Abstract
The flavodiiron proteins (FDPs) are involved in the detoxification of oxidative compounds, such as nitric oxide (NO) or O2 in Archaea and Bacteria. In cyanobacteria, the FDPs Flv1 and Flv3 are essential in the light-dependent reduction of O2 downstream of PSI. Phylogenetic analysis revealed that two genes (flvA and flvB) in the genome of Chlamydomonas reinhardtii show high homology to flv1 and flv3 genes of the cyanobacterium Synechocystis sp. PCC 6803. The physiological role of these FDPs in eukaryotic green algae is not known, but it is of a special interest since these phototrophic organisms perform oxygenic photosynthesis similar to higher plants, which do not possess FDP homologs. We have analyzed the levels of flvA and flvB transcripts in C. reinhardtii cells under various environmental conditions and showed that these genes are highly expressed under ambient CO2 levels and during the early phase of acclimation to sulfur deprivation, just before the onset of anaerobiosis and the induction of efficient H2 photoproduction. Importantly, the increase in transcript levels of the flvA and flvB genes was also corroborated by protein levels. These results strongly suggest the involvement of FLVA and FLVB proteins in alternative electron transport.
Keywords: Alternative electron transport, Chlamydomonas reinhardtii, Flavodiiron proteins, O2 photoreduction, Photosynthesis, Sulfur deprivation
Introduction
Chlamydomonas reinhardtii is a soil-dwelling green alga with great flexibility in its photosynthetic machinery and metabolism, which are employed to cope with changing light, carbon and nutrient supplies and oxic/anoxic conditions. During photosynthesis, specialized antenna complexes harvest and transfer light energy to the PSII and PSI reaction centers, where primary charge separation initiates photosynthetic linear electron flow by oxidizing water at PSII and reducing NADP+ to NADPH downstream of PSI. These electron transfer reactions are coupled with proton pumping across the thylakoid membrane, and the resulting proton gradient, ΔpH, drives the ATP synthesis. Photosynthetic organisms have developed different photoprotective mechanisms and alternative electron transport pathways to prevent the over-reduction of the photosynthetic electron transport chain and to maintain an optimal NAD(P)H/ATP ratio under different environmental conditions (reviewed in Peltier et al. 2010, Cardol et al. 2011, Shikanai 2014).
In cyanobacteria, flavodiiron proteins (FDPs, also called A-type flavoproteins, Flvs) function as a strong electron sink, redirecting excess electrons to O2 in a non-harmful way (reviewed in Allahverdiyeva et al. 2015a, Allahverdiyeva et al. 2015b). Since C. reinhardtii possesses two genes with high homology to Synechocystis sp. strain PCC 6803 (hereafter, Synechocystis) flv genes, it is highly conceivable that the proteins encoded by these genes are also involved in photosynthetic electron transport in C. reinhardtii.
FDPs are a family of enzymes with nitric oxide (NO)/O2-reductase activity and have a modular structure with a N-terminal metallo-β-lactamase-like domain and a C-terminal flavodoxin-like domain as core units (Vicente et al. 2002). The metallo-β-lactamase module harbors a non-heme di-iron center with histidine and carboxylate residues as ligands; this is the active site of NO/O2 reduction (Silaghi-Dumitrescu et al. 2003). At the C-terminus, the FMN prosthetic group is embedded and acts as the electron donor for the di-iron domain. In FDP monomers, these two redox centers are too distant from each other to perform electron transfer (Vicente et al. 2008). However, the monomers can build a ‘head-to-tail’ dimer structure for efficient electron transfer. This arrangement brings the di-iron center of each monomer in close contact with the FMN moiety from the other monomer (Vicente et al. 2008).
In organisms that conduct oxygenic photosynthesis, including cyanobacteria, green algae, mosses and lycophytes, an additional NAD(P)H:flavinoxidoreductase module is fused at the C-terminus of the FDPs. These oxygenic photosynthetic organisms always possess at least two different FDPs, which are grouped into the two clusters A and B (Zhang et al. 2009). It is noteworthy that genes encoding FDP homologs have not been detected in the sequenced genomes of diatoms, haptophytes or higher plants, Picea sitchensis being an exception. An ancient plant, P. sitchensis possesses a single gene with homology to flv; however, the enzyme encoded by this gene lacks the additional C-terminal domain that is typical of all other oxygenic photosynthetic organisms (Allahverdiyeva et al. 2015a).
Most studies conducted so far on the function of FDPs in photosynthetic organisms have been focused on cyanobacteria. The genome of Synechocystis, a non-N2-fixing, unicellular cyanobacterium, contains four genes (sll1521, sll0219, sll0550 and sll0217) encoding a family of FDPs: Flv1, Flv2, Flv3 and Flv4, respectively. A reverse genetics approach applied to Synechocystis has demonstrated the essential function of Flv1 and Flv3 proteins in the light-dependent reduction of O2, also known as the Mehler-like reaction (Helman et al. 2003). Recently, it has been found that Flv1 and Flv3 proteins are crucial for safeguarding the photosynthetic apparatus, particularly the PSI complex, under fluctuating light intensities, mimicking natural light conditions (Allahverdiyeva et al. 2013, Allahverdiyeva et al. 2015b). The other two FDPs, Flv2 and Flv4, are not involved in O2 photoreduction (Helman et al. 2003, Allahverdiyeva et al. 2015a). Instead, these proteins function as a heterodimer in the photoprotection of PSII under CO2-limiting and high light conditions by releasing excess excitation pressure at the acceptor side of PSII to a currently unknown electron acceptor (Zhang et al. 2009, Zhang et al. 2012), in co-operation with phycobilisomes (Bersanini et al. 2014, Chukhutsina et al. 2015).
The filamentous heterocystous N2-fixing cyanobacterium, Anabaena sp. strain PCC 7120 (hereafter Anabaena), possesses six FDPs. Flv1A and Flv3A proteins are specific to vegetative cells and probably function in the Mehler-like reaction, whereas Flv2 and Flv4 proteins presumably mediate photoprotection of PSII, similar to their role in Synechocystis (Ermakova et al. 2013, Ermakova et al. 2014). The additional set of two FDPs in Anabaena, Flv1B and Flv3B, are heterocyst specific (Ermakova et al. 2013). It has been shown that Flv3B protects nitrogenase by performing light-induced O2 uptake and maintaining micro-oxic conditions inside of the heterocysts, while the role of Flv1B remains unknown (Ermakova et al. 2014).
In the eukaryotic green alga C. reinhardtii, two flv genes have been identified as paralogs in each cluster: flvA (Cre12.g531900) and flvB (Cre16.g691800). Despite a lack of sufficient experimental data, the high homology between the cyanobacterial and algal FDP proteins makes the involvement of FDPs in O2 photoreduction highly likely (Zhang et al. 2009, Peltier et al. 2010, Cardol et al. 2011, Dang et al. 2014).
In this work, we analyzed the expression patterns of C. reinhardtii flvA and flvB at the transcript and protein levels under different environmental conditions, including acclimation to different light intensities, CO2 concentrations and sulfur deprivation. Our results strongly support the involvement of the FLVA and FLVB proteins in alternative electron transfer.
Results
Selection of the appropriate reference genes
Before analyzing the transcript level of flvA and flvB with real-time quantitative reverse transcription–PCR (RT-qPCR), we performed a selection of the most suitable reference genes for the environmental conditions applied here (for more details, see the Materials and Methods). The selection of putative reference genes was based on previous studies in Arabidopsis thaliana (Hong et al. 2010). The putative reference genes included Mu1-adaptin (ap1m1), eukaryotic translation elongation factor 1α (eef1), the protein phosphatase 2A subunit B (pp2a), a TIP41-like protein (tip41), β-tubulin 1 (tub1), a ubiquitin ligase (ubc8) and commonly used reference genes, such as actin (act) and a receptor of the activated protein kinase C (cblp) (Table 1).
Table 1.
