A quantitative in vitro assay based on isolated yeast microsomes reveals that SNARE-mediated membrane fusion is involved in atlastin-initiated homotypic ER fusion.
Abstract
Dynamin-like GTPases of the atlastin family are thought to mediate homotypic endoplasmic reticulum (ER) membrane fusion; however, the underlying mechanism remains largely unclear. Here, we developed a simple and quantitative in vitro assay using isolated yeast microsomes for measuring yeast atlastin Sey1p-dependent ER fusion. Using this assay, we found that the ER SNAREs Sec22p and Sec20p were required for Sey1p-mediated ER fusion. Consistently, ER fusion was significantly reduced by inhibition of Sec18p and Sec17p, which regulate SNARE-mediated membrane fusion. The involvement of SNAREs in Sey1p-dependent ER fusion was further supported by the physical interaction of Sey1p with Sec22p and Ufe1p, another ER SNARE. Furthermore, our estimation of the concentration of Sey1p on isolated microsomes, together with the lack of fusion between Sey1p proteoliposomes even with a 25-fold excess of the physiological concentration of Sey1p, suggests that Sey1p requires additional factors to support ER fusion in vivo. Collectively, our data strongly suggest that SNARE-mediated membrane fusion is involved in atlastin-initiated homotypic ER fusion.
Introduction
The ER mediates a variety of essential processes in eukaryotic cells: it synthesizes lipids and provides membranes for various endomembrane organelles and vesicles, it stores calcium ions in its lumen and thereby regulates intracellular calcium homeostasis, and it is the site where nearly all secretory and integral membrane proteins are synthesized and folded. The unique structure of the ER, with its highly dynamic network of sheets and tubules that spreads throughout the cytoplasm, is thought to be critical for these functions (Shibata et al., 2006; Friedman and Voeltz, 2011).
ER tubules and networks are generated and maintained by transmembrane ER-shaping proteins, such as the reticulons and DP1/Yop1p (Voeltz et al., 2006; Hu et al., 2008). These proteins physically interact with each other to introduce positive curvature into the ER membrane, thereby forming the highly curved regions of the ER. In addition, homotypic fusion of ER membranes plays a critical role in the establishment and maintenance of the unique shape of the ER network (Hu et al., 2009; Orso et al., 2009). Members of several distinct protein families have been suggested to mediate homotypic ER fusion. First, dynamin-like GTPases of the atlastin family and their functional orthologues (Sey1p in yeast and RHD3 in plants) are believed to mediate homotypic membrane fusion between ER tubules to form the polygonal ER network (Rismanchi et al., 2008; Orso et al., 2009; Anwar et al., 2012; Chen et al., 2012; Zhang and Hu, 2013). Atlastin molecules in different ER tubules form homodimers in trans in a GTP-dependent manner, thereby bringing these two membranes into close apposition (Orso et al., 2009). Upon GTP hydrolysis and Pi release, the cytosolic domain (CD) of the atlastin homodimers undergoes a dramatic conformational change, pulling the apposed membranes into close proximity and inducing membrane fusion (Bian et al., 2011; Byrnes and Sondermann, 2011). Second, ER-associated SNARE proteins are involved in homotypic ER fusion (Patel et al., 1998; Anwar et al., 2012). SNARE proteins, characterized by their heptad-repeat SNARE motif, mediate most endomembrane fusion events by forming a four-helical bundle between four SNARE motifs provided by one R-SNARE protein and two or three Q-SNARE proteins. Finally, Rab GTPases have been implicated in ER membrane fusion (Turner et al., 1997; English and Voeltz, 2012), with recent studies suggesting that Rab10 and Rab18 regulate ER structure in mammalian cells (English and Voeltz, 2012; Gerondopoulos et al., 2014). Although Rab proteins usually function together with SNARE proteins to support membrane fusion, it remains unclear whether Rab10 mediates homotypic ER fusion through a SNARE-mediated fusion pathway. The Dsl1 complex, which binds and regulates the assembly of ER SNAREs, and the ER SNARE syntaxin-18 were recently found to be Rab18 effectors in Drosophila melanogaster (Gillingham et al., 2014), suggesting that Rab18 is involved in ER fusion via an ER SNARE-mediated mechanism.
Although atlastins, SNAREs, and Rab GTPases appear to play important roles in homotypic ER fusion, it is still unknown how these proteins might communicate with one another to support ER fusion in the same pathway or whether they mediate ER fusion via mutually exclusive pathways. Rab GTPases are often required for SNARE-mediated membrane fusion, acting by mediating membrane docking before fusion or by regulating the assembly of trans-SNARE complexes via their effectors (McBride et al., 1999; Grosshans et al., 2006; Collins and Wickner, 2007). A recent study suggests that ER-associated SNAREs are involved in ER fusion in the absence of atlastins (Anwar et al., 2012). Interestingly, however, whether SNAREs and Rab GTPases are involved in atlastin-mediated homotypic ER membrane fusion has never been examined. Here, we developed a simple and quantitative in vitro assay for investigating homotypic ER fusion that employs isolated yeast ER microsomes. Using this assay, we demonstrated that ER-associated SNARE proteins, but not Rab GTPases, are required for Sey1p-mediated homotypic ER fusion.
Results
Establishment of an in vitro assay for Sey1p-dependent ER membrane fusion using isolated yeast microsomes
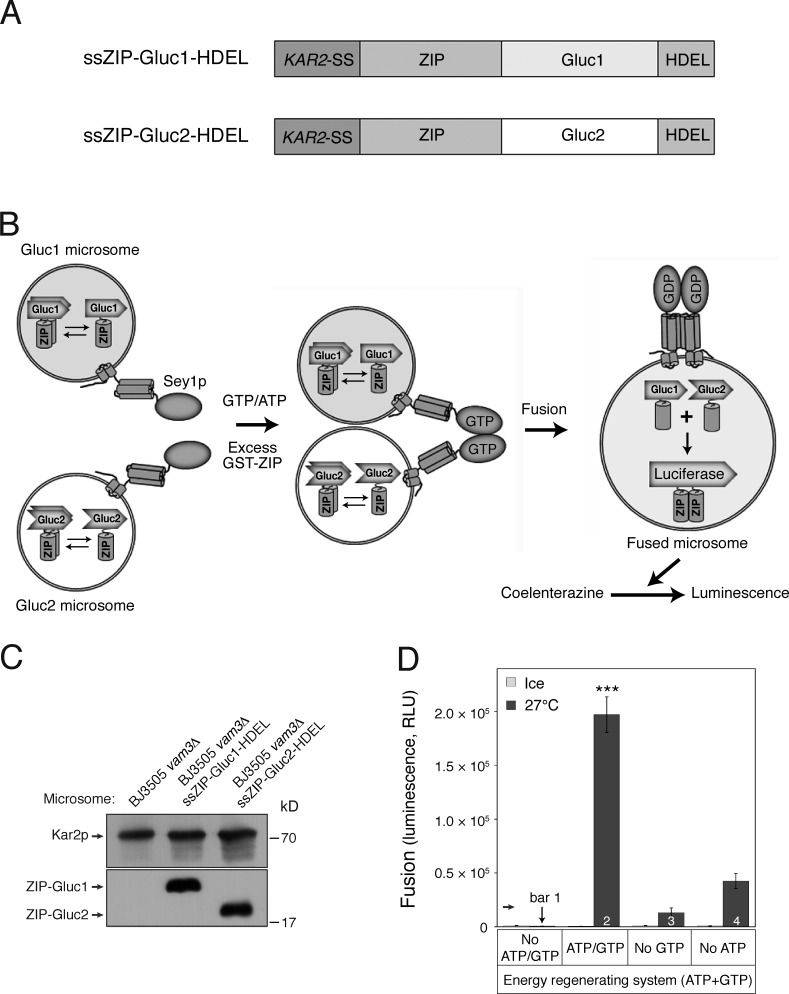
