag 1974). Details, including whether edge set E is allowed to include directed edges, that is, ordered pairs of nodes, or only undirected edges, that is, unordered pairs, depend on the specific graphical model and are discussed below.
(Besag 1974). Details, including whether edge set E is allowed to include directed edges, that is, ordered pairs of nodes, or only undirected edges, that is, unordered pairs, depend on the specific graphical model and are discussed below.
Regulatory relationships between proteins may include causal loops and dynamic reciprocal regulation (see Figure 2). This motivates the assumption that the set of conditional dependence relationships characterizing protein interactions is well represented by a class of graphical models known as reciprocal graphs (Koster 1996). These graphs allow for directed (e.g., a → b) and undirected edges (e.g., a − b), loops (e.g., a → b, b → c, c → a), and reciprocal relationships (e.g., a → b, b → a), where a, b, c ∈ V. Provided there are no directed edges between vertices of the same undirected path (e.g., a − b, b − c, c → a), this class of models has a clear causal interpretation and equivalent Markov classes may be defined via graphical moralization (Koster 1996; Lauritzen 1996).
Figure 2.
A protein interaction pathway produced by combining known protein–protein interactions from the literature. This network wiring diagram shows the connectivity of the receptor tyrosine kinase to the MAPK (mitogen-activated protein kinase), Akt, STAT, Bcl-2 (B-cell lymphoma 2), and p53 signaling proteins. (We are able to measure a large percentage of the molecules using the RPPA for AML patients. The relationships between proteins suggested in this diagram will be considered as prior information for the proposed probability model.) The online version of this figure is in color.
Our notation
 = {V, E} defines a reciprocal graph. Each
= {V, E} defines a reciprocal graph. Each
 corresponds to one and only one moral (undirected) graph
corresponds to one and only one moral (undirected) graph
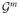 = {V, Em}, where Em represents the set of undirected edges moralized from E. While we use a directed graph
= {V, Em}, where Em represents the set of undirected edges moralized from E. While we use a directed graph
 to interpret the pathway diagram, in the absence of a time-course experiment, the data can only inform about an equilibrium distribution. For the development of the sampling model, it therefore suffices to consider the implied conditional independence structure of the equilibrium distribution. We will represent this by Gm. We proceed in the following way. We start with a prior on G; this induces a prior on Gm. Conditional on Gm, we define a sampling model for the observed data. Finally, posterior updating provides the desired posterior inference on G. In this process, it is important to note that we only learn about the features of G that would change Gm. This process parallels inference in an analysis of variance (ANOVA) model that can only inform about identifiable contrasts. The prior on the directed graph can be thought of as prior regularization.
to interpret the pathway diagram, in the absence of a time-course experiment, the data can only inform about an equilibrium distribution. For the development of the sampling model, it therefore suffices to consider the implied conditional independence structure of the equilibrium distribution. We will represent this by Gm. We proceed in the following way. We start with a prior on G; this induces a prior on Gm. Conditional on Gm, we define a sampling model for the observed data. Finally, posterior updating provides the desired posterior inference on G. In this process, it is important to note that we only learn about the features of G that would change Gm. This process parallels inference in an analysis of variance (ANOVA) model that can only inform about identifiable contrasts. The prior on the directed graph can be thought of as prior regularization.
The mapping
 →
→
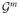 is via the moralization procedure. Henceforth, notation with superscript m will always refer to the moralized graph. The mapping is not one to one, as the same undirected graph may correspond to different directed structures, (Markov equivalence, Lauritzen 1996). In
is via the moralization procedure. Henceforth, notation with superscript m will always refer to the moralized graph. The mapping is not one to one, as the same undirected graph may correspond to different directed structures, (Markov equivalence, Lauritzen 1996). In
 , standard MRF properties apply (Besag 1974). In particular, if we denote ne(g) as the set of neighbors of protein g (a ∈ ne(b) if and only if {a, b} ∈ Em), then it is assumed that the abundance of protein g is conditionally independent of any other protein, given its neighbors ne(g).
, standard MRF properties apply (Besag 1974). In particular, if we denote ne(g) as the set of neighbors of protein g (a ∈ ne(b) if and only if {a, b} ∈ Em), then it is assumed that the abundance of protein g is conditionally independent of any other protein, given its neighbors ne(g).
We specify a joint distribution over the latent indicators et = (e1t, …, eGt)′, (t = 1, …, T), which is consistent with the Markov structure spanned by a given reciprocal graph
 , through its moral representation
, through its moral representation
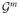 . We first reduce the trinary egt to a binary indicator zgt. We do so because in RPPA studies, researchers usually focus on protein activations. We thus define an auxiliary indicator zgt = 2I {egt = 1} − 1 ∈ {−1, 1}, which we will use to describe dependence across proteins.
. We first reduce the trinary egt to a binary indicator zgt. We do so because in RPPA studies, researchers usually focus on protein activations. We thus define an auxiliary indicator zgt = 2I {egt = 1} − 1 ∈ {−1, 1}, which we will use to describe dependence across proteins.
Next, we construct a joint probability model for zt = (zgt, g = 1, …, G). The reduction to the multivariate binary vector zt greatly simplifies this modeling step. Following the arguments by Hammersley and Clifford (1971) and Besag (1974), given G and the neighborhood structure encoded in a moral graph
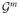 , the joint distribution p(zt) can be defined in its full generality through the complete conditional probabilities p(zgt | z(\g,t)). More precisely, let exp[Q(zt)] ∝ p(zt), then there exists an expansion of Q(zt), unique on Ω = {−1, 1}G and of the form:
, the joint distribution p(zt) can be defined in its full generality through the complete conditional probabilities p(zgt | z(\g,t)). More precisely, let exp[Q(zt)] ∝ p(zt), then there exists an expansion of Q(zt), unique on Ω = {−1, 1}G and of the form:
| (3) |
The Hammersley–Clifford theorem (Besag 1974) postulates that while the H-functions may be chosen arbitrarily, for any set of labels (1 ≤ g, h, …, s ≤ G), Hg,h,…,s may be nonnull if and only if the set of proteins labeled (g, h, …, s) forms a clique in the moral graph
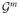 . A clique is a subset of proteins in which each pair of proteins is connected by an edge.
. A clique is a subset of proteins in which each pair of proteins is connected by an edge.
The binary nature of zgt allows us to, without loss of generality, replace any nonnull H-function with a single arbitrary parameter (Cox 1972; Besag 1974) and we can therefore write
| (4) |
The foregoing formulation is a representation result for any multivariate distribution p(z). Later, conditioning on a particular graphical model
 will substantially reduce the number of terms appearing in (4). In fact, when considering p(zt |
will substantially reduce the number of terms appearing in (4). In fact, when considering p(zt |
 ), the function Q(zt |
), the function Q(zt |
 ) will only include sets of vertices forming cliques in the moralized graph
) will only include sets of vertices forming cliques in the moralized graph
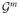 . Finally, the definition of a joint distribution over the original trinary indicators et is completed with the conditional probability p(egt = −1 | zgt = −1) = exp(γg)/{1 + exp(γg)}, (g = 1, …, G).
