Abstract
In mammalian cells, mitochondria are the only organelles besides the nucleus that house genomic DNA. The mammalian mitochondrial genome is represented by prokaryotic-type, circular, highly compacted DNA molecules. Today, more than a half-century after their discovery, the biology of these small and redundant molecules remains much less understood than that of their nuclear counterparts. One peculiarity of the mitochondrial genome that emerged in recent years is its disposable nature, as evidenced by cells abandoning a fraction of their mitochondrial DNA (mtDNA) in response to various stimuli with little or no physiological consequence. Here, we review some recent developments in the field of mtDNA biology and discuss emerging questions on the disposability and indispensability of mtDNA.
Keywords: Mitochondrial DNA, mtDNA maintenance, mtDNA copy number, mtDNA degradation, extramitohondrial mtDNA
Introduction
In mammalian cells, mitochondria are cellular organelles that generate the bulk of ATP required to sustain a plethora of diverse cellular processes. Besides generating ATP, mitochondria also play important roles in intracellular calcium signaling, apoptosis, reactive oxygen species (ROS) production and detoxification, and biosynthesis of heme and iron-sulfur clusters.
Mitochondria are encapsulated by two membranes which create four compartments: 1) the outer membrane, 2) the intermembrane space, 3) the inner membrane and 4) the matrix. The inner mitochondrial membrane houses five complexes of the oxidative phosphorylation (OXPHOS) system, of which complexes I-IV comprise an electron transport (respiratory) chain. The mitochondrial matrix houses numerous anabolic and catabolic enzymes, most prominently enzymes involved in the Krebs cycle and β-oxidation of fatty acids. The matrix is also the compartment that houses mitochondrial DNA (mtDNA).
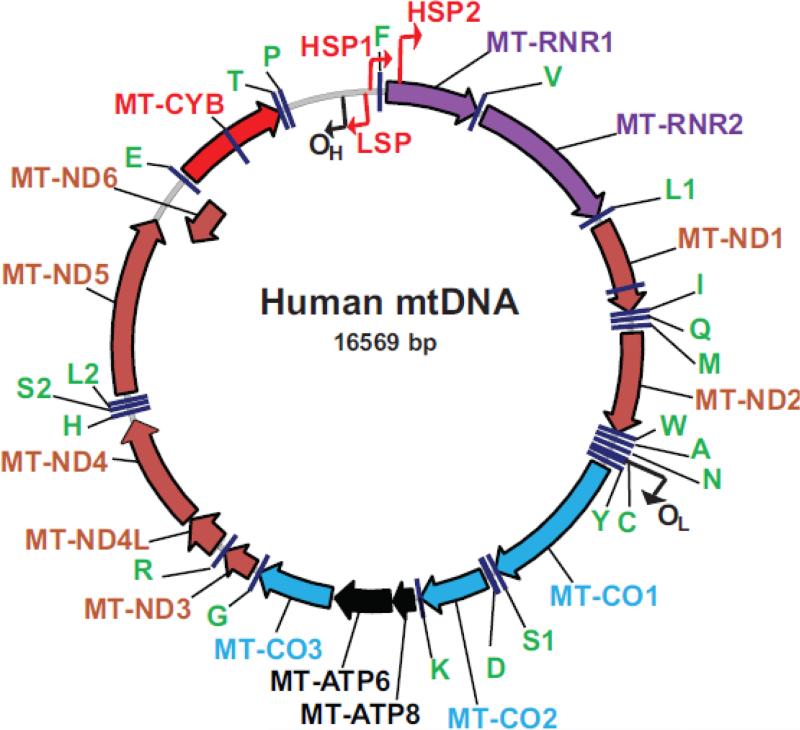
In humans, mtDNA is a circular 16,569 bp molecule (Figure 1) which encodes 37 “formal” genes: 13 polypeptides and 24 RNA components of the mitochondrial translational apparatus (2 rRNAs and 22 tRNAs). mtDNA is characterized by a tight packaging of genetic information: mitochondrial genes in mammals lack introns, and intergenic sequences are either absent or limited to a few bases. In some instances, genes overlap (MTATP8/6 and MTND4L/4), and translational termination codons of six mitochondrial genes are not encoded in the mitochondrial genome, but are completed by the addition of a polyadenine (polyA) tail to a terminal uracil encoded in mtDNA (thus generating sequence (UAA)TERAn). At the same time, mtDNA harbors a large ~1100 bp region that does not code for any genes, a so-called control region, which overlaps with a D-loop, a ~1000 nucleotide triple-helical region, which is formed by abortive initiation of replication.
Figure 1.
The map of human mtDNA. The orientation is according to the revised Cambridge Reference Sequence (rCRS, GenBank NC_012920.1). Black bent arrows designate origins and direction of new H-strand (OH) and L-strand (OL) synthesis during replication (or, conversely, origins of L-strand and H-strand replication), respectively. Red bent arrows designate mitochondrial promoters that transcribe either H-strand (HSP1 and HSP2) or L-strand (LSP), respectively. Brown arrows: MT-ND1-MT-ND6, NADH dehydrogenase subunits 1 through 6. Blue arrows: MTCO1-MTCO3, cytochrome oxidase subunits 1 through 3. Black arrows: MTATP6 and MT-ATP8, subunits 6 and 8 of the mitochondrial ATPase, Red arrow: MT-CYB, cytochrome b. tRNAs are designated by green letters using single-letter codes for corresponding amino acids.