Cell-free in vitro assays offer several advantages for studying molecular mechanisms involved in a variety of biological processes; however, a rapid and quantitative in vitro assay for measuring homotypic ER membrane fusion is limited. To establish a cell-free in vitro assay for investigating homotypic ER membrane fusion, we used a protein fragment complementation assay (PCA) using the Gaussia princeps luciferase (Gluc; Remy and Michnick, 2006). In this assay, ER microsomes isolated from two yeast strains were mixed. One strain (Gluc1) contains a chimeric protein (ssZIP-Gluc1-HDEL) with four parts: (1) the signal sequence of the ER resident protein Kar2p, which directs the chimeric protein to the ER lumen; (2) the leucine zipper helix from Gcn4p (ZIP); (3) the amino-terminal half of Gluc (Gluc1); and (4) the four–amino acid sequence HDEL, which functions as an ER-retrieval signal. The other strain (Gluc2) carries a fusion protein (ssZIP-Gluc2-HDEL) with the same signal sequence as Kar2p, the ZIP leucine zipper helix, the other half of Gluc (Gluc2), and the HDEL ER-retrieval signal (Fig. 1 A). As depicted in the assay scheme shown in Fig. 1 B, microsomes isolated from either of these yeast strains lacked luciferase activity, but membrane fusion between the two microsome populations allowed luminal content mixing, resulting in the ZIP dimerization-induced interaction between the PCA fragments, ssZIP-Gluc1-HDEL and ssZIP-Gluc2-HDEL, and reconstitution of a functional luciferase (Fig. S1). Although microsome preparations are generally contaminated by the plasma membrane and membranes of other organelles, such as endosomes and vacuoles, generation of luciferase activity is likely to result from fusion between ER membranes because the PCA fragments should reside predominantly in the ER, owing to the presence of the ER-retrieval signal (Dean and Pelham, 1990). Nevertheless, because microsomal preparations are often highly contaminated with vacuolar membranes (Shimoni and Schekman, 2002; Malkus et al., 2004) and a portion of overexpressed PCA fragments escape from the ER-retrieval system, which can be saturated (Dean and Pelham, 1990), and erroneously traffics to vacuoles, some luciferase activity may result from homotypic vacuole fusion. To minimize this vacuole fusion–derived signal, VAM3, which encodes a vacuolar SNARE essential for vacuole fusion (Nichols et al., 1997), was deleted from the tester strains. In these in vitro homotypic ER fusion reactions, microsomes isolated from the yeast strains BJ3505 vam3Δ ssZIP-Gluc1-HDEL (BJ-Gluc1) and BJ3505 vam3Δ ssZIP-Gluc2-HDEL (BJ-Gluc2) were mixed and incubated at 27°C in the presence of ATP, GTP, and an energy-regenerating system. Excess GST-ZIP was added to block extraluminal reconstitution of functional Gluc caused by membrane destabilization or rupture during incubation. After 90–120 min, coelenterazine, a luminogenic substrate of Gluc, was added, and luminescence was measured. Addition of an energy-regenerating system with ATP and GTP gave the optimal fusion signal (Fig. 1 D, bar 2), whereas GTP omission largely abolished the signal, indicating that in vitro ER fusion requires GTP (Fig. 1 D, bar 3). Furthermore, GTP hydrolysis is required for ER fusion because the nonhydrolyzable GTP analogue GTPγS did not support ER fusion (Fig. 2 A). Consistently, significant fusion signal was observed with 1 mM GTP alone (Fig. 1 D, bar 4; and Fig. 2 A), but not with GTPγS alone (Fig. 2 A). The atlastin-like GTPase Sey1p mediates homotypic ER fusion in Saccharomyces cerevisiae (Anwar et al., 2012); therefore, we first examined whether the requirement of the in vitro ER fusion reaction for GTP reflected a requirement for Sey1p in the process. To this end, we deleted SEY1 from the tester strains (Fig. 2 B) and used microsomes isolated from these sey1Δ strains for in vitro fusion reactions. GTP-dependent luciferase signals were completely abolished in fusion reactions containing sey1Δ microsomes (Fig. 2 C, black bar in the middle panel). Consistent with the suggested mode of action of Sey1p and metazoan atlastins (Orso et al., 2009; Byrnes and Sondermann, 2011; Bian et al., 2011; Anwar et al., 2012), Sey1p was required at both sides of fusing microsomes, as indicated by the fact that sey1Δ microsomes failed to fuse with wild-type microsomes (Fig. 2 C, right). In contrast, the GTP-independent fusion signal (Fig. 2 C, gray bar), although extremely low, was not affected by SEY1 deletion, suggesting that this GTP-independent signal represents the operation of Sey1p-independent ER fusion pathways, as previously reported (Anwar et al., 2012). Loss of fusion was specific to deletion of SEY1 because deletion of RTN1 or YOP1, both of which play a critical role in shaping the ER (Voeltz et al., 2006; Hu et al., 2008), did not significantly influence fusion (see Fig. S2), which is consistent with a previous study (Anwar et al., 2012).
Figure 1.
Development of an in vitro homotypic ER fusion assay. (A) Schematic representation of chimeric proteins for the Gluc PCA. (B) Assay scheme; see Results for details. Microsomes isolated from BJ-Gluc1 yeast cells overexpressing ssZIP-Gluc1-HDEL under the control of an ADH1 promoter (Gluc1 microsomes) were mixed with microsomes isolated from BJ-Gluc2 yeast cells overexpressing ssZIP-Gluc2-HDEL (Gluc2 microsomes) and then incubated at 27°C in the presence of GTP and ATP. After 90 min, the luciferase substrate coelenterazine was added, and luciferase activity was measured. Excess GST-ZIP was added to block extra-luminal reconstitution of functional Gluc caused by membrane destabilization or rupture during incubation. (C) Expression of Gluc PCA fragments in isolated microsomes. The expression of Gluc PCA fragments was analyzed by immunoblotting using the indicated antibodies. Kar2p, an ER-resident protein, was used as a loading control. (D) GTP-driven homotypic ER fusion is markedly enhanced by ATP. Gluc1 and Gluc2 microsomes were mixed and incubated on ice or at 27°C in the presence of ATP and/or GTP for 90 min. Data represent means ± SEM (error bars; n = 3). RLU, relative luminescence unit. ***, P < 0.001, Tukey’s test between experiments performed at 27°C.
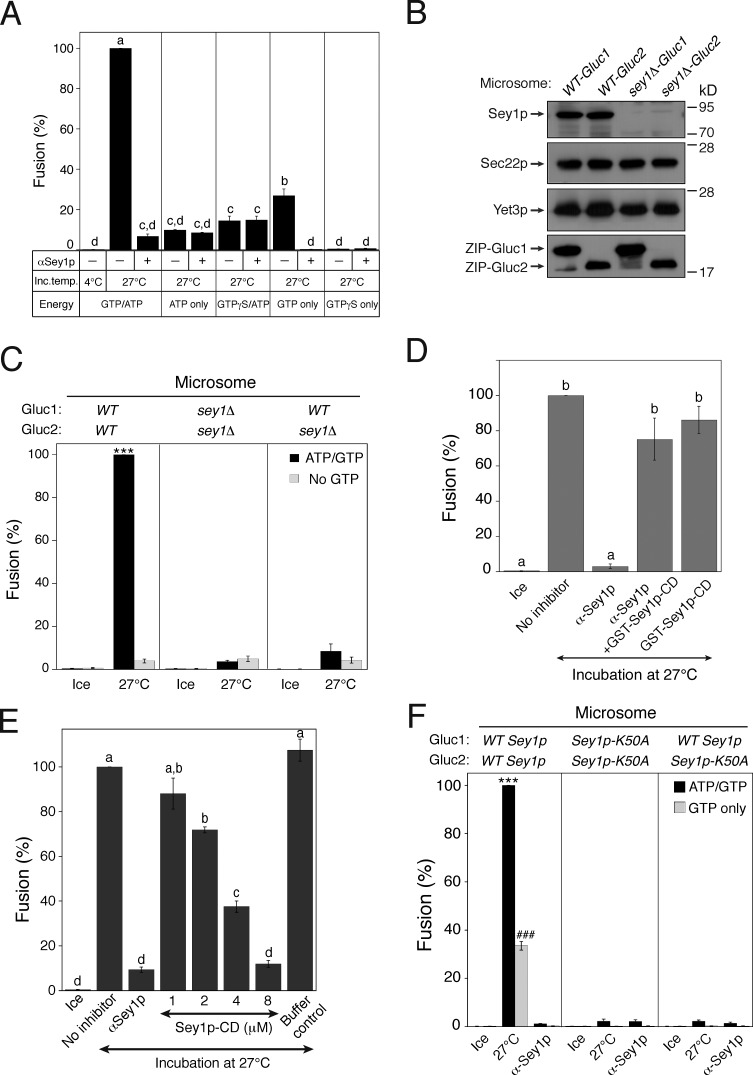
Figure 2.
