Abstract
Huntington's disease (HD) is a hereditary and devastating neurodegenerative disorder caused by a mutation in the huntingtin protein. Understanding the functions of normal and mutant huntingtin protein is the key to reveal the pathogenesis of HD and develop therapeutic targets. Huntingtin plays an important role in vesicular and organelle trafficking. Lysosomes are dynamic organelles that integrate several degradative pathways and regulate the activity of mammalian target of rapamycin complex 1 (mTORC1). In the present study, we show that the perinuclear accumulation of lysosomes is increased in a cellular model of HD derived from HD knock-in mice and primary fibroblasts from a HD patient. This perinuclear lysosomal accumulation can be reversed when normal huntingtin is overexpressed in HD cells. To further investigate the functional significance of the increased perinuclear lysosomal accumulation in HD cells, we demonstrate subsequently that basal mTORC1 activity is increased in HD cells. In addition, autophagic influx is also increased in HD cells in response to serum deprivation, which leads to a premature fusion of lysosomes with autophagosomes. Taken together, our data suggest that the increased perinuclear accumulation of lysosomes may play an important role in HD pathogenesis by altering lysosomal-dependent functions.
Keywords: vesicular trafficking, Lamp1, autophagy, autophagic influx, mTORC1
Graphical Abstract
INTRODUCTION
Huntington's disease (HD) is an inherited neurodegenerative disease caused by an abnormal polyglutamine (polyQ) expansion in the huntingtin (Htt) protein. Through molecular interactions with different proteins, Htt plays important roles in a variety of cellular functions including, but are not limited to, vesicular trafficking (Caviston & Holzbaur, 2009) and transcriptional regulation (Zuccato et al., 2003; Cui et al., 2006). Accumulating data suggest that the polyQ expansion in mutant Htt (mHtt) confers a toxic gain-of-function on mHtt and results in interference with normal functions of Htt.
Full length Htt is a cytosolic 350KDa protein that is shown to be associated with different cellular compartments, such as mitochondria (Orr et al., 2008), ER membrane/Golgi apparatus (Rockabrand et al., 2006; Atwal et al., 2007) and endosomal-lysosomal system (Kegel et al., 2000). Given the role of Htt in vesicular trafficking, it is expected that Htt can affect organelle dynamics and their functions. For example, mHtt directly interacts with mitochondrial fission protein, dynamin-related protein 1 (Drp-1) and increases its activity, leading to enhanced mitochondrial fragmentations in HD cells (Song et al., 2011). Moreover, while normal Htt promotes both anterograde and retrograde mitochondrial transport, mHtt impairs mitochondrial movements by disrupting the transport complex formation, suggesting the involvement of defective mitochondria trafficking in the pathogenesis of HD (Li et al., 2010; Sheng & Cai, 2012).
Lysosomes are dynamic organelles with crucial functions in digesting macromolecules and cellular components for normal cellular functions. Failure to do so leads to accumulation of protein aggregates, which are commonly present in a number of neurodegenerative diseases, including HD (Jeong et al., 2009). Additionally, lysosomes are the primary activation sites for mammalian target of rapamycin complex 1 (mTORC1), the master regulator of cell growth (Sancak et al., 2010; Demetriades et al., 2014; Menon et al., 2014). mTORC1 is recruited to the lysosomal membrane in the presence of amino acids through Rag GTPase, where it is then activated by its modulator, Ras homology enrich in brain (Rheb), in response to growth factor stimulation. During starvation, mTORC1 is released from lysosomes and becomes inactivated (Demetriades et al., 2014; Menon et al., 2014). Interestingly, the importance of lysosome positioning in modulating lysosomal functions and coordinating cellular nutrient responses was reported (Korolchuk et al., 2011).
Compared to mitochondria, little is known about the role of mHtt in lysosomal positioning and functions. In this study, we provide evidence showing that the subcellular lysosomal distribution is altered in HD cells. In healthy cells, lysosomes are distributed throughout the cells with moderate accumulation in the perinuclear regions. In contrast, lysosomes are highly accumulated in the perinuclear regions of HD cells. Importantly, expression of normal Htt in HD cells can redirect lysosomes to the peripheral region. Furthermore, increased perinuclear accumulation of lysosomes is associated with increased basal mTORC1 activity and accelerated autophagic influx in response to serum deprivation in HD cells. These data suggest that altered lysosomal positioning in HD contributes to the pathogenesis of HD.
MATERIALS AND METHODS
Cell culture and starvation
STHdhQ7 and STHdhQ111 cells were obtained from HD Community BioRepository Collection (New Jersey, USA) and cultured in Dulbecco's modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Mediatech, Inc. VA, USA), 1% glutamine and 1% penicillin/streptomycin at 33°C. Early passages of STHdh (less than 20 passages) were used in the studies. Primary fibroblasts, GM04693 from a HD individual (onset at age 41 years) and AG02222 from a normal subject, were obtained from the Coriell Institute for Medical Research and cultured in DMEM supplemented with 20% FBS, 1% glutamine and 1% penicillin/streptomycin at 37°C. Phoenix-Ampho cells were purchased from ATCC (Manassas, VA, USA) and cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C. For serum starvation, cells were washed three times with pre-warmed DMEM medium and then incubated in DMEM medium for 18 hours. In some experiments, 50 μM hydroxychloroquine (CQ, Sigma) was present in serum-deprived and control cells to block the fusion of lysosomes with autophagosomes. For amino acid and serum starvation, cells were washed three times with pre-warmed Hank's buffered saline solution (HBSS) and then incubated in HBSS for 1 hour. After starvation, cells were re-supplemented with complete medium for 1 hour before harvest.
Plasmids and transfection
The following plasmids were obtained from Addgene (Cambridge, MA USA) and directly used in the study: EGFP- microtubule-associated protein light chain 3 (EGFP-LC3) (Plasmid #11546), pBabe-puro-mCherry-EGFP-LC3 (plasmid #22418) and mouse kif5B (pKin1B, plasmid #31604). Kif5B were PCR amplified and subcloned into pEGFP-C1 vector. pcDNA3.1-full length htt23Q (fhtt23Q) and pcDNA3.1-fhtt145Q were originally obtained from HD Community BioRepository Collection. EGFP was engineered to the C-terminus of fhtt23Q (fhtt23Q-EGFP) and fhtt145Q (fhtt145Q-EGFP). Cells were transiently transfected with different plasmids using Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA).
Retrovirus infection and generation of mCherry-EGFP-LC3 stable cell lines
Phoenix-Ampho cells were transiently transfected with pBabe-puro-mCherry-EGFP-LC3 plasmid using Lipofectamine 2000. Seventy-two hours after transfection, viral supernatant were collected, filtered and directly used to infect STHdhQ7 and STHdhQ111 cells with 8 μg/ml polybrene (Sigma). After infection, STHdhQ7 and Q111 cells stably expressing mCherry-EGFP-LC3 were selected in the presence of 5 μg/ml puromycin (Sigma) until single colonies developed. Stable cell lines were maintained in complete medium containing 2.5 μg/ml puromycin.
