Abstract
Maf1 was first identified in yeast and studies in metazoans have primarily focused on examining its role in the repression of RNA pol III-dependent transcription. Recent work has revealed a novel and conserved function for Maf1 in the maintenance of intracellular lipid pools in C. elegans, mice, and cancer cell lines. Although additional Maf1 targets are likely, they have not been identified, and these recent findings begin to define specific activities for Maf1 in multicellular organisms beyond the regulation of RNA polymerase III transcription and suggest Maf1 plays a more diverse role in organismal physiology. We will discuss these newly defined physiological roles of Maf1, that point to its placement as an important new player in lipid metabolism with implications in human metabolic diseases such as obesity and cancer, which display prominent defects in lipid homeostasis.
Keywords: RNA polymerase III, transcription, MAF1, MAFR-1, lipid metabolism, FASN, Acc1, Caenorhabditis elegans, cancer, stress, nutrient-signaling, reproduction, Drosophila melanogaster, mouse, human
Graphical abstract
Introduction
One mechanism utilized by organisms in response to cytotoxic stress, such as DNA damage and starvation, is regulation of the transcriptional activities of RNA polymerase (pol) I, II, and III [1, 2]. RNA pol I is involved in ribosomal RNA (rRNA) transcription and regulation of its activity is complex; but is in part controlled at transcription initiation and elongation and is also sensitive to rDNA copy number variation [3, 4]. RNA pol II produces messenger RNAs (mRNAs) and many noncoding RNAs. One level of regulation is through changes in the phosphorylation status of its C-terminal repeat domain (CTD), a tandem repeat of the heptapeptide sequence YSPTSPS [5], which alters its binding affinity to a variety of nuclear factors [6, 7]. RNA pol III generates rRNA, transfer RNAs (tRNAs), 5S rRNA and the remaining complement of small noncoding RNAs [8]. RNA pol III transcripts comprise essential components of the cellular protein synthesis machinery, which must be synthesized in high-copy to fulfill the cell’s biosynthetic demand during growth. At the same time, RNA pol III activity must be amenable to negative regulation in order to impede growth during unfavorable conditions. This level of regulatory precision is mediated by the activities of multiple factors that when deregulated, influence cell cycle progression and cellular transformation; [9] they include: c-Myc [10], tumor suppressors pRB (retinoblastoma protein) [11] and p53 [12].
Not all species have clear orthologs of these well-studied regulatory factors of RNA pol III [13] mentioned above. Saccharomyces cerevisiae, for example, utilizes other well-conserved transcriptional regulators, like Maf1, to regulate RNA pol III and coordinate cellular growth and responses to stress [13]. Maf1 is a phosphoprotein that has been characterized in Saccharomyces cerevisiae [14], Drosophila melanogaster [15], Caenorhabditis elegans [16], and mammals [17] and shown to integrate stress-signaling and cellular growth and proliferation pathways by regulating RNA polymerase III-dependent transcription. The yeast, worm, mouse, and human research communities use different nomenclature for genes and proteins. For clarity, we use “Maf1” when referring to Maf1 in general terms. Specific references to C. elegans, RNA and proteins are written as mafr-1 and MAFR-1, respectively.
Maf1 may have evolved as an early mediator of cellular growth while other factors surfaced later to enhance regulation in more complex eukaryotes that require more sophisticated layers of control. The presence of Maf1 across species is suggestive of its necessity to modulate essential cellular functions, which facilitate its persistence to withstand evolutionary pressures. The study of Maf1 across multiple species has uncovered insightful information regarding Maf1 function (Table 1); but also raised new questions. This piece will summarize existing and newly identified roles that Maf1 plays in cellular homeostasis – across organisms of diverse complexity - and will give insights into the physiological impacts that Maf1 activity has in regards to nutrient signaling and cellular growth pathways.
Table 1.
Summary of experimental findings of altering Maf1 across species.
| Manipulation | Organism | Tissue/ Type | Phenotype | Ref. |
|---|---|---|---|---|
| C. elegans | All mafr-1
expressing tissues |
RNA pol II, III Lipid homeostasis Reproduction |
16 | |
| Overexpression |
|
|||
| RNA pol I, II, III | 23, 38 | |||
| Glioblastoma |
|
|||
| Human cell |
Anchorage independent growth |
38 | ||
|
|
||||
| Liver | Triglyceride levels | 23 | ||
|
| ||||
| Human cell |
Glioblastoma | RNA pol I, II, III |
38 | |
| Cellular morphology | ||||
| RNA pol II, III | ||||
| RNAi | Ubiquitous |
|
||
| C. elegans | Lipid homeostasis | 16 | ||
|
|
||||
| Intestine | Reproduction | |||
|
|
||||
| Drosophila | Fat body | Organismal growth | 15, 42 | |
|
| ||||
| Knockout | Mouse | Ubiquitous | Food intake Body weight Reproduction |
41 |
Cellular growth is an energy intensive process and under favorable conditions non-transformed cells preferentially utilize carbohydrate metabolism for ATP generation [18]. When dietary sources of sugar become scarce, cells switch to lipolysis of cellular lipid stores and generate energy through mitochondrial and peroxisomal beta-oxidation[19]. In times of nutrient excess, surplus carbohydrates can be utilized to synthesize cellular lipids through lipogenesis [5], which when left unchecked, can lead to obesity, diabetes, and other metabolic syndromes [20]. Cellular growth requires lipogenesis pathways, not only as a maintenance component of energy homeostasis, but as a vital system for the synthesis of cellular membranes and signaling molecules such as steroid hormones and prostaglandins [21]. Whereas cancer cells have upregulated lipid synthesis to drive deregulated cell division, mutation of lipid biosynthesis genes has been shown to severely impair growth and development in multiple species [22].