The base distribution between two strands of human mtDNA (44% vs. 56% purine content, GenBank NC_012920.1) is asymmetric enough to allow for their separation in denaturing buoyant density gradients of cesium chloride. It is frequently stated that the ‘Heavy’ (H) strand encodes the majority of mitochondrial genes (12 polypeptides, 2 rRNAs and 14 tRNAs), and the ‘Light’ (L) strand encodes the rest. This notation is historical [1], and according to present-day convention (and the convention used in the original report on the sequence of human mtDNA [2]), the L-strand is the coding strand for the majority of mitochondrial genes. All of the mitochondrially encoded polypeptides are components of the OXPHOS complexes I (MTND1-6 plus MTND4L), III (MTCYB), IV (MTCO1-3) and V (MTATP6 and 8). Notably, all subunits of the mitochondrial complex II, all proteins responsible for mtDNA replication and repair, as well as all proteins responsible for transcription and translation of the mitochondrial genome are encoded in the nucleus.
Non-canonical mitochondrial genes
In recent years, it became increasingly apparent that in addition to its 37 “formal” genes, mtDNA may encode short open reading frames (ORFs) which can be translated into peptides with important biological functions. The first such peptide, humanin, was identified fourteen years ago in an unbiased functional screen for clones that protect neuronal cells from death induced by amyloid precursor protein (APP) mutants, which are associated with early-onset familial Alzheimer's disease [3]. Humanin is encoded by a 75 bp ORF within the gene for 16S rRNA, and was independently isolated in a yeast two-hybrid screen as a partner of the insulin-like growth factor-binding protein-3 (IGFBP-3). Humanin has since been shown to exert cytoprotective effects against not only mutant APPs, but also against neuronal cell death induced by other stimuli such as mutant presenilins 1 and 2, cytotoxic Aβ peptides, Aβ1-42, Aβ1-43, and Aβ25-35. It has also been shown to protect against IGFBP-3 induced apoptosis. Another short ORF encoding the 16 amino-acid-long peptide mitochondrial open reading frame of the 12S rRNA-c ( MOTS-c) has been recently discovered within the gene for mitochondrial 12S rRNA. This peptide targets skeletal muscle, and its cellular actions inhibit the folate cycle and de novo purine biosynthesis, leading to activation of the AMP-activated protein kinase. MOTS-c treatment in mice prevented age-dependent and high-fat-diet-induced insulin resistance, as well as diet-induced obesity [4].
mtDNA organization
mtDNA in mitochondria is organized into compact structures called nucleoids. Nucleoids can be visualized by labelling with various DNA stains, including anti-DNA antibodies, BrdU, or fluorescent intercalators, such as DAPI and Pico Green followed by microscopy. Therefore, the number of nucleoids detected per cell (and thus, estimates of the number of mtDNA molecules per nucleoid) depends on the properties of the optical system used, such as optical resolution and signal-to-noise ratio. This may explain why values for mtDNA content of nucleoids vary greatly in the literature. The lowest reported estimate of 1.45 mtDNA molecules per nucleoid, obtained with the help of the most advanced microscopy techniques [5], is likely be the most accurate, although we cannot exclude the possibility that this number is variable and/or depends on the cell type.
Nucleoids are ovoid structures diverse in size with an average diameter of about 100 nm [5, 6]. They are associated with the mitochondrial inner membrane and are often wrapped around cristae or cristae-like membrane invaginations [6]. Experimental evidence suggest that there is little, if any, exchange of mtDNA between nucleoids. The packaging density of DNA in mitochondrial nucleoids is greater than that in the E. coli nucleoid or human nucleus, and is comparable to the packaging density in the papillomavirus capsid [7]. This high degree of compaction is achieved with the help of the mitochondrial transcription factor A (TFAM), a high mobility group (HMG)-box DNA binding protein with functions in mtDNA packaging, replication and transcription. This protein binds mtDNA with a footprint of 23 bp or 30 bp [8], and is present in mitochondria in 1,000-fold molar excess with respect to mtDNA molecules, which is sufficient for the complete coating of mtDNA [5]. TFAM can bind mtDNA specifically, at the H-strand promoter 1 (HSP1) and at the L-strand promoter (LSP) promoters to facilitate transcription and replication, and non-specifically, all over the mitochondrial genome [9], to induce mtDNA compaction. mtDNA compaction by TFAM depends on TFAM dimerization, which is mediated by HMGA domains [10]. Upon both specific and non-specific binding, TFAM imposes a sharp U-turn on its substrate mtDNA, which is essential for both transcription and packaging [10]. Interestingly, mitochondrial TFAM is predominantly DNA-bound. The release of TFAM from complexes with mtDNA is mediated by phosphorylation on Ser55 and Ser56 by PKA followed by Lon-mediated degradation [11].
