Summary
We investigated the impact of belowground and aboveground environmental heterogeneity on the ecology and evolution of a natural plant–pathogen interaction.
We combined field measurements and a reciprocal inoculation experiment to investigate the potential for natural variation in abiotic and biotic factors to mediate infection outcomes in the association between the fungal pathogen Melampsora lini and its wild flax host, Linum marginale, where pathogen strains and plant lines originated from two ecologically distinct habitat types that occur in close proximity (‘bog’ and ‘hill’).
The two habitat types differed strikingly in soil moisture and soil microbiota. Infection outcomes for different host–pathogen combinations were strongly affected by the habitat of origin of the plant lines and pathogen strains, the soil environment and their interactions. Our results suggested that trade-offs play a key role in explaining the evolutionary divergence in interaction traits among the two habitat types.
Overall, we demonstrate that soil heterogeneity, by mediating infection outcomes and evolutionary divergence, can contribute to the maintenance of variation in resistance and pathogenicity within a natural host-pathogen metapopulation.
Keywords: aboveground–belowground interactions, coevolution, ecotype, environmental heterogeneity, habitat type, host–pathogen, genotype × genotype × environment (G × G × E), trade-offs
Introduction
Turesson’s (1922) and Clausen et al’s (1940) classic transplant experiments inspired a large number of studies that demonstrated plant and animal adaptation to their local abiotic environment (Bradshaw, 1952, 1984; Linhart & Grant, 1996). Such adaptive processes may also have pronounced consequences for ecological dynamics (Ford, 1975), a view that has recently received increasing empirical support (Schoener, 2011). However, despite the realization that ecology and evolution can be strongly intertwined, we still lack fundamental insights as to how environmental heterogeneity may affect the ecoevolutionary dynamics between species, especially in natural systems. From an ecological perspective, the outcome of species interactions can be strongly mediated by the abiotic and biotic environment (Chamberlain et al., 2014). Such environmental mediation of interaction outcomes then provides the blueprint for spatial and temporal variation in evolutionary and coevolutionary dynamics (Thompson, 2005), which may feedback to further shape the ecology of species interactions. For example, with respect to host–parasite interactions, theoretical studies have demonstrated that the environment may affect coevolutionary trajectories and promote the long-term maintenance of variation in resistance and infectivity (Gavrilets & Michalakis, 2008; Mostowy & Engelstädter, 2011; Tellier & Brown, 2011; Poisot et al., 2012).
Pathologists have long realized that the environment (Pasteur et al., 1878), host genotype (Biffen, 1905), parasite genotype (Barrus, 1911) and their interactions (Flor, 1956; McNew, 1960; Wolinska & King, 2009) jointly determine the outcome of host–parasite interactions. More recently, there is increasing evidence for geographical variation in coevolutionary dynamics and patterns of local adaptation (Thompson, 2005, 2013). Microcosm (Forde et al., 2004; Vogwill et al., 2009; Lopez Pascua et al., 2012) and field studies (Laine, 2006, 2008) have assessed how a range of ecologically-relevant variables such as local encounter rates, productivity and temperature may drive spatial variation in coevolutionary dynamics and strengthen patterns of local adaptation. Nonetheless, several critical gaps in our knowledge remain. First, the majority of studies have focused on aboveground spatial heterogeneity. Given widespread documentation of spatial variation in soil abiotic and biotic conditions (Ettema & Wardle, 2002; Tedersoo et al., 2014), there is a need to investigate how belowground heterogeneity affects aboveground species interactions (Bonte et al., 2010; Fones et al., 2010). Second, identifying the environmental factors shaping species interactions in natural studies remains a challenge, as species exist within a complex and changing set of environmental conditions. Ultimately, we need to assess spatiotemporal variation in environmental conditions, as well as their direct and interactive effects on the ecology and evolution of species interactions.
Using the flax rust (Melampsora lini)–wild flax (Linum marginale) system, we have previously demonstrated rapid coevolutionary dynamics (Thrall et al., 2012) and strong pathogen adaptation at the host ecotype level (Laine et al., 2014). In this study, we use an inoculation experiment to assess the contribution of (nonhost) environmental variation in shaping infection outcomes and evolutionary divergence. More specifically, we aim to determine the impact of natural environmental variation on infection outcomes and evolutionary dynamics of L. marginale and its specialized rust pathogen M. lini across two distinct habitat types (‘bogs’ and ‘hills’), which frequently occur in close proximity within the subalpine region in Southeastern Australia. Following an initial assessment of environmental factors, we identified that soil moisture and soil microbial community structure showed the most pronounced differences among the habitat types. We then used these key factors in a multifactorial cross-inoculation experiment to investigate how environmental differentiation among habitat types might affect infection outcomes and coevolutionary trajectories. Our findings demonstrate the key role of environment on the outcome of ecological interactions, and the maintenance of variation in resistance and pathogenicity in host and parasite associations.
Materials and Methods
Study system
The native flax Linum marginale Cunn. is a perennial herbaceous plant growing throughout Southern Australia. Within the Kosciuszko National Park in southern New South Wales the plant mainly grows in montane areas and subalpine frost hollows. In these areas, flax grows as two distinct ecotypes: a hill ecotype that is found on well-drained hill sites and a bog ecotype that occurs in boggy areas and along stream courses (Carlsson-Granér et al., 1999). Morphologically, plants from the two habitat types can be distinguished by their overall size, the number of basal shoots produced and the length and adherence of sepals to buds and capsules (Carlsson-Granér et al., 1999).
The flax rust Melampsora lini (Ehrenb.) Lév. is an obligate fungal pathogen that, in Australia, is a specialist on L. marginale (Lawrence, 1989; Lawrence & Burdon, 1989). Spores are wind-dispersed, and when infection is successful, the first symptoms can be detected by highly localized light-green flecks on the leaves which turn into orange-coloured lesions (uredinia or pustules) in which urediniospores are produced asexually. This part of the lifecycle takes c. 12–14 d during the peak growing season, and multiple pathogen generations take place during a season. In contrast to the lowlands, the sexual cycle has not been observed in montane areas like Kosciuszko National Park. Hence, the pathogen overwinters on sporadic green leaves on which a few uredinia may survive during winter when the majority of plants die back to their root stocks (Jarosz & Burdon, 1992).
Significant genetic divergence and fixed allelic differences have been documented between the host plants originating from the bog and hill habitat types (Thrall et al., 2001). It has also been shown that there is extensive among-habitat phenotypic (Carlsson-Granér et al., 1999; Thrall et al., 2001) and genetic differentiation (Laine et al., 2014) for the pathogen. We therefore refer to plant lines and pathogen strains from the two habitat types as ‘ecotypes’. Previous studies on the wild flax–flax rust system have documented considerable variability in the incidence and severity of epidemics across populations and years, with high levels of disease reducing host survival (Jarosz & Burdon, 1992). Furthermore, marked differences within and among populations within and among years in pathogen infectivity (Burdon & Jarosz, 1992) and host resistance (Burdon & Thompson, 1995; Thrall et al., 2001) are associated with strong local adaptation of the pathogen to its local host ecotype (Laine et al., 2014) and rapid coevolutionary dynamics (Thrall et al., 2012).