Sample preparation and Western blot
After treatment or transfection, cells were quickly lysed in 1X SDS sample buffer containing 1X complete protease inhibitor cocktail from Sigma and 1X phosphatase inhibitor cocktail from Pierce and boiled at 95°C for 5 min. Protein concentration was determined using Bradford method. About 20 μg of sample lysates were separated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked in blocking buffer for fluorescent Western blotting (Rockland, Limerick, PA, USA) for 2 hours at room temperature and then incubated with primary antibodies overnight at 4°C. The following primary antibodies were used: rabbit anti-LC3B (1:500, Cat. #3868), rabbit anti-phospho-p70S6K (Thr389) (1:500, Cat. #9234), rabbit anti-phospho-mTORC1(Ser2448) (1:500, Cat. #5536) and rabbit anti-mTORC1 (1:500, Cat. #2983) were purchased from Cell Signaling Technology (Danvers, MA, USA). Mouse anti-actin (1:1000, Cat. sc-47778), rabbit anti-GFP (1:500, sc-8334) and mouse anti-p70S6K (1:500, Cat. sc-8418) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Rabbit p62/SQSTM1 (1:500, Cat. 18420-1-AP) was purchased from Proteintech (Chicago, IL, USA) and rat anti-lysosomal associated membrane protein 1 (Lamp1, 1:500, antibody name 1D4B) was obtained from Developmental Studies Hybridoma Bank (Iowa city, IA, USA). After primary antibodies incubation, the membranes were washed three times with Tris-buffered saline containing 0.1% Tween-20 (TBS-T) and incubated with secondary antibodies for 1 hour at room temperature. Secondary antibodies used were goat IRDye 800-conjugated anti-rabbit (1:5,000) and IRDye 680 conjugated anti-mouse (1:10,000) antibodies (LI-COR Biosciences, Lincoln, NE, USA). Fluorescent signals were detected with a LI-COR Odyssey Fc system and the images were quantified with the provided Image Studio 2.0 software.
Immunofluorescence and quantification
Cells were grown on coverslips coated with poly-D-lysine. Cells transfected with fluorescent probes were fixed in 2% paraformaldehyde in PBS for 5 min at 33°C to preserve fluorescent signals and then permeablized in 0.25% Triton X-100 in PBS for 10 min. For endogenous LC3 and Lamp1 staining, cells were fixed and permeablized in cold methanol at −20°C for 1 min. After fixation and permeablization, cells were blocked in blocking buffer [10% normal goat serum (NGS), 1% bovine serum albumin (BSA) in PBS] for 1 hour. Cells were then incubated with primary antibodies diluted in dilution buffer (1% NGS, 1% BSA in PBS) for 1 hour. The following antibodies were used: rabbit anti-LC3B (1:500) and rat anti-Lamp1 (1:500). Following primary antibody incubations, cells were further incubated with respective goat Alexa Fluor 488 or 594 secondary antibodies (Invitrogen, 1:2000 dilution in 1% NGS, 1% BSA in PBS) for 1 hour. Between steps, cells were extensively washed with PBS (3 X 5min) to remove any unbound antibodies. All incubations were carried out at the room temperature. The coverslips were mounted in Prolong® Gold antifade reagent (Invitrogen) and immunofluorescence was detected using an immunofluorescence microscope (Zeiss AxioImager D1) or a laser confocal microscope (Zeiss LSM700).
The number of LC3-positive puncti per cell was quantified using the ‘analyze particle’ plugin of the NIH ImageJ program after Background subtraction and Binary. Puncti with sizes above 0.15 μm2 were counted. Over 20 randomly selected cells from each group were analyzed for each condition.
Quantification of lysosomal distribution-perinuclear lysosomal clustering index
To quantitatively estimate the extent of lysosome clustering at the perinuclear region of a cell, we adopted a method described by Falcón-Pérez, et al. (Falcón-Pérez et al., 2005) with modifications (Supplementary Fig. S1). Specifically, mid-plane images of cells with Lamp1 and DAPI staining were taken with a Zeiss LSM700 confocal microscope [Plan-Apochromat 63x oil lens, numerical aperture (NA) 1.40] and transferred to NIH Image J for analysis. In Image J, the nuclear region of a cell was first outlined with the “freehand selection tool” based on DAPI staining. The perinuclear outline was defined as 2 μm away from the nuclear outline. This is achieved with the “Enlarge” function in Image J. Next, Lamp1 signal was adjusted to saturation in order to outline the whole cell using the “freehand selection tool”. The average Lamp1 intensity in the nuclear (FN), perinuclear (FP) and whole cell (FWC) and the areas of these regions (SN, SP and SWC, respectively) were calculated using the “measure” function. Lastly, an area of the background region outside of the cell was randomly selected and the average Lamp1 intensity in the background (FB) was measured. The average intensity in the perinuclear region was calculated according to the following formula: FPN=[(FP × SP-FN × SN)/(SP-SN)]-FB. Similarly, the average cytosolic Lamp1 intensity was calculated: FC=[(FWC × SWC-FN × SN)/(SWC-SN)]-FB. The perinuclear lysosomal clustering index was expressed as the ratio of FPN to FC (FPN/FC).
Intracellular pH measurement
Intracellular pH was determined using a pH-sensitive fluorescent dye, 2′ 7′ -bis-(2-carboxyethyl)-5-(and-6)- carboxyfluorescein-acetoxymethyl (BCECF) (Invitrogen). Cells were harvested by trypsinization and resuspended in HBSS. BCECF was added to the cell suspension at a final concentration of 1 μM and incubated for 30 min at 33°C. Cells were then washed with complete culture medium twice and then resuspended in HBSS. To establish the pH calibration curve, cells after washing were incubated with intracellular pH calibration buffer containing 10 μM nigericin and 10 μM valinomycin (pH=4.5, 5.5, 6.5 and 7.5, Invitrogen) for 15 min. All samples were aliquoted at 100 μl into a black-wall 96-well plate. The emission at 535 nm in response to excitation at 505 nm and 440 nm was measured in a SpectraMax microplate reader (Molecular Devices, Sunnyvale, CA). The ratio of emissions in response to excitation at 505 nm and 440 nm was calculated for each sample and plotted against pH values. The pH was determined from the linear standard curve generated with the pH calibration samples.