Information on the accession numbers, function and primers for eight potential reference genes and the two genes of interest: flvA and flvB
| Symbol | Accession No. | Description | Primer (5′→3′) | Amplicon length (bp) |
|---|---|---|---|---|
| act | Cre13.g603700 | Actin | for-CGCTGGAGAAGACCTACGAG | 134 |
| rev-CGCTGGAGAAGACCTACGAG | ||||
| ap1m1 | Cre06.g262250 | Mu1-Adaptin | for-ATGGTGGATGTGTTCAAGCA | 145 |
| rev-TGTACTCGGCCAGGATTTTC | ||||
| cblp | g6364 | Receptor of activated protein kinase C | for- CTTCTCGCCCATGACCAC | 105 |
| rev- CCCACCAGGTTGTTCTTCAG | ||||
| eef1 | Cre12.g498600 | Eukaryotic translation elongation factor 1α | for-GAGCTGGAGAAGCTGAAGGA | 164 |
| rev-GCGTCGATGATGGTGTAGTG | ||||
| pp2a | g1227 | Ser/Thr-specific protein phosphatase 2A, subunit B | for-GACTATGATGGCCTGCACCT | 230 |
| rev-CACCGGACCTTGTTGATTTT | ||||
| tip41 | Cre17.g734150 | TIP41-like protein | for-GTCTTCCACCTGAACCGTGT | 209 |
| rev-GGTGTGGTGAAGGTCCAGTC | ||||
| tub1 | Cre12.g542250 | β-Tubulin 1 | for-GCCCTGTACGACATCTGCTT | 75 |
| rev-GCTGATCAGGTGGTTCAGGT | ||||
| ubc8 | Cre03.g159200 | Ubiquitin-protein ligase | for-GGATCCGGAACTTCACAAGA | 184 |
| rev-CAGCAGGTGGCAGTACAGAA | ||||
| flvA | Cre12.g531900 | Flavodiiron protein A | for-CAAGTGGTGCTGTCCAACC | 246 |
| rev-GGAGAAGAGCTTGGAGCTGA | ||||
| flvB | Cre16.g691800 | Flavodiiron protein B | for-CGAGGACACCATCACCATC | 95 |
| rev-GGTAGGCGTTGTAGGTGGTG |
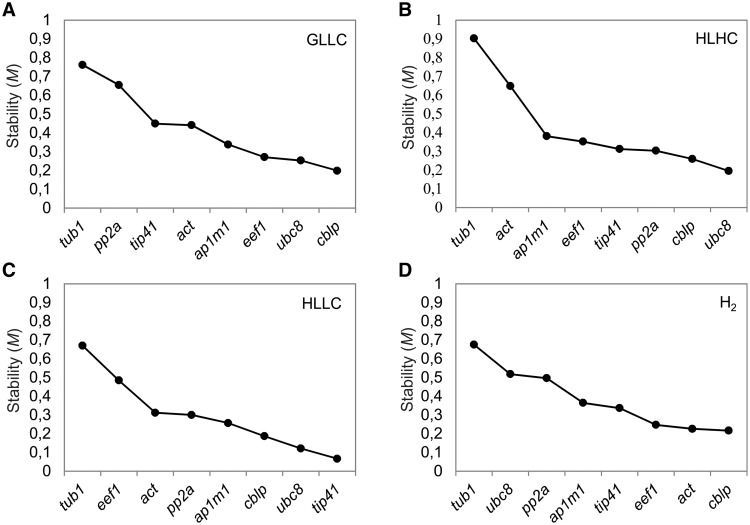
Under growth light and low (ambient level) CO2 (GLLC) conditions, the cblp and ubc8 genes showed the lowest M values and, therefore, the highest expression stabilities (Fig. 1A). Under the same conditions, tub1 and pp2a were the least stable genes. For cultures under high light and high CO2 (HLHC) conditions, ubc8 and cblp were the most stable, while tub1 and act were the least stable genes (Fig. 1B). Under the combined stress of high light and low CO2 (HLLC), the most stable reference genes were tip41, ubc8 and cblp, whereas tub1 and eef1 could not be considered stable (Fig. 1C). The cblp, act and eef1 genes showed the most stable expression pattern under the long-term H2 photoproduction condition caused by sulfur deprivation, while tub1 and ubc8 were the least stable genes (Fig. 1D).
Fig. 1.
The average expression stability (M) of eight tested potential reference genes. Chlamydomonas reinhardtii cultures were exposed for 0, 2, 12 and 24 h to GLLC (A), HLHC (B) and HLLC (C). The samples during sulfur deprivation for H2 photoproduction (D) were collected at 0, 2, 40 and 150 h from the beginning of sulfur deprivation.
Consequently, cblp and ubc8 were considered suitable reference genes under GLLC, HLHC and combined HLLC conditions. In line with these results, the cblp gene has previously been used as a reference gene in different studies of C. reinhardtii (Mus et al. 2007, Brzezowski et al. 2012, Pape et al. 2012). The least stable gene in GLLC, HLHC and HLLC cultures was tub1, which also corresponds well to previous results (Hong et al. 2010, Rosic et al. 2011, Liu et al. 2012). The cblp and act genes were selected as suitable reference genes for sulfur deprived experiments.
Expression of FLVA and FLVB under different environmental conditions
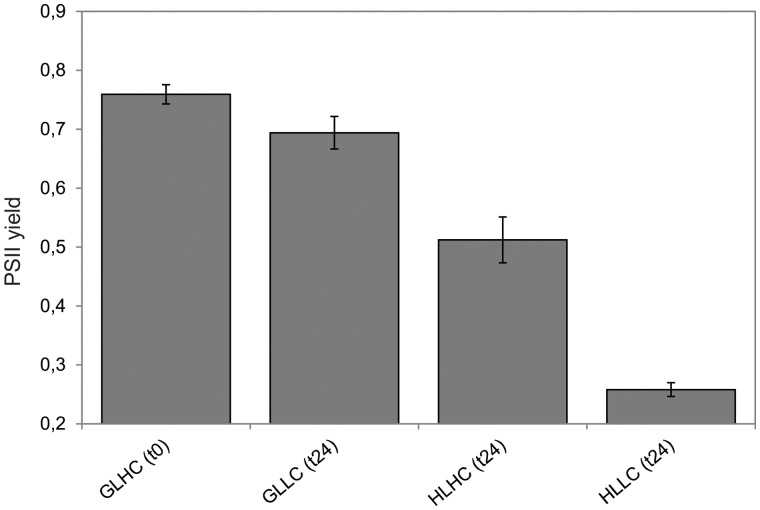
To evaluate the possible physiological role of FDPs in autotrophically-grown C. reinhardtii, flvA and flvB transcript levels were studied during the shift from growth light and high CO2 (GLHC) condition to GLLC, HLHC and, finally, to HLLC condition. The shift of the cultures from moderate growth light to high light and/or low CO2 should lead to a more reduced state of the photosynthetic electron transport chain in the cells. Indeed, the effective yield of PSII dramatically decreased after the 24 h shift from GLHC (0.76) to HLLC (0.26), whereas the shifts to GLLC or HLHC demonstrated a somewhat milder effect (0.69 and 0.51, respectively) on the photosynthetic activity (Fig. 2).
Fig. 2.
The effective PSII yield monitored directly (t0) and 24 h after the shift from GLHC to GLLC, HLHC or HLLC. The values are the mean of three biological replicates (±SD).