Study sites
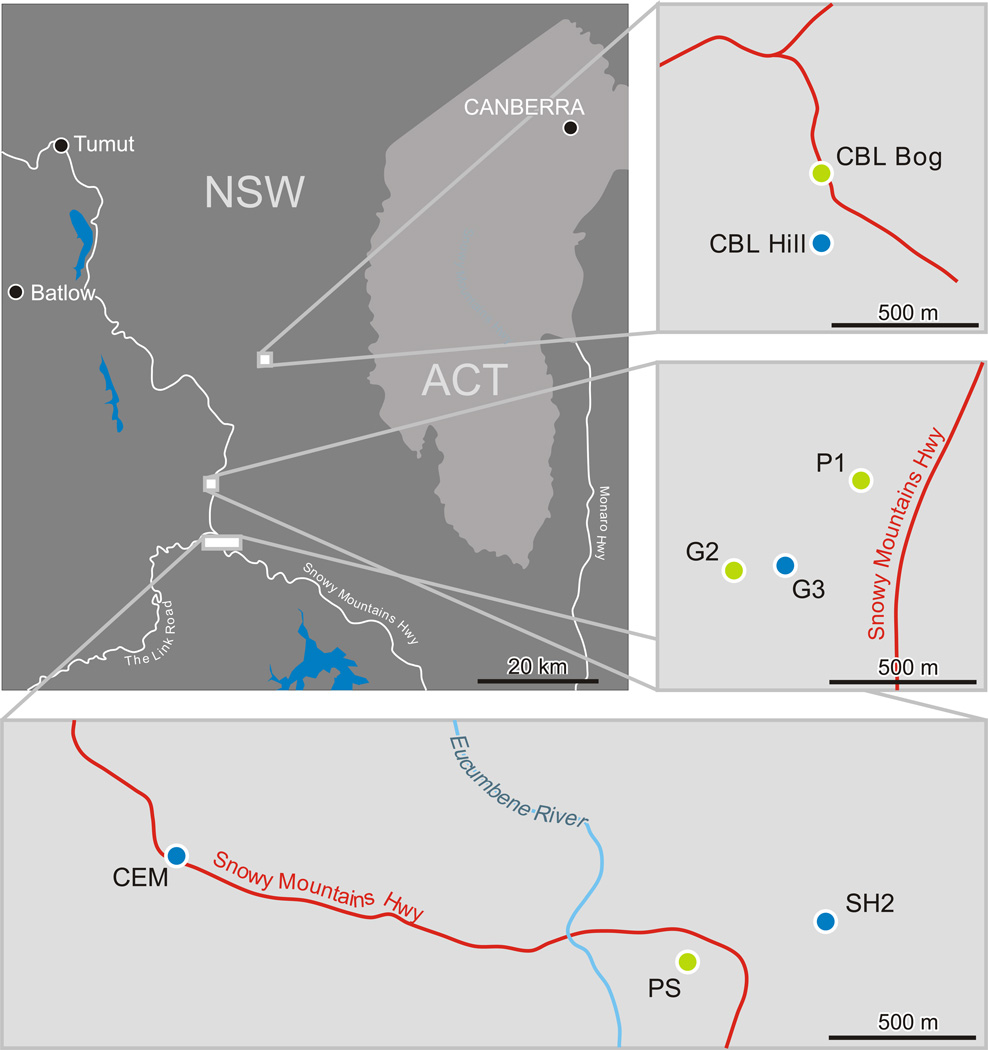
For this study, we focused on the same eight populations of L. marginale used by Laine et al. (2014; Fig. 1). Four of these populations (CBL_H, CEM, G3 and SH2) occurred in characteristic hill habitat, whereas the other four (CBL_B, G2, P1, PS) occurred in typical bog habitats. To account for spatial variation, each habitat type was represented within distinct areas (separated by >10 km) within the Kosciuszko region (Fig. 1).
Fig. 1.
Map of the study area. The large map illustrates New South Wales (NSW) and the Australian Capital Territory (ACT) in Southeast Australia. Within NSW, three areas were selected with at least one bog and one hill population. Green circles, bog populations; blue circles, hill populations.
Identification of key environmental differences among the habitat types
We first identified the key environmental factors that differed among the habitat types. For this, we assessed soil moisture, ambient temperature, soil chemistry, and soil microbial biota.
Soil moisture
In each of the eight plant populations, we randomly collected five 10 cm deep soil cores with field weights between 30–80 g into plastic bags that were then sealed. Collections were made at two time points: at the onset of epidemics on 14 December 2009 and again during the peak of epidemics on 11 January 2010. Percentage soil moisture was determined by weighing the soil core mass before and after drying in an oven at 65°C for 24 h.
Ambient temperature
Temperature loggers (HOBO Pendant® Temperature/Alarm Data Logger 8K – UA-001-08; Onset Computer Corporation, Bourne, MA, USA) were placed at the study sites between 12 January–23 March 2010. The loggers measured temperature every 30 min and were placed 25 cm above the ground to measure temperature similar to that experienced by Linum plants and developing rust infections. At site CBL-B the logger was stolen halfway through the growing season, and hence temperature data is available for the remaining seven sites only.
Soil chemistry
To test for differences in soil abiotic conditions between the habitat types, we analysed a single pooled soil sample for each of the eight populations. Each pooled soil sample consisted of three 10 cm deep soil cores taken from random locations within each of the respective populations. Samples were collected on 25 February 2010 and, following air-drying and homogenisation of the cores from each population, analysed by Incitec Pivot Ltd, Southbank, Australia (based on 500 g samples) for pH, electric conductivity, organic carbon, soil texture, nitrate, phosphorus, potassium, calcium, magnesium, sodium, aluminium, chloride, copper, iron, manganese, zinc and sulphate (Supporting information Table S1).
Biotic component of the soil
Soil community composition (bacteria and fungi) at the bog and hill sites was assessed using terminal restriction fragment length polymorphism (T-RFLP). T-RFLP is a polymerase chain reaction-based community fingerprinting method that is commonly used for comparative microbial community analyses (van Dorst et al., 2014). The method assumes that individual terminal restriction fragment lengths are representative of unique operational taxonomic units (OTUs) present within the sampled community.
DNA was extracted in duplicate from 0.25 g aliquots of soil and pooled, using a Power Soil DNA Isolation kit (MO BIO Laboratories Inc., Solana Beach, CA, USA) according to the manufacturer’s instructions and quantified spectrophotometrically (NanoDrop ND-1000, Thermo Scientific, Wilmington, DE, USA). Multiplex T-RFLP targeting bacteria and fungi was performed using primers 27f–519r (bacteria) and ITS1f–ITS4r (fungi) as described in Singh et al. (2006). Each 50 µl PCR reaction contained c. 10 ng template, and was completed with Platinum Taq DNA polymerase (Invitrogen, Mulgrave, Australia) according to the manufacturer’s instructions: 1.5 mM MgCl2, 0.2 mM each HPLC-purified primer, 1× PCR buffer, 1 U of Taq DNA polymerase, 50 µM of each dNTP (Promega, Sydney, Australia). PCR comprised 1 cycle of 94°C for 3 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s, and a final extension step of 72°C for 10 min. Triplicate PCRs were pooled, isopropanol precipitated, quantified as above and 70 ng digested independently with the restriction endonucleases MspI, AliI and HinfI. Digests were isopropanol precipitated, resuspended in 10 µl formamide containing LIZ600 size standard (Applied Biosystems, Foster City, CA, USA), denatured at 95°C (3 min) and separated by capillary electrophoresis (ABI PRISM 3130×l Genetic 110 Analyzer, Applied Biosystems). T-RFLP profiles were first checked for stable baselines, voltage and calibration and peaks in the range 50–500 bp. Absolute peak areas were initially determined with GeneMapper software v4.0 (Applied Biosystems) using a minimum peak height of five fluorescence units. Final minimum peak height threshold was determined with the T-REX T-RFLP online analysis tool (http://trex.biohpc.org/; Culman et al., 2009) removing peaks with heights less than twice the SD computed over all peaks. Data comprising the area of these ‘true’ peaks was exported for conversion to samples-by-fragments tables and subsequently to samples-by-binned-OTUs tables by the custom R script ‘interactive binner’ (Ramette, 2009) using a relative fluorescence intensity (RFI) cut-off of 0.09%, a window size of 1 and a shift size of 0.1. T-RFLP profiles were finally analysed as relative abundance (RFI) after being fourth-root transformed.