Fluorescence Recovery After Photobleaching (FRAP)
Cells were seeded on a 35mm glass bottom dish. Before FRAP experiments, cells were incubated with 50 nM LysoTracker Red DND-99 (Invitrogen) for 30 min at 33°C, followed by brief washes with the complete medium. FRAP experiments were carried out with a Zeiss LSM700 confocal microscope at room temperature. Images were taken with a Zeiss Plan-Apochromat 63x oil lens (NA 1.40). Three pre-bleached images were first acquired to record the original intensity of lysosome labeling of the foci. Photobleaching in the designated ROI area was performed with a 555 nm solid-state laser at 100% intensity for 20 iterations. The bleaching time lasted for 4 seconds. Subsequently, images were acquired every second for 2 min at a maximum scanning speed. FRAP analysis was performed using “FRAP profiler” plugin of Image J. Curve fitting method was “single exponential recovery” and the equation used was p[0]*(1-exp(-p[1]*x). The half-time (τ1/2) to reach to maximum recovery level and mobile fractions were also calculated.
Statistical analysis
All data were expressed as means ± S.E.M. To establish significance, data were subjected to unpaired student's t-tests or one-way ANOVA followed by the Tukey's multiple comparison test using the GraphPad Prism software statistical package 6.0 (GraphPad Software). The criterion for significance was set at P ≤ 0.05.
RESULTS
Increased accumulation of lysosomes in the perinuclear region of cells expressing mHtt
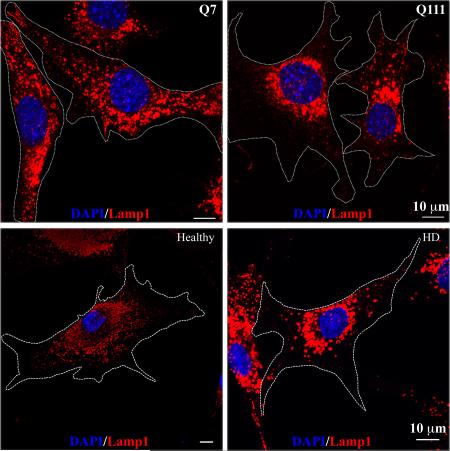
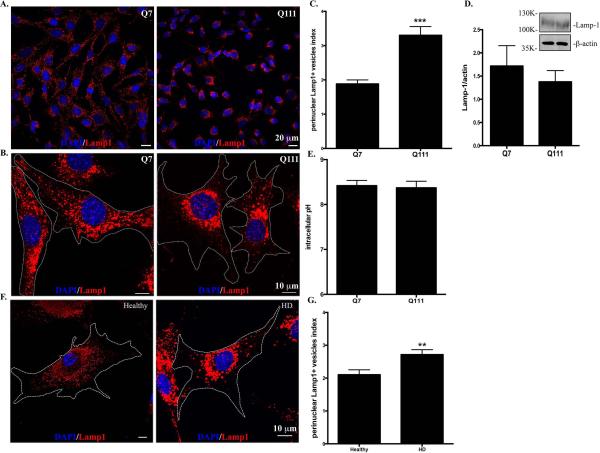
We first investigated the subcellular distribution of lysosomes in two clonal striatal cell lines derived from wild-type (STHdhQ7/Q7, hereafter referred as STHdhQ7) and mHtt (STHdhQ111/Q111, hereafter referred as STHdhQ111) knock-in mice (Trettel et al., 2000). Using antibodies against Lamp1, a specific lysosomal marker, we found that lysosomes were distributed throughout the cytosol of STHdhQ7 cells, including lamellipodia (Fig. 1A, for lower magnification, Fig. 1B, for higher magnification). In contrast, this distribution pattern was changed in STHdhQ111 cells with more lysosomes accumulated in the perinuclear regions (Fig. 1A-B). To further quantify the differences, we calculated the perinuclear lysosomal clustering index, which is the intensity ratio of perinuclear to cytosolic Lamp1 staining (Supplementary Fig. S1). An index of 1 should indicate that lysosomes are evenly distributed in the cytosol and perinuclear region of the cell. The average index number for STHdhQ7 cells is 1.882±0.118 (Fig. 1C), suggesting a moderate accumulation of lysosomes in the perinuclear regions, which is consistent with the current understanding of the lysosomal distribution in a normal cell. In contrast, STHdhQ111 cells had a much higher index number (3.302±0.256), indicating an increased lysosomal distribution in the perinuclear region (Fig. 1C, t98 = 4.62, P < 0.0001,). Notably, we did not find a significant change in Lamp1 protein expression between these two cell lines (Fig. 1D, t16 = 0.68, P = 0.51), ruling out the possibility that the observed differences in lysosomal positioning is due to changes in Lamp1 protein levels. It was reported that intracellular pH (pHi) controls lysosomal positioning (Heuser, 1989), we therefore measured pHi and found no significant differences between STHdhQ7 and STHdhQ111 cells (Fig. 1E, t6=0.26, P = 0.80). We further examined the lysosomal distribution in primary fibroblasts from a healthy individual and a HD patient. Similarly, more lysosomes were accumulated in the perinuclear regions of HD fibroblasts compared to normal fibroblasts (Fig. 1F-1G, t44=2.80, P = 0.0077). To exclude the possibility that an artifact in the immunostaining procedure might cause the differences in lysosomal positioning, we also stained lysosomes with LysoTracker Red DND-99 in live cells. Consistently, we observed an increased perinuclear accumulation of lysosomes in STHdhQ111 cells compared to STHdhQ7 cells (Supplementary Fig. S2). Taken together, our data suggest that the perinuclear accumulation of lysosomes is increased in HD cells.
Figure 1.
Lysosomes are accumulated in the perinuclear regions of HD cells. Cells were methanol-fixed and immunostained with Lamp1 (red) and counterstained with DAPI (blue). A. Representative images of Lamp1 staining in STHdhQ7 and Q111 cells at lower magnification (EC Plan-Neofluar 40x oil, NA 1.30). B. Representative images of Lamp1 staining in STHdhQ7 and Q111 cells at higher magnification (Plan-Apochromat 63x oil lens, NA 1.40). Cell boundaries were outlined. C. Quantitative analysis of perinuclear lysosomal clustering index in STHdh cells. A total of 44 cells for STHdhQ7 and 56 cells for STHdhQ111 were analyzed. ***P < 0.001, compared to STHdhQ7 cells, student's t-test. D. Western blot analysis of Lamp1 protein expression levels in STHdh cells. The graph represents the quantification of Lamp1 levels relative to β-actin in four independent replicates. E. Radiometric measurement of intracellular pH using an intracellular pH indicator, BCECF, in STHdh cells. The value represents four independent measurements. F. Representative images of Lamp1 staining in human primary fibroblasts from a healthy individual and a HD patient. Cells were methanol-fixed and immunostained with Lamp1 (red) and counterstained with DAPI (blue). G. Quantitative analysis of perinuclear lysosomal clustering index in human primary fibroblasts (a total of 22 for healthy and 27 for HD primary fibroblasts were analyzed). **P < 0.01, compared to the healthy control, student's t-test.