In vitro homotypic ER fusion requires the atlastin GTPase Sey1p. (A) GTP hydrolysis is required for ER fusion in vitro. Gluc1 and Gluc2 microsomes were incubated on ice or at 27°C in the presence of the indicated nucleotide for 90 min. Fusion values were normalized to those obtained using an ATP/GTP-driven reaction incubated at 27°C. Data represent the means ± SEM (error bars; n = 3). Lowercase letters indicate statistically different groups (P < 0.001, Tukey’s test). (B) Protein profiles of wild-type and sey1Δ microsomes. The expression of Gluc PCA fragments and ER-associated proteins was analyzed by immunoblotting using the indicated antibodies. The ER-associated proteins Sec22p and Yet3p were used as loading controls. (C) Sey1p in both of the fusing microsomes is required to support fusion. The indicated microsomes were incubated on ice or at 27°C in the presence of ATP and/or GTP for 90 min. Data represent the means ± SEM (error bars; n = 3). ***, P < 0.001, Tukey’s test between experiments performed at 27°C. (D) Anti-Sey1p antibodies specifically inhibit the in vitro ER fusion reaction. Gluc1 and Gluc2 microsomes were incubated on ice or at 27°C in the presence of anti-Sey1p antibodies (88 nM) and/or GST-Sey1p (1 µM). Data represent the means ± SEM (error bars; n = 3). Lowercase letters indicate statistically different groups (P < 0.001, Tukey’s test). (E) Addition of the cytoplasmic domain of Sey1p inhibits ER fusion in vitro. Gluc1 and Gluc2 microsomes were incubated on ice or at 27°C in the presence of the Sey1p CD (Sey1p-CD). Data represent the means ± SEM (error bars; n = 3). Lowercase letters indicate statistically different groups (P < 0.001, Tukey’s test). (F) Sey1p-K50A, a GTP-binding-defective mutant, did not support in vitro ER fusion. The indicated microsomes were incubated on ice or at 27°C in the presence of ATP and/or GTP for 90 min. Data represent the means ± SEM (error bars; n = 3). ***, P < 0.001, between ATP/GTP-driven reactions; ###, P < 0.001, between GTP-only-driven reactions, Tukey’s test.
Although sey1Δ microsomes failed to fuse, this loss of fusion could be caused by indirect effects of SEY1 deletion, such as the defective ER localization of other proteins that are required for ER membrane fusion. Thus, to examine whether Sey1p is directly required for the fusion of wild-type microsomes, we added affinity-purified antibodies against recombinant Sey1p to fusion reactions. Antibodies to Sey1p effectively prevented the in vitro fusion of wild-type microsomes, confirming that Sey1p is essential for in vitro homotypic ER fusion (Fig. 2 D). Co-incubation of anti-Sey1p antibodies with the GST-fused CD of Sey1p (Sey1p-CD) at a subinhibitory concentration (1 µM) largely relieved this inhibition (Fig. 2 D), indicating that the inhibitory effect of anti-Sey1p antibodies was specific. Furthermore, the addition of Sey1p-CD at higher concentrations effectively inhibited in vitro ER fusion, presumably by competing with membrane-bound Sey1p molecules (Fig. 2 E). To confirm that the requirement of in vitro ER fusion for GTP reflected the essential role of the GTPase Sey1p in this process, we examined whether Sey1p-K50A, a GTP-binding mutant of Sey1p (Hu et al., 2009; Anwar et al., 2012), supported in vitro ER fusion. Sey1p-K50A did not support ER fusion (Fig. 2 F). Collectively, these data clearly establish the functional validity of our in vitro assay for Sey1p-dependent yeast ER fusion.
Rab GTPases are not involved in Sey1p-dependent ER fusion
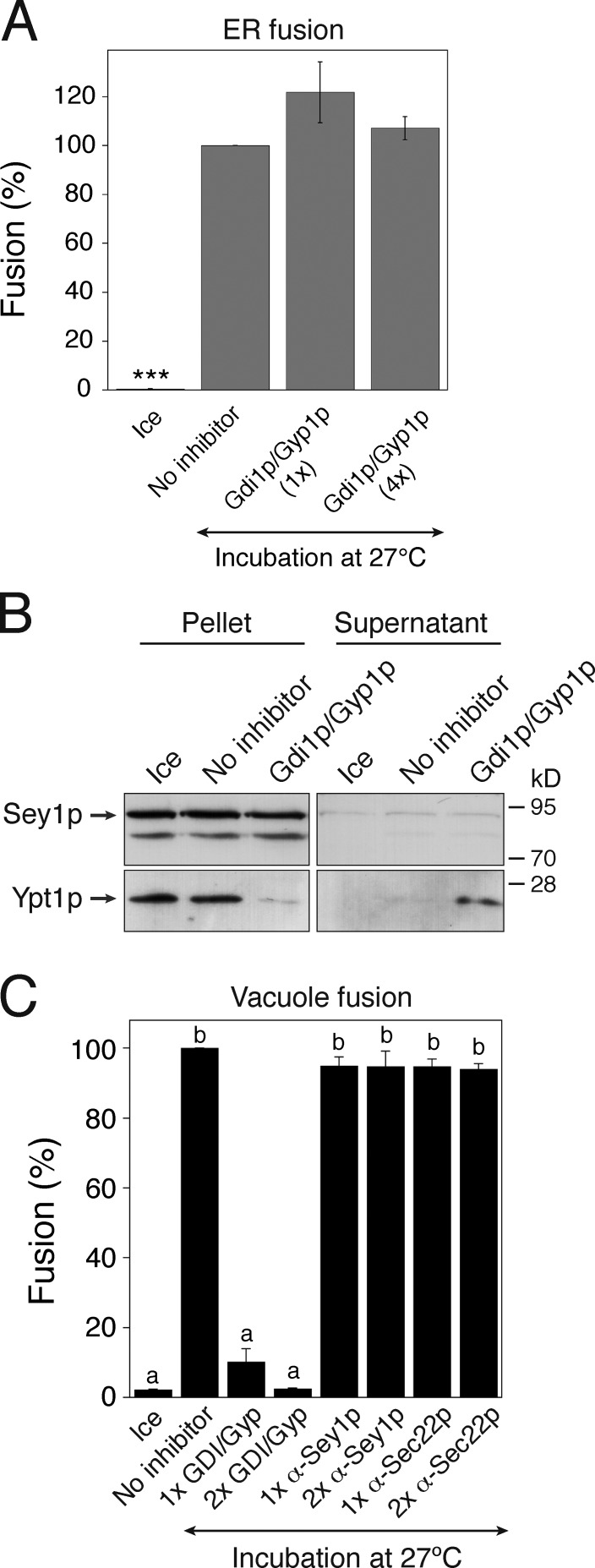
Rab GTPases have been implicated in ER membrane fusion in metazoan cells (Turner et al., 1997; Audhya et al., 2007; English and Voeltz, 2012; Gerondopoulos et al., 2014). To determine whether a Rab GTPase is involved in atlastin-dependent ER fusion in yeast, we used Gdi1p and Gyp1p. Gdi1p is the only GDP dissociation inhibitor (GDI) identified in S. cerevisiae and can probably act on all yeast Rab (YPT) proteins and remove their GDP-bound forms from the membrane (Garrett et al., 1994; Haas et al., 1995; Kamena et al., 2008). Gyp1p is a Rab GTPase-activating protein (GAP) that preferentially inactivates Ypt1p (a Rab protein involved in ER-to-Golgi trafficking) and other yeast Rab proteins (e.g., Ypt51p, Ypt7p, and Sec4p; Du et al., 1998; Albert et al., 1999). In vitro ER fusion was fully resistant to the addition of a fourfold excess of the amount of Gdi1p/Gyp1p sufficient to completely inhibit in vitro yeast vacuole fusion (compare Fig. 3 A and Fig. 3 C). The concentration of Gdi1p/Gyp1p used in this experiment (1×) was sufficient to extract nearly all Ypt1p molecules from ER membranes (Fig. 3 B). Thus, Sey1p-dependent ER fusion likely occurs independently of the function of Rab GTPases.