Statistical analyses
We first investigated differences in abiotic conditions among the two habitat types. We modelled log-transformed soil moisture as a function of the fixed variables ‘Habitat type’ and ‘Date’ (including the interaction), and specified a compound symmetric covariance structure for the subject ‘Population’ (nested under ‘Habitat type’) to account for sampling the same populations twice. To analyse differences in ambient temperature among the two habitat types, we first calculated the average, minimum and maximum daily temperature for each population (n = 71 d). We then modelled each response variable as a function of the fixed factors ‘Habitat type’ and ‘Day’ (including the interaction), and specified the first-order autoregressive covariance structure for the subject ‘Population’ (nested under ‘Habitat type’) to account for repeatedly sampling the same populations through time. To derive the degrees of freedom we used the Kenward-Roger adjustment (Littell et al., 2006). Repeated-measures analyses were implemented in SAS 9.3. Soil abiotic characteristics were analysed using linear discriminant analysis (function lda in package vegan in R 2.15.1) (R Core Team, 2012; Oksanen et al., 2013), where we reduced the initial set of soil variables using variance inflation factors following Neter (1996) with a threshold of five. Significance was evaluated using a chi-square test on a contingency table with the number of correct and incorrect predictions for each habitat type using jackknife-based classification (Borcard et al., 2011). Multivariate analyses of the microbial soil community were conducted on Bray–Curtis similarities calculated from relative abundance data. The multivariate approach used in this study was similar to that advocated by Clarke & Warwick (2001), namely the following steps: (1) a visual representation of the community, for which we employed nonmetric multidimensional scaling (nMDS) (Clarke, 1993) on three axes and (2) discrimination of samples using significance testing. The effect of ‘treatment’ (bog or hill) on community structure was tested with one-way analysis of similarity (ANOSIM) (Clarke, 1993) using Primer v6 (http://www.primer-e.com/).
Environmental differentiation among habitat types
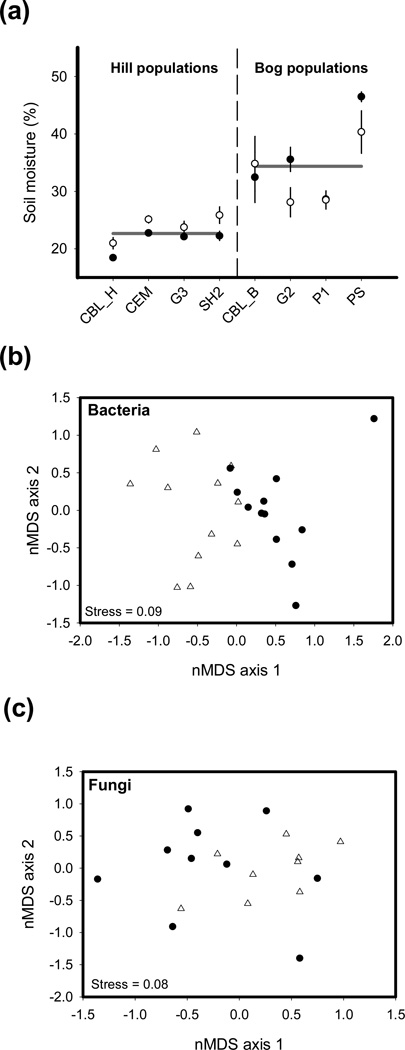
Soil moisture was significantly higher in bog than in hill populations (Fig. 2a; F1,6 = 15.59, P = 0.008). While we detected no difference in soil moisture content between the sample dates (F1,70 = 0.16, P = 0.89), there was some evidence for an interaction between sampling date and habitat type (Fig. 2a; F1,70 = 8.78, P = 0.004). In contrast to soil moisture, the mean, minimum and maximum daily temperature varied strongly among days (P < 0.001 for all three response variables), but did not vary among habitat types (P > 0.05 for all three response variables). Likewise, temporal changes in temperature did not differ among the habitat types (interaction among ‘Habitat type’ and ‘Day’: P > 0.8 for all three response variables).
Fig. 2.
Variation among bog and hill sites in the abiotic and biotic environment. (a) Differences in soil moisture among habitat types. Closed symbols depict soil samples collected in December at the start of epidemics and open symbols samples collected in January at the peak of epidemics, where vertical lines reflect ± SE of the mean and horizontal grey lines represent ecotypic averages of soil moisture in bog and hill populations. (b, c) Nonmetric multidimensional plot of the (b) bacterial and (c) fungal soil communities, respectively; triangles, samples from bog populations; circles, samples from hill populations.
The structure of the abiotic soil environment did not differ significantly between bog and hill sites (, P = 0.48). When testing differences in individual compounds, potassium (hill > bog), and to a lesser extent sodium (hill < bog), differed significantly among the two habitat types (F1,6 = 17.68, P = 0.006 and F1,6 = 7.74, P = 0.03, respectively). The bog and hill sites contained distinct bacterial communities (Fig. 2b, ANOSIM Rho = 0.26, P < 0.001), but such a clear distinction was not found for soil fungal communities (Fig. 2c, ANOSIM Rho = 0.09, P = 0.10).
Large-scale inoculation experiment
Origin of host plant lines and pathogen isolates and experimental design
For the Linum marginale–Melampsora lini study system we have recently shown strong patterns of pathogen local adaptation (measured as infectivity) among the ecotypes (Laine et al., 2014). However, how abiotic and biotic nonhost variables might affect infection outcomes and evolutionary dynamics has not yet been investigated. We therefore investigated the potential for the host–pathogen interaction to be modified by the environmental variables that were most clearly differentiated among bog and hill sites. To do this, we selected plant lines and rust pathogen isolates from three bog and four hill populations (Fig. 1; see also Laine et al., 2014). Seed was collected directly from individual plants in the field noting that the low level of outcrossing in mountain populations of L. marginale (≤3%; Burdon et al., 1999) ensured relative within-line uniformity. Due to limited seed set in some plant lines, we deviated from a fully reciprocal cross-inoculation design. Instead, we reciprocally inoculated seven plant lines with seven pathogen isolates (one randomly selected plant line and pathogen isolate from each of three bog and four hill populations) and repeated this for three sets of plant lines and pathogen isolates (Table S2). This design amounted to 147 plant line × pathogen strain combinations. Each combination of plant lines and pathogen strains was tested in four experimental treatments (see the Experimental treatments subsection), resulting in a total of 588 pairwise inoculations. Seeds were missing or did not germinate for 44 plants, and an additional three plants died during the experiment (see Table S2).