mtDNA replication
Only one replicative DNA polymerase, DNA polymerase γ (PolG), has been described in mammalian mitochondria. This holoenzyme mediates both replication and repair of mtDNA, and consists of a large catalytic subunit and two accessory subunits. The catalytic subunit possess the polymerase, 3′→5′ exonuclease (proofreading), and 5′ - deoxyribose phosphate (dRP) lyase activities, while the accessory subunits enhance DNA binding and processivity. Also, a novel DNA polymerase/primase, PrimPol, has been recently identified in mitochondria. This enzyme is believed to facilitate replication fork progression by acting as a translesion DNA polymerase or as a specific DNA primase reinitiating downstream of lesions that block synthesis during both mitochondrial and nuclear DNA replication. Unlike PolG−/−, PrimPol−/− mice are viable indicating accessory role for the enzyme [12].
The mitochondrial replisome consists of PolG, the mitochondrial single-stranded DNA binding protein (mtSSB), mitochondrial DNA helicase, topoisomerases and RNaseH [13]. The lack of dedicated primase, an enzyme that generates an RNA primer for replication, is a unique feature of mtDNA replication. Instead of being synthesized by a dedicated enzyme, the primer for H-strand replication is generated by transcription initiated by mitochondrial RNA polymerase on an L-strand promoter (see below). Thus, the RNA polymerase serves to generate not only transcripts, but also the primers needed for mtDNA replication.
The mode of mtDNA replication remains controversial, and at least three models of replication have been described (recently reviewed in [14]). The original asynchronous strand-displacement model suggests that mtDNA replication is primed by an L-strand transcript. Interaction of the transcription elongation factor TEFM with POLRMT and the nascent transcript prevents the generation of replication primers and increases transcription processivity to enable synthesis of a near-genomic length L-strand transcript. Therefore, TEFM serves as a molecular switch between replication and transcription [15]. The primer generated at LSP is extended by the mitochondrial replisome and displaces H-strand over ~70% of mtDNA length. Then H-strand replication exposes the origin of the L-strand replication (OL), and the synthesis of a new L-strand is initiated in the opposite direction. The asymmetry of strand synthesis leaves one segregated daughter molecule with an incompletely synthesized L-strand, called a gapped circle (GpC).
In the alternative strand-coupled (synchronous) model, there is thought to be a zone of replication initiation within a broad area beyond the D-loop. Within this zone, both strands are synthesized bidirectionally as the conventional double-stranded replication forks advance through continuous synthesis of leading and discontinuous synthesis (through Okazaki fragments) of lagging strands. However, this model relies on continuous ligation of Okazaki fragments during lagging strand synthesis and appears to be inconsistent with recent findings that 100-fold reduction in mitochondrial DNA ligase III does not appreciably affect the rate of mtDNA replication or copy number [16]. A third model is based on the observation of RNA incorporation throughout the lagging strand (RITOLS) [17]. According to this model, replication proceeds as in the strand-displacement model, except displaced H-strand is present not as single-strand DNA, but rather as a DNA/RNA hybrid sensitive to RNase H, until it is made duplex by PolG. It is not clear whether RITOLS serve as primers for Okazaki fragments, but it appears unlikely due to the low response of mtDNA replication to DNA ligase III (see above). Recently, it was demonstrated that the in vivo occupancy profile of mtSSB displays a distinct pattern, with the highest levels of mtSSB close to the mitochondrial control region and with a gradual decline towards OriL. This pattern correlates with the replication products expected for the strand displacement mode of mtDNA synthesis thus lending strong in vivo support [18].
mtDNA copy number control
mtDNA is maintained at different copy numbers in different tissues, the two extremes of this spectrum being mammalian erythrocytes and sperm which have no mtDNA and ~5 copies of mtDNA per cell [19], respectively, and oocytes, which may contain >500,000 copies [20]. In Drosophila, mtDNA is eliminated from sperm in the male genital tract prior to fertilization, so that fertilizing sperm may contain no mtDNA at all [21]. Curiously, human oocyte quality directly correlates with mtDNA copy number, whereas this correlation is inverse for the human sperm [20].
It is often overlooked that normal mtDNA copy number in a given tissue is not a set figure, but is rather a range. Therefore, in most population studies mtDNA copy number in apparently healthy individuals varies over a 2-10 fold range [22], and mtDNA content between 40-150% of the average is considered clinically normal [23]. Apart from normal variation in mtDNA copy number, mtDNA content can be altered under various pathologic scenarios. For example, mtDNA depletion syndromes [24] are associated with a profound loss of mtDNA, down to as low as 2% of normal [25]. Even though mtDNA depletion in these patients is frequently associated with perinatal lethality, long survival has been reported in a 29 year old patient with 76% depletion, who was observed for this condition since early childhood [26]. Also, a profound (91%) loss of mtDNA in a 47 year old patient was associated with relatively mild symptoms such as daytime sleepiness, exercise intolerance, and myalgias in the lower-limb muscles [27]. Paradoxically, both increased [28] and decreased skeletal muscle mtDNA content has been reported in patients with mutations in mitofusin 2 (MFN2). Changes in mtDNA content in tissues of aged individuals have been widely reported, although the direction of these changes remains controversial. Some studies report increased mtDNA copy number in the elderly [29], while others report a decrease and associate frailty with either lower [30] or higher [31] mtDNA copy number.