Changes in lysosomal mobility in cells expressing mHtt
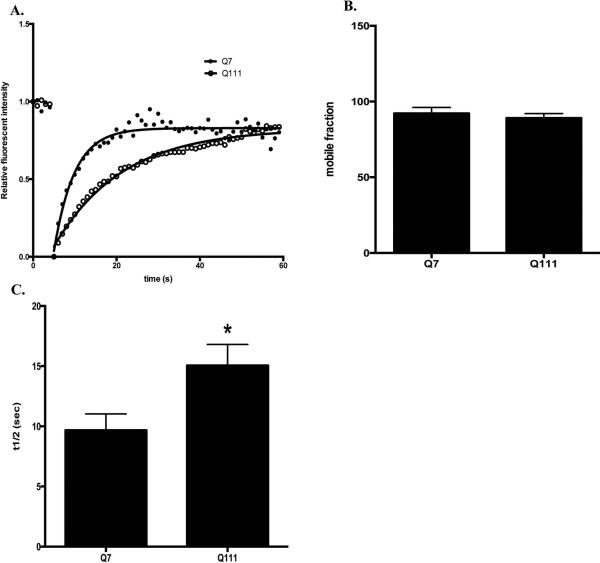
We next investigated whether lysosomal dynamics is affected in STHdhQ111 cells with FRAP analysis. A designated area of lysosomes labeled with LysoTracker Red DND-99 were subjected to photobleaching and the dynamic fluorescent recovery after bleaching is shown in Fig. 2A (also see Supplementary Fig. S3 and S4 for the representative time-lapse images before and after bleaching in STHdhQ7 and STHdhQ111 cells, respectively). No difference in the percentage of mobile lysosomes was observed in these two groups (Fig. 2B, t26 = 0.64, P = 0.53). However, it took a longer time for fluorescent recovery of labeled lysosomes in STHdhQ111 cells, suggesting that lysosomes in STHdhQ111 cells moved slower (Fig. 2A). Indeed, the half-time to reach to maximum fluorescent recovery increased from 9.7±1.4 seconds in STHdhQ7 cells to 15.1±1.7 seconds in STHdhQ111 cells (Fig. 2C, t26 = 2.38, P = 0.025).
Figure 2.
Lysosomal mobility is reduced in STHdhQ111 cells. Lysosomes in live cells were labeled with LysoTracker Red DND-99 and subjected to FRAP analysis. A. Representative traces of time-dependent LysoTracker fluorescent recovery after bleach in STHdhQ7 and STHdhQ111 cells. B. Quantitative analysis of mobile fractions of lysosomes in STHdhQ7 (n=13) and STHdhQ111 cells (n=15). C. Quantitative analysis of the half-time for full fluorescence recovery in STHdhQ7 (n=13) and STHdhQ111 cells (n=15). *P < 0.05, student's t-test.
Mutant huntingtin causes increased perinuclear accumulation of lysosomes in HD cells
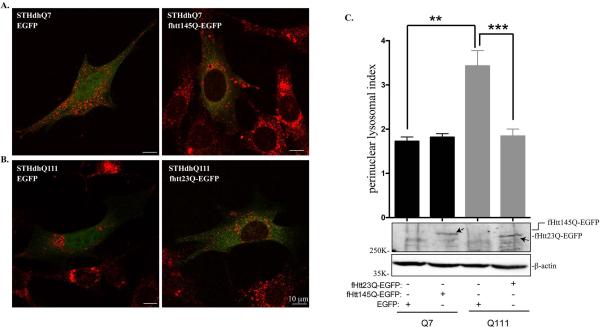
Normal Htt has been reported to coordinate retrograde transport of lysosomes in HeLa cells (Caviston et al., 2011). The effect of mHtt on lysosomal trafficking has not been investigated. We then examined whether there is a causal relationship between perinuclear lysosomal positioning and mHtt expression. To address this, we transiently transfected EGFP or fHtt145Q-EGFP in STHdhQ7 cells and fHtt23Q-EGFP in STHdhQ111 cells. Lysosomal positioning in the transfected cells was then analyzed with Lamp1 immunostaining. If mHtt is responsible for perinuclear accumulation of lysosomes in STHdhQ111 cells, we expected that expression of normal Htt in STHdhQ111 cells would reverse this phenomenon and vice versa. Expression of EGFP alone has no effect on lysosome positioning in both cell lines (Fig. 3A-B, left panels). Expressing fHtt145Q-EGFP in STHdhQ7 cells caused lysosomes to accumulate in the perinuclear region as indicated by Lamp1 staining (Fig. 3A, right panel). Quantitative analysis from a large number of transfected cells showed an increase in the perinuclear lysosomal index, but did not reach to significance (1.73±0.10 in STHdhQ7 cells transfected with EGFP alone vs. 1.82±0.08 in STHdhQ7 cells expressing fHtt145Q-EGFP, Fig. 3C, t63 = 0.66, P = 0.51). The underlying cause needs to be further determined. One possibility is that over-expressed fHtt145Q-EGFP formed perinuclear aggregates in some of the transfected STHdhQ7 cells (Supplementary Fig. S5), and the aggregates may have no effect on lysosomal trafficking. On the contrary, expressing fHtt23Q-EGFP in STHdhQ111 cells apparently redirected lysosomes to the peripheral regions (Fig. 3B, right panel). Moreover, perinuclear lysosomal index was significantly decreased in STHdhQ111 cells overexpressing normal Htt compared to STHdhQ111 cells expressing EGFP alone (1.84±0.16 vs. 3.43±0.35, Fig. 3C, t35 = 4.47, P < 0.0001). Collectively, these data suggest that mHtt interferes with normal distribution of lysosomes and overexpressing normal Htt can redirect lysosomes to the periphery in HD cells.
Figure 3.
Normal Htt expression reverses abnormal perinuclear lysosomal positioning in STHdhQ111 cells. A-B. Representative confocal images of Lamp1 staining in (A) STHdhQ7 cells transfected with EGFP alone or fHtt145Q-EGFP and in (B) STHdhQ111 cells transfected with EGFP alone or fHtt23Q-EGFP. Cells were fixed in 2% PFA and stained with Lamp1 (red). C. Top panel, quantitative analysis of perinuclear lysosomal clustering index in STHdhQ7 cells transfected with EGFP (n=21) or Htt145Q-EGFP (n=47) and STHdhQ111 cells transfected with EGFP (n=8) or htt23Q-EGFP (n=29). **P < 0.01, STHdhQ111 vs. STHdhQ7 transfected with EGFP; ***P < 0.001, STHdhQ111 transfected with htt23Q-EGFP vs. STHdhQ111 transfected with EGFP, student's t-test. Bottom panel, western blot analysis of ectopically expressed fHtt23Q-EGFP and fHtt145Q-EGFP in STHdh cells with GFP antibodies.