Experimental treatments
The key environmental differences among the bog and hill habitat types were soil moisture and soil bacterial community structure (Fig. 2), and we therefore manipulated the watering regime and the soil community in a multifactorial design.
Soil was collected from each of the four bog and four hill sites. Soils from each ecotype were then bulked and homogenised to form a single representative bog or hill soil. Seeds from each plant line were grown in cylindrical tubes (ø = 8 cm, depth = 15 cm; c. 700 ml of soil). For the glasshouse experiment, pots were two thirds filled with a 1 : 1 sterilized vermiculite : sand mixture. For each pot, 50 g of either a bog or hill soil was layered over this mixture and then covered with the vermiculite : sand mixture to within 1 cm from the top. The multifactorial design consisted of four experimental treatments: high moisture and soil biota from the bog habitat type; high moisture and soil biota from the hill habitat type; low moisture and bog soil biota; and low moisture and hill soil biota. To mimic natural variation in moisture between bog and hill soils, we established different watering regimes: in the high moisture treatment (representing the bog sites) plants were watered daily with 100 ml, whereas plants in the dry treatment were watered twice a week with 20 ml.
Eight weeks after planting, plants were inoculated with isolates of M. lini in a settling tower (10 plants per tower; 10 mg of rust spores mixed with 10 parts of talc per inoculation) according to the inoculation matrix (Table S2).
Measured responses variables
As life-history stages may be differentially affected by environmental conditions, we measured multiple plant and pathogen life-history traits. From day 6 (postinoculation) onwards we checked plants daily for the first appearance of flecking (representing the first visible stage in pustule formation and henceforth referred to as ‘incubation period’) and the appearance of orange-coloured pustules (representing the first possible time point at which pathogen transmission could occur, and henceforth referred to as ‘latent period’). At 12 d post inoculation we scored pathogen infection type using a categorical scoring system with five classes: (1) fully susceptible (S), large full-sized sporulating pustules (uredinia) on all leaves; (2) partial resistance (P2), large full-sized sporulating pustules on the younger leaves, grading down to no pustules on the oldest leaves; (3) partial resistance (P3), large full-sized pustules on only the youngest one or two leaves; (4) partial resistance (P4), no sporulation, but with necrotic flecks on older leaves; (5) fully resistant (R), no macroscopic evidence of infection. A frequently observed phenomenon was leaf drop, particularly after the initiation of disease (leaf drop occurred in roughly half of the inoculations). This was recorded as a binary variable (0 = no leaf abscission; 1 = leaf abscission).
Analysis
We used the framework of generalized linear mixed models (GLMMs) to analyse the data in SAS 9.3 (Littell et al., 2006). To investigate the role of genotype and environment in pathogen performance and plant resistance, we analysed each of the response variables as a function of ‘Host ecotype’, ‘Pathogen ecotype’, ‘Soil humidity’, ‘Soil biota’ and their two-, three- and four-way interactions. To account for variation among host and pathogen populations, we included the random factors ‘Host population’ and ‘Pathogen population’ (nested within host and pathogen ecotype, respectively). Likewise, we included the random factors ‘Plant line’ and ‘Pathogen genotype’ (nested within host and pathogen population, respectively) to account for variation among plant lines and pathogen strains from the same population.
To determine the factors with a significant impact on the observed response, we reduced each maximal GLMM to a minimum adequate GLMM by sequentially removing non-significant fixed factors (P > 0.10) from the model using backward selection (Crawley, 2012).
Results
The impact of plant and pathogen ecotype and environmental variation on host resistance and pathogen performance
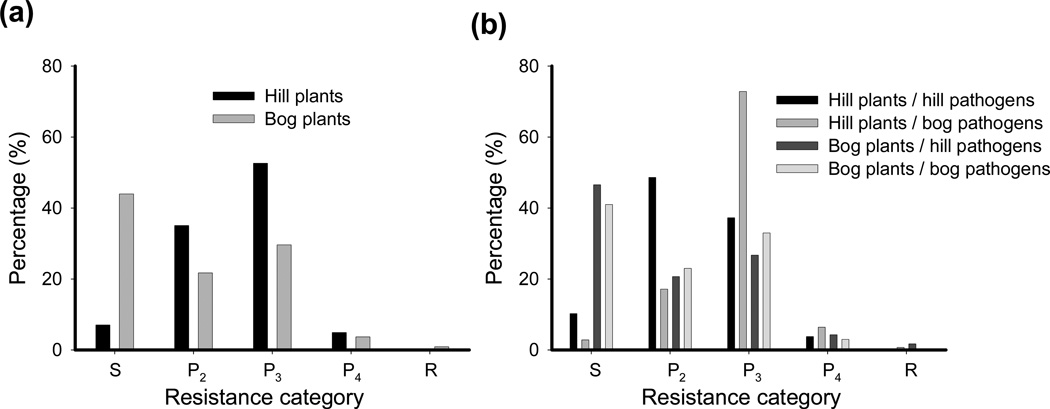
The expression of resistance strongly differed among plant ecotypes, with the majority of hill plants expressing partial resistance (Figs 3a, 4a; Tables 1, S3). By contrast, the bog plants were fully susceptible (S) to roughly half of the pathogen strains (Figs 3a, 4a). When accounting for the habitat of origin of both the plant and the pathogen, it further becomes apparent that bog pathogens induce higher levels of partial resistance in hill plants (mostly P3), whereas hill pathogens induce relatively low levels of partial resistance on hill plants (mostly P2; Fig. 3b). As a result, the average resistance score was affected by the interaction between plant and pathogen genotype (Table 1). Notably, full resistance (R) was nearly absent and occurred only in a few nonnative host–pathogen interactions (Fig. 3b). As shown in earlier studies (Laine et al., 2014), hill pathogens induced the least resistance on their local plant ecotype (Fig. 4b). However, bog pathogens and hill pathogens performed equally well on plants from the bog ecotype (Fig. 4b).
Fig. 3.
Histogram showing the resistance type expressed by the bog and hill plants (Linum marginale) in response to either (a) the full set of pathogens (Melampsora lini) or (b) separately for pathogens originating from the bog and hill habitat type.
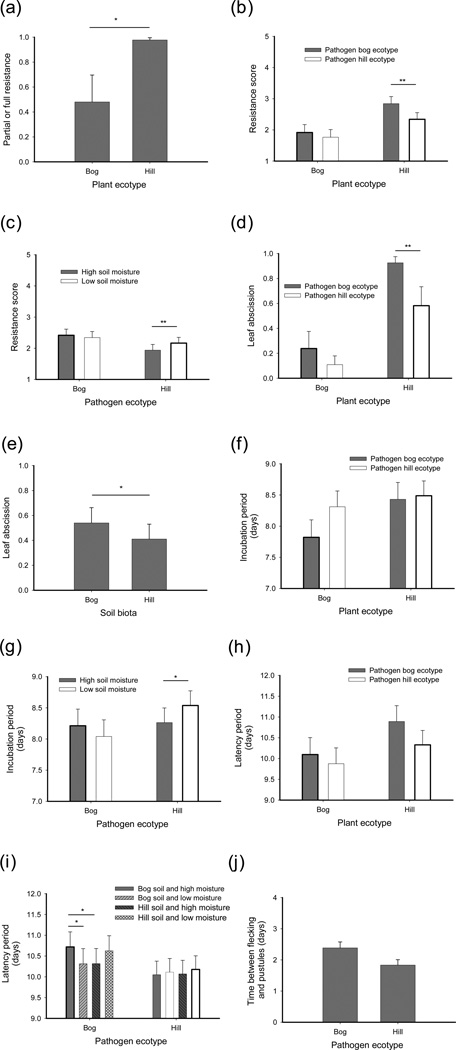
Fig. 4.