Findings by Palian et. al [23] and Khanna et. al [16] have highlighted a non-canonical role for Maf1 in the regulation of intracellular lipid stores. These two papers demonstrated that in addition to RNA pol III-dependent regulation of cellular biosynthetic capacity, Maf1 also regulates select RNA pol II genes, such as Acc1 and Fasn, which encode enzymes for the first two steps in de novo lipogenesis (discussed below). While some of the molecular mechanisms that regulate Maf1 function have been identified, it is clear that other post-translational modifications of Maf1 that influence cellular functions remain to be uncovered. However, the extent of diversity in modifications to specific residues of the Maf1 protein, which are that are essential for Maf1 functions, remains to be elucidated.
Maf1 structure, function, and regulation
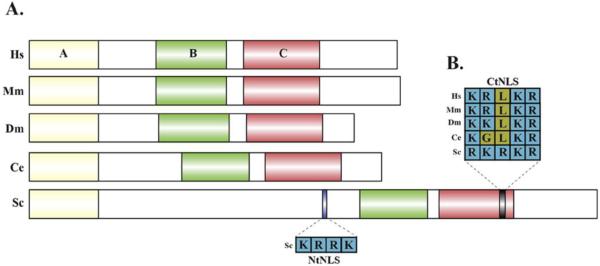
A large body of work has characterized the role of yeast Maf1 as a repressor of RNA pol III transcription in a TFIIIB (Transcription Factor for polymerase III B)-dependent manner [24]. Subsequent research has demonstrated biochemical evidence of human Maf1 interaction with RPAC2 (alpha like subunit of RNA pol III), RPC1 (the largest subunit of RNA pol III) and Brf1 (subunit of TFIIIB)[17]. It is believed that this direct interaction with the RNA pol III machinery facilitates the ability for Maf1 to regulate transcriptional output. Maf1 proteins in all species share three regions of high similarity, named A, B, and C box. The domains defined in these regions are unique to Maf1 and do not contain sequence motifs of known function. By using mutant alleles of human Maf1, it was shown that the A box is required for Maf1 to interact with the large RNA pol III subunits, and the B box is required for interaction with Brf1 [17].
Across all species examined, Maf1 has been shown to localize to both the cytoplasm and nuclear cellular compartments. As the canonical role of Maf1 is as a negative regulator of transcription, it is expected that its primary function will be in the nucleus. This suggests that nuclear/cytoplasmic transport is at least one of the mechanisms of regulating Maf1 activity. Yeast Maf1 contains two nuclear localization sequences (NLS): one near the N-terminal (NtNLS) end and another near the C-terminal (CtNLS) end of the protein [24]. Although there is no predicted NLS in Maf1 of higher eukaryotes, the yeast CtNLS shows high conservation across different species (Fig.1). The crystal structure of human Maf1 revealed that the N- and C-terminal domains of Maf1 (the location of the two NLS sequences in yeast) are exposed at the surface of the Maf1 globular protein structure [25]. The linker regions that separate the Maf1 A and B boxes and the C-terminal acidic tail were missing in the RNA pol III-Maf1 complex crystal structure, which suggests that these two regions do not play a role in Maf1 binding to RNA pol III [8]. Further analysis of the functional relevance of these individual regions that were missing from the crystal structure are of great interest to the field and will help to further refine the functional domains that influence Maf1 activity.
Fig. 1. Maf1 protein structure.
A. Maf1 protein sequences in different species share regions of high similarity - A, B and C box regions. Putative and defined NLS sequences are labeled. B. Alignment of CtNLS sequences. Hs- Homo sapiens, Mm- Mus musculus, Dm-Drosophila melanogaster, Ce- Caenorhabditis elegans, Sc- Saccharomyces cerevisiae.
Maf1 has been shown to repress RNA pol III transcription by blocking the assembly of the RNA pol III complex in response to DNA damage, nutrient limitation, oxidative stress and defects in the secretory pathway [14]. Different stress conditions have been shown to alter the phosphorylation status of Maf1. In yeast, Roberts et al. [26] and Oficjalska-Pham et al. [27] provided biochemical evidence that Maf1 is phosphorylated under favorable conditions and is rapidly dephosphorylated under stressful growth environments[14]. In contrast, a phosphorylated form of Maf1 has been shown to localize to the nucleus of HeLa cells where it retains the ability to suppress RNA pol III target genes [28]. These findings should be taken with caution, as HeLa cells are a transformed cell line derived from a cervical cancer biopsy and may not reflect normal physiological regulation of Maf1 [29].