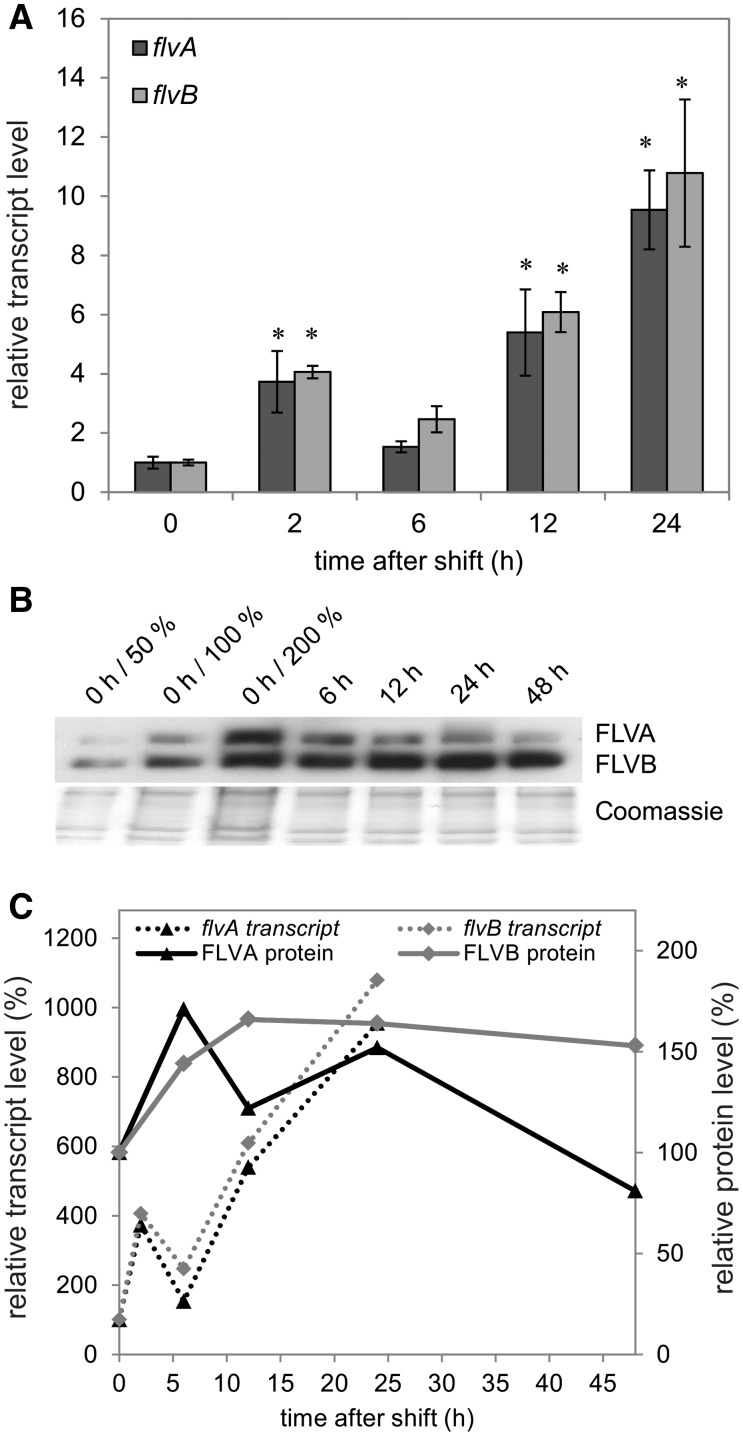
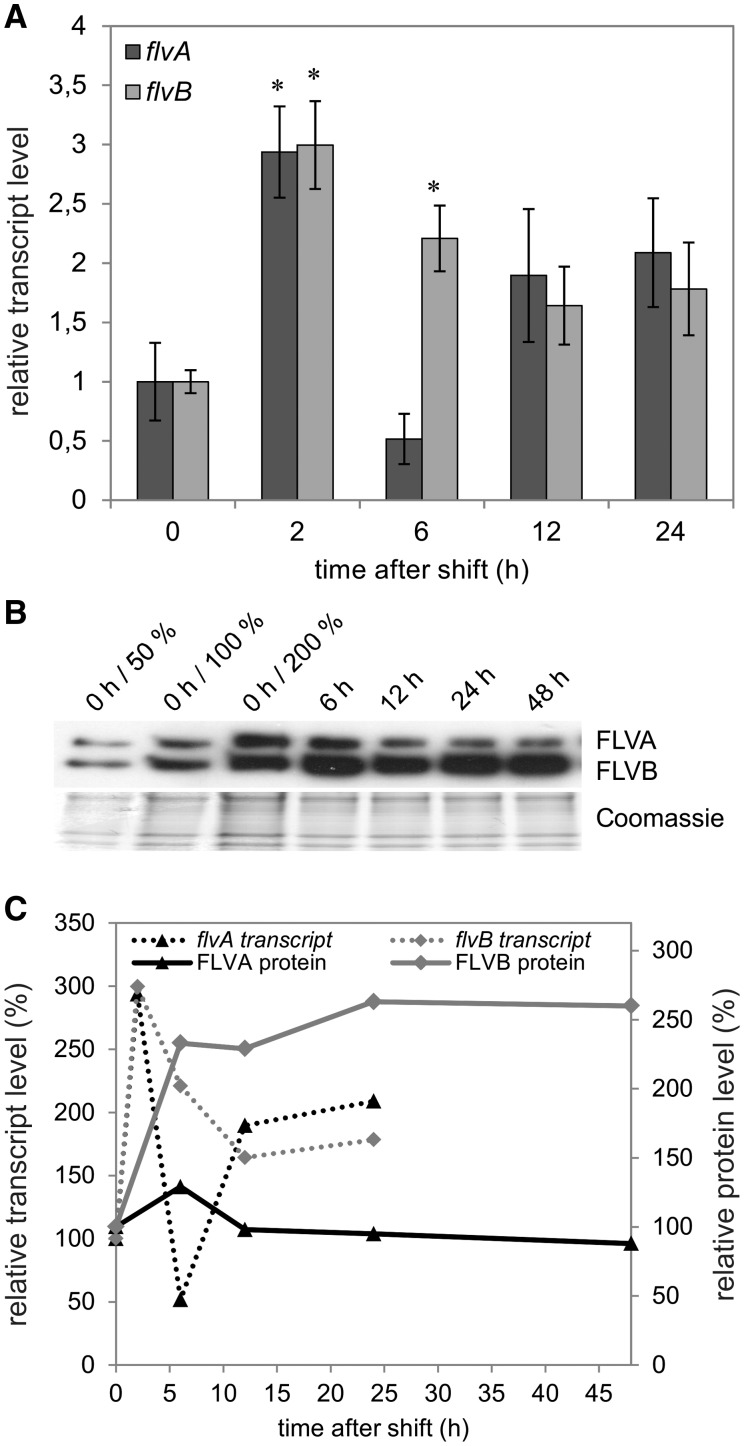
The shift of the cells from GLHC to GLLC conditions led to an approximately 4-fold increase in flvA transcript abundance after 2 h and up to approximately 9-fold after 24 h acclimation to GLLC (Fig. 3A). The flvB transcript level was also significantly up-regulated under the GLLC condition (Fig. 3A), approximately 4-fold after 2 h and approximately 11-fold after 24 h.
Fig. 3.
Changes in flv transcript (A) and FDP protein (B) levels upon the shift from GLHC to GLLC. RNA was isolated after 0, 2, 6, 12 and 24 h. The values are the mean of three biological replicates (± SD). The significance was evaluated with Student’s t-test (an asterisk represents P ≤ 0.05). The FLVA and FLVB protein accumulation was probed at 0, 6, 12, 24 and 48 h after the shift by a specific antibody. Correlation between transcript (dashed line) and protein (solid line) accumulation level (C). The protein levels were determined by densitometric analysis of three Western blots, performed with Gene Tools (Perkin Elmer).
In order to corroborate the transcript level results obtained by RT–qPCR on a protein expression level, we generated an antibody against FLVB. The antibody raised against FLVB showed two strong bands around 70 and 58 kDa. The respective bands were cut from the SDS–PAGE gel and submitted to liquid chromatography-tandem mass spectrometry (LC-MS/MS) for further analysis of the functionality of the antibodies. FLVB was identified at approximately 58 kDa and FLVA at approximately 70 kDa. This correlated well with the two strongest bands detected by immunoblotting. The analysis of FDPs under the different environmental conditions demonstrated up-regulation (∼160%) of the FLVA and FLVB proteins 6–48 h after the shift from GLHC to GLLC (Fig. 3B, C).