Figure 3.
In vitro homotypic ER fusion is Rab independent. (A) Homotypic ER fusion is insensitive to Rab inhibition by Gdi1p/Gyp1p. Gluc1 and Gluc2 microsomes were incubated on ice or at 27°C with ATP/GTP in the absence or presence of his6-Gdi1p and his6-Gyp1p for 90 min. For 1×Gdi1p/Gyp1p, the concentrations of his6-Gdi1p and his6-Gyp1p were 1.5 and 2.5 µM, respectively; the corresponding concentrations for 4×Gdi1p/Gyp1p were 6 and 10 µM, respectively. Data represent the means ± SEM (error bars; n = 3). ***, P < 0.001, Tukey’s test. (B) Ypt1p is efficiently removed from microsomes by the addition of Gdi1p/Gyp1p. ER fusion reactions were performed as described in A in the absence or presence of his6-Gdi1p (1.5 µM) and his6-Gyp1p (2.5 µM), and then centrifuged. Ypt1p partitioning between the soluble (supernatant) and insoluble (pellet) fractions was assessed by immunoblotting using anti-Ypt1p and anti-Sey1p antibodies. (C) In vitro yeast vacuole fusion reactions were efficiently prevented by Gdi1p/Gyp1p, but were completely resistant to anti-Sey1p and anti-Sec22p antibodies. Vacuoles isolated from BJ3505 and DKY6281 were mixed and incubated in fusion reaction buffer (see Materials and methods for details) at 27°C. After 90 min, alkaline phosphatase activity was measured, and fusion values (%) were normalized to those obtained in reactions performed without an inhibitor at 27°C. Data represent the means ± SEM (error bars; n = 3). Lowercase letters indicate statistically different groups (P < 0.001, Tukey’s test).
ER-associated SNAREs are involved in Sey1p-dependent ER fusion
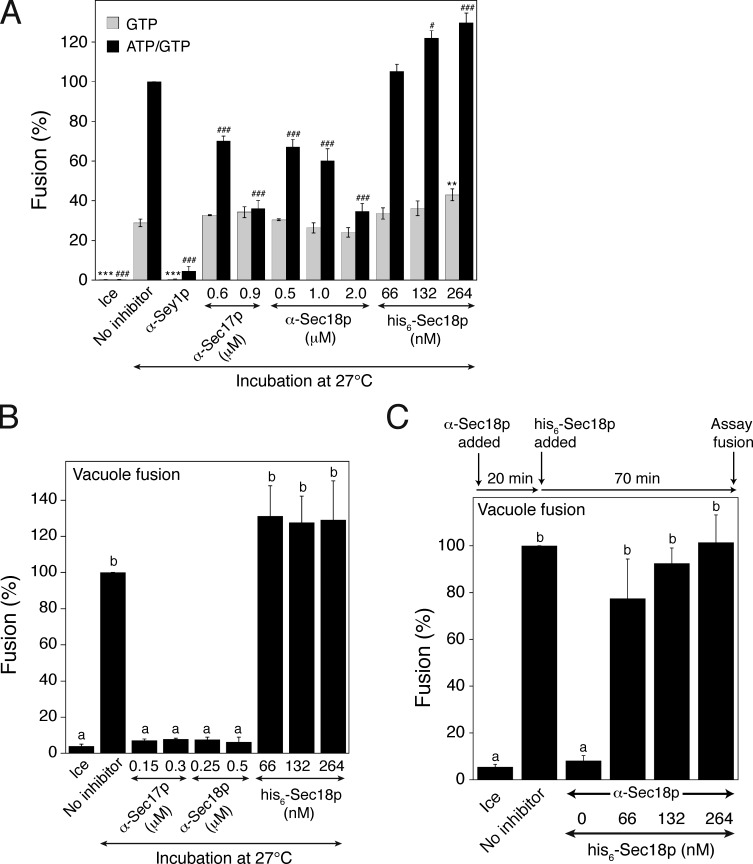
Previous studies have indicated that ER-associated SNARE proteins are involved in ER fusion (Patel et al., 1998; Anwar et al., 2012; Rogers et al., 2013), although this SNARE-mediated ER fusion seems to occur independently of Sey1p and mediate residual fusion events. Interestingly, however, whether ER SNAREs are involved in Sey1p-mediated ER fusion has never been directly tested. During SNARE-mediated membrane fusion in yeast, the cytosolic ATPase Sec18p (the yeast orthologue of mammalian NSF [N-ethylmaleimide-sensitive factor]) and its cochaperone Sec17p (the yeast orthologue of mammalian α-SNAP) use ATP energy to disassemble SNARE complexes, formed from previous membrane fusion events, into individual SNARE proteins for the next rounds of fusion (Mayer et al., 1996). To examine whether Sec18p is involved in Sey1p-dependent ER fusion, we added affinity-purified anti-Sec18p antibodies or recombinant his6-Sec18p to the in vitro ER fusion reactions (Fig. 4 A). ER fusion was markedly blocked by anti-Sec18p antibodies and enhanced by his6-Sec18p, suggesting that Sec18p, although not essential, is involved in Sey1p-dependent ER fusion. This result also suggests that ATP in the energy-regenerating system used in our in vitro assay is consumed not only for regenerating GTP but also for the action of Sec18p. Consistent with the involvement of Sec18p in Sey1p-dependent ER fusion, anti-Sec17 antibodies inhibited the in vitro ER fusion reactions to an extent similar to inhibition by anti-Sec18p antibodies. Inhibition by anti-Sec18p or anti-Sec17p antibodies seems to be specific because ER fusion driven by GTP alone was largely resistant to inhibition by these antibodies (Fig. 4 A, gray bars). As previously reported, these antibodies effectively inhibited Sec18p/Sec17p-dependent homotypic yeast vacuole fusion, while his6-Sec18p stimulated vacuole fusion and rescued the fusion between vacuoles pretreated with anti-Sec18p antibodies (Fig. 4, B and C).
Figure 4.
The SNARE chaperones Sec17p and Sec18p are involved in ATP/GTP-driven ER fusion. (A) Sec17p and Sec18p are involved in ATP/GTP-driven ER fusion, but not in GTP-driven fusion. Gluc1 and Gluc2 microsomes were incubated on ice or at 27°C in the presence of GTP or ATP/GTP for 90 min. Some reactions were treated with anti-Sec17p antibodies, anti-Sec18p antibodies, or recombinant his6-Sec18p at the indicated concentrations. Data represent the means ± SEM (error bars; n = 3). **, P < 0.01; ***, P < 0.001, between GTP-only-driven reactions; #, P < 0.05; ###, P < 0.001, between ATP/GTP-driven reactions, Tukey’s test compared with the “no inhibitor” group. (B) In vitro yeast vacuole fusion reactions were efficiently prevented by anti-Sec17p or anti-Sec18p antibodies. Data represent the means ± SEM (error bars; n = 3). Lowercase letters indicate statistically different groups (P < 0.001, Tukey’s test). (C) Recombinant his6-Sec18p can rescue the fusion of anti-Sec18p–treated vacuoles. BJ3505 and DKY6281 vacuoles were incubated in the presence of anti-Sec18p antibodies at 27°C. After 20 min, recombinant his6-Sec18p was added at the indicated concentrations, and the mixture was further incubated for 70 min at 27°C. Data represent the means ± SEM (error bars; n = 3). Lowercase letters indicate statistically different groups (P < 0.01, Tukey’s test).