Regulation of mtDNA copy number is complex and remains incompletely understood. Nitric oxide, PGC1α, NCOR1 and their coactivators have been implicated as regulators through their effects on mitochondrial biogenesis. A systematic analysis in yeast revealed 102 ORFs whose deletion led to the loss of mtDNA. Remarkably, 55% of those ORFs were associated with biosynthesis of mitochondrial proteins [32]. In humans, sex-specific quantitative trait loci for mtDNA content have been identified on chromosomes 1, 2 and 3 [33]. Recently, epigenetic modification of exon 2 of the gene for the catalytic subunit of PolG has been implicated in mtDNA copy number regulation [34].
mtDNA repair and abandonment
Mitochondria contain a reduced complement of DNA repair pathways as compared to those present in the nucleus (reviewed in [35]). As mtDNA encodes only 13 out of ~1500 polypeptides that comprise mitochondria, all components of mtDNA repair machinery are encoded in the nucleus, translated on cytoplasmic ribosomes, and post-translationally imported into mitochondria. Mitochondria are proficient in both short-patch and long-patch subpathways of the Base Excision Repair (BER) pathway, which is responsible for the repair of oxidative and alkylating lesions as well as single strand breaks [35]. Importantly, some oxidized base lesions are repaired in mitochondria more efficiently than they are in the nucleus [35]. The presence of any other complete DNA repair pathway in mitochondria has not been demonstrated yet, although some components of the mismatch repair pathway have been identified in these organelles. How, then, do mitochondria cope with the mutagenic effects of DNA lesions that they are unable to repair? It turns out that in mammalian cells, the high redundancy of mtDNA enables a unique, mitochondria-specific pathway for the preservation of DNA integrity through degradation of damaged molecules. This pathway is nonspecific to the type of lesion and is mobilized not only in response to lesions that mitochondria are unable to repair, but also in response to the presence of an overwhelming amount of lesions that mitochondria are normally able repair in moderate quantities, such as oxidative lesions [36], abasic sites, or gapped duplexes [37]. The kinetics of this process is different in different cell lines, and in some cell lines mtDNA loss can be detected as soon as 5-10 min after challenge with H2O2 [38]. Strikingly, almost no information exists on the mechanism of induced mtDNA degradation or nucleases involved in the process, although it appears unlikely that mitophagy can account for the rapid mtDNA turnover observed in some experiments [38].
Extramitochondrial mtDNA
Circulating cell-free mtDNA (mtDNAcf) was described in blood plasma 15 years ago by Zhong et al. [39], and has been suggested to have prognostic value in cancer, cardiac arrest and severe sepsis [40, 41]. Recently, mtDNAcf emerged as a danger-associated molecular pattern (DAMP), a major mediator of innate immunity and systemic inflammatory response syndrome (SIRS). Upon tissue damage (e.g., blunt-force trauma), blood levels of mtDNAcf dramatically increase leading to toll-like receptor 9 (TLR-9)-mediated neutrophil activation and systemic inflammation [42]. Curiously, there appears to be discordance between the reported activation of TLR-9 exclusively by DNA containing unmethylated CpG islands and the well-documented cytosine methylation in mtDNA, which is mediated by an organellar isoform of the nuclear methyltransferase DNMT1 [43]. Since both nuclear and mitochondrial CpG islands can be methylated, one possible explanation for the selective activation of the immune system by mtDNA is in the greater content of unmethylated CpGs in mtDNA.
Extramitochondrial mtDNA has also been reported in the cytosol. Cytosolic release of mtDNA can be mediated by strong insults like oxidative stress, or by subtle changes like altered compaction of mitochondrial nucleoids secondary to TFAM haploinsufficiency [44]. This intracellular release of mtDNA has been implicated in cell-intrinsic innate immune responses [45, 46].