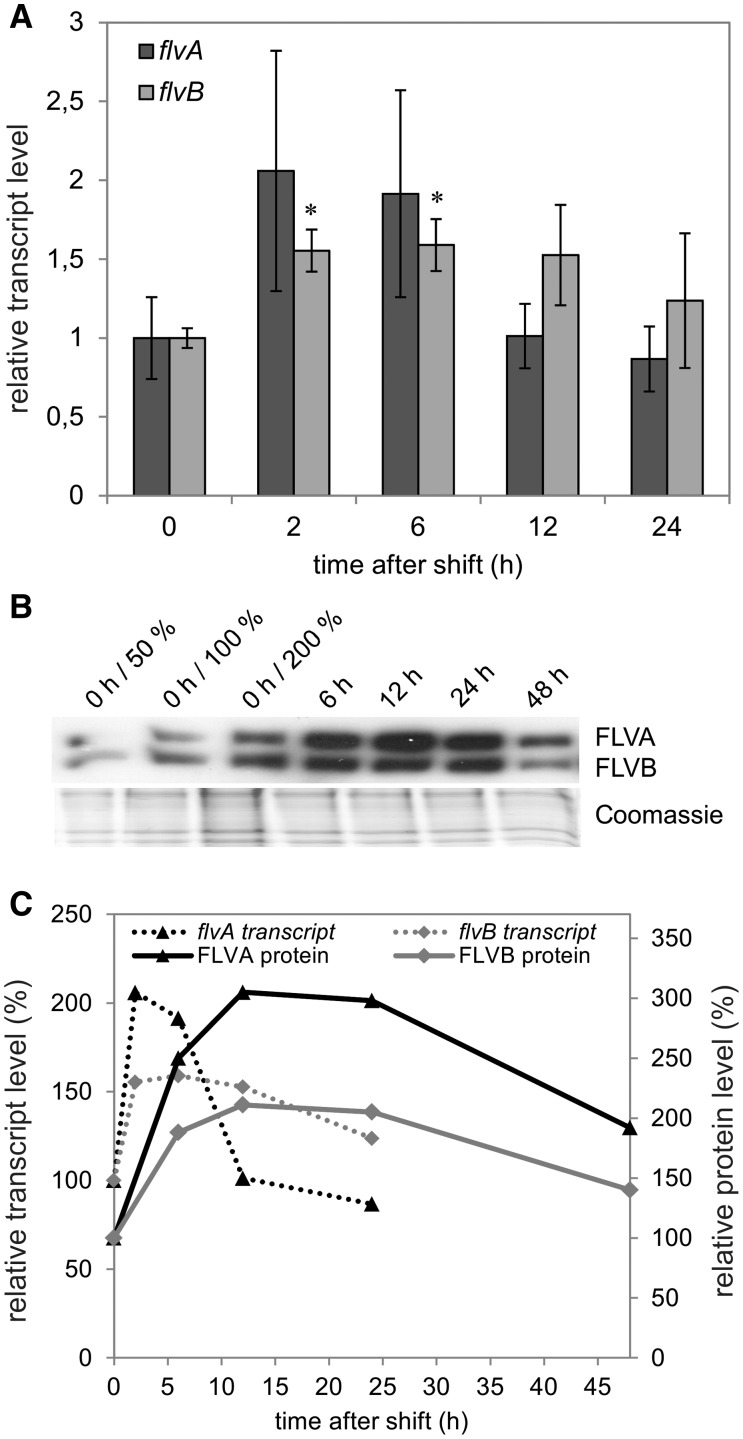
The shift from growth (GLHC) to high light (HLHC) conditions resulted in a small but significant increase in the transcript abundance of flvB (∼1.5-fold) (Fig. 4A). This was reflected at the protein level by immunoblotting experiments showing an increase in the FLVB protein (∼210%) during the first 24 h of the shift to HLHC (Fig. 4B, C). Despite the absence of statistically significant change in flvA transcript levels (Fig. 4A), at the protein level FLVA was strongly up-regulated (∼300%) during the first 24 h after the shift to HLHC (Fig. 4B, C). However, 48 h after the shift to HLHC, the abundance of FDPs decreased to approximately 200% for FLVA and to 140% for FLVB.
Fig. 4.
Changes in flv transcript (A) and FDP protein (B) levels upon the shift from GLHC to HLHC. RNA was isolated after 0, 2, 6, 12 and 24 h. The values are the mean of three biological replicates ( ± SD). The significance was evaluated with Student’s t-test (an asterisk represents P ≤ 0.05). The FLVA and FLVB protein accumulation was probed at 0, 6, 12, 24 and 48 h after the shift by a specific antibody. Correlation between transcript (dashed line) and protein (solid line) accumulation level (C). The protein levels were determined by densitometric analysis of three Western blots, performed with Gene Tools (Perkin Elmer).
The combined stress caused by the shift from GLHC to HLLC led to a small (∼3-fold), but significant, up-regulation of both flvA and flvB transcripts 2 h after the shift (Fig. 5A). The immunoblot analysis showed a slight up-regulation of FLVA (∼130%) and a strong up-regulation of FLVB (∼260%) 6 h after the shift (Fig. 5B, C). Furthermore, after 48 h at HLLC conditions, the FLVB level did not change further, while the FLVA content decreased to initial levels.
Fig. 5.
Changes in flv transcript (A) and FDP protein (B) levels upon the shift from GLHC to HLLC. RNA was isolated after 0, 2, 6, 12 and 24 h. The values are the mean of three biological replicates (± SD). The significance was evaluated with Student’s t-test (an asterisk represents P ≤ 0.05). The FLVA and FLVB protein accumulation was probed at 0, 6, 12, 24 and 48 h after the shift by a specific antibody. Correlation between transcript (dashed line) and protein (solid line) accumulation level (C). The protein levels were determined by densitometric analysis of three Western blots, performed with Gene Tools (Perkin Elmer).
Long-term H2 photoproduction and FDPs
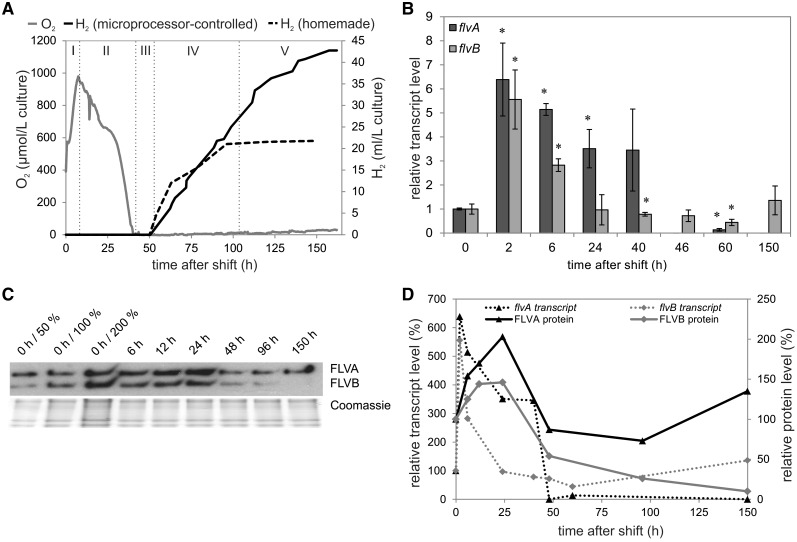
Next, we investigated the gene transcription and protein expression levels of FDPs throughout acclimation to the H2 photoproduction condition, which was triggered by applying a two-stage sulfur deprivation protocol (Melis et al. 2000). Initial experiments were performed in a photobioreactor system that allowed the detection of H2 photoproduction yields and continuous monitoring of dissolved O2 levels in the media containing sulfur-deprived C. reinhardtii cells (Fig. 6A). During acclimation to sulfur deprivation, C. reinhardtii cultures pass through five consecutive phases: photosynthetic (I), O2 consumption (II), anaerobic (III), H2 production (IV) and termination (V) (Kosourov et al. 2002). As shown in Fig. 6A, after the transfer to sulfur deprived photoheterotrophic conditions, the C. reinhardtii cells continued to evolve O2 intensively during the first 10 h (phase I). In the second phase of sulfur deprivation, the cells undergo strong metabolic changes, PSII activity drops down strongly and respiratory activity increases, inducing a transition to anaerobic conditions. This phase is followed by a complete anaerobic phase (Fig. 6A, phase III), where two [Fe–Fe]-hydrogenases are expressed, finally leading to H2 photoproduction (phase IV). Similar experiments were also performed in sealed flasks, where H2 photoproduction was monitored regularly (Fig 6A, dashed line) and samples were collected from different time points of sulfur deprivation for further investigation by RT–qPCR and Western blotting experiments.