Post-translational modifications (PTMs), such as phosphorylation, are established mechanisms of regulating protein localization [30]. The phosphorylation status of residues between the A and B boxes of Maf1 have been linked to Maf1 distribution between the nuclear and cytoplasmic compartments [31] but not all of these sites are conserved (Fig. 1 and Table 2). Early work in yeast alluded to a mechanism by which the regulation of Maf1 activity was linked to its phosphorylation status; the dephosphorylated form would be active in repressing RNA pol III-dependent transcription[26]. Using immunoprecipitation Roberts et al. reported that Maf1 dephosphorylation greatly increases its binding affinity for the RNA pol III subunit Rpc82 when under stress induced by exposure to Chlorpromazine (membrane stretching), methyl methanesulfonate (DNA damage), or nutrient limitation. However, conflicting studies have also demonstrated that phosphorylated Maf1 can be found in the nucleus of HeLa cells. Moreover, this phosphorylated Maf1 is localized at the promoters of tRNA and 5s rRNA genes [28]. Taken together, the studies linking phosphorylation, Maf1 localization, and activity are contradictory and require further attention.
Table 2.
Maf1 protein with conserved A, B, and C box regions across different species.
| Species | Protein Size |
Box |
||
|---|---|---|---|---|
| A | B | C | ||
| Human | 257 | 1 - 49 | 89 - 138 | 150 - 203 |
| Mouse | 259 | 1 - 49 | 89 - 138 | 150 - 203 |
| Drosophila | 227 | 1 - 49 | 91 - 140 | 152 - 205 |
| C. elegans | 246 | 1 - 49 | 107 - 154 | 165 - 218 |
| Yeast | 396 | 1 - 49 | 231 - 278 | 286 - 338 |
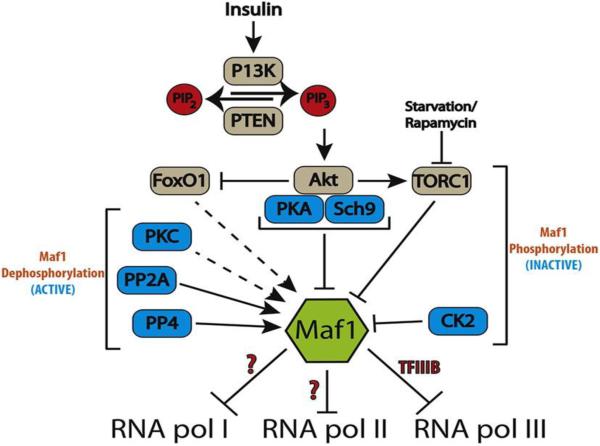
TORC1 (target of rapamycin complex 1) has been shown to phosphorylate Maf1 in yeast [32], drosophila [15] and humans [33]. However, the specific sites of TORC1 phosphorylation have only been identified for human Maf1: Ser 60, Ser 68, Ser 75. The TOR signaling pathway has been shown to inactivate Maf1 in yeast cells [32] as well as in IMR90hTert, a human lung cell line [33]. However, rapamycin treatment in HeLa cells did not result in any changes in Maf1 localization [28], suggesting the regulatory role of Maf1 by TOR is aberrant in malignant cells. Nevertheless, these studies are a clear example of the complex role that Maf1 plays in the regulation of RNA pol III transcription and clearly reveals the sensitivity of Maf1-dependent activity to both cell type and cellular transformation status. Moir et al. identified cAMP-dependent protein kinase A (PKA) sites near the NtNLS of yeast Maf1 that are phosphorylated under normal conditions and lead to sequestration in the cytoplasm [34]. However, the canonical PKA sites identified near the NtNLS in yeast are not apparent in worms, flies, mice, or humans. In addition, Protein Kinase C1 (Pkc1) activity was identified as crucial for yeast Maf1 dephosphorylation under nutrient deprivation [26], but a role for this kinase in other species has not been investigated. Caesin Kinase 2 (CK2) stimulates RNA pol III transcription via phosphorylation of TFIIIB (CK2 forms a stable complex with TFIIIB) and has also been reported to directly phosphorylate Maf1 in yeast and mammalian cell culture [35]. There are contradicting reports however on whether CK2 phosphorylation of Maf1 is required for derepression of RNA pol III transcription in yeast[35, 36] and a role for CK2 has yet to be investigated in other species. Taken together, phosphorylation clearly plays an important regulatory role in Maf1 function, but the details underlying this control need further refinement (Fig. 2).
Fig. 2. Model of Maf1 signaling for inhibition of RNA pol I, II, and III.
Maf1 integrates nutrient and stress signals to growth by regulating RNA pol III activity. Insulin signaling activates PI3K, which in turn activates members of the AGC kinase gene family. PKA (Protein kinase A) and Sch9 (Serine/threonine-protein kinase) phosphorylate Maf1 to inhibit Maf1 activity. Nutrient deprivation or treatment with Rapamycin inhibits TORC1, which leads to the activation of Maf1. PKC (Protein kinase C) is required for Maf1 dephosphorylation by an unknown mechanism. PP4 (Protein phosphates 4) and PP2A (Protein phosphates 2A) can activate Maf1 by facilitating its dephosphorylating. FoxO1 transcriptionally regulates Maf1 (only observed in C. elegans) and post-translationally by an unknown mechanism. Regulation in mammals is illustrated in grey and cases only seen in yeast are colored blue. Dotted lines and “?” denote documented regulation of Maf1 that occur through unknown mechanisms.
SUMOylation has recently been shown to alter human Maf1 activity without changing Maf1 subcellular localization [37], indicating that different residues and modifications can regulate Maf1 activity and localization. But more importantly, Maf1 activity and subcellular localization are distinctly regulated. An investigation of whether mutating residues that facilitate Maf1 SUMOylation can alter the ability of Maf1 to respond to stress is still needed. While these studies on the impact of post-translational modifications of Maf1 and its influence on activity are compelling and informative, the totality of PTMs on Maf1 remains to be uncovered and is of critical importance to the field.