Increased perinuclear lysosomal positioning promotes basal mTORC1 activity in STHdhQ111 cells
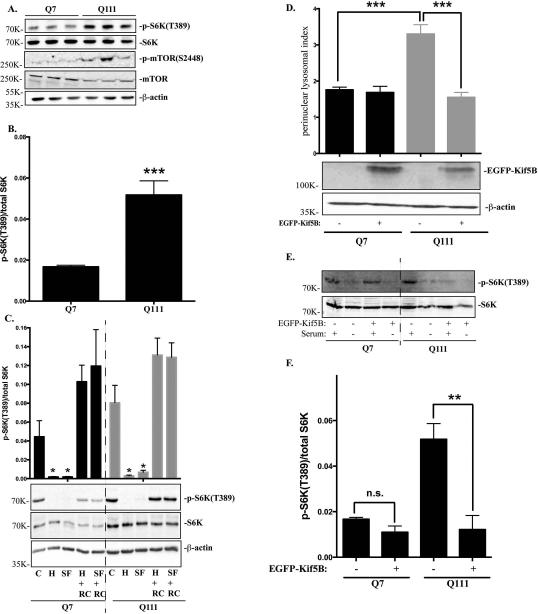
Lysosomal positioning has been closely related to nutrient response in non-neuronal cells by regulating mTORC1 activity (Korolchuk et al., 2011). We then asked whether the observed differences in lysosomal positioning affects mTORC1 activity in STHdhQ7 and STHdhQ111 cells. We first investigated mTORC1 activation as reflected by the phosphorylation status of p70S6 kinase (S6K) at Threonine 389, an mTORC1-specific phosphorylation site. The level of phospho-S6K was significantly increased in STHdhQ111 cells, suggesting that basal mTORC1 activity was elevated in STHdhQ111 cells (Fig. 4A-B, t19 = 4.82, P = 0.0001). Consistently, phosphorylation of mTORC1 at Serine 2448, another indicator of mTORC1 activation, was also up-regulated in STHdhQ111 cells (Fig. 4A). mTORC1 activity is quickly inhibited when cells are under nutrient deprivation and activated when cells are re-supplemented with nutrients (Demetriades et al., 2014; Menon et al., 2014). Changes in mTORC1 activity in response to nutrients appears to correlate with changes in lysosomal positioning during nutrient challenges (Korolchuk et al., 2011). Therefore, we next investigated whether this sharp regulation of mTORC1 activity is impaired in HD cells. Cells were subjected to two types of starvation: serum starvation, in which cells were incubated in DMEM medium (SF), or amino acid and serum deprivation, in which cells were incubated in HBSS for one hour. Under both starvation conditions, mTORC1 activity was greatly decreased in both cell types (Fig. 4C). When cells were re-supplemented with nutrients for one hour, mTORC1 were quickly activated in both cell types, indicating that the lysosome nutrient sensing machinery is not impaired in HD cells (Fig. 4C). Lastly, we investigated whether increased perinuclear lysosomal positioning contributes to the elevated basal mTORC1 activity in STHdhQ111 cells. Kinesin motor protein appears to have a key role in the positioning of lysosomes. It was reported that knocking out kinesin family member 5B (Kif5B) causes perinuclear accumulation of lysosomes in neurons (Tanaka et al., 1998), suggesting that Kif5B is important for anterograde transport of lysosomes. Overexpressing Kif5B redirected lysosomes to the peripheral region and significantly reduced the perinuclear lysosomal index in STHdhQ111 cells (Fig. 4D, ***P < 0.001, one-way ANOVA followed by Tukey's post hoc test). Moreover, p-S6K levels were also greatly reduced in STHdhQ111 cells overexpressing Kif5B compared to STHdhQ111 cells alone under basal conditions, indicating that overexpression of Kif5B can reduce mTORC1 activity in STHdhQ111 cells (Fig. 4E-F, **P < 0.01, one-way ANOVA followed by Tukey's post hoc test).
Figure 4.
mTORC1 basal activity is increased in STHdhQ111 cells. A. Western blot analysis of total p70S6K (S6K), phospho-S6K(T389), phospho-mTOR(S2448) and total mTOR expression in three independent STHdhQ7 and Q111 cell lysates. β-actin was used as the loading control. B. mTORC1 activation as quantified by the ratio of phospho-p70S6K to total p70S6K in STHdhQ7 and Q111 cells (***P < 0.001, student's t-test, n=3). C. Changes of mTORC1 activity during nutrient starvation and recovery. Cells were incubated with HBSS (for amino acid and serum starvation) or DMEM (for serum starvation only) for 1 hour. After starvation, cells were re-supplemented with complete medium for another one hour before harvest. C, control; H, HBSS; SF, serum free (DMEM only); H+RC, HBSS treatment followed by medium recovery. SF+RC: serum deprivation followed by medium recovery. Top panel, quantification of changes in p-S6K/S6K changes from three independent experiments (*P < 0.05, student's t-test, n=3, compared to their respective controls). Bottom panel, a representative western blot image is shown. D. Overexpressing Kif5B reduces perinuclear lysosomal clustering index in STHdhQ111 cells. STHdh cells transfected with EGFP-Kif5B or EGFP alone were immunostained with Lamp1 and confocal images were taken. Top panel, perinuclear lysosomal clustering indexes from STHdhQ7 cells transfected with EGFP (n=36) or EGFP-Kif5B (n=22) and STHdhQ111 cells transfected with EGFP (n=56) or EGFP-Kif5B (n=21). ***P < 0.001, student's t-test. Bottom panel, western blot analysis of EGFP-Kif5B overexpression. E. Western blot analysis of total p70S6K (S6K), phospho-S6K(T389) in STHdh cells overexpressing EGFP-Kif5B in the presence or absence of serum deprivation. F. Quantitative analysis of mTORC1 activation as indicated by the ratio of phospho-p70S6K to total p70S6K in STHdhQ7 and Q111 cells overexpressing EGFP-Kif5B (*** P < 0.01, student's t-test, n=3).
Perinuclear lysosomal positioning accelerates autophagic influx in STHdhQ111 cells under serum deprivation
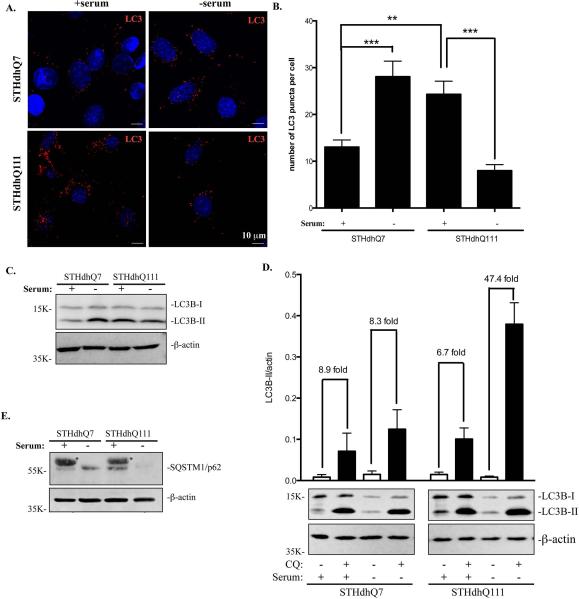
Another important function of lysosomes is to degrade cytoplasmic materials and organelles delivered by autophagosomes. It is known that autophagosomes are retrogradely transported to the perinuclear region to fuse with lysosomes. We thus hypothesized that changes in lysosomal positioning could have an impact on autophagic influx, i.e., the fusion of autophagosomes with lysosomes in STHdhQ111 cells. Upon induction of autophagy, LC3 is conjugated to phosphatidylethanolamine (LC3-II) and targeted to autophagosomal membranes. Therefore, changes in LC3-II expression has been used as a reliable marker to monitor autophagy (Kabeya et al., 2000). We first performed immunofluorescent studies in STHdhQ7 and Q111 cells with LC3 antibodies to label autophagosomes and autophagolysosomes. Interestingly, the number of LC3-positive puncti was increased in STHdhQ111 cells when compared to STHdhQ7 cells at basal conditions (Fig. 5A-B). We next examined serum starvation-induced autophagy responses in these two cell lines. Serum deprivation caused an increase in the number of LC3-positive puncti in STHdhQ7 cells (Fig 5A-B). However, the number of LC3-positive puncti was significantly reduced in STHdhQ111 cells under serum deprivation (Fig. 5A-B). This change correlated with a decrease in LC3-II conversion in STHdhQ111 cells as analyzed by Western blot (Fig. 5C).