The impact of host ecotype, pathogen ecotype, environmental factors, and their interactions on host resistance and pathogen performance of flax rust Melampsora lini on the wild flax Linum marginale. (a) Host resistance is affected by plant ecotype. The resistance level is affected by (b) the interaction between plant and pathogen ecotype and (c) the interaction among pathogen ecotype and soil moisture. Leaf abscission in response to pathogen infection is affected by (d) the interaction among plant and pathogen ecotype and (e) the soil biota. The incubation period is affected by the interactions between (f) plant and pathogen ecotype and (g) pathogen ecotype and soil moisture. Latent period is affected by (h) the interaction between plant and pathogen ecotype and (i) the three-way interaction between pathogen ecotype, soil biota and soil moisture. (j) The time between the first appearance of flecking and pustules is weakly affected by pathogen ecotype. The bold outline of vertical bars indicates (where relevant) the sympatric (local) combinations of plants, parasites and environmental variables. Shown are (back-transformed) least-squares means and + SE; significant pair-wise differences within each ecotype (as indicated on the x-axis) or among main factors (a, e, j) are indicated: *, P < 0.05; **, P < 0.01.
Table 1.
The effect of host ecotype, pathogen ecotype, soil humidity, soil biota and their interactions on host resistance and pathogen performance for the interaction between flax rust (Melampsora lini) and wild flax (Linum marginale)
| Response variable | Host ecotype |
Pathogen ecotype |
Soil moisture |
Soil biota |
Host ecotype × pathogen ecotype |
Host ecotype × soil humidity |
Host ecotype × soil biota |
Pathogen ecotype × soil humidity |
Pathogen ecotype × soil biota |
Soil humidity × soil biota |
Host ecotype × pathogen ecotype × soil humidity |
Host ecotype × soil humidity × soil biota |
Pathogen ecotype × soil humidity × soil biota |
Host ecotype × pathogen ecotype × soil humidity × soil biota |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Infectivity (n = 541) | 0.019 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Resistance level (n = 541) | 0.045 | 0.120 | 0.188 | - | 0.004 | - | - | 0.009 | - | - | - | - | - | - |
| Leaf abscission (n = 541) | 0.009 | 0.067 | - | 0.024 | 0.021 | - | - | - | - | - | - | - | - | - |
| Incubation period (n = 531) | 0.061 | 0.447 | 0.532 | 0.010 | - | - | 0.007 | - | - | - | - | - | - | |
| Latent period (n = 517) | 0.141 | 0.373 | 0.814 | 0.988 | 0.035 | - | - | 0.394 | 0.572 | 0.015 | - | - | 0.032 | - |
| Time between flecking and pustules (n = 517) | - | 0.052 | - | - | - | - | - | - | - | - | - | - | - | - |
Shown are P-values of the fixed effects as estimated with generalized linear mixed models, where significant P-values are in bold typeface.
Pathogens from the hill ecotype induced higher resistance responses when plants were grown in low soil moisture (Fig. 4c; Table 1). Leaf abscission in response to pathogen infection took place in roughly half of the inoculations and occurred much more frequently on hill ecotype plants (Fig. 4d; Table 1). Leaf abscission was closely related to the degree of plant resistance: leaf abscission was nearly absent when plants were either fully susceptible or resistant (<1%), but increased with partial resistance of the plant (P2: 60.2%; P3: 69.8%; P4: 87.5%). The incidence of leaf abscission was also strongly affected by the interaction between plant and pathogen ecotype; leaf abscission occurred in >90% of the encounters between bog pathogens and hill plants, whereas hill pathogens induced leaf abscission on <60% of the hill plants (Fig. 4d; Table 1). Leaf abscission was somewhat lower when plants were grown in soil inoculated with soil biota from the hill ecotype (Fig. 4e; Table 1).
The incubation period (time to appearance of first disease symptoms) was affected by the interaction between plant and pathogen ecotype. The incubation period was shortest for bog pathogens infecting bog plants, whereas hill pathogens developed at the same pace regardless of plant ecotype (Fig. 4f; Table 1). Incubation period was also affected by the interaction between pathogen ecotype and soil moisture, with hill pathogens developing relatively slowly on plants growing in low soil moisture (Fig. 4g; Table 1). Latent period (time to when disease transmission could first occur) was weakly affected by the interaction between plant and pathogen genotype (Fig. 4h; Table 1) and further affected by a three-way interaction between pathogen ecotype, soil biota and soil moisture (Fig. 4i; Table 1). Notably, latent period was relatively long for bog pathogens in their sympatric (local) environmental conditions (Fig. 4i). Finally, there was a trend for pathogen ecotype to affect the number of days between the first flecking and pustule eruption (Fig. 4j; Table 1). Overall, the majority of quantitative life-history traits were affected by the interaction between host and pathogen ecotype. Moreover, several of the traits were affected by the interaction between pathogen ecotype and the environment; by contrast, host ecotype did not interact with the environment to shape infection outcomes.
Discussion
Previous work on the Linum–Melampsora interaction has shown strong local adaptation between adjacent habitat types (bogs and hills) occurring within a natural metapopulation (Laine et al., 2014). These findings beg the question of what factors might maintain such strong differentiation at these local spatial scales. Here we used an experimental approach to explicitly investigate the potential for environmental heterogeneity to impact on the ecology of the interaction. We first showed that bog and hill habitat types were clearly differentiated with respect to soil moisture and soil biota. We then manipulated these belowground environmental factors in a multifactorial cross-inoculation experiment. Our findings highlight that environmental heterogeneity – here in the form of two distinct habitat types frequently occurring in close proximity – can drive ecological interactions and spatial divergence in resistance and aggressiveness, thereby maintaining genetic variation in interaction traits within a natural host–pathogen metapopulation.
The impact of genetic differentiation and environmental variation on infection outcomes
While interactions between plant and pathogen genotypes have been a major focus of research since the seminal studies of Flor (1956; Thompson & Burdon, 1992), our results indicate that the direct impact of the environment and genotype-by-environment interactions are also important predictors of infection outcomes. Moreover, the existence of environment-by-environment (E × E) and G × E × E interactions illustrate the need for multifactorial designs to understand variation in pathogen performance and how it might relate to patterns of local adaptation. Interestingly, while Gpath × E interactions were common, the host genotype did not interact with the environment; however, future research in this and other pathosystems involving a range of environmental factors is needed to assess the generality of this phenomenon. Overall, these results provide evidence that genotype × environment interactions are not merely an interesting laboratory phenomenon, but have high ecological relevance in natural systems. By contrast, we did not detect G × G × E interactions, even though our design had enough resolution to detect those. Interestingly, the low amount of variation explained by G × G × E interactions is supported by the few studies currently available in wild host–parasite systems (Laine, 2007; Hall & Ebert, 2012). If this turns out to be a general rule, it has pronounced implications for theoretical modelling, which increasingly assumes the existence of spatially or temporally variable G × G interactions (e.g. Gavrilets & Michalakis, 2008; Mostowy & Engelstädter, 2011).