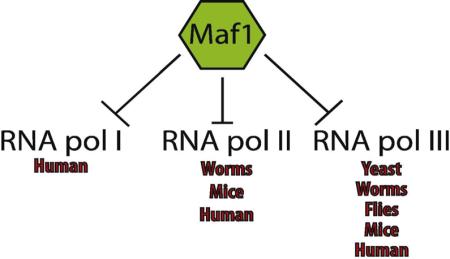
Maf1 can regulate the expression of targets of all three RNA polymerase
Recently, three studies have documented important roles for Maf1 beyond the negative regulation of RNA pol III transcription [16, 23, 38]. These non-canonical roles of Maf1 are just beginning to be explored, but reveal that Maf1 plays other important functions in the maintenance of cellular and organismal homeostasis. Johnson et al. [38] discovered that altered levels of human Maf1 can affect the activity of all the three RNA polymerases. These effects were mediated partially through the ability of Maf1 to suppress transcription of TATA binding protein (TBP) by blocking the binding of Elk-1 to the TBP promoter in glioblastoma cell lines [38]. TBP is an important transcription factor needed by all the RNA polymerases in order to form an active transcription complex, which is considered to be the rate-limiting step in transcription initiation [39]. TBP expression levels are crucial for the maintenance of cell proliferation, and deregulated TBP expression can promote oncogenesis [40]. In C. elegans, Maf1 levels can also influence TBP expression in addition to canonical RNA pol III targets [16]. In contrast, a whole-body knockout of Maf1 in mice showed no significant changes in the steady state transcript levels of TBP in liver [41]. At the present time we can only speculate about the reasons for the differences in these results. One possible explanation could be due to the partial reduction of Maf1 in cell culture and C. elegans versus the complete ablation of Maf1 in mice. The methodological difference might be significant and further supports the notion that cells/organisms are sensitive to even small changes in Maf1 levels; 50% less Maf1 and single copy overexpression of Maf1 in worms result in opposing physiological responses. Secondly, there are clear examples of cell non-autonomous effects of Maf1 [15, 16] but we are only beginning to appreciate, in which tissues Maf1 is required and which tissues are responsive to changes in Maf1 levels. Lastly, two studies investigated the role of Maf1 in Drosophila melanogaster [15, 42] and confirmed its ability to regulate RNA pol III transcription, these reports did not directly test for any RNA pol I or RNA pol II transcriptional targets. These questions are of particular interest, as the differences in Maf1 function across species could reveal how and when Maf1 evolved to regulate all RNA polymerases, which will then provide mechanistic insight toward regulation of Maf1 activity.
The finding that Maf1 can regulate all RNA polymerases was surprising and although there is evidence for the necessity of the A and B-box regions of Maf1 to repress RNA pol III transcription, it remains to be seen whether this region of Maf1 also regulates the newly reported RNA pol II-dependent functions [16, 23]. As such, the underlying mechanisms by which Maf1 can influence RNA pol I and RNA pol II activities requires further investigation. Maf1 has been found on chromatin at the promoters of RNA pol II genes [23, 43]; however, the spectrum and diversity of Maf1 targets has yet to be determined. Microarray analysis in C. elegans with altered levels of Maf1 display deregulated expression of genes involved in a variety of essential cellular pathways, including protein synthesis, growth, stress response, reproduction, and development [16]. Although RNAseq of epidydymal white adipose tissue from the Maf1 knockout mice showed no significant changes in the RNA pol II transciptome 41] an exploration of the importance of Maf1 in all tissues will be of great importance to understand the diverse Maf1 function. Further studies of the impact Maf1 has on gene expression under nutrient deprivation, DNA damage, or oxidative stress will further help identify potential direct targets of Maf1 and perhaps reveal mechanisms of target selection.
Strikingly, the Maf1 protein does not contain any known DNA binding motifs. As such, the molecular basis for Maf1 recruitment to chromatin remains a mystery, but the absence of a clear DNA binding domain could possibly explain why Maf1 overexpression does not repress all RNA pol III-dependent transcripts, as seen in yeast, C. elegans and mammalian cell culture [16, 38, 44]. Regardless of whether Maf1 associates with DNA directly through a novel domain or if its recruitment is facilitated by other transcriptional mediators, these studies reveal the role of Maf1 in the regulation of metazoan RNA biology to be more complex than initially expected.
Maf1 and metabolic homeostasis
Two studies from the Curran and Johnson laboratories discovered the capacity of Maf1 to influence lipid homeostasis in C. elegans and human cells, respectively [16, 23] (Table 1). This regulation is in part accomplished by the capacity of Maf1 to negatively regulate the expression of the lipid biosynthesis genes Fasn and Acc1. Mechanistically, Palian et al. discovered Maf1 occupancy at the Fasn promoter, which functionally inhibited Srebp1-mediated transcription at that locus [23]. This study also revealed that reducing Maf1 levels in Huh-7 cells increased intracellular lipids and that Maf1 levels in mouse liver are repressed by dietary carbohydrates. This finding functionally links Maf1 levels with lipogenesis from excess sugars. Importantly, studies of the Maf1 knockout mouse revealed a significant increase in the levels of newly synthesized palmitate in Maf1(−/−) livers [41], which suggests the negative regulation of de novo lipogenesis by Maf1 is functionally conserved. However, Acc1 and Fasn transcripts were unaltered, and surprisingly the Maf1(−/−) animals were phenotypically lean. One interesting difference between these studies is that while the Maf1(−/−) mouse is constitutively lacking Maf1, the Maf1 RNAi studies in worms are conducted on adolescent animals; thus Maf1 levels are maintained at normal levels during embryogenesis. Further study of the temporal requirements of Maf1 will be highly impactful.