Figure 5.
Autophagic influx is increased in STHdhQ111 cells under serum deprivation. For serum deprivation, cells were incubates in DMEM medium for 24 hours. A. Representative images of endogenous LC3 staining under different conditions. Cells were stained with LC3 antibodies and counterstained with DAPI. B. Quantitative analysis of the number of LC3-positive puncti per cell in SThdhQ7 (n=27), STHdhQ7 with serum deprivation (n=15), STHdhQ111 (n=19) and STHdhQ111 cells with serum deprivation (n=12). ***P < 0.001, **P < 0.01, student's t-test. C. Western blot analysis of endogenous LC3-II expression in STHdhQ7 and STHdhQ111 cells in the presence or absence of serum. β-actin was used as the loading control. D. Quantitative analysis of the fold changes in LC3-II expression under different conditions by densitometry. Cells were incubated with 50 μM CQ in the presence or absence of serum for 24 hours. A representative image of Western blot analysis of endogenous LC3-II expression in STHdhQ7 and STHdhQ111 cells under different conditions is shown below the graph. Data represent statistical analysis of four independent Western blots. E. Western blot analysis of p62 levels in STHdhQ7 and STHdhQ111 cells in the presence or absence of serum. β-actin was used as the loading control. * denotes non-specific bands from serum.
Decreased LC3-II levels are associated with either decreased autophagosomal synthesis or increased autophagosomal turnover (autophagic influx). To further test which pathway is responsible for the decreased LC3-II levels (Fig. 5A-C), we incubated cells with hydroxychloroquine (CQ), which blocks lysosome acidification and thereby autophagic influx and degradation of LC3-II. As expected, CQ treatment greatly increased LC3-II levels in all conditions as analyzed by Western blot (Fig. 5D). Further quantitative analysis revealed that CQ treatment caused similar folds increase in LC3-II levels in STHdhQ7 cells under normal or serum-free conditions (Fig. 5D), suggesting that serum deprivation did not significantly affect the rate of autophagic influx in STHdhQ7 cells. Interestingly, CQ treatment caused a ~47-fold increase in LC3-II levels under serum deprivation compared to a ~7-fold increase under normal conditions in STHdhQ111 cells (Fig. 5D), indicating that the rate of autophagosomal fusion with lysosomes was increased in STHdhQ111 cells in response to serum deprivation. To further confirm this, we also measured changes in p62 levels in response to serum starvation. P62 binds to LC3 and is itself degraded by autophagy (Bjørkøy et al., 2005; Pankiv et al., 2007). Therefore, changes in p62 levels should reflex autophagic influx. Indeed, p62 levels were greatly reduced in STHdhQ111 cells in response to serum starvation (Fig. 5E).
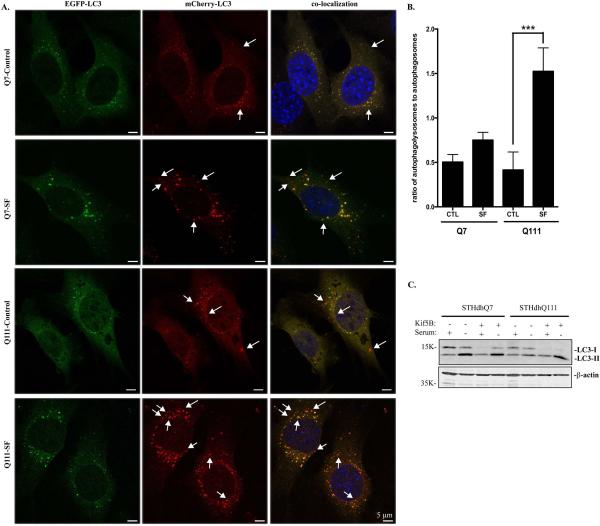
We then employed the tandem mCherry-EGFP-LC3 fluorescence analysis in which mCherry is more stable in acidic compartments whereas EGFP fluorescence is rapidly degraded. Under normal condition, the LC3 staining appears as yellow as the result of detecting both mCherry and GFP signals. If serum deprivation causes a rapid fusion of lysosomes and autophagosomes, we then expect more red-only signal after serum deprivation. Consistent with Fig. 5, we detected an increased autophagic influx in STHdhQ111 cells compared to STHdhQ7 cells, as indicated by more red-only autophagolysosomes in STHdhQ111 cells under serum deprivation (Fig. 6A-B). Collectively, our data indicate that autophagic influx is increased in STHdhQ111 cells in response to serum deprivation.
Figure 6.
A-B. Analysis of autophagic influx by tandem LC3 expression. A. Representative images showing the expression pattern of mCherry-EGFP-LC3 in STHdhQ7 and Q111 cells under different conditions. STHdhQ7 and Q111 cells stably expressing mCherry-EGFP-LC3 were subjected to serum deprivation for 18 hours. Arrows indicate red-only puncti. B. Statistical analysis of the ratio of autophagolysosomes to autophagosomes. About 20 cells were analyzed in each group. ***P < 0.001, student's t-test. SF, serum free. C. Overexpression of Kif5B in STHdhQ111 cells increased LC3-II expression under serum deprivation. Cells were transfected with EGFP-Kif5B for 24 hours and serum-starved for another 24 hours before harvest for Western blot analysis of LC3 levels.
Lastly, we asked whether serum starvation-induced increase in autophagic influx in STHdhQ111 cells is related to the perinuclear lysosomal positioning. EGFP-Kif5B was overexpressed in STHdhQ111 cells to redirect lysosomes to the perinuclear region. LC3-II levels were increased in STHdhQ111 cells overexpressing Kif5B under serum deprivation (Fig. 6C), indicating that accumulation of lysosomes in the perinuclear region of STHdhQ111 cells facilitates autophagosomal fusion with lysosomes under serum deprivation.