. Finally, the definition of a joint distribution over the original trinary indicators et is completed with the conditional probability p(egt = −1 | zgt = −1) = exp(γg)/{1 + exp(γg)}, (g = 1, …, G).
In summary, we have mapped
 to
to
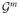 and then indexed all possible joint probability models on et that respect the conditional independence structure represented in
and then indexed all possible joint probability models on et that respect the conditional independence structure represented in
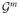 by (4).
by (4).
2.3 Priors Over Interaction Parameters and Graphical Structures: p(β |
 ) p(
) p(
 )
)
We introduce an informative class of priors for the unknown graph
 . In words, our prior model is based on a pathway diagram that summarizes substantive prior information about the biochemical pathway of interest. First, we interpret the pathway diagram as a known and fixed reciprocal graph
. In words, our prior model is based on a pathway diagram that summarizes substantive prior information about the biochemical pathway of interest. First, we interpret the pathway diagram as a known and fixed reciprocal graph
 = {V, E0}. Here, we assume that the biochemical pathway is displayed as a set of nodes corresponding to proteins and edges between nodes. We assume that all edges are directed. In general, a reciprocal graph could also include undirected edges, subject only to the constraint that there be no directed edges between nodes of the same (undirected) pathway component (Koster 1996). We do not make use of this feature in our implementation, as all edges in the original pathway are indeed directed.
= {V, E0}. Here, we assume that the biochemical pathway is displayed as a set of nodes corresponding to proteins and edges between nodes. We assume that all edges are directed. In general, a reciprocal graph could also include undirected edges, subject only to the constraint that there be no directed edges between nodes of the same (undirected) pathway component (Koster 1996). We do not make use of this feature in our implementation, as all edges in the original pathway are indeed directed.
Conceptually, we write p(
 ) ∝ f {d(
) ∝ f {d(
 ,
,
 )}, where d(·, ·) is a discrepancy measure and f′ ≤ 0, so that graphs that deviate from
)}, where d(·, ·) is a discrepancy measure and f′ ≤ 0, so that graphs that deviate from
 are assigned a lower prior probability. A similar approach was introduced by Mukherjee and Speed (2008) for Gaussian DAGs.
are assigned a lower prior probability. A similar approach was introduced by Mukherjee and Speed (2008) for Gaussian DAGs.
Let Ac denote the complement of set A and define
, δ > 1. Here, |E| identifies the number of edges in a graph
 = {V, E}. This defines a discrepancy measure d(
= {V, E}. This defines a discrepancy measure d(
 ,
,
 ) as the weighted sum of edges dropped from
) as the weighted sum of edges dropped from
 and edges added to
and edges added to
 . If we assume for a moment δ = 1, then d(
. If we assume for a moment δ = 1, then d(
 ,
,
 ) reduces to the number of changed edges relative to
) reduces to the number of changed edges relative to
 . A weight δ > 1 allows us to include a notion of parsimony by using an increased penalty for adding edges compared with dropping edges. For a given d(·, ·), we assume that
. A weight δ > 1 allows us to include a notion of parsimony by using an increased penalty for adding edges compared with dropping edges. For a given d(·, ·), we assume that
| (5) |
where we assume φ~
 (aφ, bφ). The prior (5), with δ = 1, is equivalent to assuming exchangeable coin flips to determine each edge, with odds of matching edges in
(aφ, bφ). The prior (5), with δ = 1, is equivalent to assuming exchangeable coin flips to determine each edge, with odds of matching edges in
 equal to φ. Similar probability models have been shown to provide in several contexts automatic multiplicity correction in the posterior (Scott and Berger 2008). For any given φ, the expected density of a graph1
equal to φ. Similar probability models have been shown to provide in several contexts automatic multiplicity correction in the posterior (Scott and Berger 2008). For any given φ, the expected density of a graph1
 is computed as
, which is a decreasing function of δ. Furthermore, the distribution of the graph density d(
is computed as
, which is a decreasing function of δ. Furthermore, the distribution of the graph density d(
 | φ) has mode at
and variance decreasing with δ. A key property of (5) is therefore the informative nature of the prior, maximized at the prior guess
| φ) has mode at
and variance decreasing with δ. A key property of (5) is therefore the informative nature of the prior, maximized at the prior guess
 , leading to posterior simulation being a local exploration of the model space. This is in sharp contrast to alternate approaches that allow for global model exploration by using prior probability models that are far more diffuse over the model space. If desired, further structural restrictions are easily incorporated in this framework by introducing changes in d(
, leading to posterior simulation being a local exploration of the model space. This is in sharp contrast to alternate approaches that allow for global model exploration by using prior probability models that are far more diffuse over the model space. If desired, further structural restrictions are easily incorporated in this framework by introducing changes in d(
 ,
,
 ). For example, this function could be defined in ways that force the inclusion of a subset of edges, say
, or in ways that introduce discrete cutoffs to the overall model size.
). For example, this function could be defined in ways that force the inclusion of a subset of edges, say
, or in ways that introduce discrete cutoffs to the overall model size.
Our model is completed by defining normal priors over the interaction parameters (βgh, βghk, …), (∀ g < h < k …) so that , for any generic index of the parameter vector β. The mean value ηu is used to represent an important feature of typical molecular pathway representations. Pathway diagrams usually include a distinction between stimulatory and inhibitory interactions. In Figure 2, for example, edges between proteins are labeled with an arrowed-tip (i → j) if interactions observed a priori are of a stimulatory nature, and with a bullet-tip (—•) if prior interactions are inhibitory.
We formalize the definition of stimulatory and inhibitory relationships, following the framework introduced in Section 2.2 and focusing on second-order interactions parameters βgh, (g < h). More precisely, indexing protein activation with binary indicators zgt ∈ {−1, 1}, we assume that protein g stimulates the activation of protein h if p(zgt = 1 | zht = 1) > p(zgt = 1 | zht = −1). Similarly, for inhibitory relationships, we assume p(zgt = −1 | zht = 1) > p(zgt = −1 | zht = −1). It is easy to verify that these definitions motivate the choice of ηu > 0 for stimulatory and ηu < 0 for prior inhibitory interactions. In the absence of prior information, ηu is simply set to 0.
For reference, we summarize the complete model. Let ; we have
where the trinary indicator egt is an elaboration of the binary indicator zgt for activation, with and egt = 1 when zgt = 1.
In our analysis, we consider a default prior setting inspired by the considerations in Jiang (2007). Specifically, we choose
, and
, where mG = G(G − 1), and ζ and ν are positive constants. This simplifies tuning and sensitivity analyses to the choice of two parameters: ζ > 0, regulating overall sparsity, and ν > 0, defining prior deviation penalties from
 .