In C. elegans, altering MAFR-1 levels in the whole animal could reciprocally affect lipid metabolism; increasing Maf1 levels lead to a reduction of lipids while Maf1 RNAi increased stored lipids. Importantly, this lipid homeostasis function of Maf1 was independent of its canonical regulation of RNA pol III-dependent transcription [16]. It is intriguing to see that overexpression of Maf1 can protect against diet-induced obesity in C. elegans and mice [16, 23], and although more research is needed, it appears that this protection is correlated to the degree of overexpression. Despite the fact that Maf1 levels are clearly linked to lipid homeostasis, it still remains to be seen whether other cell non-autonomous effects of Maf1 exist.
The identification of Maf1 as an inhibitor of fatty acid synthesis and regulator of intracellular lipid abundance opens up the exciting possibility of exploring Maf1 as a target for new therapeutic strategies in order to study the incidence and progression of obesity, diabetes, non-alcoholic fatty liver disease, and other pathologies that feature deregulated lipid homeostasis. Beyond its importance in metabolic diseases, Maf1 has emerged as a significant regulator that interlinks lipid homeostasis and cellular growth. This highlights Maf1 as an interesting new target to look at in the progression of cancer where both of these processes are deregulated [23].
Maf1 in cell cycle, cancer and reproduction
Maf1 clearly functions as a transcriptional repressor that can inhibit cell proliferation [23] and lipogenesis [16, 23], but other other potential roles of Maf1 have not been fully explored in metazoans. Maf1 has become an emerging player in the progression of cancer and oncogenic transformation as its expression has been shown to induce changes in cell morphology and can influence anchorage-independent growth [23, 38]. In addition to suppressing cellular growth by inhibiting RNA pol III and TBP, as discussed above, Maf1 inhibits de-novo lipid synthesis, an important process that supports cancer progression. Taken together, these findings point to a potential role for Maf1 as a tumor suppressor. However, unlike other tumor suppressors, such as pRB, p53 and PTEN (phosphatase and tensin homolog) the molecular mechanisms that control Maf1 activity are largely unknown. PTEN is a tumor suppressor that inhibits the transcriptional activity of RNA pol III [45]. Mutations in PTEN have been identified in multiple human cancers and are linked to increased PI3K signaling and activation of AKT [46]. The targets of AKT are established mediators that are important for tumor cell growth and survival [47]. Importantly, Maf1 has recently emerged as a downstream target of PTEN and PI3K signaling[16, 23]. PTEN-mediated positive regulation of Maf1 appears to be important checkpoint for a variety of cell lines in controlling cell proliferation. Given the intersection of Maf1 with the insulin/IGFI pathway and its impact on growth, metabolism, and cellular status, it is of particular interest to investigate the role of Maf1, whether similar or different, in PTEN(+) cancer cells, as well as aberrant Maf1 activity in cancer progression and specificity. Moreover, in light of recent evidence supporting the idea that Maf1 functions as a tumor suppressor, it would be useful to examine the frequency of Maf1 mutations across a broader spectrum of cancer types.
MAFR-1 overexpression in C. elegans results in a significant reduction in reproductive capacity through a mechanism independent of its capacity to regulate RNA pol III-dependent transcription (Table 1). Surprisingly, increased Maf1 did not impact embryogenesis, as all laid eggs hatched; there were simply fewer progeny produced. Similar to the role of C. elegans MAFR-1 in lipid homeostasis, levels of MAFR-1 were inversely correlated with the expression of the vitellogenin genes, which function in the transport of lipids from the intestine (where they are stored) to the germline (where they fuel oocyte development) [48]. Increased Maf1 expression reduced vitellogenesis, which decreased oocyte maturation [16]. Importantly, these studies revealed the cell non-autonomous role that Maf1 plays in an intact organism. The increased expression of Maf1 – specifically in the intestine – was causal for reproductive decline, which points to the necessity of investigating the tissue-specific differences in Maf1 activity and regulation in mammals. Interestingly, ablation of Maf1 in mice also results reduced fecundity [41] by a currently unknown mechanism. These findings raise an intriguing question of whether Maf1 could play a role in mammalian reproductive diseases, including polycystic ovarian syndrome (PCOS), premature ovarian aging (POA) and testicular dysgenesis (TDS), which are also influenced by metabolism [49-51]. Further examination of mechanisms of the effect of Maf1 on vertebrate reproduction deserves particular attention.