Fig. 6.
Phases during sulfur deprivation in C. reinhardtii cultures in a microprocessor-controlled photobioreactor system (A). H2 photoproduction in C. reinhardtii cultures was induced by transferring the cells at 0 h into TAP-S medium and the produced H2 (full line, microprocessor-controlled photobioreactor system; dashed line, home-made photobioreactor) was gathered by water displacement in the upside-down graduated cylinder filled with water. The O2 level was monitored by the microprocessor-controlled photobioreactor system. The relative transcript levels of flvA and flvB during the shift to H2 photoproduction conditions are shown for 0, 2, 6, 24, 40, 46, 60 and 150 h after the beginning of sulfur deprivation (B). The values are the mean of three biological replicates (± SD) and the significance was evaluated with Student’s t-test (an asterisk represents P ≤ 0.05). The protein levels during H2 photoproduction are shown for 0, 6, 12, 24, 48, 96 and 150 h after the shift to TAP-S medium (C). Correlation between transcript (dashed line) and protein (solid line) accumulation level (D). The protein levels were determined by densitometric analysis of three Western blots, performed with Gene Tools (Perkin Elmer).
RT–qPCR experiments demonstrated that the transcript levels of flvA and flvB were significantly up-regulated (∼6.5-fold and ∼5.5-fold, respectively) 2 h after the shift to TAP-S (Tris-acetate-phosphate without sulfur) medium (Fig. 6B), when PSII is still active and a significant increase in the level of dissolved O2 is observed (Fig. 6A). The flvA transcript level remained up-regulated (∼3.5-fold) while the flvB level decreased to approximately 0.7-fold of the initial level at 40 h of sulfur deprivation, when complete anaerobiosis was established and H2 photoproduction began. During the H2 photoproduction phase (46–60 h after the shift), when cultures had established a long-term anaerobiosis in the medium (Fig. 6A), both flvA and flvB transcript levels were significantly down-regulated to 0.1-fold (or not detectable) and approximately 0.5-fold, respectively (Fig. 6B). During the termination phase, 150 h after the transfer to sulfur deprivation conditions when cultures cease producing H2 and, instead, start to show residual O2 evolution activity (Fig. 6A), flvB returned to its initial transcript level and flvA was no longer detectable.
Immunoblotting revealed a strong up-regulation in the amounts of FLVA (∼200%) and FLVB (∼150%) proteins from 6 to 24 h after the shift to TAP-S medium (Fig. 6C, D). The later time points showed that the FLVA protein had returned to its initial level by the start of H2 photoproduction and FLVB declined until it was undetectable at the end of the experiment. Although the protein levels did not strictly follow the trend of the transcript abundance towards the termination phase, it is clear that the FLVA and FLVB proteins were up-regulated during the photosynthetic phase of adaption to sulfur deprivation and remained high until anaerobiosis was established in the culture.
Discussion
Analysis of putative reference genes
As a first approach to obtain information about the function of FDPs in C. reinhardtii, we applied RT–qPCR to determine the response of flv transcript levels to varying environmental conditions. The determination of appropriate reference genes for each organism under particular environmental conditions is crucial to employing the correct normalization strategy to transcript analysis (Huggett et al. 2005, Guenin et al. 2009). In this study, we analyzed several candidate reference genes, and the cblp and ubc8 genes were determined to be the most suitable for the interpretation of the transcript data obtained after the shift of algal cultures from high CO2 and standard growth light conditions to low CO2 and/or high light conditions (Fig. 1A–C).
The situation was different when the sulfur deprivation protocol was applied (Melis et al. 2000) for the initiation of long-term H2 photoproduction. Under nutrient deprivation conditions, the sealed algal cultures pass through several physiological stages, resulting in massive changes in cellular metabolism from oxygenic photosynthesis to anaerobic photo-fermentation (Atteia et al. 2013, Catalanotti et al. 2013, Allahverdiyeva et al. 2014). In this case, the cblp and act genes were the most stable reference genes (Fig. 1D). This study confirmed that there are no universal reference genes, and the choice of appropriate reference genes varies depending on the environmental conditions and the nature of the analyzed target genes.
FDPs possibly work as an alternative electron sink in C. reinhardtii
FDPs are known to function in alternative electron transport routes in cyanobacteria (reviewed in Allahverdiyeva et al. 2015a, Allahverdiyeva et al. 2015b). The presence of homologs of the genes coding for FDPs in C. reinhardtii suggests a possible involvement of their products in photosynthetic electron transport. However, the function of FDPs in C. reinhardtii has not been addressed thoroughly and needs to be elucidated. RT–qPCR and Western blot analysis demonstrated that flvA and flvB were significantly up-regulated on both the transcript and protein levels after the change in CO2 (shifting the cultures from HC to LC) (Figs. 3, 5) and/or light (shifting the cultures from GL to HL) regimes (Figs. 4, 5). The strongest up-regulation of the FLVA protein was observed after the shift to high light, whereas the FLVB protein was up-regulated under all three different environmental conditions tested in the present study.
The treatment of cells with both high light and/or limited CO2 concentrations led to a decrease in photosynthetic activity (Fig. 2). The exposure of cells to high light causes an increase in NAD(P)H levels, while the lower CO2 availability led to a higher ATP demand (Kramer and Evans 2011). During evolution, photosynthetic organisms have developed sophisticated mechanisms to dissipate excess reducing power in harmless ways and to balance possible mismatches in production and demand of ATP and NAD(P)H, the ratio of which changes upon environmental cues through the regulation of linear and alternative electron transport pathways (Peltier et al. 2010, Cardol et al. 2011, Kramer and Evans 2011). Recent studies with cyanobacteria have demonstrated the function of FDPs as powerful electron sinks under stress conditions: Flv2 and Flv4 are strongly up-regulated and involved in the photoprotection of PSII under ambient CO2 and high light conditions (Zhang et al. 2009, Zhang et al. 2012, Bersanini et al. 2014), whereas Flv1 and Flv3 proteins can release electron pressure after PSI, thus safeguarding PSI under fluctuating light intensities (Allahverdiyeva et al. 2013). Flv1 and Flv3 proteins act as a strong electron sink, redirecting about 20–60% of electrons originating from PSII to O2 during illumination under air-level CO2 and under strong Ci deprivation, respectively (Allahverdiyeva et al. 2011). Importantly, O2 photoreduction by the FDP pathway generates water without the formation of reactive oxygen species (ROS) (Vicente et al. 2002), thus also contributing to ATP synthesis.
The data obtained with C. reinhardtii flvA and flvB genes strongly resembles the gene expression pattern of Anabaena flv1A and flv3A, where both genes were strongly up-regulated at low CO2 and moderately up-regulated at high light conditions (Ermakova et al. 2013). Moreover, accumulation of the flv3 transcript and a strong up-regulation of the Flv3 protein have been observed in Synechocystis cells under low CO2 conditions, whereas the flv1 and flv3 transcripts in Synechocystis did not show a remarkable induction under high light conditions (Zhang et al. 2009). The limited information about the response of flv1 transcripts to different environmental cues is probably due to low transcript abundance of this gene in Synechocystis (Zhang et al. 2009, Allahverdiyeva et al. 2015a).