While many studies only measure a single pathogen trait, our data highlight that the impact of environmental and genetic variation differs strikingly among plant and pathogen life-history traits. In particular, infectivity was not affected by the environment. This mirrors comparable findings in both plant and animal pathosystems that infectivity may be relatively insensitive to genotype-by-environment interactions (Laine, 2007; Duneau et al., 2011). The traditional focus on this qualitative trait (infectivity) may then have relegated genotype-by-environment interactions as minor drivers of host–parasite interactions. By contrast, all quantitative traits were affected by the environment, even though the relevant environmental factor(s) varied among the life-history stages.
The impact of environmental heterogeneity on (co)evolutionary dynamics
Despite the potential for strong gene flow to occur among populations belonging to different habitat types, especially for an aerially-dispersed rust pathogen such as M. lini, we found clear environmentally-driven differentiation in interaction traits among plant and pathogen populations from the two habitat types. Some traits were clearly adaptive for the pathogen, where hill pathogens were more infective and induced less leaf abscission on the hill plants than the bog pathogens. The adaptation of hill pathogens to their local plants may be driven by strong selection pressures due to higher average level of plant resistance in hill populations; by contrast, the low resistance of bog pathogens may preclude strong natural selection. The high leaf drop of the hill plants may have evolved as a means of getting rid of infection that disrupts the plant cuticle and makes plants more vulnerable to water loss, which is likely more of a problem in the drier hill habitat. However, despite spatial consistency in the impact of the environment on the host–parasite interaction, the adaptive nature of specific plant and pathogen responses to local environmental conditions was not always apparent: Why would plants in the bog environment not evolve higher resistance? Why do hill pathogens induce higher resistance and develop more slowly in their native (low) moisture conditions? Why do bog pathogens develop relatively slowly in their local environmental conditions? These results illustrate the notion that coevolutionary dynamics may result in adaptive mismatches, or natural selection – in this case in response to the belowground abiotic and biotic environment – can be evolutionarily constrained (Dargent et al., 2013; Garland Jr, 2014).
Based on previous studies within the same and related pathosystems, we postulate that trade-offs and limited gene flow play a key role in explaining the distinct coevolutionary dynamics among the two habitat types. Carlsson-Granér et al. (1999) demonstrated very low survival of hill plants in the bog habitat in a 2-yr transplant experiment and that reciprocal crosses between bog and hill host lines were far less successful when the bog plant was the maternal parent. This supports the idea that a trade-off between high levels of resistance and survival in the bog habitat, as well as reduced offspring survival, may select against the introgression of resistance genes from hill populations into those growing in bogs and supports the role of the environment as a potential driving force for the large spatial and temporal variation in resistance levels within and among host species (Laine et al., 2011; Tack et al., 2012). Equally, the high level of resistance and leaf abscission of the hill plants in response to the bog pathogen will reduce pathogen gene flow from the bog to the hill habitat, whereas a trade-off between high infectivity levels and spore production (Thrall & Burdon, 2003) may prevent pathogen migration from the hill to the bog habitat. The rapid purging of immigrant pathogen genotypes is further supported by absence of full resistance in local host–parasite interactions, whereas full resistance was detected in a few cross-ecotype inoculations. Overall, these results shed light on how spatial divergence in host resistance and pathogen aggressiveness can persist in the face of high dispersal rates among habitat types that can be as close as several 100 m. Further, a positive relationship between developmental time and pustule spore load may explain the slower development of pathogen strains in their local environment. Given the omnipresence of trade-offs and evolutionary constraints in host–parasite systems (Laine & Barrès, 2013; Susi & Laine, 2013; Bruns et al., 2014), we argue that the apparent inconsistency in adaptive signals across qualitative and quantitative infection traits will be mirrored in the majority of host–parasite systems. This is evidenced by an emerging number of local adaptation studies measuring a range of life-history traits (Lemoine et al., 2012; Tack et al., 2014).
Providing a feedback between ecology and evolution, environmentally-mediated spatial evolutionary divergence in resistance may in turn affect epidemiological dynamics: thus susceptible bog populations sustain higher levels of infection that the more resistant hill populations (Laine et al., 2014).
Belowground drivers of host–parasite dynamics
Studies of evolutionary responses of single species to below-ground heterogeneity – in particular of plants to serpentine and heavy-metal soils – have provided some of the most convincing examples of rapid evolution and local adaptation (Bradshaw, 1952, 1984; Brady et al., 2005). Our study demonstrates that belowground abiotic and biotic heterogeneity may also drive the ecological outcome of aboveground species interactions and thereby exert selection pressures on those interactions. Illustrating the evolutionary potential of pathogens in response to the soil environment, Fones et al. (2010) showed that naturally colonizing bacteria growing on hyperaccumulating Noccea caerulans had a higher zinc tolerance compared with bacteria isolated from nonaccumulating plants, suggesting local adaptation of the bacteria to high metal concentrations. As another example, a recent artificial selection experiment by Bonte et al. (2010) demonstrated that spider mites may adapt to plants infested with either mycorrhizae or nematodes (as compared with control plants) within the relatively short time span of 15 spider mite generations.
Conclusion
Overall, our study highlights that environmental heterogeneity has the potential to shape the outcome of species interactions and spatial divergence in interaction traits despite the potential for frequent dispersal among neighbouring populations. While environmental heterogeneity has previously been identified as a key determinant of species diversity (Stein et al., 2014), we here show that environmental heterogeneity can also maintain variation at the genetic level relevant for species interactions. The high level of environmental heterogeneity in natural settings – as compared with agricultural fields – may then provide at least a partial explanation for the classic observation that genetic variation in natural pathosystems is higher in natural than agricultural systems (Burdon, 1987; Tack et al., 2012). Although we did not detect G × G × E interactions, the general importance of a complex set of gene-by-environment interactions matches the framework of the geographic mosaic of coevolution, which emphasizes that environmental heterogeneity plays a major role in the ecology and evolution of species interactions across spatial scales (Thompson, 2005). Importantly, belowground variation in the abiotic and biotic environment is likely to represent a major but unrecognized ecological and evolutionary force in natural communities and contribute to the long-term maintenance of variation in resistance and pathogenicity.
Supplementary Material
Acknowledgements
We thank Caritta Eliasson for help with the glasshouse experiment and Guillaume Blanchet for advice on the multivariate analysis of the abiotic soil environment. A-L.L. and A.J.M.T. were supported by funding from the Academy of Finland (Grants 250444 and 136393 to A-L.L.). This research was supported by the National Institutes of Health (NIH grant 5RO1 GM074265-01A2).
Footnotes
Supporting Information
Additional supporting information may be found in the online version of this article.