Pathways that regulate Maf1 expression
Recent studies of Maf1 utilize reduction of function models, but a true null mutant has yet to be characterized. In C. elegans, Maf1 transcript and protein levels are highest in embryos [16], but its role in early development has not been studied. Taken together, it will be interesting to examine changes in development and adult physiology in Maf1 knockout models in other systems as well. Although previous studies have demonstrated effects of ectopically modulating Maf1 levels in different model systems, there is a dearth of data to shed light on the regulation of steady state levels of endogenous Maf1 protein, at any level of expression. Recent studies using C. elegans and mammalian cells have identified FoxO1, a component of the insulin signaling pathway, as an important regulator of Maf1 protein levels. This regulation by DAF-16/FoxO is in part at the level of transcription in C. elegans [16], but appears to be regulated specifically at the protein level in mammalian cells [23]. These differences are intriguing and perhaps reveal important differences in the models used for study, which need to be taken into consideration. Regardless, these studies identify FoxO1 as a potential upstream regulator of Maf1 in both mammalian cell lines and multicellular organisms.
DAF-16/FoxO is the major downstream effector of insulin signaling [52]. In C. elegans, regulation by this pathway is particularly important, as loss of wither daf-16/Foxo or daf-18/PTEN (DAF-16/FoxO regulator), directly impact the capacity of MAFR-1 to modulate levels of stored lipids, in part by altering MAFR-1 levels [16]. The intersection of Maf1 with the insulin signaling pathway has also been documented in D. melanogaster, where Maf1 has been shown to cell non-autonomously coordinate with the insulin-like signaling pathway in order to regulate organismal growth when raised on nutrient-rich conditions[15] (Table 1). Specifically, loss of Maf1 in the Drosophila fat body induces the secretion of insulin-like peptides from the brain to promote growth of peripheral tissues [15]. Similar findings were reported in mice that were fed a high-fat diet, which led to reduced steady-state levels of Maf1 protein [23]. The mechanisms underlying the response of Maf1 protein to changes in available nutrients requires further investigation. Collectively, these studies highlight that in addition to the nutrient signaling pathways that influence Maf1 phosphorylation, there exists other nutrient responsive pathways, like the insulin/IGF-I pathway, that can alter levels of Maf1 protein. It remains to be determined if this regulation occurs in a cell autonomous or non-autonomous manner, and to what extent the effects that Maf1 imparts on RNA pol III activity, growth, and reproduction are independent or a part of established insulin/IGF-I signaling mechanisms.
Future perspectives
The Maf1 research field is relatively new, as the initial discovery of Maf1 was reported in 1997 - less than 20 years ago. Recent studies have significantly accelerated our understanding about the importance of Maf1 on cellular and organismal function. However, before we can complete the puzzle and reveal a comprehensive picture of the functional capacity of Maf1 in cellular and organismal physiology, we need to find all the pieces (Fig. 3)
Fig. 3. Future perspectives on Maf1.
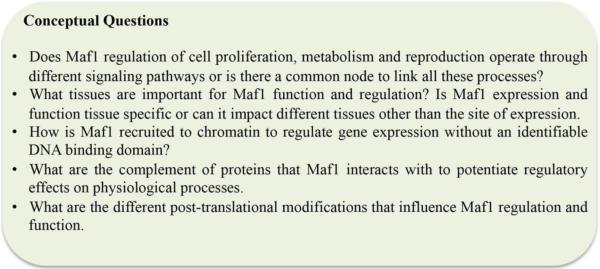
The multitude of roles for Maf1 in organismal physiology are only starting to be fully uncovered. The figure identifies key questions that will significantly improve our understanding of the diversity of Maf1 functions at the molecular level and organismal levels.
Organismal pieces
Comparative biology approach to explore the similarities and differences in Maf1 function across species. Are there life periods – from conception to death - where Maf1 function is more critical than others? What other physiological factors are influenced by Maf1?
Tissue/Cell pieces
In what organ(s) is Maf1 most influential? Is the regulation of each RNA polymerase by Maf1 variable in different tissues? From studies in flies, what is the fat derived signal that originates in the fat body and signals the brain to regulate insulin signaling? Is this signal also present in other metazoans? To that end, what are the cell autonomous and cell-non autonomous impacts of Maf1 activity?
Regulatory pieces
Multiple studies, across different model systems, have identified a role for nutrient signaling in the control of Maf1 function. Specifically, TOR and insulin signaling have been shown to have the largest impacts, albeit to varying degrees. It has yet to be determined: how, where, and when this regulation is of critical importance. Moreover, an investigation of the impact of other signaling pathways on Maf1 activity has not been exhausted. Although DAF-16/FoxO appears to regulate Maf1 transcript levels in C. elegans [16]; this regulation is not essential, as genetic ablation of daf-16/FoxO null mutants still express Maf1 at about 50% of wild type levels. As such the transcription factors that drive Maf1 expression need identification.
Molecular pieces
To what extent is Maf1 post-translationally modified and what enzymes are responsible for these modifications? In addition, how does decorating specific residues on Maf1 alter stability, localization, target selection and/or function. Evidence supports the idea that Maf1 does not act alone, and aside from factors that regulate target selection, the identification of Maf1-associated proteins that mediate its subcellular localization and activity are needed; perhaps there are protein factors that negatively regulate this negative regulator. Future structure/function studies on the Maf1 protein will help to identify domains that are either important for post-translational modifications, regulating its subcellular localization, or that are required for association with other components that regulate the activities of RNA pols I, II, and III.
Our capacity to address these fascinating questions will require the coordinated use of multiple biological models. In addition, as new roles for Maf1 undoubtedly emerge, collaborations among researchers with expertise across diverse disciplines will be essential.
Highlights.