A recent study showing an up-regulation of FLVA and FLVB proteins in the pgrl1 mutant of C. reinhardtii under low CO2 as well as high light conditions indicates that FDPs could function as an electron valve to compensate for the lack of, or impaired, cyclic electron flow (Dang et al. 2014). Interestingly, under the combined HLLC stress condition used in that study, the up-regulation of FDPs in the pgrl1 mutant was transient and disappeared after 48 h. Instead, the elevated H2O2 level indicated a replacement of the FDP pathway by the true Mehler reaction and the formation of ROS (Dang et al. 2014). Similarly, our results with C. reinhardtii wild type during HLLC stress showed an up-regulation of both FDPs during the first 48 h (Fig. 5). This implies an important function for these proteins upon changes in environmental conditions. Our expression analysis of FLVA and FLVB, together with previous results, suggests that FDPs in C. reinhardtii also play an important role as alternative electron sinks in order to prevent redox poise at the photosynthetic electron transport chain.
The possible electron donor of FDPs in C. reinhardtii is not known yet. Based on in vitro studies on recombinant Synechocystis Flv3 proteins, it was concluded that FDPs function as an NAD(P)H-O2-oxidoreductase (Vicente et al. 2002). However, this is not the case for Synechocystis Flv2 and Flv4 proteins functioning at the PSII acceptor side. Recently, ferredoxin 1 (FDX1) was found to interact with FLVB, thus opening up a new discussion about the possibility of FDX1 as an electron donor to the FDPs proteins in C. reinhardtii (Peden et al. 2013).
FDPs participate in photosynthetic acclimation of C. reinhardtii to sulfur deprivation
During acclimation to sulfur deprivation, algae experience a strong metabolic shift from oxygenic photosynthesis, where CO2 is assimilated and starch accumulated, towards anaerobic photo-fermentation, where starch reserves are metabolized to produce ATP and NAD(P)H. The anaerobic re-oxidation of NAD(P)H involves several fermentative pathways that produce organic acids (acetate, formate, lactate, malate and succinate), ethanol, H2 and CO2 (reviewed in Atteia et al. 2013, Catalanotti et al. 2013). Some enzymes of fermentative metabolism, such as the [Fe–Fe]-hydrogenases and pyruvate formate-lyase, are sensitive to O2 remaining in the chloroplast (Atteia et al. 2013).
The acclimation to sulfur deprivation that triggers H2 photoproduction in algae can be divided into several phases (Kosourov et al. 2002) (Fig. 6A). During the photosynthetic stage (phase I) of acclimation to sulfur deprivation (0 to ∼10 h) the O2 concentration in the bioreactor rises until respiratory processes take over (phase II). Anaerobiosis is established at approximately 40 h after the shift to sulfur deprivation (phase III). The up-regulation of both FLVA and FLVB proteins demonstrates a correlation with the presence of O2 in the culture, with the maximum FDP amount observed approximately 24 h after the shift (Fig 6C, D). The FDP up-regulation during the photosynthetic and respiratory phase of H2 photoproduction indicates that O2 photoreduction via FDPs is important in the acclimation to these conditions. It has been postulated that the decrease of O2 after a shift to sulfur deprived medium is mainly due to an increase in mitochondrial respiration (Melis et al. 2000, Melis 2007, Ghirardi et al. 2010). Our results indicate that FDPs contribute to the establishment of anaerobiosis by functioning in light-induced O2 uptake. The increased levels of FDPs in the chloroplast during the first phases of sulfur deprivation may accelerate the establishment of anaerobiosis and therefore help to ensure the function of the fermentative pathways within a shorter time period. In the later phase IV, while H2 is produced in anaerobiosis, the FDPs are down-regulated (Fig. 6).
Taken together, we propose that FDPs in C. reinhardtii function as an alternative electron sink during oxygenic photosynthesis by actively assisting to decrease the O2 level inside the chloroplast at the onset of anaerobiosis and are replaced by [Fe–Fe]-hydrogenases later on, when anaerobiosis is fully established. In both cases, FDPs and [Fe–Fe]-hydrogenases support electron flow in thylakoids for the production of ATP at the expense of reducing power accumulated downstream of PSI and, thus, also protect the photosynthetic electron transport chain from over-reduction. This rapid acclimation to anaerobiosis is likely to be advantageous for the soil-dwelling C. reinhardtii, which regularly faces anoxic or micro-oxic conditions in nature.
Materials and Methods
Strains and culture conditions
The wild-type C. reinhardtii, strain CC406, was maintained photoheterotrophically in TAP medium (Gorman and Levine 1965) at ambient air under a continuous light intensity of 50 µmol photons m−2 s−1 photosynthetically active radiuiation (PAR) under agitation (90 r.p.m.) at 25°C. For preparing experimental C. reinhardtii cultures, the cells were harvested at OD750 = approximately 1.2, transferred to high salt medium (HSM; Sueoka 1960), diluted to OD750 = approximately 0.2 and cultivated photoautotrophically by bubbling the cultures with sterile air containing 3% CO2 (high CO2, HC) under a continuous light intensity of 50 µmol photons m−2 s−1 PAR (GL), at 25°C for 48 h. To perform the shift to different environmental conditions, the cells were harvested at OD750 = 1.2 by centrifugation (2,500 r.p.m., 2 min), resuspended in fresh HSM and adjusted to OD750 = 1.0. High light conditions were achieved by illuminating the cells at 150 µmol photons m−2 s−1 with 3% CO2 (HLHC) and low CO2 conditions were obtained by bubbling the cells with ambient air under 150 µmol photons m−2 s−1 (HLLC), or under standard growth light, 50 µmol photons m−2 s−1 (GLLC). Cells for transcript analysis were collected at 0, 2, 6, 12 and 24 h, and for protein analysis at 0, 6, 12, 24 and 48 h after the shift and stored at –80°C until further use.
Chl fluorescence measurement
The Chl a fluorescence was analyzed with a Dual-PAM-100 fluorometer (Walz). The cultures were adjusted to a final Chl (a+b) concentration of 10 µg ml−1. Red actinic light (630 nm) intensity of 54 µmol photons m−2 s−1 (cells grown under standard growth light) or 217 µmol photons m−2 s−1 (cells treated with high light) was applied. The PSII effective yield was calculated as Y(II) = (Fm′ – F)/Fm′ after illumination of the cells with an actinic light for 5 min. A saturating pulse (4,000 µmol photons m−2 s−1, 500 ms) was fired to probe Fm′, the maximum fluorescence level under the actinic light. F is the steady-state fluorescence level under actinic light.
H2 photoproduction
For the long-term H2 photoproduction, a sulfur deprivation protocol was applied (Melis et al. 2000). Cultures grown in standard TAP medium (Gorman and Levine, 1965) under ambient CO2 and continuous light intensity of 50 µmol photons m−2 s−1 PAR were harvested by centrifugation at a cell density of approximately 25 µg ml−1 Chl (a+b) and transferred into TAP medium without sulfur (TAP-S). After a series of centrifugations (twice at 2,500 r.p.m. for 2 min) and re-suspensions in TAP-S medium, the cells were adjusted to a Chl (a+b) concentration of 20 µg ml−1. Home-made cylindrical photobioreactors with an inner diameter of 60 mm were filled with 550 ml of culture, placed under continuous illumination of 75 µmol photons m−2 s−1 PAR from cool white fluorescent lamps (Mitsubishi/Osram) at 25°C and kept sealed with threaded rubber stoppers and attached tubing for gas collection. H2 production was monitored by collecting the gas in an upside-down graduated cylinder filled with water. For transcriptional analysis, cells were taken at 0, 2, 6, 24, 40, 46, 60 and 150 h after the shift to TAP-S medium. Cells for the protein analysis were collected at 0, 6, 12, 24, 48, 96 and 150 h after the shift. For continuous monitoring of the O2 level in the cultures, the experiment was repeated in a microprocessor-controlled photobioreactor system, described in Tsygankov et al. (2006).
RNA extraction
Total RNA was extracted using TRIsure (Bioline) reagent. The cells were broken via heating at 65°C. RNA was further purified by extraction with phenol/chloroform/isoamylalcohol (25 : 24 : 1) and precipitated by isopropanol, followed by removal of genomic DNA (Ambion Turbo DNase kit) with 0.5 µl of DNase (2 U µl−1). The RNA concentration was determined using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific) and the quality was checked by RNA-gel electrophoresis.
cDNA synthesis
Purified RNA (2 µg) was used for cDNA synthesis. Reverse transcription was performed with poly(dT)(20) primers and SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s protocol. Synthesized cDNA was diluted 5-fold and used as a template for RT–qPCR.