mtDNA is disposable but indispensable
The prevailing evidence indicates that mtDNA is disposable and has a limited life span. In unstressed cells and tissues, it turns over with relatively short half-lives [47]. The substantial variability of mtDNA content in the same tissue of different healthy individuals revealed by epidemiological studies suggests that, within limits, alterations in mtDNA copy number do not affect normal mitochondrial function [22, 30, 31, 48]. In support of this notion, it has been observed that reduced mtDNA copy number has no major effect on mitochondrial transcript levels or enzyme activities in various tissues [49, 50]. It has also been shown that some cells can survive after losing their mtDNA. For example, in response to stimulation, eosinophils abandon their mtDNA by catapulting it into extracellular milieu, while remaining viable [51, 52]. In response to a heat shock and upon antibody-induced clustering of GPI-anchored antigens, ciliated protozoa jettison mitochondria out of the cells without losing viability [53]. In transgenic mice, doxycycline-induced expression of the mutant UNG1 resulted in hippocampal mtDNA depletion [54], which was accompanied by relatively mild behavioral changes such as increased locomotor activity, impaired cognitive abilities, and lack of anxietylike responses [55]. Lastly, mitochondria are normally shed together with enclosed mtDNA by retinal ganglion cell axons for degradation by surrounding astrocytes [56]. Reversible loss of mtDNA in experimental animals in response to treatments also supports the notion that mtDNA molecules are disposable. In mice, intragastric administration of ethanol was accompanied by a ~50% depletion of mtDNA in all organs studied, which reversed 24h after discontinuation of treatment [57]. Degradation of mtDNA was also observed in a rat model of cerebral ischemia/reperfusion [58]. Similarly to mtDNA levels following depletion induced by intragastric ethanol administration, mtDNA levels following cerebral ischemia returned to normal within 24h of reperfusion [58]. Unaccustomed resistance exercise can induce up to 32% mtDNA depletion in mouse muscle [59], whereas prolonged endurance training results in a 50% increase in mtDNA copy number in human muscle [60]. These examples demonstrate that a partial mtDNA loss is relatively common in some cells and tissues and that this loss, at least in some instances, 1) can be accompanied by either mild or no consequences and 2) can be transient and accompanied by compensatory adaptive mtDNA resynthesis.
While the evidence reviewed above suggests that individual mtDNA molecules are disposable, it does not answer the question of whether or not mtDNA in general is superfluous for cell viability: in other words, is mtDNA dispensable? For mammalian erythrocytes, which contain no mtDNA, it is. Also, cultured animal cells can be induced to undergo a complete loss of mtDNA. The resulting ρ0 cells become auxotrophic, but retain viability. Interestingly, however, these cells fail to form tumors in experimental animals unless they capture mtDNA from surrounding tissues [61]. Yet, successful attempts to induce global mtDNA loss in multicellular organisms have not been reported. As noted above, infants with mtDNA depletion syndromes, which are accompanied by severe loss of mtDNA, usually die in the first year of life. Also, whole-body ablation of TFAM, a protein required for mtDNA replication, results in severe mtDNA depletion and is embryonically lethal in mice [62]. These observations suggest that mtDNA is indispensable for the viability of multicellular organisms.
Conclusion
More than 50 years of research have led to dramatic progress in our understanding of mtDNA biology. Our views have evolved from dismissing the mitochondrial genome as vestigial to appreciating the key role of mtDNA and its alterations in cellular bioenergetics and mitochondrial disease. Yet it appears that mammalian cells have a relaxed reliance on mtDNA, which is perpetually destroyed and resynthesized. Under physiological and pathophysiological conditions, the balance between these two processes can be shifted and restored, often with mild or no consequences. The absence of a direct correlation between mtDNA copy number and levels of mitochondrially encoded transcripts or proteins, which may remain unaltered during mtDNA depletion [49, 50], provides a mechanistic explanation for this relaxed reliance. Yet, some key questions remain unanswered. What are the triggers and mechanism(s) of mtDNA degradation? What enzymes are involved in the process? Is mtDNA degraded down to nucleotides, or to oligonucleotides? Is degrading mtDNA contained inside mitochondria or released intra/extracellularly? Does degrading mtDNA contribute to innate immune responses? These and other questions will be addressed in the coming years.
Highlights.
The loss of mtDNA in mammals is incompatible with life
However, broad variations in mtDNA copy number are observed and appear inconsequential
Mitochondria can dispose of severely damaged mtDNA and resynthesize new
mtDNA released into the cytosol or extracellularly plays an important role in innate immunity