Maf1 regulate de novo lipognesis
Species specific effects of altering Maf1 levels
Future perspectives of Maf1 research
Acknowledgements
This work was supported by the American Federation for Aging Research; the Ellison Medical Foundation; National Institutes of Health grant R01GM109028 and American Heart Association grant 14GRNT20380731 to SPC. We thank D. Johnson for vibrant discussions, Jacqueline Lo and H. Dalton for careful review of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ghavidel A, Schultz MC. TATA binding protein-associated CK2 transduces DNA damage signals to the RNA polymerase III transcriptional machinery. Cell. 2001;106:575–84. doi: 10.1016/s0092-8674(01)00473-1. [DOI] [PubMed] [Google Scholar]
- [2].White RJ. RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol. 2005;6:69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- [3].Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- [4].Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P. RNA polymerase I structure and transcription regulation. Nature. 2013;502:650–5. doi: 10.1038/nature12712. [DOI] [PubMed] [Google Scholar]
- [5].Hellerstein MK. No common energy currency: de novo lipogenesis as the road less traveled. Am J Clin Nutr. 2001;74:707–8. doi: 10.1093/ajcn/74.6.707. [DOI] [PubMed] [Google Scholar]
- [6].Hirose Y, Ohkuma Y. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J Biochem. 2007;141:601–8. doi: 10.1093/jb/mvm090. [DOI] [PubMed] [Google Scholar]
- [7].Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–36. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- [8].Boguta M. Maf1, a general negative regulator of RNA polymerase III in yeast. Biochim Biophys Acta. 2013;1829:376–84. doi: 10.1016/j.bbagrm.2012.11.004. [DOI] [PubMed] [Google Scholar]
- [9].White RJ. RNA polymerase III transcription and cancer. Oncogene. 2004;23:3208–16. doi: 10.1038/sj.onc.1207547. [DOI] [PubMed] [Google Scholar]
- [10].Poortinga G, Hannan KM, Snelling H, Walkley CR, Jenkins A, Sharkey K, et al. MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J. 2004;23:3325–35. doi: 10.1038/sj.emboj.7600335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Woitach JT, Zhang M, Niu CH, Thorgeirsson SS. A retinoblastoma-binding protein that affects cell-cycle control and confers transforming ability. Nat Genet. 1998;19:371–4. doi: 10.1038/1258. [DOI] [PubMed] [Google Scholar]
- [12].Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–58. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Willis IM, Moir RD. Integration of nutritional and stress signaling pathways by Maf1. Trends Biochem Sci. 2007;32:51–3. doi: 10.1016/j.tibs.2006.12.001. [DOI] [PubMed] [Google Scholar]
- [14].Upadhya R, Lee J, Willis IM. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell. 2002;10:1489–94. doi: 10.1016/s1097-2765(02)00787-6. [DOI] [PubMed] [Google Scholar]
- [15].Rideout EJ, Marshall L, Grewal SS. Drosophila RNA polymerase III repressor Maf1 controls body size and developmental timing by modulating tRNAiMet synthesis and systemic insulin signaling. Proc Natl Acad Sci U S A. 2012;109:1139–44. doi: 10.1073/pnas.1113311109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Khanna A, Johnson DL, Curran SP. Physiological roles for mafr-1 in reproduction and lipid homeostasis. Cell Rep. 2014;9:2180–91. doi: 10.1016/j.celrep.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Reina JH, Azzouz TN, Hernandez N. Maf1, a new player in the regulation of human RNA polymerase III transcription. PLoS One. 2006;1:e134. doi: 10.1371/journal.pone.0000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Furuta E, Okuda H, Kobayashi A, Watabe K. Metabolic genes in cancer: their roles in tumor progression and clinical implications. Biochim Biophys Acta. 2010;1805:141–52. doi: 10.1016/j.bbcan.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lafontan M. Advances in adipose tissue metabolism. Int J Obes (Lond) 2008;32(Suppl 7):S39–51. doi: 10.1038/ijo.2008.237. [DOI] [PubMed] [Google Scholar]
- [20].Lodhi IJ, Wei X, Semenkovich CF. Lipoexpediency: de novo lipogenesis as a metabolic signal transmitter. Trends Endocrinol Metab. 2011;22:1–8. doi: 10.1016/j.tem.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Baenke F, Peck B, Miess H, Schulze A. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis Model Mech. 2013;6:1353–63. doi: 10.1242/dmm.011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chirala SS, Chang H, Matzuk M, Abu-Elheiga L, Mao J, Mahon K, et al. Fatty acid synthesis is essential in embryonic development: fatty acid synthase null mutants and most of the heterozygotes die in utero. Proc Natl Acad Sci U S A. 2003;100:6358–63. doi: 10.1073/pnas.0931394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Palian BM, Rohira AD, Johnson SA, He L, Zheng N, Dubeau L, et al. Maf1 is a novel target of PTEN and PI3K signaling that negatively regulates oncogenesis and lipid metabolism. PLoS Genet. 2014;10:e1004789. doi: 10.1371/journal.pgen.1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pluta K, Lefebvre O, Martin NC, Smagowicz WJ, Stanford DR, Ellis SR, et al. Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:5031–40. doi: 10.1128/MCB.21.15.5031-5040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Vannini A, Ringel R, Kusser AG, Berninghausen O, Kassavetis GA, Cramer P. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell. 2010;143:59–70. doi: 10.1016/j.cell.2010.09.002. [DOI] [PubMed] [Google Scholar]