Table S1 Methods and units for quantification of the soil structure and chemistry
Table S2 Experimental inoculation matrix
Table S3 F-values and associated degrees of freedom for the statistical results reported in Table 1
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
References
- Barrus MF. Variation of varieties of beans in their susceptibility to anthracnose. Phytopathology. 1911;1:190–195. [Google Scholar]
- Biffen RH. Mendel’s laws of inheritance and wheat breeding. The Journal of Agricultural Science. 1905;1:4–48. [Google Scholar]
- Bonte D, De Roissart A, Vandegehuchte ML, Ballhorn DJ, Van Leeuwen T, de la Peña E. Local adaptation of aboveground herbivores towards plant phenotypes induced by soil biota. PLoS ONE. 2010;5:e11174. doi: 10.1371/journal.pone.0011174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borcard D, Gillet F, Legendre P. Numerical ecology with R. New York, NY, USA: Springer; 2011. [Google Scholar]
- Bradshaw AD. Populations of Agrostis tenuis resistant to lead and zinc poisoning. Nature. 1952;169:1098–1098. doi: 10.1038/1691098a0. [DOI] [PubMed] [Google Scholar]
- Bradshaw AD. Ecological significance of genetic variation between populations. In: Dirzo R, Sarukhán J, editors. Perspectives in plant population ecology. Sunderland, MA, USA: Sinauer; 1984. pp. 213–228. [Google Scholar]
- Brady KU, Kruckeberg AR, Bradshaw HD., Jr Evolutionary ecology of plant adaptation to serpentine soils. Annual Review of Ecology, Evolution, and Systematics. 2005;36:243–266. [Google Scholar]
- Bruns E, Carson ML, May G. The jack of all trades is master of none: a pathogen’s ability to infect a greater number of host genotypes comes at a cost of delayed reproduction. Evolution. 2014;68:2453–2466. doi: 10.1111/evo.12461. [DOI] [PubMed] [Google Scholar]
- Burdon JJ. Diseases and plant population biology. Cambridge, UK: Cambridge University Press; 1987. [Google Scholar]
- Burdon JJ, Jarosz AM. Temporal variation in the racial structure of flax rust (Melampsora lini) populations growing on natural stands of wild flax (Linum marginale): local versus metapopulation dynamics. Plant Pathology. 1992;41:165–179. [Google Scholar]
- Burdon JJ, Thompson JN. Changed patterns of resistance in a population of Linum marginale attacked by the rust pathogen Melampsora Lini. Journal of Ecology. 1995;83:199–206. [Google Scholar]
- Burdon JJ, Thrall PH, Brown AHD. Resistance and virulence structure in two Linum marginale–Melampsora lini host–pathogen metapopulations with different mating systems. Evolution. 1999;53:704–716. doi: 10.1111/j.1558-5646.1999.tb05365.x. [DOI] [PubMed] [Google Scholar]
- Carlsson-Granér U, Burdon JJ, Thrall PH. Host resistance and pathogen virulence across a plant hybrid zone. Oecologia. 1999;121:339–347. doi: 10.1007/s004420050937. [DOI] [PubMed] [Google Scholar]
- Chamberlain SA, Bronstein JL, Rudgers JA. How context dependent are species interactions? Ecology Letters. 2014;17:881–890. doi: 10.1111/ele.12279. [DOI] [PubMed] [Google Scholar]
- Clarke KR. Non-parametric multivariate analyses of changes in community structure. Australian Journal of Ecology. 1993;18:117–143. [Google Scholar]
- Clarke KR, Warwick RM. Change in marine communities: an approach to statistical analysis and interpretation. Plymouth, UK: PRIMER-E; 2001. [Google Scholar]
- Clausen J, Keck DD, Hiesey WM. Experimental studies on the nature of species. I. Effect of varied environments on western North American plants. Washington DC, USA: Carnegie Institution of Washington Publication; 1940. [Google Scholar]
- Crawley MJ. The R Book. Chichester, UK: John Wiley & Sons Ltd.; 2012. [Google Scholar]
- Culman SW, Bukowski R, Gauch HG, Cadillo-Quiroz H, Buckley DH. T-REX: software for the processing and analysis of T-RFLP data. BMC Bioinformatics. 2009;10:171. doi: 10.1186/1471-2105-10-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dargent F, Scott ME, Hendry AP, Fussmann GF. Experimental elimination of parasites in nature leads to the evolution of increased resistance in hosts. Proceedings of the Royal Society B: Biological Sciences. 2013;280:20132371. doi: 10.1098/rspb.2013.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duneau D, Luijckx P, Ben-Ami F, Laforsch C, Ebert D. Resolving the infection process reveals striking differences in the contribution of environment, genetics and phylogeny to host–parasite interactions. BMC Biology. 2011;9:11. doi: 10.1186/1741-7007-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettema CH, Wardle DA. Spatial soil ecology. Trends in Ecology & Evolution. 2002;17:177–183. [Google Scholar]
- Flor HH. The complementary genic systems in flax and flax rust. Advances in Genetics. 1956;8:29–54. [Google Scholar]
- Fones H, Davis CAR, Rico A, Fang F, Smith JAC, Preston GM. Metal hyperaccumulation armors plants against disease. PLoS Pathog. 2010;6:e1001093. doi: 10.1371/journal.ppat.1001093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford EB. Ecological genetics. London, UK: Chapman & Hall; 1975. [Google Scholar]
- Forde SE, Thompson JN, Bohannan BJM. Adaptation varies through space and time in a coevolving host–parasitoid interaction. Nature. 2004;431:841–844. doi: 10.1038/nature02906. [DOI] [PubMed] [Google Scholar]
- Garland T., Jr Trade-offs. Current Biology. 2014;24:R60–R61. doi: 10.1016/j.cub.2013.11.036. [DOI] [PubMed] [Google Scholar]
- Gavrilets S, Michalakis Y. Effects of environmental heterogeneity on victim–exploiter coevolution. Evolution. 2008;62:3100–3116. doi: 10.1111/j.1558-5646.2008.00513.x. [DOI] [PubMed] [Google Scholar]
- Hall MD, Ebert D. Disentangling the influence of parasite genotype, host genotype and maternal environment on different stages of bacterial infection in Daphnia magna. Proceedings of the Royal Society B: Biological Sciences. 2012;279:3176–3183. doi: 10.1098/rspb.2012.0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz AM, Burdon JJ. Host–pathogen interactions in natural populations of Linum marginale and Melampsora lini. Oecologia. 1992;89:53–61. doi: 10.1007/BF00319015. [DOI] [PubMed] [Google Scholar]
- Laine A-L. Evolution of host resistance: looking for coevolutionary hotspots at small spatial scales. Proceedings of the Royal Society B: Biological Sciences. 2006;273:267–273. doi: 10.1098/rspb.2005.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine A-L. Pathogen fitness components and genotypes differ in their sensitivity to nutrient and temperature variation in a wild plant–pathogen association. Journal of Evolutionary Biology. 2007;20:2371–2378. doi: 10.1111/j.1420-9101.2007.01406.x. [DOI] [PubMed] [Google Scholar]
- Laine A-L. Temperature-mediated patterns of local adaptation in a natural plant–pathogen metapopulation. Ecology Letters. 2008;11:327–337. doi: 10.1111/j.1461-0248.2007.01146.x. [DOI] [PubMed] [Google Scholar]
- Laine AL, Barrès B. Epidemiological and evolutionary consequences of life-history trade-offs in pathogens. Plant Pathology. 2013;62:96–105. [Google Scholar]