RT–qPCR
RT–qPCR was performed with a Bio-Rad IQ5 system using iQ SYBR Green Supermix (Bio-Rad) in 96-well plates. The PCR protocol was 3 min initial denaturation at 95°C, followed by 50 cycles of 95°C for 15 s, 60°C for 30 s and 72°C for 35 s. At the end, a melting curve was performed. Relative changes in the gene expression level were calculated using the qbase+ software by Biogazelle.
Selection of putative reference genes and primer design
For calculation of the expression levels of target transcripts, most popular methods, such as the 2–ΔΔCT method (Schmittgen and Livak 2008), apply reference genes for normalization. Eight putative reference genes (Table 1) were selected based on previous studies of reference genes in A. thaliana (Hong et al. 2010) and some commonly used reference genes in C. reinhardtii. The specific primers were designed by using Primer3plus software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) and are shown in Table 1.
For correct comparison, it is recommended to select reference genes with a similar expression level compared with the target genes (Hruz et al. 2011). As shown in Supplementary Fig. 1, the Cq values of the studied putative reference genes varied between 24 and 35, and those of the target genes varied between 28 and 40. Each putative reference gene showed a relatively stable expression, with Cq values that differed in just 1–2 cycles. The stability of the putative reference genes was validated by the geNorm algorithm (Vandesompele et al. 2002) included in the qbase+ software. The minimum number of reference genes needed for normalization was determined by calculation of the pairwise variation (V). A threshold of 0.15 was applied and the lowest V values were obtained at V2/3, which means that a minimum of 2–3 reference genes are sufficient for normalization of the expression data under all studied conditions (Supplementary Fig. 2).
Protein analysis
Total protein extracts were isolated by resuspending the sample cell pellet in lysis buffer (50 mM Tris pH 8, 2% SDS, 10 mM EDTA, protease inhibitors; Sigma) and freezing. After thawing the samples, the total protein extracts were separated by 14% SDS–PAGE without urea, transferred to a polyvinylidene difluoride membrane (Millipore) and blocked with 5% blotting grade blocker (Bio-Rad). The samples were loaded on an equal protein basis determined with a Direct Detect™ Spectrometer (Millipore) and visualized with Coomassie brilliant blue (Bio-Rad). The FLVA and FLVB proteins were detected using a purified rabbit antibody prepared against an FLVB peptide antigen mix (CKVVIAESYGGRDEP and CARKKAAMSGEVAKA) conjugated with keyhole limpet hemocyanin. The high homology between FLVB and FLVA allows this antibody to recognize both proteins.
The specificity of the antibody was verified via LC-MS/MS. As a secondary antibody, anti-rabbit horseradish peroxidase (HRP) was used and visualized with ECL. The protein levels were determined by densitometric ananlysis of three Western blots, performed with Gene Tools (Perkin Elmer)
Identification of proteins by LC-MS/MS
Samples for LC-MS/MS analysis were prepared according to the protocol of Shevchenko et al. (1996). Silver-stained protein bands were excised from the SDS–PAGE gel, reduced, alkylated and in-gel digested with Trypsin Gold (Promega). The peptides were extracted by repeated incubation with 5% formic acid and50% acetonitrile, lyophilized and desalted on C18 resin. Samples were analyzed by LC-MS/MS using a QExactive mass spectrometer (Thermo Fisher Scientific, Inc.) connected in line with an Easy-nLC II HPLC system (Thermo Fisher Scientific, Inc.). Peptides were dissolved in 18 µl of 2% formic acid. From the sample, 5 µl were loaded onto a pre-column (2 cm × 100 µm inner diameter) packed with a Magic C18 AQ 200-Å resin (Michrom Bioresources) and subjected to reverse-phase chromatography on a 15 cm × 75 µm inner diameter nanoscale LC column packed with the same resin. A gradient of 2–40% acetonitrile in 0.2% formic acid was applied for 28 min followed by a gradient of 40–100% acetonitrile in 0.2% formic acid for 2 min with a flow rate of 300 nl min−1. MS data acquisition of positively charged precursor ions was performed in a data-dependent mode, with the 10 most intensive ions sent to MS/MS analysis in each duty cycle. The data were processed with the Mascot search engine (version 2.4; Matrix Science) through the Protein Discoverer software, version 1.4 (Thermo Fisher Scientific, Inc.). Database searches were performed against a database of C. reinhardtii proteins supplemented with sequences of common protein contaminants and with a reverse decoy database. The search criteria allowed for one miscleavage of trypsin, oxidation of methionine, and 5 and 10 p.p.m. mass accuracies for MS and MS/MS modes, respectively.
Supplementary data
Supplementary data are available at PCP online.
Funding
This research was financially supported by the Kone foundation [to Y.A.]; the Academy of Finland and Russian Academy of Science [Research exchange program (2011–2013) mobility (grant # 267409 to Y.A.)]; the Academy of Finland [FCoE program (271832 to E-M.A.)]; the Maj and Tor Nessling Foundation [2014050 to S.K.] the Nordic Energy Research AquaFEED project.
Supplementary Material
Acknowledgments
We grateully acknowledge the National Doctoral Programme in Informational and Structural Biology (ISB).
Disclosures
The authors have no conflicts of interest to declare.
Glossary
Abbreviations
- FDP
flavodiiron protein
- FDX1
ferredoxin 1
- GL
growth light
- HC
high CO2
- HL
high light
- LC
low CO2
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- PAR
photosynthetically active radiation
- ROS
reactive oxygen species
- RT–qPCR
real-time quantitative reverse transcription–PCR
- TAP-S
Tris-acetate-phosphate without sulfur
References
- Allahverdiyeva Y., Ermakova M., Eisenhut M., Zhang P., Richaud P., Hagemann M., et al. (2011) Interplay between flavodiiron proteins and photorespiration in Synechocystis sp. PCC 6803. J. Biol. Chem. 286: 24007–24014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allahverdiyeva Y., Aro E.M., Kosourov S.N. (2014) Recent developments on cyanobacteria and green algae for biohydrogen photoproduction and its importance in CO2 reduction. In Bioenergy Research: Advances and Applications. Edited by Gupta V.K., Tuohy M., Kubicek C.P., Saddler J., Xu F. pp. 367–387. Elsevier, Amsterdam. [Google Scholar]
- Allahverdiyeva Y., Isojärvi J., Zhang P., Aro E.M. (2015a) Cyanobacterial oxygenic photosynthesis is protected by flavodiiron proteins. Life 5: 716–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allahverdiyeva Y., Mustila H., Ermakova M., Bersanini L., Richaud P., Ajlani G., et al. (2013) Flavodiiron proteins Flv1 and Flv3 enable cyanobacterial growth and photosynthesis under fluctuating light. Proc. Natl Acad. Sci. USA 110: 4111–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allahverdiyeva Y., Suorsa M., Tikkanen M., Aro E.M. (2015b) Photoprotection of photosystems in fluctuating light intensities. J. Exp. Bot. 66: 2427–2436. [DOI] [PubMed] [Google Scholar]
- Antal T.K., Krendeleva T.E., Laurinavichene T.V., Makarova V.V., Ghirardi M.L., Rubin A.B., et al. (2003) The dependence of algal H2 production on Photosystem II and O2 consumption activities in sulfur-deprived Chlamydomonas reinhardtii cells. Biochim. Biophys. Acta 1607: 153–160. [DOI] [PubMed] [Google Scholar]
- Atteia A., van Lis R., Tielens A.G., Martin W.F. (2013) Anaerobic energy metabolism in unicellular photosynthetic eukaryotes . Biochim. Biophys. Acta 1827: 210–223. [DOI] [PubMed] [Google Scholar]