- [26].Roberts DN, Wilson B, Huff JT, Stewart AJ, Cairns BR. Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol Cell. 2006;22:633–44. doi: 10.1016/j.molcel.2006.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Oficjalska-Pham D, Harismendy O, Smagowicz WJ, Gonzalez de Peredo A, Boguta M, Sentenac A, et al. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol Cell. 2006;22:623–32. doi: 10.1016/j.molcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- [28].Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ. mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc Natl Acad Sci U S A. 2010;107:11823–8. doi: 10.1073/pnas.1005188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gey GO, Coffman WD, Kubicek MT. Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Cancer Res. 1952;12:264–5. [Google Scholar]
- [30].Cohen P. The regulation of protein function by multisite phosphorylation – a 25 year update. Trends Biochem Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- [31].Wei Y, Zheng XS. Maf1 regulation: a model of signal transduction inside the nucleus. Nucleus. 2010;1:162–5. doi: 10.4161/nucl.1.2.11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wei Y, Tsang CK, Zheng XF. Mechanisms of regulation of RNA polymerase III-dependent transcription by TORC1. EMBO J. 2009;28:2220–30. doi: 10.1038/emboj.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Michels AA, Robitaille AM, Buczynski-Ruchonnet D, Hodroj W, Reina JH, Hall MN, et al. mTORC1 directly phosphorylates and regulates human MAF1. Mol Cell Biol. 2010;30:3749–57. doi: 10.1128/MCB.00319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Moir RD, Willis IM. Regulation of pol III transcription by nutrient and stress signaling pathways. Biochim Biophys Acta. 2013;1829:361–75. doi: 10.1016/j.bbagrm.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Graczyk D, Debski J, Muszynska G, Bretner M, Lefebvre O, Boguta M. Casein kinase II-mediated phosphorylation of general repressor Maf1 triggers RNA polymerase III activation. Proc Natl Acad Sci U S A. 2011;108:4926–31. doi: 10.1073/pnas.1010010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Moir RD, Lee J, Willis IM. Recovery of RNA polymerase III transcription from the glycerol-repressed state: revisiting the role of protein kinase CK2 in Maf1 phosphoregulation. J Biol Chem. 2012;287:30833–41. doi: 10.1074/jbc.M112.378828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rohira AD, Chen CY, Allen JR, Johnson DL. Covalent small ubiquitin-like modifier (SUMO) modification of Maf1 protein controls RNA polymerase III-dependent transcription repression. J Biol Chem. 2013;288:19288–95. doi: 10.1074/jbc.M113.473744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Johnson SS, Zhang C, Fromm J, Willis IM, Johnson DL. Mammalian Maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Mol Cell. 2007;26:367–79. doi: 10.1016/j.molcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- [39].Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–83. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- [40].Johnson SA, Dubeau L, Kawalek M, Dervan A, Schonthal AH, Dang CV, et al. Increased expression of TATA-binding protein, the central transcription factor, can contribute to oncogenesis. Mol Cell Biol. 2003;23:3043–51. doi: 10.1128/MCB.23.9.3043-3051.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bonhoure N, Byrnes A, Moir RD, Hodroj W, Preitner F, Praz V, et al. Loss of the RNA polymerase III repressor MAF1 confers obesity resistance. Genes Dev. 2015;29:934–47. doi: 10.1101/gad.258350.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Marshall L, Rideout EJ, Grewal SS. Nutrient/TOR-dependent regulation of RNA polymerase III controls tissue and organismal growth in Drosophila. EMBO J. 2012;31:1916–30. doi: 10.1038/emboj.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rollins J, Veras I, Cabarcas S, Willis I, Schramm L. Human Maf1 negatively regulates RNA polymerase III transcription via the TFIIB family members Brf1 and Brf2. Int J Biol Sci. 2007;3:292–302. doi: 10.7150/ijbs.3.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ciesla M, Towpik J, Graczyk D, Oficjalska-Pham D, Harismendy O, Suleau A, et al. Maf1 is involved in coupling carbon metabolism to RNA polymerase III transcription. Mol Cell Biol. 2007;27:7693–702. doi: 10.1128/MCB.01051-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Woiwode A, Johnson SA, Zhong S, Zhang C, Roeder RG, Teichmann M, et al. PTEN represses RNA polymerase III-dependent transcription by targeting the TFIIIB complex. Mol Cell Biol. 2008;28:4204–14. doi: 10.1128/MCB.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–50. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–74. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Grant B, Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol Biol Cell. 1999;10:4311–26. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sukur YE, Kivancli IB, Ozmen B. Ovarian aging and premature ovarian failure. J Turk Ger Gynecol Assoc. 2014;15:190–6. doi: 10.5152/jtgga.2014.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Virtanen HE, Rajpert-De Meyts E, Main KM, Skakkebaek NE, Toppari J. Testicular dysgenesis syndrome and the development and occurrence of male reproductive disorders. Toxicol Appl Pharmacol. 2005;207:501–5. doi: 10.1016/j.taap.2005.01.058. [DOI] [PubMed] [Google Scholar]
- [51].Gambineri A, Pelusi C, Vicennati V, Pagotto U, Pasquali R. Obesity and the polycystic ovary syndrome. Int J Obes Relat Metab Disord. 2002;26:883–96. doi: 10.1038/sj.ijo.0801994. [DOI] [PubMed] [Google Scholar]
- [52].Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–7. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]