Acknowledgements
This work was supported by the National Institutes of Health grants PO1 HL66299 and OD010944.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 2.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 3.Hashimoto Y, Niikura T, Tajima H, Yasukawa T, Sudo H, Ito Y, Kita Y, Kawasumi M, Kouyama K, Doyu M, Sobue G, Koide T, Tsuji S, Lang J, Kurokawa K, Nishimoto I. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer's disease genes and Abeta. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6336–6341. doi: 10.1073/pnas.101133498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, Wan J, Kim SJ, Mehta H, Hevener AL, de Cabo R, Cohen P. The Mitochondrial-Derived Peptide MOTS-c Promotes Metabolic Homeostasis and Reduces Obesity and Insulin Resistance. Cell metabolism. 2015;21:443–454. doi: 10.1016/j.cmet.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kukat C, Wurm CA, Spahr H, Falkenberg M, Larsson NG, Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown TA, Tkachuk AN, Shtengel G, Kopek BG, Bogenhagen DF, Hess HF, Clayton DA. Super-resolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol. Cell. Biol. 2011 doi: 10.1128/MCB.05694-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogenhagen DF. Mitochondrial DNA nucleoid structure. Biochim. Biophys. Acta. 2012;1819:914–920. doi: 10.1016/j.bbagrm.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Farge G, Laurens N, Broekmans OD, van den Wildenberg SM, Dekker LC, Gaspari M, Gustafsson CM, Peterman EJ, Falkenberg M, Wuite GJ. Protein sliding and DNA denaturation are essential for DNA organization by human mitochondrial transcription factor A. Nature communications. 2012;3:1013. doi: 10.1038/ncomms2001. [DOI] [PubMed] [Google Scholar]
- 9.Wang YE, Marinov GK, Wold BJ, Chan DC. Genome-wide analysis reveals coating of the mitochondrial genome by TFAM. PLoS One. 2013;8:e74513. doi: 10.1371/journal.pone.0074513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ngo HB, Lovely GA, Phillips R, Chan DC. Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nature communications. 2014;5:3077. doi: 10.1038/ncomms4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu B, Lee J, Nie X, Li M, Morozov YI, Venkatesh S, Bogenhagen DF, Temiakov D, Suzuki CK. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol. Cell. 2013;49:121–132. doi: 10.1016/j.molcel.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Gomez S, Reyes A, Martinez-Jimenez MI, Chocron ES, Mouron S, Terrados G, Powell C, Salido E, Mendez J, Holt IJ, Blanco L. PrimPol, an archaic primase/polymerase operating in human cells. Mol. Cell. 2013;52:541–553. doi: 10.1016/j.molcel.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasiviswanathan R, Collins TR, Copeland WC. The interface of transcription and DNA replication in the mitochondria. Biochim. Biophys. Acta. 2012;1819:970–978. doi: 10.1016/j.bbagrm.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKinney EA, Oliveira MT. Replicating animal mitochondrial DNA. Genet. Mol. Biol. 2013;36:308–315. doi: 10.1590/S1415-47572013000300002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agaronyan K, Morozov YI, Anikin M, Temiakov D. Mitochondrial biology. Replication-transcription switch in human mitochondria. Science. 2015;347:548–551. doi: 10.1126/science.aaa0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shokolenko IN, Fayzulin RZ, Katyal S, McKinnon PJ, Wilson GL, Alexeyev MF. Mitochondrial DNA ligase is dispensable for the viability of cultured cells but essential for mtDNA maintenance. J. Biol. Chem. 2013;288:26594–26605. doi: 10.1074/jbc.M113.472977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yasukawa T, Reyes A, Cluett TJ, Yang MY, Bowmaker M, Jacobs HT, Holt IJ. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25:5358–5371. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miralles Fuste J, Shi Y, Wanrooij S, Zhu X, Jemt E, Persson O, Sabouri N, Gustafsson CM, Falkenberg M. In vivo occupancy of mitochondrial single-stranded DNA binding protein supports the strand displacement mode of DNA replication. PLoS genetics. 2014;10:e1004832. doi: 10.1371/journal.pgen.1004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabriel MS, Chan SW, Alhathal N, Chen JZ, Zini A. Influence of microsurgical varicocelectomy on human sperm mitochondrial DNA copy number: a pilot study. J. Assist. Reprod. Genet. 2012;29:759–764. doi: 10.1007/s10815-012-9785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynier P, May-Panloup P, Chretien MF, Morgan CJ, Jean M, Savagner F, Barriere P, Malthiery Y. Mitochondrial DNA content affects the fertilizability of human oocytes. Mol. Hum. Reprod. 2001;7:425–429. doi: 10.1093/molehr/7.5.425. [DOI] [PubMed] [Google Scholar]
- 21.DeLuca SZ, O'Farrell PH. Barriers to male transmission of mitochondrial DNA in sperm development. Dev. Cell. 2012;22:660–668. doi: 10.1016/j.devcel.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stringer HA, Sohi GK, Maguire JA, Cote HC. Decreased skeletal muscle mitochondrial DNA in patients with statin-induced myopathy. J. Neurol. Sci. 2013;325:142–147. doi: 10.1016/j.jns.2012.12.023. [DOI] [PubMed] [Google Scholar]