.
3. POSTERIOR COMPUTATION AND INFERENCE
3.1 Stochastic Search and MCMC Computation
We are interested in identifying patterns of molecular interactions, as informed by experimental data and guided by a prior pathway. The full model is determined by mixture membership indicators e, a conditional dependence configuration summarized in a graph
 , MRF parameters β, and a collection of parameters θ, which we assume contains all remaining random quantities with the exception of Y. Our inference centers on the posterior distribution p(e,
, MRF parameters β, and a collection of parameters θ, which we assume contains all remaining random quantities with the exception of Y. Our inference centers on the posterior distribution p(e,
 , β, θ |Y), which fully summarizes the available evidence on protein abundance and protein expression profiles similarities.
, β, θ |Y), which fully summarizes the available evidence on protein abundance and protein expression profiles similarities.
The posterior probability model is not available in closed form. However, it is conceptually straightforward to define MCMC simulation from this target distribution. In particular, we could proceed as usual and draw the parameters of interest sequentially or in random order from the full conditional posterior distributions of θ, e, β, and
 . Updating of e and θ can be implemented via standard Gibbs sampling.
. Updating of e and θ can be implemented via standard Gibbs sampling.
Unfortunately, updating β and
 remains problematic (Green and Richardson 2002). The complete conditional posterior for β and
remains problematic (Green and Richardson 2002). The complete conditional posterior for β and
 depends on e only indirectly through z. Recall that we further summarized the trinary indicators e into binary indicators z ∈ {−1, 1}G of protein activation. The conditional posterior of β is given by
depends on e only indirectly through z. Recall that we further summarized the trinary indicators e into binary indicators z ∈ {−1, 1}G of protein activation. The conditional posterior of β is given by
| (6) |
where
and the sum Σz is over all 2G possible realizations of z.
The full conditional distribution of β is therefore defined in terms of a partition function g(β), which requires summation over all possible 2G realizations of zt, for any t = 1, …, T. This quantity can be evaluated efficiently only for a very small G. This is often not the case in RPPA studies, which leaves us with the problem of devising a feasible strategy to update β. Similar considerations apply for updates involving
 , with the further complication that changes in
, with the further complication that changes in
 define changes in the dimensionality of the parameter vector β.
define changes in the dimensionality of the parameter vector β.
This problem is well known in the literature on MRFs and several solutions have been proposed, based on ad-hoc approximation to the partition function of interest (Green and Richardson 2002; Friel et al. 2009). In the following, we propose a solution based on MCMC ingenuity and show that exploiting a transition kernel that updates mixture indicators e and dependence parameters β in a joint fashion is possible without calculation of g(β).
3.2 Updating β
To update β, we consider a Metropolis–Hastings (MH) transition kernel that involves also changes in z and consequently in e. For any generic element of β, say βu (u ⊂ {1, …, M}), we consider a proposal distribution
, where q(·) is an arbitrary proposal density and possibly dependent on the previous values βu, whereas p(e | β,
 , θ) is the conditional prior defined in Section 2.2. In our implementation, for all i > j = 1, …, G, we consider a random-walk proposal
, where sβ is calibrated at burn-in to achieve acceptance rates between 30% and 70%.
, θ) is the conditional prior defined in Section 2.2. In our implementation, for all i > j = 1, …, G, we consider a random-walk proposal
, where sβ is calibrated at burn-in to achieve acceptance rates between 30% and 70%.
Draws from e′| β′,
 , θ may then be obtained easily via a short Gibbs run, based on full conditionals p(egt | e\g, β′,
, θ may then be obtained easily via a short Gibbs run, based on full conditionals p(egt | e\g, β′,
 , θ). It is easy to verify that for the construction of a standard MH transition kernel the newly proposed values (e′,
) must be accepted with probability
, θ). It is easy to verify that for the construction of a standard MH transition kernel the newly proposed values (e′,
) must be accepted with probability
| (7) |
The structure of the proposal distribution allows for the elimination of the partition function g(β), at the cost of having to simulate G × T mixture indicators for each βij. Gains in efficiency could be further achieved by considering block updates for the elements of β. Our approach is similar to that of Møller et al. (2006).
In short, we replace the problem of evaluating the partition function with a problem of approximate prior simulation. The resulting MCMC provides an approximation to the desired posterior only to the extent to which the prior Gibbs simulation can be considered a draw from the prior. A systematic discussion of related MCMC schemes appears in the work of Andriew and Roberts (2009), who also discussed several more elaborate variations of this strategy. Further precision may be achieved by considering exact simulation from e | β,
 , θ (Propp and Wilson 1996), but this would come at a higher computational cost. In fact, the asymptotic validity of the proposed computation and a favorable comparison with exact sampling methods have recently been discussed by Liang (2010). A comparative review of alternate computational strategies associated with similar probability models (Ising and Potts models) has been compiled by Zhou and Schmidler (2009).
, θ (Propp and Wilson 1996), but this would come at a higher computational cost. In fact, the asymptotic validity of the proposed computation and a favorable comparison with exact sampling methods have recently been discussed by Liang (2010). A comparative review of alternate computational strategies associated with similar probability models (Ising and Potts models) has been compiled by Zhou and Schmidler (2009).
3.3 Updating
 via Reversible-Jump MCMC (RJ-MCMC)
via Reversible-Jump MCMC (RJ-MCMC)
Updating
 involves changes in β and its dimensionality. We are therefore faced with two complications, one associated with the evaluation of the partition function g(β), as explained above, and the other associated with the need to maintain detailed balance across dimensions. To solve this problem, we combine the reversible-jump algorithm of Green (1995) with the approach described in Section 3.2. More precisely, we follow Giudici and Green (1999) and propose the following transitions:
involves changes in β and its dimensionality. We are therefore faced with two complications, one associated with the evaluation of the partition function g(β), as explained above, and the other associated with the need to maintain detailed balance across dimensions. To solve this problem, we combine the reversible-jump algorithm of Green (1995) with the approach described in Section 3.2. More precisely, we follow Giudici and Green (1999) and propose the following transitions:
Select an edge (i, j) at random. If (i, j) ∈ E, propose its elimination; otherwise, if (i, j) ∉ E, propose its birth. This corresponds to moving from the current graph
 to a new graph
to a new graph
 , with moral representation
, with moral representation
 .
.Assume the move in Step 1 involves a birth. Propose a set of, say k, new interaction parameters in correspondence to new undirected edges in the moral graph
 . In our implementation, we consider a proposal density based on local logistic regressions. More precisely, if we propose the birth of an interaction parameter βij involving zi and zj, we define: a = Σt I{zit = −1, zjt = −1}, b = Σt I{zit = −1, zjt = 1}, c = Σt{zit = 1, zjt = −1}, and d = Σt I{zit = 1, zjt = 1}. We calculate
and
. Here, β̂ij estimates the log odds ratio for a logistic regression involving zi and zj The parameter δβ is a standard large-sample approximation to SE(β̂ij). The proposal density is then constructed as a normal approximation to the conditional likelihood surface so that
.