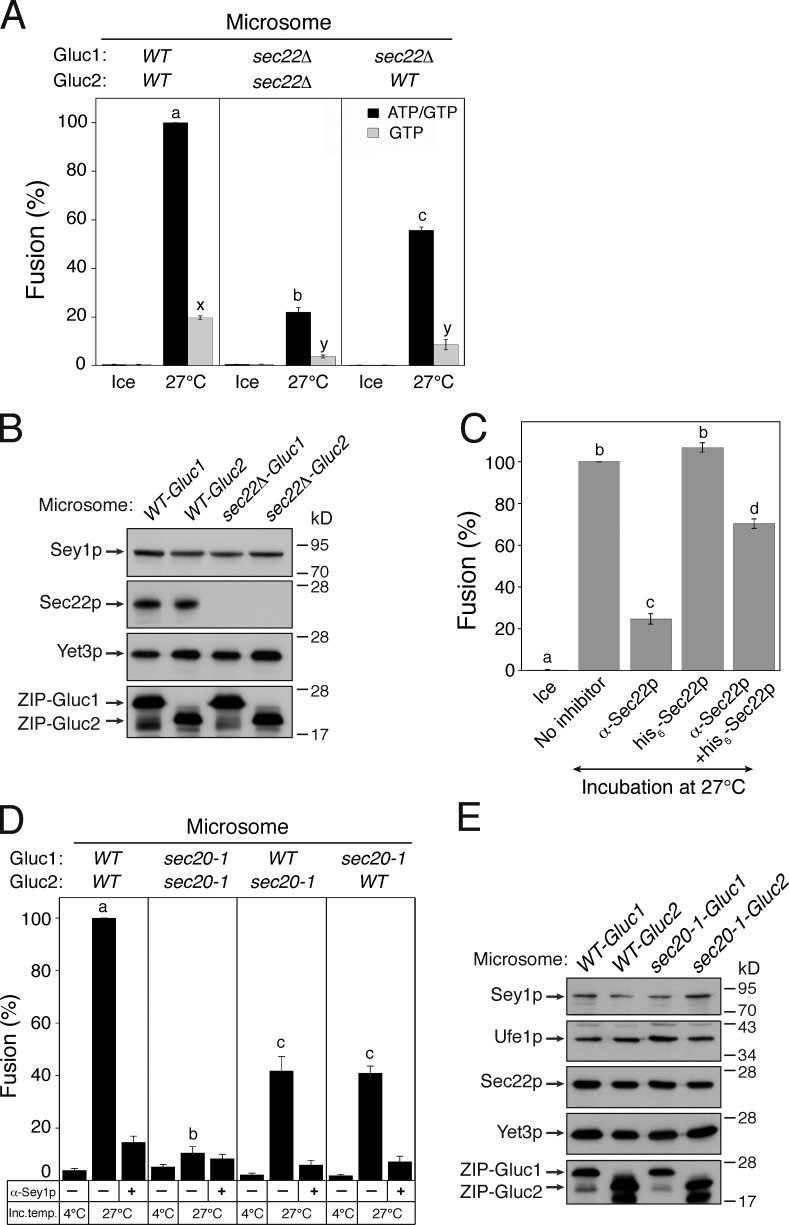
The involvement of Sec17p and Sec18p in Sey1p-mediated ER fusion indicates that SNARE proteins play a crucial role in the process. To determine which SNARE proteins are involved in homotypic ER membrane fusion in yeast, we first focused on the SNARE proteins that are localized to the ER or vesicles that traffic between the ER and the Golgi apparatus. Among genes encoding ER-associated SNAREs, SEC22 is the only one that is not essential for cell survival and can thus be genetically deleted. Whereas deletion of SEC22 did not affect the amount of luciferase reporter constructs or Sey1p in isolated ER microsomes (Fig. 5 B), it markedly reduced fusion (Fig. 5 A, middle panel). Thus, Sec22p is likely involved in Sey1p-mediated ER fusion. Unexpectedly, fusion in the presence of GTP alone was also inhibited by deletion of SEC22 (Fig. 5 A, gray bars), suggesting that Sec22p is generally required for efficient Sey1p-mediated ER fusion. How is Sec22p involved in the ATP-independent ER fusion driven by GTP alone? One plausible scenario is that significant portions of Sec22p molecules on isolated microsomes were not complexed with other SNAREs because Sec18p had catalyzed their disassembly from SNARE complexes in vivo, before microsome isolation (Thorngren et al., 2004). Thus, these uncomplexed Sec22p molecules may have participated in Sey1p-dependent ER fusion, even in the absence of ATP. The degree of fusion between wild-type microsomes and sec22Δ microsomes was nearly half of that between wild-type microsomes, suggesting that Sec22p (R-SNARE) molecules in one membrane interact with ER-associated Q-SNARE proteins in the other to support Sey1-dependent ER fusion.
Figure 5.
ER SNAREs are involved in Sey1p-dependent ER fusion. (A) Sec22p is involved in Sey1p-mediated ER fusion. The indicated microsomes were incubated on ice or at 27°C in the presence of GTP or ATP/GTP for 90 min. Data represent the means ± SEM (error bars; n = 3). Lowercase letters indicate statistically different groups. A Tukey’s test between reactions was performed at 27°C. P < 0.001 between groups a–c and P < 0.01 between groups x and y. (B) Protein profiles of wild-type and sec22Δ microsomes. Yet3p was used as a loading control. (C) Affinity-purified anti-Sec22p antibodies specifically inhibit in vitro ER fusion. Gluc1 and Gluc2 microsomes were incubated on ice or at 27°C with GTP/ATP in the absence or presence of anti-Sec22p antibodies and/or his6-Sec22p for 90 min. Data represent the means ± SEM (error bars; n = 3). Lowercase letters indicate statistically different groups (P < 0.001, Tukey’s test). (D) The function of Sec20p is required for Sey1p-dependent ER fusion. The indicated microsomes were mixed with or without anti-Sey1p antibodies and incubated on ice or at 27°C in the presence of GTP/ATP for 90 min. Lowercase letters indicate statistically different groups. Tukey’s test between reactions was performed without anti-Sey1p antibodies at 27°C (P < 0.001). (E) Protein profiles of wild-type and sec20-1 microsomes.
The involvement of Sec22p in Sey1p-mediated ER fusion was reexamined using anti-Sec22p antibodies in in vitro ER fusion reactions containing wild-type microsomes. In vitro ER fusion was efficiently prevented by affinity-purified anti-Sec22p antibodies, an inhibitory effect that was largely eliminated by the addition of recombinant his6-Sec22p (Fig. 5 C). Furthermore, homotypic yeast vacuole fusion was completely resistant to anti-Sec22p antibodies (Fig. 3 B), indicating that the inhibition of fusion by anti-Sec22p antibodies was specific.