- Laine A-L, Burdon JJ, Dodds PN, Thrall PH. Spatial variation in disease resistance: from molecules to metapopulations. Journal of Ecology. 2011;99:96–112. doi: 10.1111/j.1365-2745.2010.01738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine A-L, Burdon JJ, Nemri A, Thrall PH. Host ecotype generates evolutionary and epidemiological divergence across a pathogen metapopulation. Proceedings of the Royal Society B: Biological Sciences. 2014;281:20140522. doi: 10.1098/rspb.2014.0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence GJ. Flax rust from Linum marginale: pathogenicity reactions on the Linum usitatissimum set of differential varieties. Canadian Journal of Botany. 1989;67:3187–3191. [Google Scholar]
- Lawrence GJ, Burdon JJ. Flax rust from Linum marginale: variation in a natural host–pathogen interaction. Canadian Journal of Botany. 1989;67:3192–3198. [Google Scholar]
- Lemoine M, Doligez B, Richner H. On the equivalence of host local adaptation and parasite maladaptation: an experimental test. The American Naturalist. 2012;179:270–281. doi: 10.1086/663699. [DOI] [PubMed] [Google Scholar]
- Linhart YB, Grant MC. Evolutionary significance of local genetic differentiation in plants. Annual Review of Ecology and Systematics. 1996;27:237–277. [Google Scholar]
- Littell RC, Milliken GA, Stroup WW, Wolfinger RD, Schabenberger O. SAS® for Mixed Models. Cary, NC, USA: SAS Institute Inc.; 2006. [Google Scholar]
- Lopez Pascua L, Gandon S, Buckling A. Abiotic heterogeneity drives parasite local adaptation in coevolving bacteria and phages. Journal of Evolutionary Biology. 2012;25:187–195. doi: 10.1111/j.1420-9101.2011.02416.x. [DOI] [PubMed] [Google Scholar]
- McNew GL. The nature, origin, and evolution of parasitism. In: Horsfall JG, Dimond AE, editors. Plant pathology: an advanced treatise. New York, NY, USA: Academic Press; 1960. pp. 19–69. [Google Scholar]
- Mostowy R, Engelstädter J. The impact of environmental change on host–parasite coevolutionary dynamics. Proceedings of the Royal Society B-Biological Sciences. 2011;278:2283–2292. doi: 10.1098/rspb.2010.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neter J, Kutner MH, Nachtsheim CJ, Wasserman W. Applied linear statistical models. Irwin, IL, USA: McGraw–Hill Higher Education; 1996. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. vegan: community ecology package. R package version 2.0–10. 2013 http://CRAN.R-project.org/package=vegan. [Google Scholar]
- Pasteur L, Joubert JF, Chamberland C. Sur le charbon des poules. Comptes Rendus des seances De L’Academie Des Sciences. 1878;87:47–48. [Google Scholar]
- Poisot T, Thrall PH, Hochberg ME. Trophic network structure emerges through antagonistic coevolution in temporally varying environments. Proceedings of the Royal Society B: Biological Sciences. 2012;279:299–308. doi: 10.1098/rspb.2011.0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. [Google Scholar]
- Ramette A. Quantitative community fingerprinting methods for estimating the abundance of operational taxonomic units in natural microbial communities. Applied and Environmental Microbiology. 2009;75:2495–2505. doi: 10.1128/AEM.02409-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoener TW. The newest synthesis: understanding the interplay of evolutionary and ecological dynamics. Science. 2011;331:426–429. doi: 10.1126/science.1193954. [DOI] [PubMed] [Google Scholar]
- Singh BK, Nazaries L, Munro S, Anderson IC, Campbell CD. Use of multiplex terminal restriction fragment length polymorphism for rapid and simultaneous analysis of dfifferent components of the soil microbial community? Applied and Environmental Microbiology. 2006;72:7278–7285. doi: 10.1128/AEM.00510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A, Gerstner K, Kreft H. Environmental heterogeneity as a universal driver of species richness across taxa, biomes and spatial scales. Ecology Letters. 2014;17:866–880. doi: 10.1111/ele.12277. [DOI] [PubMed] [Google Scholar]
- Susi H, Laine A-L. Pathogen life-history trade-offs revealed in allopatry. Evolution. 2013;67:3362–3370. doi: 10.1111/evo.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tack AJM, Horns F, Laine A-L. The impact of spatial scale and habitat configuration on patterns of trait variation and local adaptation in a wild plant parasite. Evolution. 2014;68:176–189. doi: 10.1111/evo.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tack AJM, Thrall PH, Barrett LG, Burdon JJ, Laine A-L. Variation in infectivity and aggressiveness in space and time in wild host–pathogen systems: causes and consequences. Journal of Evolutionary Biology. 2012;25:1918–1936. doi: 10.1111/j.1420-9101.2012.02588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R, Ruiz LV, Vasco-Palacios AM, Thu PQ, Suija A, et al. Global diversity and geography of soil fungi. Science. 2014;346:1256688. doi: 10.1126/science.1256688. [DOI] [PubMed] [Google Scholar]
- Tellier A, Brown JKM. Spatial heterogeneity, frequency-dependent selection and polymorphism in host–parasite interactions. BMC Evolutionary Biology. 2011;11:319. doi: 10.1186/1471-2148-11-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JN. The geographic mosaic of coevolution. Chicago, IL, USA: University of Chicago Press; 2005. [Google Scholar]
- Thompson JN. Relentless evolution. Chicago, IL, USA: University Of Chicago Press; 2013. [Google Scholar]
- Thompson JN, Burdon JJ. Gene-for-gene coevolution between plants and parasites. Nature. 1992;360:121–125. [Google Scholar]
- Thrall PH, Burdon JJ. Evolution of virulence in a plant host–pathogen metapopulation. Science. 2003;299:1735–1737. doi: 10.1126/science.1080070. [DOI] [PubMed] [Google Scholar]
- Thrall PH, Burdon JJ, Young A. Variation in resistance and virulence among demes of a plant host–pathogen metapopulation. Journal of Ecology. 2001;89:736–748. [Google Scholar]
- Thrall PH, Laine A-L, Ravensdale M, Nemri A, Dodds PN, Barrett LG, Burdon JJ. Rapid genetic change underpins antagonistic coevolution in a natural host–pathogen metapopulation. Ecology Letters. 2012;15:425–435. doi: 10.1111/j.1461-0248.2012.01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turesson G. The genotypical response of the plant species to the habitat. Hereditas. 1922;3:211–350. [Google Scholar]
- Van Dorst J, Bissett A, Palmer AS, Brown M, Snape I, Stark JS, Raymond B, McKinlay J, Ji M, Winsley T, et al. Community fingerprinting in a sequencing world. FEMS Microbiology Ecology. 2014;89:316–330. doi: 10.1111/1574-6941.12308. [DOI] [PubMed] [Google Scholar]
- Vogwill T, Fenton A, Buckling A, Hochberg ME, Brockhurst MA. Source populations act as coevolutionary pacemakers in experimental selection mosaics containing hotspots and coldspots. The American Naturalist. 2009;173:E171–E176. doi: 10.1086/597374. [DOI] [PubMed] [Google Scholar]
- Wolinska J, King KC. Environment can alter selection in host–parasite interactions. Trends in Parasitology. 2009;25:236–244. doi: 10.1016/j.pt.2009.02.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.