. In our implementation, we consider a proposal density based on local logistic regressions. More precisely, if we propose the birth of an interaction parameter βij involving zi and zj, we define: a = Σt I{zit = −1, zjt = −1}, b = Σt I{zit = −1, zjt = 1}, c = Σt{zit = 1, zjt = −1}, and d = Σt I{zit = 1, zjt = 1}. We calculate
and
. Here, β̂ij estimates the log odds ratio for a logistic regression involving zi and zj The parameter δβ is a standard large-sample approximation to SE(β̂ij). The proposal density is then constructed as a normal approximation to the conditional likelihood surface so that
.Propose new values for the mixture indicators e′ ~ p(e′| β′,
 , θ). Draws are made from the conditional prior defined in Section 2.2.
, θ). Draws are made from the conditional prior defined in Section 2.2.- Accept Steps 1–3 with probability ρb(
 ,
,
 ):
):
The reverse move, involving the death of a random edge (i, j), is implemented by setting to 0 the elements of β disappearing from
 and it is accepted with probability ρd (
and it is accepted with probability ρd (
 ,
,
 ) = 1/ρb(
) = 1/ρb(
 ,
,
 ).
).
The transition probability for
 includes a proposal for elements of β. One could therefore drop the transition probability described in Section 3.1.1, and still maintain irreducibility. However, without Section 3.1.1, the resulting MCMC demonstrates far slower mixing.
includes a proposal for elements of β. One could therefore drop the transition probability described in Section 3.1.1, and still maintain irreducibility. However, without Section 3.1.1, the resulting MCMC demonstrates far slower mixing.
We conclude by summarizing the overall MCMC. We use notation x−i to indicate the vector x without the element xi, and [x | y, z] to generically indicate a transition probability that changes x and depends on the currently imputed values of y, z. Each iteration in the proposed posterior simulation algorithm involves the following transition sequence:
4. SIMULATED DATA
We carried out a small simulation experiment to validate inference under the proposed model, to investigate the impact of prior choices and to compare with alternate approaches. In brief, we simulated data under a model that differs from the proposed sampling model. Hypothetical data were sampled as:
, where the mixing proportions depend on the realizations of a matrix normal random quantity W ~
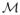
 (M, Σ, IT) through a multivariate probit link (Chib and Greenberg 1998) so that
and
. The concentration matrix Σ−1 was structured via structural equations to encode a notion of true dependence in a reciprocal graph. We fitted our model using a misspecified network prior. Given the true set of edges E* (in our example, | E* |= 50), we consider
, with
omitted from the prior edge set (in our example,
) and Ẽ serving as a random misspecification set including false edges (in our example, |Ẽ| = 30). Ideally, this prior would mimic what is often the case in practice, where a prior pathway may include edges that do not play a relevant role in the biological process under investigation (Ẽ) as well as previously unidentified but relevant interactions (
).
(M, Σ, IT) through a multivariate probit link (Chib and Greenberg 1998) so that
and
. The concentration matrix Σ−1 was structured via structural equations to encode a notion of true dependence in a reciprocal graph. We fitted our model using a misspecified network prior. Given the true set of edges E* (in our example, | E* |= 50), we consider
, with
omitted from the prior edge set (in our example,
) and Ẽ serving as a random misspecification set including false edges (in our example, |Ẽ| = 30). Ideally, this prior would mimic what is often the case in practice, where a prior pathway may include edges that do not play a relevant role in the biological process under investigation (Ẽ) as well as previously unidentified but relevant interactions (
).
Figure 3(a) explores robustness of protein activation inference to data arising from misspecified sampling models, by comparing inference on protein activation with the simulation truth. Figure 3(b) shows receiver operating characteristic (ROC) curves for selecting edges in the graph and compares inference under the proposed model, under GeneNet (Schäfer and Strimmer 2005) and under a model with exchangeable Bernoulli priors on all edges. Figure 3(c) shows similar ROC curves under alternate priors with the proposed model, comparing the proposed informative prior (5) and two relatively noninformative priors. Details and sensitivity analyses are reported in the supplementary materials to this article, available online. Overall, we found that a sampling model mimicking a potential data generative process is likely to provide some advantages over standard multivariate Gaussian calculations. Furthermore, we remark how using prior information as a source of structural constraints on the model space can provide a substantial advantage in the identification of meaningful interactions.
Figure 3.

Simulated data. Panel (a) shows ROC curves for the correct classification of into activated versus nonactivated proteins. Panels (b) and (c) show ROC curves for reporting edges as present or not. In panel (b), we compare different models. In panel (c), we compare alternate priors under the proposed model (δ = 50 and 100, corresponding to ζ = 1 and 2, respectively). The online version of this figure is in color.
5. EXAMPLE: ANALYSIS OF RPPA DATA
5.1 A Study in Acute Myeloid Leukemia
The current classification of AML uses the French–American–British system based on morphological features, along with flow cytometric analysis of surface marker expression, cytogenetics, and assessment of recurrent molecular abnormalities. These classification schemes have prognostic relevance, but with the exception of acute promyelocytic leukemia, they generally do not alter therapeutic recommendations (Mrozek et al. 2007). Furthermore, the predictive abilities of prognostic models based on current clinical and laboratory features are generally low, with less than half of the outcomes explained by these features. Researchers have recently started to investigate genomic and proteomic features for prognosticating AML. Proteins within signaling pathways that exhibit heterogeneous expression in AML are often prognostic, a characteristic that has been studied by researchers such as Kornblau et al. (2006) and Tanner et al. (2001). These authors find that distinct molecular abnormalities and patterns of pathway activation in leukemic cells collectively suggest potential targets for therapeutic intervention. Consequently, knowledge allowing rational evaluation of targeted therapies on an individualized basis in AML is sorely needed. As part of a knowledge-learning process, expression levels and activation process of a single protein or a limited number of proteins have been studied by Kornblau et al. (1994), Kornblau et al. (2000), and Kreuter et al. (2006).
The natural next step is formal inference on different patterns of pathway activation and modification. This requires inference on the joint probability distribution for multiple proteins in a pathway. The proposed model and the inference approach are built to facilitate such inference.
We use data from a large AML study based on RPPA. Specifically, we have probed protein samples from 531 newly diagnosed, primary, refractory, and relapsed AML patients. The objective of this experiment was to investigate interactions of important protein markers related to AML. We selected 51 proteins involved in signal transduction, apoptosis, and cell cycle regulatory pathways (Figure 2) and studied their expression pro-files in all 531 samples. An important feature of the AML data under study is that the number of samples (n = 531) is much greater than the number of proteins (p = 51). This feature facilitates principled model-based inference and model assessment and contrasts inference for RPPA data with inference for many other high-throughput genomic platforms.