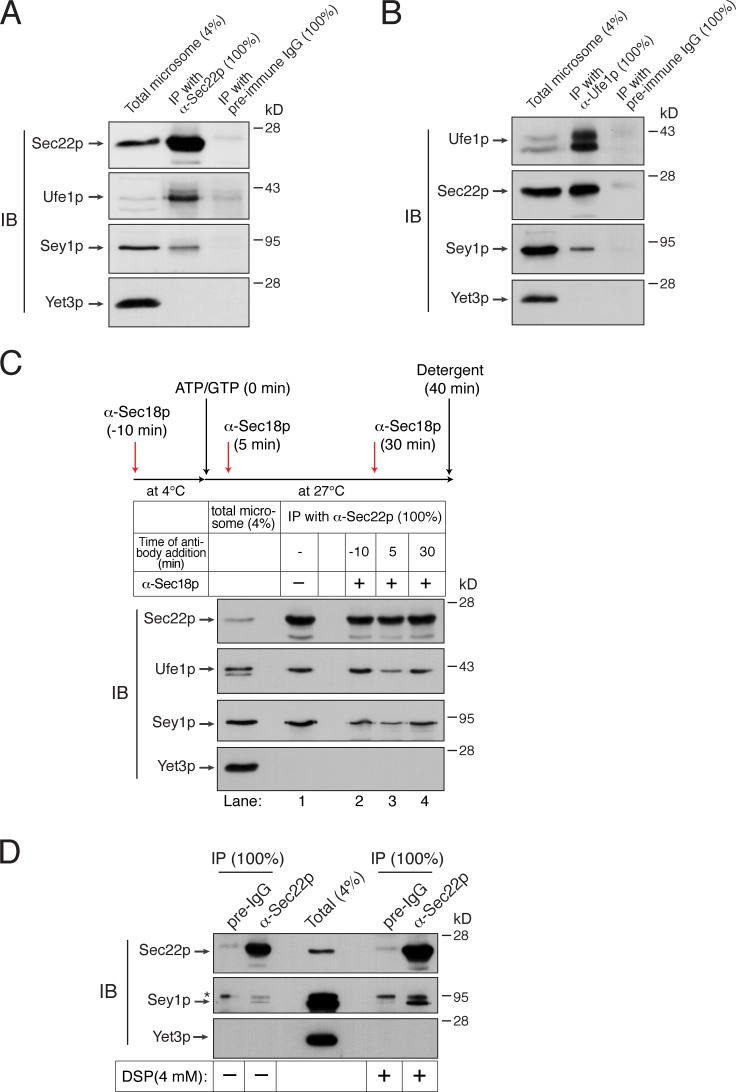
The ER-associated Q-SNAREs Sec20p, Ufe1p, and Use1p bind Sec22p to form a SNARE complex on the ER (Lewis et al., 1997; Burri et al., 2003; Dilcher et al., 2003). To examine whether these ER Q-SNAREs also participate in Sey1p-dependent ER fusion, ER microsomes isolated from sec20-1 (a SEC20 temperature-sensitive mutant) cells (Lewis et al., 1997) carrying ssZIP-Gluc1-HDEL or ssZIP-Gluc2-HDEL (Fig. 5 E) were tested for their fusion capability (Fig. 5 D). Interestingly, even without incubation at the nonpermissive temperature, sec20-1 microsomes failed to support ER fusion, whereas a significant fusion signal was observed with microsomes isolated from the parental wild-type strains. Because sec20-1 cells grow poorly at 30°C and a strong karyogamy defect was seen in sec20-1 cells at the semipermissive temperature (30°C; Rogers et al., 2013), the function of the mutant Sec20p might have been impaired during microsome preparation, which involves the incubation of yeast cells at 30°C. Together, these data strongly suggest that ER SNARE proteins participate in Sey1-mediated ER membrane fusion in yeast. This interpretation is further supported by the physical interaction of Sey1p with Sec22p and Ufe1p, although Sey1p and ER SNAREs appear to interact only transiently, rather than assembling into a stable complex, during ER fusion (Fig. 6, A and B). To test whether the interaction between Sey1p and ER SNAREs is regulated by the SNARE chaperone Sec18p, we blocked the function of Sec18p by adding anti-Sec18p antibodies (2 µM) to ER fusion reactions at different times and analyzed the degree of interactions between Sey1p and Sec22p by coimmunoprecipitation (Fig. 6 C). To preserve nearly all preexisting SNARE complexes, which can be immediately disrupted by ATP-loaded Sec18p before antibodies act, one reaction was preincubated with anti-Sec18p antibodies for 10 min before the addition of ATP and GTP (lane 2). While comparable amounts of Ufe1p were precipitated with Sec22p in reactions with or without anti-Sec18p antibodies (compare lanes 1 and 2), significantly less Ufe1p was copurified with Sec22p in reactions that had received anti-Sec18p antibodies 5 min after ATP/GTP addition (lane 3). These results indicate that preexisting SNARE complexes were initially broken up by Sec18p, but the resulting individual SNAREs reassembled into SNARE complexes, mediating membrane fusion. Interestingly, the amount of Sey1p bound to Sec22p was similarly affected by the addition of anti-Sec18p antibody (compare lanes 2–4 between Sey1p and Ufe1p); the amount of Sey1p precipitated with Sec22p was directly proportional to the amount of ER SNARE complexes. Thus, these data suggest that Sey1p preferentially binds to SNARE complexes over individual SNARE proteins.
Figure 6.
Sey1p physically interacts with ER SNAREs. (A and B) Sey1p physically interacts with the ER SNARE proteins Sec22p and Ufe1p. Microsomes isolated from BJ3505 were detergent-solubilized and incubated with anti-Sec22p antibodies (A), anti-Ufe1p antibodies (B), or preimmune IgG (control) in the presence of protein A Sepharose. Protein A Sepharose–bound material was then analyzed by immunoblotting using the indicated antibodies. (C) Sec18p regulates the interaction between Sec22p and Sey1p. BJ3505 microsomes were preincubated in the absence or presence of anti-Sec18p antibodies for 10 min at 4°C. After ATP/GTP was added, microsomes were further incubated for 40 min. During incubation, some samples received anti-Sec18p antibodies at the indicated time points. Sec22p was precipitated using anti-Sec22p antibody–conjugated Dynabeads, and bound proteins were analyzed by immunoblotting using the indicated antibodies. (D) Detection of an in vivo interaction between Sec22p and Sey1p by DSP cross-linking. BJ3505 spheroplasts were incubated in the absence or presence of 4 mM DSP for 30 min at 4°C. After DSP was quenched, detergent-solubilized spheroplasts were subjected to immunoprecipitation using anti-Sec22p antibodies or preimmune IgG (preIgG). Cross-links were cleaved using β-mercaptoethanol in SDS sample buffer, and proteins cross-linked with Sec22p were analyzed by immunoblotting using the indicated antibodies. The asterisk indicates nonspecific bands. All experiments were performed multiple times with similar results, and the data shown are representative of all results.
Finally, to examine whether Sey1p interacts with Sec22p in vivo, we attempted to purify Sec22p from yeast cells using anti-Sec22p antibodies and probing copurifying proteins with anti-Sey1p antibodies (Fig. 6 D). Although very weak, but specific, interaction between the two proteins was detected without cross-linking, we were able to recover significant amounts of Sey1p bound to Sec22p by treating spheroplasts with the cleavable cross-linker dithiobis-succinimidyl-propionate (DSP), followed by detergent extraction of membranes. This cross-linking between Sey1p and Sec22p seems to be specific because no cross-linking was detected between Sec22p and Yet3p, an abundant ER-resident protein.
The physiological concentration of Sey1p may not be high enough to drive ER fusion by itself
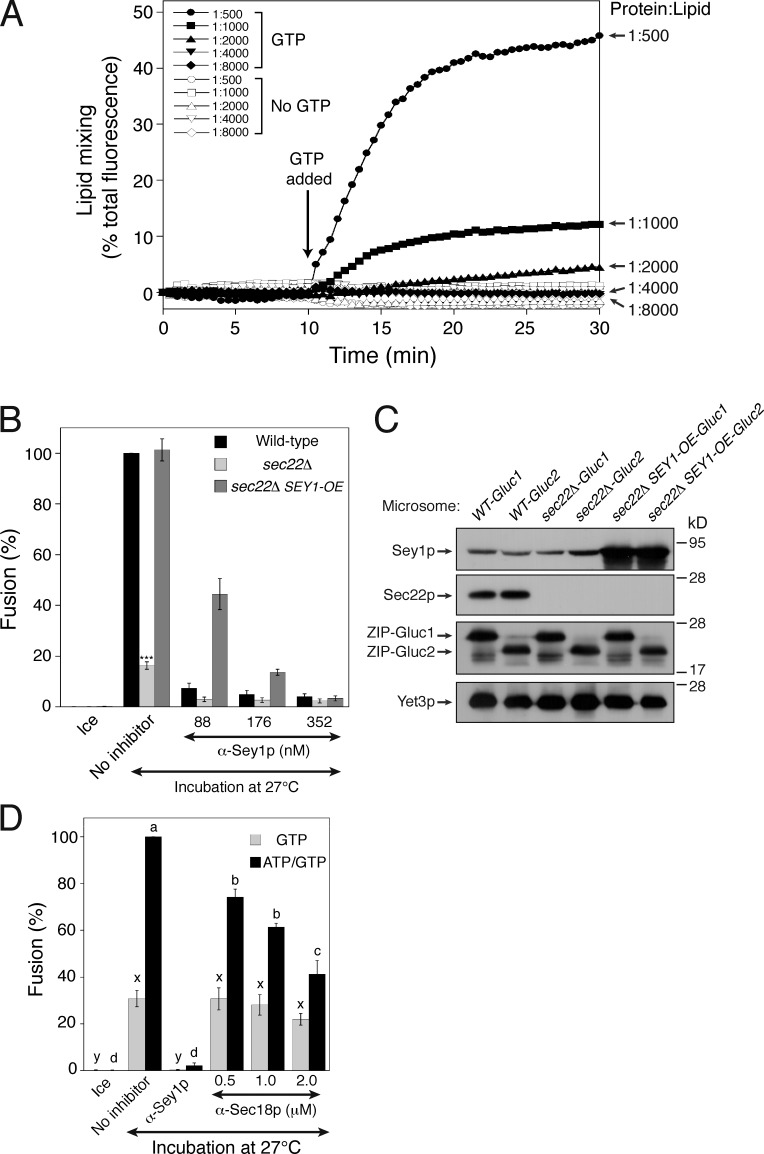