- Bersanini L., Battchikova N., Jokel M., Rehman A., Vass I., Allahverdiyeva Y., et al. (2014) Flavodiiron protein Flv2/Flv4-related photoprotective mechanism dissipates excitation pressure of PSII in cooperation with phycobilisomes in Cyanobacteria. Plant Physiol. 164: 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezowski P., Wilson K.E., Gray G.R. (2012) The PSBP2 protein of Chlamydomonas reinhardtii is required for singlet oxygen-dependent signaling. Planta 236: 1289–1303. [DOI] [PubMed] [Google Scholar]
- Cardol P., Forti G., Finazzi G. (2011) Regulation of electron transport in microalgae. Biochim. Biophys. Acta 1807: 912–918. [DOI] [PubMed] [Google Scholar]
- Catalanotti C., Yang W., Posewitz M.C., Grossman A.R. (2013) Fermentation metabolism and its evolution in algae. Front. Plant Sci. 4: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukhutsina V., Bersanini L., Aro E.M., van Amerongen H. (2015) Cyanobacterial flv4-2 operon-encoded proteins optimize light harvesting and charge separation in photosystem II. Mol. Plant 8: 747–761. [DOI] [PubMed] [Google Scholar]
- Dang K.V., Plet J., Tolleter D., Jokel M., Cuiné S., Carrier P., et al. (2014) Combined increases in mitochondrial cooperation and oxygen photoreduction compensate for deficiency in cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell 26: 3036–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermakova M., Battchikova N., Allahverdiyeva Y., Aro E.M. (2013) Novel heterocyst-specific flavodiiron proteins in Anabaena sp. PCC 7120. FEBS Lett. 587: 82–87. [DOI] [PubMed] [Google Scholar]
- Ermakova M., Battchikova N., Richaud P., Leino H., Kosourov S., Isojärvi J., et al. (2014) Heterocyst-specific flavodiiron protein Flv3B enables oxic diazotrophic growth of the filamentous cyanobacterium Anabaena sp. PCC 7120. Proc. Natl Acad. Sci. USA 111: 11205–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardi M.L., Kosourov S., Maness P., Smolinski S., Seibert M., Flickinger M.C. (2010) Algal hydrogen production. In Encyclopedia of Industrial Biotechnology. Edited by Flickinger, M.C. pp. 184–198. John Wiley & Sons, Inc. [Google Scholar]
- Gorman D.S., Levine R.P. (1965) Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA 54: 1665–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenin S., Mauriat M., Pelloux J., Van W.O., Bellini C., Gutierrez L. (2009) Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J. Exp. Bot. 60: 487–493. [DOI] [PubMed] [Google Scholar]
- Helman Y., Tchernov D., Reinhold L., Shibata M., Ogawa T., Schwarz R., et al. (2003) Genes encoding A-type flavoproteins are essential for photoreduction of O2 in cyanobacteria. Curr. Biol. 13: 230–235. [DOI] [PubMed] [Google Scholar]
- Hong S.M., Bahn S.C., Lyu A., Jung H.S., Ahn J.H. (2010) Identification and Testing of Superior Reference Genes for a Starting Pool of Transcript Normalization in Arabidopsis. Plant Cell Physiol. 51(10): 1694–1706. [DOI] [PubMed] [Google Scholar]
- Hruz T., Wyss M., Docquier M., Pfaffl M., Masanetz S., Borghi L., et al. (2011) RefGenes: identification of reliable and condition specific reference genes for RT–qPCR data normalization. BMC Genomics 12: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J., Dheda K., Bustin S., Zumla A. (2005) Real-time RT–PCR normalisation; strategies and considerations. Genes Immun. 6: 279–284. [DOI] [PubMed] [Google Scholar]
- Kosourov S., Tsygankov A., Seibert M., Ghirardi M.L. (2002) Sustained hydrogen photoproduction by Chlamydomonas reinhardtii: effects of culture parameters. Biotechnol. Bioeng. 78: 731–740. [DOI] [PubMed] [Google Scholar]
- Kramer D.M., Evans J.R. (2011) The importance of energy balance in improving photosynthetic productivity . Plant Physiol. 155: 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Wu G., Huang X., Liu S., Cong B. (2012) Validation of housekeeping genes for gene expression studies in an ice alga Chlamydomonas during freezing acclimation. Extremophiles 16: 419–425. [DOI] [PubMed] [Google Scholar]
- Melis A., Zhang L., Forestier M., Ghirardi M.L., Seibert M. (2000) Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol. 122: 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis A. (2007) Photosynthetic H2 metabolism in Chlamydomonas reinhardtii (unicellular green algae). Planta 226: 1075–1086. [DOI] [PubMed] [Google Scholar]
- Mus F., Dubini A., Seibert M., Posewitz M.C., Grossman A.R. (2007) Anaerobic acclimation in Chlamydomonas reinhardtii. Anoxic gene expression, hydrogenase induction and metabolic pathways. J. Biol. Chem. 282: 25475–25486. [DOI] [PubMed] [Google Scholar]
- Pape M., Lambertz C., Happe T., Hemschemeier A. (2012) Differential expression of the Chlamydomonas [FeFe]-hydrogenase-encoding HYDA1 gene is regulated by the COPPER RESPONSE REGULATOR1. Plant Physiol. 159: 1700–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden E.A., Boehm M., Mulder D.W., Davis R., Old W.M., King P.W., et al. (2013) Identification of global ferredoxin interaction networks in Chlamydomonas reinhardtii. J. Biol. Chem. 288: 35192–35209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier G., Tolleter D., Billon E., Cournac L. (2010) Auxiliary electron transport pathways in chloroplasts of microalgae. Photosynth. Res. 106: 19–31. [DOI] [PubMed] [Google Scholar]
- Rosic N., Pernice M., Rodriguez-Lanetty M., Hoegh-Guldberg O. (2011) Validation of housekeeping genes for gene expression studies in Symbiodinium exposed to thermal and light stress. Mar. Biotechnol. 29: 1–11. [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008) Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68: 850–858. [DOI] [PubMed] [Google Scholar]
- Shikanai T. (2014) Central role of cyclic electron transport around photosystem I in the regulation of photosynthesis. Curr. Opin. Biotechnol. 26: 25–30. [DOI] [PubMed] [Google Scholar]
- Silaghi-Dumitrescu R., Coulter E.D., Das A., Ljungdahl L.G., Jameson G.N., Huynh B.H., et al. (2003) A flavodiiron protein and high molecular weight rubredoxin from Moorella thermoacetica with nitric oxide reductase activity. Biochemistry 42: 2806–2815. [DOI] [PubMed] [Google Scholar]
- Sueoka N. (1960) Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA 46: 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsygankov A., Kosourov S.N., Tolstygina I.V., Ghirardi M.L., Seibert M. (2006) Hydrogen production by sulfur-deprived Chlamydomonas reinhardtii under photoautotrophic conditions. Int. J. Hydrogen Energ. 31: 1574–1584. [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002) Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: research0034–research0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente J.B., Gomes C.M., Wasserfallen A., Teixeira M. (2002) Module fusion in an A-type flavoprotein from the cyanobacterium Synechocystis condenses a multiple-component pathway in a single polypeptide chain. Biochem. Biophys. Res. Commun. 294: 82–87. [DOI] [PubMed] [Google Scholar]
- Vicente J.B., Justino M.C., Gonçalves V.L., Saraiva L.M., Teixeira M. (2008) Biochemical, spectroscopic, and thermodynamic properties of flavodiiron proteins. Methods Enzymol. 437: 21–45. [DOI] [PubMed] [Google Scholar]
- Zhang P., Allahverdiyeva Y., Eisenhut M., Aro E.M. (2009) Flavodiiron proteins in oxygenic photosynthetic organisms: Photoprotection of photosystem II by Flv2 and Flv4 in Synechocystis sp. PCC 6803. PLoS One 4: e5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Eisenhut M., Brandt A.M., Carmel D., Silén H.M., Vass I., et al. (2012) Operon flv4–flv2 provides cyanobacterial photosystem II with flexibility of electron transfer. Plant Cell 24: 1952–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.