The targeted interactions are illustrated in Figure 2, where we present a comprehensive signaling network map for the selected proteins based on the three pathways. The signaling network map was developed to show signaling interactions based on published articles from PubMed searches as well as from the connections map in Signal Transduction Knowledge Environment (http://www.stke.org).
5.2 Data Analysis
Using a desktop scanner at an optical resolution of 1200 dpi, we scanned the hybridized RPPA slides and saved them as TIFF files. The protein expression intensity of each spot was measured with an automated software program, MicroVigene™ (VigeneTech, Inc., North Billerica, MA). The dilution series of the samples provided a dilution–concentration–expression curve, providing relative expression intensities that were read off in the linear part of the curve. We used these numbers for data preprocessing and calculation after standardization and topographical normalization (for details, refer to the SuperCurve software at http://bioinformatics.mdanderson.org/Software/OOMPA). Good preprocessing of data from any high-throughput experiments in general, and RPPA data in particular, is critically important. We used SuperCurve as it is the best currently available model-based and principled implementation of inference to quantify RPPA data.
We estimate protein pathway structure by combining prior knowledge of protein interaction and RPPA measurements. Specifically, given the prior consensus pathway in Figure 2, we use the proposed probability model to update our prior knowledge, combining the qualitative information about the consensus pathway with quantitative information from RPPA data (Figure 1). We fit the model presented in Section 2 to this set of 51 proteins. Reported inference is based on 500,000 MCMC samples (thinned by 10) after discarding 100,000 samples for burn-in.
In Figure 4 (left panel), we illustrate the fit of the proposed model to the AML protein expression data reported in Figure 1. Defining and , we follow Parmigiani et al. (2002) and define a probability of expression (POE) scale . This quantity represents a univariate posterior summary, designed to relate directly to the mixture indicator labels egt ∈ {−1, 0, 1} and may be considered as the normalized scale for raw intensities ygt. Here, we show how the original abundance measurement is translated into a POE on the new scale (p*). Plotting the raw correlation coefficients versus the the simple correlation estimated in the p* scale (Figure 4, right panel), we note that the two measures of association are clearly correlated, with a tendency of stronger agreement toward large absolute values of the correlation coefficients.
Figure 4.

RPPA study: (Left panel) Centered protein abundance ygt versus POE scale intensities ( ). (Right panel) Raw simple correlation estimates versus simple correlations in the POE scale.
Our inference on molecular interaction is based on posterior edge inclusion probabilities, P{(i, j) ∈ E or (j, i) ∈ E | Y)}. These quantities are estimated directly from the RJ-MCMC output as the percentage of iterations where the edge is included in the current graphical structure
 . In Figure 5, we represent the posterior edge inclusion probabilities for all possible protein interactions, against expected posterior interactions E(βij | Y). Solid diamonds denote edges originally included in the prior pathway
. In Figure 5, we represent the posterior edge inclusion probabilities for all possible protein interactions, against expected posterior interactions E(βij | Y). Solid diamonds denote edges originally included in the prior pathway
 . Our differential penalization scheme favors edges in the target pathway, but still allows for the model to explore interactions that are not included in the prior. Reporting a final pathway may be based on several criteria (Telesca et al. 2009). Here, we consider the median model (Barbieri and Berger 2004; Scott and Berger 2008). That is, we select edges with posterior inclusion probability greater than 0.5.
. Our differential penalization scheme favors edges in the target pathway, but still allows for the model to explore interactions that are not included in the prior. Reporting a final pathway may be based on several criteria (Telesca et al. 2009). Here, we consider the median model (Barbieri and Berger 2004; Scott and Berger 2008). That is, we select edges with posterior inclusion probability greater than 0.5.
Figure 5.
Panel (a): Posterior expected interactions E(βu | Y) versus posterior edge inclusion probabilities p((i, j ) or (j, i) ∈ E | Y). Solid diamonds correspond to edges originally included in the prior pathway. Panel (b): Median model identified selecting edges with posterior inclusion probability greater than 0.5. Arrows define stimulatory relationships, whereas dotted arrowheads define inhibitory relationships. Edge thickness is proportional to the absolute size of the posterior expected interaction parameters.
The selected posterior network is reported in Figure 5. The edge thickness is proportional to the absolute value of the associated expected posterior interaction parameter E(βij | Y). We distinguish between stimulatory and inhibitory relationships. Following the argument introduced in Section 2.3, we base our inference on the sign of the posterior mean E(βij | Y ). For a generic edge index u, a positive posterior mean is interpreted as stimulation, whereas a negative sign is interpreted as inhibition. In the figure, we represent stimulation with arrows and inhibition with dotted arrowheads.
Our analysis identifies two main subpathways [Akt/mTOR (protein kinase B/mammalian target of rapamycin) and STATs (signal transducers and activators of transcription)] as groups of proteins exhibiting significant similarities in their overabundance patterns across samples. The Akt/mTOR pathway is known to be a key player in cell growth, proliferation, and survival. Recent literature identifies this pathway as a strong contributor to proliferation and drug resistance in AML [a comprehensive review is provided by Martelli et al. (2009)]. The second active pathway involves the family of STAT proteins (STAT3/STAT5). These molecules are known to act as important regulators in hematopoiesis (blood cell formation), and their upregulation via β-catenin (BCAT) has been documented by several authors (Boeuf et al. 2001; Hao et al. 2006). In the same pathway, we also recover key interactions between the STAT proteins and extracellular signal-regulated kinase (ERK) (Jain et al. 1998). The important role of STAT5 in AML was also recently noted by Kornblau et al. (2010).
Some of our results are surprising. For example, pPTEN (phosphatase and tensin homolog) is inactivated when phosphorylated and therefore cannot inhibit Akt phosphorylation. The negative relationship between pPTEN and pAkt-308 seems, therefore, contrary to canonical expectations (Wan and Helman 2003). The PTEN–Akt pathway is however of great interest in oncology, and this finding may prove useful in the search for possible feedback regulatory interactions or latent oncogenes [see, e.g., the discussion in Palomero, Dominguez, and Ferrando (2008)]. Likewise, one would expect the pAkts to have an edge with pGSK-3 (glycogen synthase kinase 3) instead of total Akt. On the other hand, the inferred diagram confirms evidence that, in active form, Akt phosphorylates a wide variety of downstream substrates. Also, the diagram highlights MTOR as a main activator of the serine p70S6K (p70S6 kinase), possibly contributing to tumor cell survival. Furthermore, the fact that MTOR signaling is confirmed to operate downstream makes it a promising therapeutic target for AML patients (Chen et al. 2010).