- 23.Nakano Y, Murayama K, Tsuruoka T, Aizawa M, Nagasaka H, Horie H, Ohtake A, Saitou K. Fatal case of mitochondrial DNA depletion with severe asphyxia in a newborn. Pediatr. Int. 2011;53:240–242. doi: 10.1111/j.1442-200X.2010.03183.x. [DOI] [PubMed] [Google Scholar]
- 24.Nogueira C, Almeida LS, Nesti C, Pezzini I, Videira A, Vilarinho L, Santorelli FM. Syndromes associated with mitochondrial DNA depletion. Ital. J. Pediatr. 2014;40:34. doi: 10.1186/1824-7288-40-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee IC, Lee NC, Lu JJ, Su PH. Mitochondrial depletion causes neonatal-onset leigh syndrome, myopathy, and renal tubulopathy. J. Child Neurol. 2013;28:404–408. doi: 10.1177/0883073812469722. [DOI] [PubMed] [Google Scholar]
- 26.Vu TH, Tanji K, Valsamis H, DiMauro S, Bonilla E. Mitochondrial DNA depletion in a patient with long survival. Neurology. 1998;51:1190–1193. doi: 10.1212/wnl.51.4.1190. [DOI] [PubMed] [Google Scholar]
- 27.Finsterer J, G GK, Ahting U. Adult mitochondrial DNA depletion syndrome with mild manifestations. Neurol. Int. 2013;5:28–30. doi: 10.4081/ni.2013.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sitarz KS, Yu-Wai-Man P, Pyle A, Stewart JD, Rautenstrauss B, Seeman P, Reilly MM, Horvath R, Chinnery PF. MFN2 mutations cause compensatory mitochondrial DNA proliferation. Brain. 2012 doi: 10.1093/brain/aws049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navarro-Sastre A, Tort F, Garcia-Villoria J, Pons MR, Nascimento A, Colomer J, Campistol J, Yoldi ME, Lopez-Gallardo E, Montoya J, Unceta M, Martinez MJ, Briones P, Ribes A. Mitochondrial DNA depletion syndrome: New descriptions and the use of citrate synthase as a helpful tool to better characterise the patients. Mol. Genet. Metab. 2012 doi: 10.1016/j.ymgme.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 30.Mengel-From J, Thinggaard M, Dalgard C, Kyvik KO, Christensen K, Christiansen L. Mitochondrial DNA copy number in peripheral blood cells declines with age and is associated with general health among elderly. Hum. Genet. 2014;133:1149–1159. doi: 10.1007/s00439-014-1458-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Leeuwen N, Beekman M, Deelen J, van den Akker EB, de Craen AJ, Slagboom PE, t Hart LM. Low mitochondrial DNA content associates with familial longevity: the Leiden Longevity Study. Age. 2014;36:9629. doi: 10.1007/s11357-014-9629-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Singh KK. Global genetic determinants of mitochondrial DNA copy number. PLoS One. 2014;9:e105242. doi: 10.1371/journal.pone.0105242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez S, Buil A, Souto JC, Casademont J, Blangero J, Martinez-Perez A, Fontcuberta J, Lathrop M, Almasy L, Soria JM. Sex-specific regulation of mitochondrial DNA levels: genome-wide linkage analysis to identify quantitative trait Loci. PLoS One. 2012;7:e42711. doi: 10.1371/journal.pone.0042711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee W, Johnson J, Gough DJ, Donoghue J, Cagnone GL, Vaghjiani V, Brown KA, Johns TG, St John JC. Mitochondrial DNA copy number is regulated by DNA methylation and demethylation of POLGA in stem and cancer cells and their differentiated progeny. Cell Death Dis. 2015;6:e1664. doi: 10.1038/cddis.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexeyev M, Shokolenko I, Wilson G, LeDoux S. The maintenance of mitochondrial DNA integrity--critical analysis and update. Cold Spring Harb. Perspect. Biol. 2013;5:a012641. doi: 10.1101/cshperspect.a012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furda AM, Marrangoni AM, Lokshin A, Van Houten B. Oxidants and not alkylating agents induce rapid mtDNA loss and mitochondrial dysfunction. DNA Repair (Amst) 2012;11:684–692. doi: 10.1016/j.dnarep.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shokolenko IN, Wilson GL, Alexeyev MF. Persistent damage induces mitochondrial DNA degradation. DNA Repair (Amst) 2013;12:488–499. doi: 10.1016/j.dnarep.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shokolenko IN, Wilson GL, Alexeyev MF. The “fast” and the “slow” modes of mitochondrial DNA degradation. Mitochondrial DNA. 2014 doi: 10.3109/19401736.2014.905829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong S, Ng MC, Lo YM, Chan JC, Johnson PJ. Presence of mitochondrial tRNA(Leu(UUR)) A to G 3243 mutation in DNA extracted from serum and plasma of patients with type 2 diabetes mellitus. J. Clin. Pathol. 2000;53:466–469. doi: 10.1136/jcp.53.6.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnalich F, Codoceo R, Lopez-Collazo E, Montiel C. Circulating cell-free mitochondrial DNA: A better early prognostic marker in patients with out-of-hospital cardiac arrest. Resuscitation. 2012 doi: 10.1016/j.resuscitation.2012.03.032. [DOI] [PubMed] [Google Scholar]