Our data strongly suggest that ER SNAREs are required for Sey1p-mediated ER fusion. Given that Sey1p alone can induce liposome fusion (Anwar et al., 2012), why are SNAREs required for Sey1p-mediated fusion of isolated microsomes? One possibility is that the physiological concentration of Sey1p is not sufficient to drive ER fusion. Non-physiologically high concentrations of SNARE proteins can induce the fusion of liposomes (Chen et al., 2006; Dennison et al., 2006) or isolated organelles (Starai et al., 2007) without additional factors, which are otherwise required for fusion in vivo. To measure the concentration of Sey1p in the biological membrane, ER microsomes were purified and analyzed for lipid phosphorus and bound Sey1p. According to our estimation, the lipid/Sey1p ratios of BJ-Gluc1 and BJ-Gluc2 microsomes were 1.19 × 105 and 1.10 × 105, respectively. These values are 100–600-fold lower in isolated microsomes than in Sey1p phosphatidylcholine/phosphatidylserine proteoliposomes capable of lipid mixing in a previous report (Anwar et al., 2012). The rate and extent of fusion are governed not only by the molar ratio of fusogenic proteins to lipid but also by the proteoliposomal lipid composition. Therefore, we generated Sey1p proteoliposomes of a mixed lipid composition mimicking that of the ER, based on the established composition of the isolated organelle (Zinser et al., 1991), bearing Sey1p at a molar ratio to lipids of 1:500, 1:1,000, 1:2,000, 1:4,000, or 1:8,000 (Fig. 7 A, and also see Fig. S3). Although these ER-mimicking mixed lipids enabled liposomes to be fused with significantly less Sey1p in comparison to phosphatidylcholine/phosphatidylserine alone (Anwar et al., 2012), fusion was hardly detectable for proteoliposomes with Sey1p at a 1:4,000 molar ratio to lipids, which is still 25-fold more abundant than in isolated microsomes (Fig. 7 A). Thus, Sey1p at the physiological concentration is unlikely to drive liposome fusion by itself. Although overexpression of Sey1p restored fusion to sec22Δ microsomes (Fig. 7, B and C), the restored fusion was still sensitive to inhibition by anti-Sec18p antibodies (Fig. 7 D), indicating that SNARE proteins are still required. Another R-SNARE, such as Ykt6p (Liu and Barlowe, 2002), may replace the function of Sec22p to mediate ER fusion in sec22Δ microsomes overexpressing Sey1p. Together, these results suggest that Sey1p-mediated ER fusion in vivo requires additional factors, such as SNAREs, to occur efficiently.
Figure 7.
Sey1p at its physiological concentration is not sufficient to induce liposome fusion. (A) Proteoliposomes with the indicated Sey1p-to-lipid ratio were generated as described in the Materials and methods. Donor and acceptor proteoliposomes were mixed and incubated at 30°C for 10 min. After GTP and Mg2+ were added, and NBD fluorescence was measured at 30-s intervals for 30 min. β-Octylglucoside was then added to determine total fluorescence. Fusion is expressed as the percentage of total fluorescence. All experiments were performed multiple times with similar results, and the data shown are representative of all results. (B) Overexpression of Sey1p restores fusion to sec22Δ microsomes. The indicated microsomes were incubated on ice or at 27°C with GTP/ATP in the absence or presence of anti-Sey1p antibodies for 90 min. SEY1-OE, SEY1 overexpressor. Data represent the means ± SEM (error bars; n = 3). ***, P < 0.001, Tukey’s test between the no inhibitor groups of the indicated genotypes. (C) Protein profiles of wild-type, sec22Δ, and sec22Δ SEY1-OE microsomes. (D) The fusion of sec22Δ SEY1-OE microsomes requires Sec18p function. Microsomes (sec22Δ SEY1-OE) were incubated on ice or at 27°C with GTP/ATP or GTP alone in the presence of anti-Sey1p antibodies or increasing concentrations of anti-Sec18p antibodies for 90 min. Data represent the means ± SEM (error bars; n = 3). Statistically different groups are indicated by different lowercase letters (a–d, between ATP/GTP-driven reactions, P < 0.001; and x–y, between GTP-only-driven reactions, P < 0.05).
The function of Sey1p precedes that of Sec22p during homotypic ER fusion
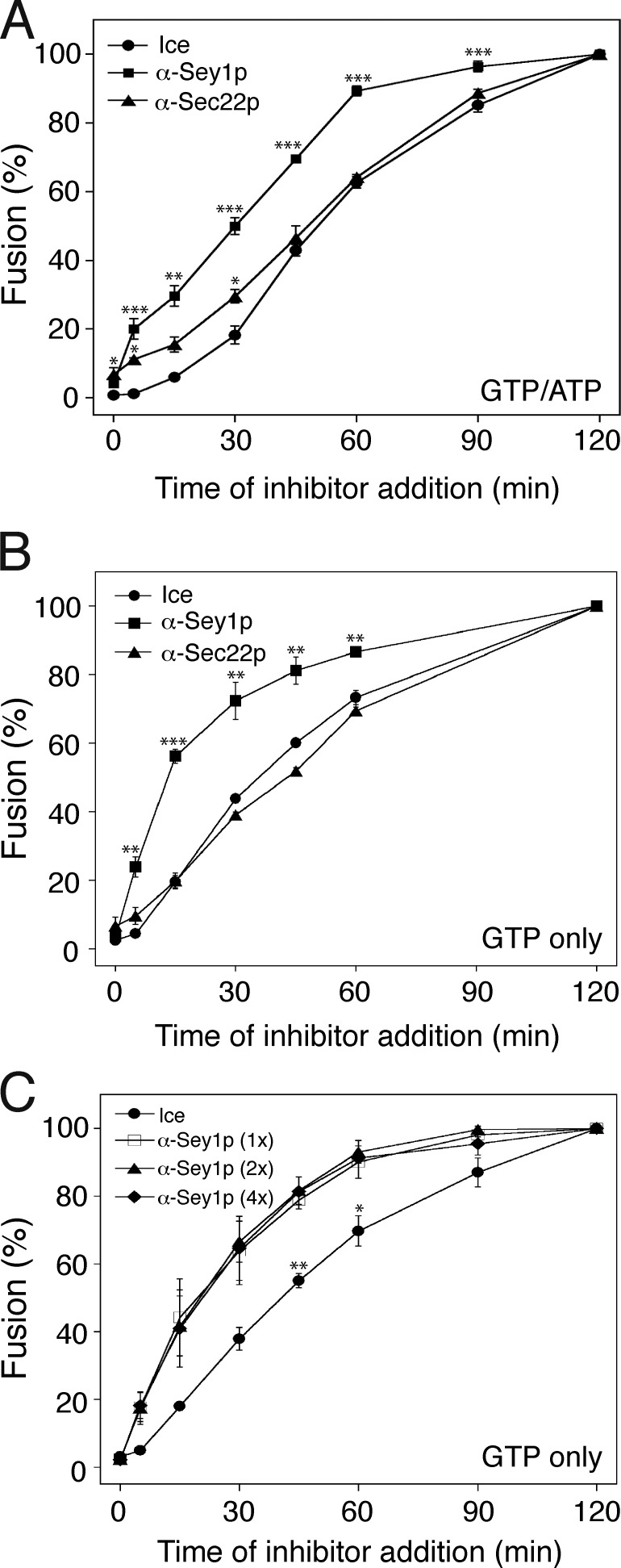
Our data strongly suggest that ER-associated SNARE proteins and Sey1p function together in the same pathway to support homotypic ER fusion. To determine whether Sey1p and Sec22p function simultaneously or act sequentially, we performed the staging experiment originally developed for homotypic yeast vacuole studies (Mayer et al., 1996). During yeast vacuole fusion in vitro, once a vacuole completes a step of the membrane fusion reaction, it becomes resistant to inhibitors of that particular step (Mayer et al., 1996; Jun et al., 2006); thus application of suitable inhibitors can reveal the sequence of events in vacuole fusion. Applying this concept to determine the order of Sey1p and Sec22p functions, we examined the kinetics of the sensitivity of fusion to anti-Sey1p and anti-Sec22 antibodies. To this end, we mixed aliquots of a fusion reaction with the indicated antibodies after incubating for different amounts of times. When added from the start, either anti-Sey1p or anti-Sec22p antibodies markedly prevented ER fusion driven by GTP and ATP (Fig. 8 A) or by GTP alone (Fig. 8 B). However, after 60 min, the reaction was almost completely resistant to anti-Sey1p antibodies, indicating that the function of Sey1p was no longer required (Figs. 8, A and B, squares). In contrast, fusion remained sensitive to anti-Sec22p antibodies throughout the incubation period; the inhibition curve for anti-Sec22p antibodies (Fig. 8, A and B, triangles) was similar to that produced by placing the reaction samples on ice (circles). Similar sensitivity kinetics were observed with a fourfold excess of the amount of anti-Sey1p antibodies sufficient for complete inhibition (Fig. 8 C), suggesting that the difference seen with antibodies to Sey1p and Sec22p was not due to different kinetics of antibody binding to the antigens. Therefore, Sec22p is required in an extremely late, post-Sey1p phase of the reaction and acts near the time of content mixing during homotypic ER fusion in yeast.