These unexpected findings may simply represent misidentified relationships or may reflect regulatory feedback within the system. At the same time, however, they may define the basis for genuinely novel discoveries. While this analysis is exploratory in nature, we believe it provides a principled hypothesis generation instrument for further experimental investigation. This instrument may prove particularly significant in a disease like AML, where standard chemotherapy is too often not fully effective and where there is a great need for new therapeutic strategies.
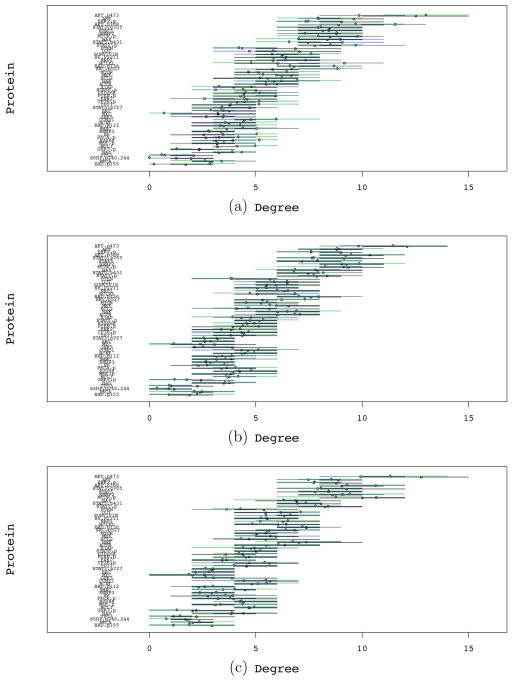
We assess the sensitivity of our inference with respect to prior settings by repeating our analysis for a range of values associated with prior hyperparameters. We summarize the posterior distribution of the proteomic pathway by analyzing the degree of each protein. As we consider reciprocal graphs, the degree of a vertex (protein) is defined as the number of edges incident to the vertex, with reciprocal relationships counted twice. A summary of our results is reported in Figure 6, where we represent the posterior mean degree associated with each protein and 95% credible intervals. Our analysis considered varying levels of prior sparsity (ζ = 0.5, 1.0, and 2.0), as well as varying degrees of prior pathway faithfulness, by considering pathway departure penalties, ν = 0.5, 1.0, and 2.0. We found that our results are robust to varying degrees of prior pathway faithfulness and overall sparsity. The proteomic degree distribution is maintained in agreement with respect to the order and magnitude of the expected number of neighbors. We observed some added instability associated with increasing levels of the sparsity parameter ζ > 2. However, posterior degree distributions are, for the large majority, significantly overlapping for most proteins over wide ranges of prior hyperparameters.
Figure 6.
Posterior mean degree and associated 95% CI by protein. Panels (a), (b), and (c) correspond to increasing sparsity penalties = 0.5, 1.0, and 2.0, respectively. For each protein, we report the posterior mean vertex degree evaluated in relation to three settings of the pathway deviation penalty parameter ν [ν = 0.5 (black), ν = 1.0 (blue), ν = 2.0 (green)]. The online version of this figure is in color.
6. DISCUSSION
We have proposed a probabilistic framework for the analysis of RPPA data. We focus on assessing the probability of protein–protein interactions by considering patterns of similarity characterized by hidden states of activation (as compared with inactive states).
Our probability model makes explicit use of prior information regarding evidence of protein interactions reported in the literature. This allows for the definition of a prior over graphical structures that explicitly anchors the model to known patterns of interaction, still allowing for a local search of new interaction patterns. Among the appealing features of the prior defined in this article is the explicit consideration of edge direction and the possibility of feedback loops between proteins.
We observed strong robustness of posterior inference about the pathway, to the setting of prior hyperparameters. This is expected as RPPA data are often characterized by large sample sizes. We however maintain that careful tuning and diagnostic checks are often required and are useful tools to assess sensitivity to the prior.
The large sample sizes characterizing RPPA data provide a unique opportunity for principled probabilistic modeling. At the same time, we are fully aware that departure from the multivariate Gaussian framework comes at a technical and computational cost. Particularly, computations involving partition functions, like the one reported in Section 3.1, characterize an entire area of research (Møller et al. 2006, Friel et al. 2009) and are known to be potentially problematic in high-dimensional settings. The strategy suggested in our article relies on the ability to simulate from a multivariate binary distribution in a fast and reliable fashion. Therefore, if the number of proteins or samples is very large, one may consider alleviating the computation burden via parallelization across samples and/or relying on alternate approximation strategies (Besag 1974; Green and Richardson 2002; Friel et al. 2009). While a full review of this problem would be perhaps too ambitious for this application, we defer the reader to Zhou and Schmidler (2009) for a comparative discussion of alternate computational strategies.
On a related subject, it is worth noting that even standard Gaussian MRF representations are not fully immune from computational difficulties, as closed-form expressions are usually only available upon assuming very stringent restrictions (decomposability) on the graph topologies admitted for inference (Giudici and Green 1999; Roverato 2002; Wong, Carter, and Khon 2003; Atay-Kays and Massam 2005, among others). In contrast, the general Hammersley–Clifford representation (Hammersley and Clifford 1971; Besag 1974) does not prescribe unrealistic restrictions on the graph of interest and provides a very flexible recipe for the definition of conditional dependence relationships.
Finally, in this article, we model dependence between protein by assuming that there are no significant alterations in patterns of covariation between different subsets of patients. While this is beyond the scope of this work, our formulation provides a straightforward basis for methodological extensions aimed at allowing for formal tests of differential pathway activation, both in a supervised and in an unsupervised fashion. These possible developments will be particularly useful in the analysis of proteomic studies of AML, since the disease is markedly heterogeneous, with numerous underlying genetic aberrations, and we still lack a strategy for the treatment of different subtypes of AML based on the current knowledge about genetic markers.
Footnotes
The density of a reciprocal graph
 is defined as
.
is defined as
.
SUPPLEMENTARY MATERIALS
The supplementary materials are intended to provide an expanded discussion of some of the aspects characterizing the methodology proposed in our article. We discuss identifiability of the sampling model’s mixture components and assess how the proposed methodology compares to current network inference techniques. We also provide full conditional distributions, when available.
Supplementary materials for this article are available online. Please go to www.tandfonline.com/r/JASA
Contributor Information
Donatello Telesca, Email: dtelesca@ucla.edu, Assistant Professor, Department of Biostatistics, UCLA School of Public Health, Los Angeles, CA.
Peter Müller, Email: pmueller@math.utexas.edu, Professor, Department of Mathematics, University of Texas at Austin.
Steven M. Kornblau, Email: skornbla@mdanderson.org, Professor, Department of Stem Cell Transplantation, University of Texas M. D. Anderson Cancer Center, Houston, TX
Marc A. Suchard, Email: msuchard@ucla.edu, Professor, Departments of Biomathematics and Human Genetics, David Geffen School of Medicine at UCLA, and Department of Biostatistics, UCLA School of Public Health, Los Angeles, CA
Yuan Ji, Email: koaeraser@gmail.com, Director of Cancer Research Informatics, North Shore University Health System.