DISCUSSION
Increasing evidence suggest that lysosomes are crucial regulators of cell homeostasis, playing important roles in degrading macromolecules and regulating nutrient response (Boya, 2012). It is known that Htt is associated with lysosomal membranes, suggesting a possible role of Htt in lysosomal functions (Kegel et al., 2000). However, whether mHtt affects lysosomal functions is not clear. In this study, we present evidences showing that the perinuclear accumulation of lysosomes is increased in HD cells and may play important roles in the pathogenesis of HD by regulating basal mTORC1 activity and autophagic influx under serum deprivation.
Htt and lysosomal trafficking
The underlying molecular mechanisms for increased perinuclear accumulation of lysosomes in HD cells remains to be further investigated. Based on the known role of Htt in protein trafficking and our data, it is reasonable to suggest that Htt is involved in lysosomal trafficking and mHtt expression may decrease the anterograde or facilitate the retrograde lysosomal transport, resulting in an increased accumulation of perinuclear lysosomes.
First, it is interesting to know whether Htt is directly associated with lysosomes and affects their trafficking. Using a clonal striatal cell line overexpressing Flag-Htt, it was shown that Htt is co-localized to vesicles positive for cathepsin D, a lysosomal marker, by immunofluorescence and electron microscopic studies (Kegel et al., 2000). However, overexpression of Flag-Htt in a cell line may cause it to mis-localize. Therefore, it is important to investigate the subcellular localization of endogenous Htt. We co-stained endogenous Htt with Lamp1 in PFA-fixed STHdhQ7 and STHdhQ111 cells and found that some of the Htt-positive puncti co-localized with Lamp-1 positive vesicles (Supplementary Fig. S6A-B) and polyQ expansion did not affect the association (Supplementary Fig. S6C, t6 = 1.55, P = 0.17). However, it is well known that there are pitfalls when defining the subcellular localization of a protein with immunocytochemistry, which includes antibodies against different immunogenic epitopes, fixation methods and detection techniques used (Hughes & Jones, 2011). Therefore, although our data suggest that Htt is associated with lysosomes, we could not rule out the possibility that it is affected by our immunostaining method. Moreover, assuming immunohistochemistry gives the correct subcellular localization of Htt, obtaining super resolution images with super-resolution microscopy, is needed to clearly examine the co-localization of Htt with certain organelles including lysosomes.
Alternatively, Htt acts as a scaffold protein by binding to huntingtin-associated protein 1 (HAP1), which mediates its association with kinesin and dynein–dynactin motor proteins and stimulates the trafficking of various membrane-bound organelles in both directions (Caviston & Holzbaur, 2009). Whereas normal Htt promotes bi-directional mitochondrial transport, mHtt disrupts the formation of transport complexes and impairs mitochondrial movement (Sheng & Cai, 2012). Several studies strongly suggest that the htt-HAP1 complex associates with Kif5 and regulate the trafficking of brain-derived neurotrophic factor (Colin et al., 2008), GABAA receptor (Twelvetrees et al., 2010) and autophagosomes (Wong & Holzbaur, 2014). In neurons, Kif5B plays a more important role in regulating vesicular trafficking (Hirokawa & Takemura, 2005). Knocking out Kif5B is embryonically lethal with abnormal perinuclear clustering of lysosomes and mitochondria (Tanaka et al., 1998). In the present study, we demonstrate that overexpressing Kif5B directs lysosomes to the peripheral region in HD cells. It is thus possible that mHtt preferentially impairs anterograde transport of lysosomes through interactions with HAP1 and Kif5B, leading to perinuclear lysosomal clustering.
Lysosomal positioning and mTORC1 activity
We found that basal mTORC1 activity is increased in STHdhQ111 cells. Consistent with our study, it was recently reported that mTORC1 activity was increased in STHdhQ111 cells and N171-Q82 transgenic HD mice (Pryor et al., 2014). Elevated mTORC1 activity appears to be important for HD pathogenesis since abnormally high mTORC1 activity promotes rapid accumulation of cytoplasmic inclusions and causes neurodegeneration (Kassai et al., 2014). The molecular basis for increased mTORC1 activity in HD cells is poorly understood. Lysosomes are the primary activation sites for mTORC1 activity (Demetriades et al., 2014; Menon et al., 2014). It is possible that mHtt has an enhanced affinity for Rheb/mTORC1 complex, which is further stabilized on perinuclear structures by amino acids signals, leading to sustained mTORC1 activity (Pryor et al., 2014). Our data alternatively suggest that lysosomal positioning is another key factor in regulating mTORC1 activity. It is possible that the perinuclear accumulation of lysosomes activates mTORC1 activity by providing more docking sites for mTORC1 complex and its co-activators. In line with this, mTOR, Rheb and Rag are mostly concentrated in the perinuclear region (Yadav et al., 2013). It is also supported by our data showing that redirecting lysosomes to peripheral area by overexpressing Kif5B in STHdhQ111 cells significantly reduced mTORC1 activity when using phospho-S6K as readout.
Lysosomal positioning and autophagic influx
We demonstrate in this study that another functional aspect of accumulated perinuclear lysosomal positioning is associated with increased autophagic influx in response to serum starvation. As fusions between lysosomes and autophagosomes primarily occur in the perinuclear region, increasing the number of perinuclear lysosomes can facilitate such a fusion. We attempted to use live cell imaging to track and quantify the fusion rate in STHdh cells stably expressing mCherry-GFP-LC3 without success. This is possibly due to the fact that fusion occurs quickly in less than 60 seconds and most of the fusions occur in a kiss-and-run pattern (Jahreiss et al., 2008). Increased autophagic influx under serum deprivation suggests that HD cells have a defective autophagic response to nutrient stress. Since autophagosomes fuse faster with lysosomes under serum deprivation, autophagosomes do not have time to collect cargos, leading to the pre-mature fusion of lysosomes with empty autophagosomes in HD cells. The presence of empty autophagosomes in HD cells has been demonstrated by electron microscopy studies (Martinez-Vicente et al., 2010). It has been reported that anterograde transport of autophagosomes is impaired in polyQ-htt expressing cells, preventing it from collecting cellular wastes (Wong & Holzbaur, 2014). In our studies, overexpression of Kif5B could redirect lysosomes away from the perinuclear region and prevent the premature autophagosome-lysosomal fusion upon serum deprivation in HD cells. Although we cannot rule out the possibility that Kif5B directly participates in anterograde transport of autophagosomes, our data support the idea that perinuclear accumulation of lysosomes facilitates their fusion with autophagosomes. Collectively, our data suggest that defective autophagic response contribute to the protein degradation failure and accumulation of damaged protein in HD cells, which makes HD cells more vulnerable to other toxic stimuli, contributing to further cellular dysfunctions and neurodegeneration in HD.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by National Institute of Neurological Disorders and Stroke (R15NS066339-02 to J.W.).