- 41.Kung CT, Hsiao SY, Tsai TC, Su CM, Chang WN, Huang CR, Wang HC, Lin WC, Chang HW, Lin YJ, Cheng BC, Su BY, Tsai NW, Lu CH. Plasma nuclear and mitochondrial DNA levels as predictors of outcome in severe sepsis patients in the emergency room. J. Transl. Med. 2012;10:130. doi: 10.1186/1479-5876-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3630–3635. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, Kaech SM, Smiley JR, Means RE, Iwasaki A, Shadel GS. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015 doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rongvaux A, Jackson R, Harman CC, Li T, West AP, de Zoete MR, Wu Y, Yordy B, Lakhani SA, Kuan CY, Taniguchi T, Shadel GS, Chen ZJ, Iwasaki A, Flavell RA. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159:1563–1577. doi: 10.1016/j.cell.2014.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White MJ, McArthur K, Metcalf D, Lane RM, Cambier JC, Herold MJ, van Delft MF, Bedoui S, Lessene G, Ritchie ME, Huang DC, Kile BT. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159:1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gross NJ, Getz GS, Rabinowitz M. Apparent turnover of mitochondrial deoxyribonucleic acid and mitochondrial phospholipids in the tissues of the rat. J. Biol. Chem. 1969;244:1552–1562. [PubMed] [Google Scholar]
- 48.Wang D, Su LY, Zhang AM, Li YY, Li XA, Chen LL, Long H, Yao YG. Mitochondrial DNA copy number, but not haplogroup, confers a genetic susceptibility to leprosy in Han Chinese from Southwest China. PLoS One. 2012;7:e38848. doi: 10.1371/journal.pone.0038848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barazzoni R, Short KR, Nair KS. Effects of aging on mitochondrial DNA copy number and cytochrome c oxidase gene expression in rat skeletal muscle, liver, and heart. J. Biol. Chem. 2000;275:3343–3347. doi: 10.1074/jbc.275.5.3343. [DOI] [PubMed] [Google Scholar]
- 50.Barthelemy C, Ogier de Baulny H, Diaz J, Cheval MA, Frachon P, Romero N, Goutieres F, Fardeau M, Lombes A. Late-onset mitochondrial DNA depletion: DNA copy number, multiple deletions, and compensation. Ann. Neurol. 2001;49:607–617. [PubMed] [Google Scholar]
- 51.Yousefi S, Gold JA, Andina N, Lee JJ, Kelly AM, Kozlowski E, Schmid I, Straumann A, Reichenbach J, Gleich GJ, Simon HU. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat. Med. 2008 doi: 10.1038/nm.1855. [DOI] [PubMed] [Google Scholar]
- 52.Yousefi S, Mihalache C, Kozlowski E, Schmid I, Simon HU. Viable neutrophils release mitochondrial DNA to form neutrophil extracellular traps. Cell Death Differ. 2009;16:1438–1444. doi: 10.1038/cdd.2009.96. [DOI] [PubMed] [Google Scholar]
- 53.Bisharyan Y, Clark TG. Calcium-dependent mitochondrial extrusion in ciliated protozoa. Mitochondrion. 2011;11:909–918. doi: 10.1016/j.mito.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lauritzen KH, Cheng C, Wiksen H, Bergersen LH, Klungland A. Mitochondrial DNA toxicity compromises mitochondrial dynamics and induces hippocampal antioxidant defenses. DNA Repair (Amst) 2011;10:639–653. doi: 10.1016/j.dnarep.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 55.Lauritzen KH, Moldestad O, Eide L, Carlsen H, Nesse G, Storm JF, Mansuy IM, Bergersen LH, Klungland A. Mitochondrial DNA toxicity in forebrain neurons causes apoptosis, neurodegeneration, and impaired behavior. Mol. Cell. Biol. 2010;30:1357–1367. doi: 10.1128/MCB.01149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis CH, Kim KY, Bushong EA, Mills EA, Boassa D, Shih T, Kinebuchi M, Phan S, Zhou Y, Bihlmeyer NA, Nguyen JV, Jin Y, Ellisman MH, Marsh-Armstrong N. Transcellular degradation of axonal mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2014;111:9633–9638. doi: 10.1073/pnas.1404651111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mansouri A, Gaou I, De Kerguenec C, Amsellem S, Haouzi D, Berson A, Moreau A, Feldmann G, Letteron P, Pessayre D, Fromenty B. An alcoholic binge causes massive degradation of hepatic mitochondrial DNA in mice. Gastroenterology. 1999;117:181–190. doi: 10.1016/s0016-5085(99)70566-4. [DOI] [PubMed] [Google Scholar]
- 58.Chen H, Hu CJ, He YY, Yang DI, Xu J, Hsu CY. Reduction and restoration of mitochondrial dna content after focal cerebral ischemia/reperfusion. Stroke. 2001;32:2382–2387. doi: 10.1161/hs1001.097099. [DOI] [PubMed] [Google Scholar]
- 59.Ogborn DI, McKay BR, Crane JD, Safdar A, Akhtar M, Parise G, Tarnopolsky MA. Effects of age and unaccustomed resistance exercise on mitochondrial transcript and protein abundance in skeletal muscle of men. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015 doi: 10.1152/ajpregu.00005.2014. ajpregu 00005 02014. [DOI] [PubMed] [Google Scholar]
- 60.Zoladz JA, Grassi B, Majerczak J, Szkutnik Z, Korostynski M, Grandys M, Jarmuszkiewicz W, Korzeniewski B. Mechanisms responsible for the acceleration of pulmonary VO2 on-kinetics in humans after prolonged endurance training. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014;307:R1101–1114. doi: 10.1152/ajpregu.00046.2014. [DOI] [PubMed] [Google Scholar]
- 61.Tan AS, Baty JW, Dong LF, Bezawork-Geleta A, Endaya B, Goodwin J, Bajzikova M, Kovarova J, Peterka M, Yan B, Pesdar EA, Sobol M, Filimonenko A, Stuart S, Vondrusova M, Kluckova K, Sachaphibulkij K, Rohlena J, Hozak P, Truksa J, Eccles D, Haupt LM, Griffiths LR, Neuzil J, Berridge MV. Mitochondrial Genome Acquisition Restores Respiratory Function and Tumorigenic Potential of Cancer Cells without Mitochondrial DNA. Cell metabolism. 2015;21:81–94. doi: 10.1016/j.cmet.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 62.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]