References
- Airoldi EM, Markowetz F, Blei DM, Troyanskaya O. Statistical Discovery of Signaling Pathways From an Ensemble of Weakly Informative Data Sources. Poster presentation at the 2007 NIPS Workshop on Statistical Models of Networks; Whistler, BC, Canada. 2007. [Google Scholar]
- Andriew C, Roberts GO. The Pseudo-Marginal Approach for Efficient Markov Chain Monte Carlo Computation. The Annals of Statistics. 2009;37:697–725. [Google Scholar]
- Atay-Kays A, Massam H. The Marginal Likelihood for Decomposable and Non-Decomposable Graphical Gaussian Models. Biometrika. 2005;92:674–659. [Google Scholar]
- Barbieri MM, Berger JO. Optimal Predictive Model Selection. The Annals of Statistics. 2004;32(3):870–897. [Google Scholar]
- Besag J. Spatial Interaction and the Statistical Analysis of Lattice Systems,” (with discussion) Journal of Royal Statistical Society, Series B. 1974;36:192–236. [Google Scholar]
- Boeuf H, Merienne K, Jacquot S, Duval D, Zeniou M, Hauss C, Reinhardt B, Huss-Garcia Y, Dierich A, Frank DA, Hanauer A, Kedinger C. The Ribosomal S6 Kinases, cAMP-Responsive Element-Binding, and STAT3 Proteins Are Regulated by Different Leukemia Inhibitory Factor Signaling Pathways in Mouse Embryonic Stem Cells. Journal of Biological Chemistry. 2001;276:46204–46211. doi: 10.1074/jbc.M106718200. [DOI] [PubMed] [Google Scholar]
- Chen W, Drakos E, Ioannis G, Schlette E, Li J, Vasiliki L, Staikou E, Patsouris E, Panayotidis P, Medeiros J, Rassidakis GZ. mTOR Signaling Is Activated by FLT3 Kinase and Promotes Survival of FLT3-Mutated Acute Myeloid Leukemia Cells. Molecular Cancer. 2010;9:292. doi: 10.1186/1476-4598-9-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chib S, Greenberg E. Analysis of Multivariate Probit Models. Biometrika. 1998;85(2):347–361. [Google Scholar]
- Cox DR. The Analysis of Multivariate Binary Data. Applied Statistics. 1972;21:113–120. [Google Scholar]
- Dean N, Raftery AE. Normal Uniform Mixture Differential Gene Expression Detection for cDNA Microarrays. Bioinformatics. 2005;6:173–179. doi: 10.1186/1471-2105-6-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobra A, Hans C, Jones B, Yao G, West M. Sparse Graphical Models for Exploring Gene Expression Data. Journal of Multivariate Analysis. 2004;90:196–212. [Google Scholar]
- Efron B, Tibshirani R. On Testing the Significance of Sets of Genes. The Annals of Applied Statistics. 2006;1:101–129. [Google Scholar]
- Friedman N. Inferring Cellular Networks Using Probabilistic Graphical Models. Science. 2004;303:799–805. doi: 10.1126/science.1094068. [DOI] [PubMed] [Google Scholar]
- Friel N, Pettitt AN, Reeves R, Wit E. Bayesian Inference in Hidden Markov Random Fields for Binary Data Defined on Large Lattices. Journal of Computational and Graphical Statistics. 2009;18(2):243–261. [Google Scholar]
- Giudici P, Green PJ. Decomposable Graphical Gaussian Model Determination. Biometrika. 1999;86(4):785–801. [Google Scholar]
- Green PJ. Reversible Jump Markov Chain Monte Carlo Computation and Bayesian Model Determination. Biometrika. 1995;82(4):711–732. [Google Scholar]
- Green PJ, Richardson S. Hidden Markov Models and Disease Mapping. Journal of the American Statistical Association. 2002;97(460):1055–1070. [Google Scholar]
- Hammersley J, Clifford P. Markov Filed on Finite Graphs and Lattices. University of Cambridge, Statistical Laboratory; 1971. unpublished manuscript. [Google Scholar]
- Hao J, Li TG, Qi X, Zhao DF, Zhao GQ. WNT/Beta-Catenin Pathway Up-Regulates Stat3 and Converges on LIF to Prevent Differentiation of Mouse Embryonic Stem Cells. Developmental Biology. 2006;290(1):81–91. doi: 10.1016/j.ydbio.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Jain N, Zhang T, Fong SL, Lim CP, Caoa X. Repression of Stat3 Activity by Activation of Mitogen-Activated Protein Kinase (MAPK) Oncogene. 1998;17(24):3157–3167. doi: 10.1038/sj.onc.1202238. [DOI] [PubMed] [Google Scholar]
- Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan N, Chung S, Emili A, Snyder J, Greenblatt M, Gerstein M. A Bayesian Networks Approach for Predicting Protein-Protein Interactions From Genomic Data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- Jiang W. Bayesian Variable Selection for High Dimensional Generalized Linear Models: Convergence Rates of the Fitted Densities. The Annals of Statistics. 2007;35:1487–1511. [Google Scholar]
- Jones B, Carvalho C, Dobra A, Hans C, West M. Experiments in Stochastic Computation for High-Dimensional Graphical Models. Statistical Science. 2005;20:388–400. [Google Scholar]
- Kornblau SM, Minden M, Rosen D, Putta S, Cohen A, Covey T, Spellmeyer D, Fantl W, Gayko U, Cesano A. Dynamic Single-Cell Network Profiles in Acute Myelogenous Leukemia Are Associated With Patient Response to Standard Induction Therapy. Clinical Cancer Research. 2010;16(14):3721–3733. doi: 10.1158/1078-0432.CCR-10-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblau SM, Vu H, Ruvolo P, Estrov Z, O’Brien S, Cortes J, Kantarjian H, Andreeff M, May W. BAX and PKC-Alpha Modulate the Prognostic Impact of BCL2 Expression in Acute Myelogenous Leukemia. Clinical Cancer Research. 2000;6:1401–1409. [PubMed] [Google Scholar]
- Kornblau SM, Womble M, Qiu Y, Jackson E, Chen W, Konopleva M, Estey E, Andreeff M. Simultaneous Activation of Multiple Signal Transduction Pathways Confers Poor Prognosis in Acute Myelogenous Leukemia. Blood. 2006;108:2358–2365. doi: 10.1182/blood-2006-02-003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblau SM, Xu H, Zhang W, Hu S, Beran M, Smith T, Hester J, Estey E, Benedict W, Deisseroth A. Levels of Retinoblastoma Protein Expression in Newly Diagnosed Acute Myelogenous Leukemia. Blood. 1994;84:256–261. [PubMed] [Google Scholar]
- Koster JTA. Markov Properties of Non-Recursive Causal Models. The Annals of Statistics. 1996;24:2148–2177. [Google Scholar]