ABBREVIATIONS
- BCECF
2′ 7′ -bis-(2-carboxyethyl)-5-(and-6)- carboxyfluorescein-acetoxymethyl
- CQ
hydroxychloroquine
- DMEM
Dulbecco's modified eagle medium
- EGFP
enhanced green fluorescent protein
- FRAP
fluorescence recovery after photobleaching
- HAP1
huntingtin-associated protein 1
- HBSS
HEPES-buffered saline solution
- HD
Huntington's disease
- Htt
huntingtin
- Kif5B
kinesin family membrane 5B
- Lamp1
lysosomal associated membrane protein 1
- LC3
microtubule-associated protein light chain 3
- LC3-II
LC3 conjugated to phosphatidylethanolamine
- mHtt
mutant Htt
- mTORC1
mammalian target of rapamycin complex 1
- NGS
normal goat serum
- pHi
intracellular pH
- polyQ
polyglutamine
- Rheb
Ras homology enrich in brain
- S6K
p70S6 kinase
Footnotes
There are no conflicts of interest to be reported.
REFERENCES
- Atwal RS, Xia J, Pinchev D, Taylor J, Epand RM, Truant R. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum. Mol. Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P. Lysosomal Function and Dysfunction: Mechanism and Disease. Antioxid. Redox Signal. 2012;17:766–774. doi: 10.1089/ars.2011.4405. [DOI] [PubMed] [Google Scholar]
- Caviston JP, Holzbaur ELF. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 2009;19:147–155. doi: 10.1016/j.tcb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston JP, Zajac AL, Tokito M, Holzbaur ELF. Huntingtin coordinates the dynein-mediated dynamic positioning of endosomes and lysosomes. Mol. Biol. Cell. 2011;22:478–492. doi: 10.1091/mbc.E10-03-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin E, Zala D, Liot G, Rangone H, Borrell-Pagès M, Li X-J, Saudou F, Humbert S. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 2008;27:2124–2134. doi: 10.1038/emboj.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional Repression of PGC-1α by Mutant Huntingtin Leads to Mitochondrial Dysfunction and Neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Demetriades C, Doumpas N, Teleman AA. Regulation of TORC1 in Response to Amino Acid Starvation via Lysosomal Recruitment of TSC2. Cell. 2014;156:786–799. doi: 10.1016/j.cell.2014.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcón-Pérez JM, Nazarian R, Sabatti C, Dell'Angelica EC. Distribution and dynamics of Lamp1-containing endocytic organelles in fibroblasts deficient in BLOC-3. J. Cell. Sci. 2005;118:5243–5255. doi: 10.1242/jcs.02633. [DOI] [PubMed] [Google Scholar]
- Heuser J. Changes in lysosome shape and distribution correlated with changes in cytoplasmic pH. J. Cell Biol. 1989;108:855–864. doi: 10.1083/jcb.108.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R. Molecular motors and mechanisms of directional transport in neurons. Nat. Rev. Neurosci. 2005;6:201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- Hughes A, Jones L. Huntingtin localisation studies - a technical review. PLoS Curr. 2011;3:RRN121. doi: 10.1371/currents.RRN1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahreiss L, Menzies FM, Rubinsztein DC. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 2008;9:574–587. doi: 10.1111/j.1600-0854.2008.00701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Then F, Melia TJ, Mazzulli JR, Cui L, Savas JN, Voisine C, Paganetti P, Tanese N, Hart AC, Yamamoto A, Krainc D. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassai H, Sugaya Y, Noda S, Nakao K, Maeda T, Kano M, Aiba A. Selective Activation of mTORC1 Signaling Recapitulates Microcephaly, Tuberous Sclerosis, and Neurodegenerative Diseases. Cell Rep. 2014;7:1626–1639. doi: 10.1016/j.celrep.2014.04.048. [DOI] [PubMed] [Google Scholar]
- Kegel KB, Kim M, Sapp E, McIntyre C, Castaño JG, Aronin N, DiFiglia M. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J. Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, O'Kane CJ, Deretic V, Rubinsztein DC. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 2011;13:453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-J, Orr AL, Li S. Impaired mitochondrial trafficking in Huntington's disease. Biochim. Biophys. Acta. 2010;1802:62–65. doi: 10.1016/j.bbadis.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat. Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. Spatial Control of the TSC Complex Integrates Insulin and Nutrient Regulation of mTORC1 at the Lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr AL, Li S, Wang C-E, Li H, Wang J, Rong J, Xu X, Mastroberardino PG, Greenamyre JT, Li X-J. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J. Neurosci. 2008;28:2783–2792. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun J-A, Outzen H, Overvatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Pryor WM, Biagioli M, Shahani N, Swarnkar S, Huang W-C, Page DT, MacDonald ME, Subramaniam S. Huntingtin promotes mTORC1 signaling in the pathogenesis of Huntington's disease. Sci Signal. 2014;7:ra103. doi: 10.1126/scisignal.2005633. [DOI] [PubMed] [Google Scholar]
- Rockabrand E, Slepko N, Pantalone A, Nukala VN, Kazantsev A, Marsh JL, Sullivan PG, Steffan JS, Sensi SL, Thompson LM. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum. Mol. Genet. 2006;16:61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z-H, Cai Q. Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat. Rev. Neurosci. 2012;13:77–93. doi: 10.1038/nrn3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Chen J, Petrilli A, Liot G, Klinglmayr E, Zhou Y, Poquiz P, Tjong J, Pouladi MA, Hayden MR, Masliah E, Ellisman M, Rouiller I, Schwarzenbacher R, Bossy B, Perkins G, Bossy-Wetzel E. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med. 2011;17:377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Kanai Y, Okada Y, Nonaka S, Takeda S, Harada A, Hirokawa N. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93:1147–1158. doi: 10.1016/s0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F, Cattaneo E, MacDonald ME. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum. Mol. Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- Twelvetrees AE, Yuen EY, Arancibia-Carcamo IL, MacAskill AF, Rostaing P, Lumb MJ, Humbert S, Triller A, Saudou F, Yan Z, Kittler JT. Delivery of GABAARs to synapses is mediated by HAP1-KIF5 and disrupted by mutant huntingtin. Neuron. 2010;65:53–65. doi: 10.1016/j.neuron.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC, Holzbaur ELF. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J. Neurosci. 2014;34:1293–1305. doi: 10.1523/JNEUROSCI.1870-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RB, Burgos P, Parker AW, Iadevaia V, Proud CG, Allen RA, O'Connell JP, Jeshtadi A, Stubbs CD, Botchway SW. mTOR direct interactions with Rheb-GTPase and raptor: sub-cellular localization using fluorescence lifetime imaging. BMC Cell Biol. 2013;14:3. doi: 10.1186/1471-2121-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, Cataudella T, Leavitt BR, Hayden MR, Timmusk T, Rigamonti D, Cattaneo E. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.