- Kreuter M, Woelke K, Bieker R, Schliemann C, Steins M, Buechner T, Berfel W, Mesters R. Correlation of Neuropilin-1 Overexpression to Survival in Acute Myeloid Leukemia. Leukemia. 2006;20:1950–1954. doi: 10.1038/sj.leu.2404384. [DOI] [PubMed] [Google Scholar]
- Lauritzen SL. Graphical Models. Oxford: Clarendon; 1996. [Google Scholar]
- Liang F. A Double Metropolis-Hastings Sampler for Spatial Models With Intractable Normalizing Constants. Journal of Statistical Computing and Simulation. 2010;80:1007–1022. [Google Scholar]
- Martelli AM, Evangelisti C, Chiarini F, Grimaldi C, Manzoli L, McCubrey JA. Targeting the pi3k/Akt/mTOR Signaling Network in Acute Myelogenous Leukemia. Expert Opinion on Investigational Drugs. 2009;18(9):1333–1349. doi: 10.1517/14728220903136775. [DOI] [PubMed] [Google Scholar]
- Møller J, Pettittt AN, Reeves R, Berthelsen KK. An Efficient Markov Chain Monte Carlo Method for Distributions With Intractable Normalizing Constants. Biometrika. 2006;93(2):451–458. [Google Scholar]
- Mrozek K, Marcucci G, Paschka P, Whitman S, Bloomfiled C. Clinical Relevance of Mutations and Gene-Expression Changes in Adult Acute Myeloid Leukemia With Normal Cytogenetics: Are We Ready for a Prognostically Prioritized Molecular Classification? Blood. 2007;109:431–448. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Speed TP. Network Inference Using Informative Priors. Proceedings of the National Academy of Sciences. 2008;105(38):1433–14318. doi: 10.1073/pnas.0802272105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton MA, Quintana F, den Boon J, Sengupta S, Ahlquist P. Random-Set Methods Identify Distinct Aspects of the Enrichment Signal in Gene-Set Analysis. The Annals of Applied Statistics. 2007;1:85–106. [Google Scholar]
- Palomero T, Dominguez M, Ferrando A. The Role of the PTEN/AKT Pathway in NOTCH1-Induced Leukemia. Cell Cycle. 2008;7(8):965–970. doi: 10.4161/cc.7.8.5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmigiani G, Garrett ES, Anbazhagan R, Gabrielson E. A Statistical Framework for Expression-Based Molecular Classification in Cancer. Journal of the Royal Statistical Society, Series B. 2002;64:717–736. [Google Scholar]
- Pittman J, Huang E, Dressman H, Horng C-F, CSH, Tsou M, Chen C-M, Bild A, Iversen E, Liao M, Huang A, Nevins J, West M. Integrated Modelling of Clinical and Gene Expression Information for Personalized Prediction of Disease Outcomes. Proceedings of the National Academy of Sciences. 2004;101:8431–8436. doi: 10.1073/pnas.0401736101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Propp JG, Wilson DB. Exact Sampling With Coupled Markov Chains and Applications to Statistical Mechanics. Random Structures & Algorithms. 1996;9:223–252. [Google Scholar]
- Roverato A. Hyper-Inverse Wishart Distribution for Non-Decomposable Graphs and Its Application to Bayesian Inference for Gaussian Graphical Models. Scandinavian Journal of Statistics. 2002;29:391–411. [Google Scholar]
- Schäfer J, Strimmer K. A Shrinkage Approach to Large Scale Covariance Matrix Estimation and Implications for Functional Genomics. Statistical Applications in Genetics and Molecular Biology. 2005;4(1):32. doi: 10.2202/1544-6115.1175. [DOI] [PubMed] [Google Scholar]
- Scott J, Berger J. Technical Report (2008–10) Department of Statistical Science, Duke University; 2008. Multiple Testing, Empirical Bayes, and the Variable Selection Problem. [Google Scholar]
- Scott J, Carvalho CM. Feature-Inclusion Stochastic Search for Gaussian Graphical Models. Journal of Computational and Graphical Statistics. 2008;17:790–808. [Google Scholar]
- Sebastiani P, Ramoni M. Normative Selection of Bayesian Networks. Journal of Multivariate Analysis. 2005;93:340–357. [Google Scholar]
- Shojaie A, Michailidis G. Analysis of Gene Sets Based on the Underlying Regulatory Network. Journal of Computational Biology. 2009;16(3):407–426. doi: 10.1089/cmb.2008.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shojaie A, Michailidis G. Network Enrichment Analysis in Complex Experiments. Statistical Applications in Genetics and Molecular Biology. 2010;9(1):Article 22. doi: 10.2202/1544-6115.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner SM, Austin JL, Leone G, Rush LJ, Plass C, Heinonen K, Mrozek K, Sill H, Knuutila S, Kolitz JE, Archer KJ, Caligiuri MA, Bloomfield CD, de la Chapelle A. BAALC, The Human Member of a Novel Mammalian Neuroectoderm Gene Lineage, Is Implicated in Hematopoiesis and Acute Leukemia. Proceedings of the National Academy of Sciences. 2001;98:13901–13906. doi: 10.1073/pnas.241525498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesca D, Inoue L, Neira M, Etzioni R, Gleave M, Nelson C. Differential Expression and Network Inferences Through Functional Data Modeling. Biometrics. 2009;65(3):793–804. doi: 10.1111/j.1541-0420.2008.01159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesca D, Müller P, Parmigiani G, Friedman RS. Technical Report. Department of Biostatistics, UCLA School of Public Health; 2010. Modeling Dependent Gene Expression. [Google Scholar]
- Tibes R, Qiu Y, Hennessy B, Andreeff M, Mills G, Kornblau S. Reverse Phase Protein Array: Validation of a Novel Proteomic Technology and Utility for Analysis of Primary Leukemia Specimens and Hematopoietic Stem Cell. Molecular Cancer Therapeutics. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- Wan X, Helman LJ. Levels of PTEN Protein Modulate Akt Phosphorylation on Serine 473, But Not on Threonine 308, in IGF-II-Overexpressing Rhabdomysarcomas Cells. Oncogene. 2003;22:8205–8211. doi: 10.1038/sj.onc.1206878. [DOI] [PubMed] [Google Scholar]
- Wong F, Carter CK, Khon R. Efficient Estimation of Covariance Selection Models. Biometrika. 2003;90:809–830. [Google Scholar]
- Zhou X, Schmidler SC. Technical Report. Duke University; 2009. Bayesian Parameter Estimation in Ising and Potts Models: A Comparative Study With Applications to Protein Modeling. [Google Scholar]