Figure 8.
Sey1p acts before Sec22p during homotypic ER fusion. Gluc1 and Gluc2 microsomes were incubated at 27°C in the presence of both ATP and GTP (A) or GTP alone (B). At the indicated times, a portion of the reaction received anti-Sey1p (squares) or anti-Sec22p (triangles) antibodies, or was placed on ice (circles). Luciferase activity was measured after 120 min. Fusion values were normalized to those obtained in reactions that received antibodies or were placed on ice at 120 min. Data represent the means ± SEM (error bars; n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001, Tukey’s test compared with the ice groups (circles). (C) Similar sensitivity kinetics was observed with increasing amounts of anti-Sey1p antibodies. Gluc1 and Gluc2 microsomes were incubated at 27°C in the presence of GTP. At the indicated times, a portion of the reaction received increasing concentrations of anti-Sey1p antibodies (88 nM for squares, 176 nM for triangles, 352 nM for diamonds) or was placed on ice (circles). Luciferase activity was measured after 120 min. Data represent the means ± SEM (error bars; n = 3). *, P < 0.05; **, P < 0.01, Tukey’s test.
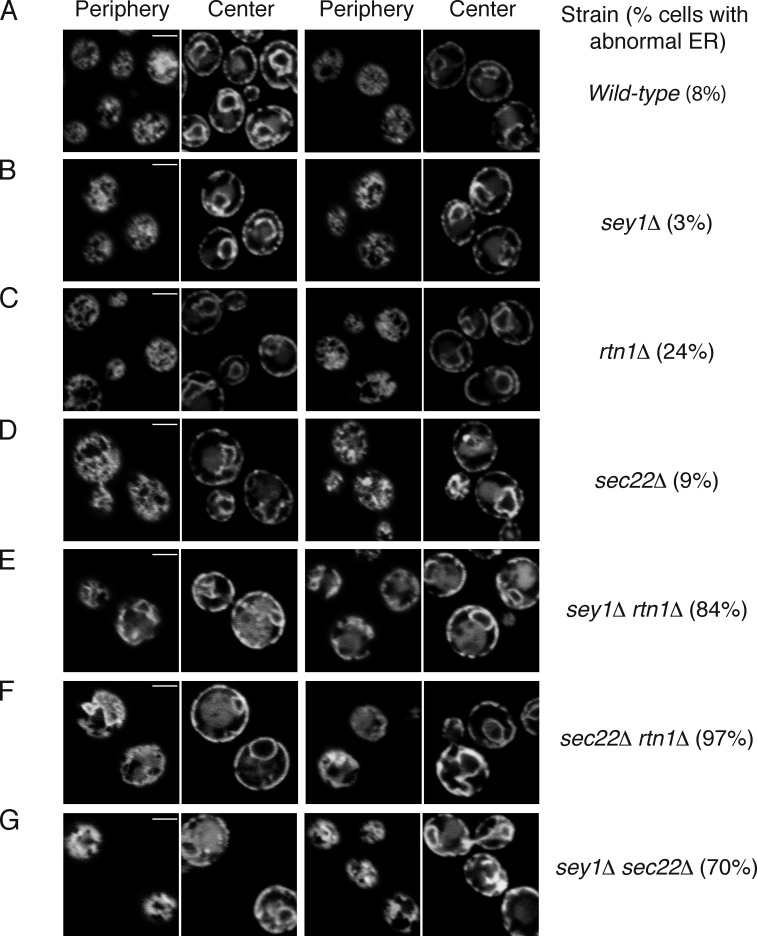
Sec22p is required to maintain ER morphology in yeast cells lacking Rtn1p or Sey1p
To address whether Sec22p is involved in ER fusion in vivo, we analyzed ER morphology in yeast cells lacking Sec22p. In yeast lacking only Sec22p, no significant defect in ER morphology was observed (Fig. 9 D) compared with wild-type cells (Fig. 9 A), which is similar to previous observations for single deletions of Sey1p or Rtn1p (Voeltz et al., 2006; Hu et al., 2009; Anwar et al., 2012; also see Fig. 9, B and C). However, cells lacking both Sec22p and Rtn1p had an abnormal ER morphology; they lacked the tubular network and instead contained expanded ER sheets (Fig. 9 F). This is reminiscent of the synergistic defect in ER morphology observed in cells lacking both Sey1p and Rtn1p (Voeltz et al., 2006; Hu et al., 2009; also see Fig. 9 E). Furthermore, a similar defect was observed in cells lacking both Sey1p and Sec22p (sey1Δ sec22Δ, Fig. 9 G). These results are consistent with the idea that Sey1p and Sec22p cooperate to mediate ER fusion.
Figure 9.
Sec22p regulates ER morphology in vivo. (A) The ER was visualized by expressing EGFP-HDEL in wild-type (BY4742) yeast cells. The localization of the fluorescent protein was determined by confocal fluorescence microscopy, with the microscope focused either at the center or periphery of the cell. (B) As in A, but with cells lacking Sey1p (BY4742 sey1Δ). (C) As in A, but with BY4742 rtn1Δ. (D) As in A, but with BY4742 sec22Δ. (E) As in A, but with BY4742 sey1Δ rtn1Δ. (F) As in A, but with BY4742 sec22Δ rtn1Δ. (G) As in A, but with BY4742 sey1Δ sec22Δ. The percentage of cells with abnormal ER was determined from 30–80 cells per strain. Bars, 2 µm.
Discussion
Here, we described an in vitro assay using isolated yeast microsomes for measuring atlastin-dependent homotypic ER fusion that exploits content mixing–dependent reconstitution of Gluc activity from two complementary fragments. Although a cell-free assay has been developed for measuring homotypic fusion of yeast ER microsomes based on glucose trimming of N-linked oligosaccharides (Latterich and Schekman, 1994), ER fusion in this assay strictly requires ATP, but not GTP. Thus, this assay may not represent atlastin-dependent ER membrane fusion events, which require GTP. Using our in vitro assay, we found that ER-associated SNARE proteins are involved in atlastin-mediated homotypic ER membrane fusion in yeast.
ER morphology (Hu et al., 2009; Anwar et al., 2012) and intermixing between different ER markers from two mating cells during yeast cell mating (Anwar et al., 2012; Chen et al., 2012; Rogers et al., 2013) have been used as measures of homotypic ER fusion in yeast. Although these measures may provide an indication of in vivo events in homotypic ER fusion, they have several limitations. First, defects in the genes required for homotypic ER fusion do not always correlate with ER morphology defects. For example, yeast cells deficient for Sey1p show a normal ER morphology, whereas defects in genes encoding ER-shaping proteins, which normally do not participate in ER fusion per se, result in abnormal ER morphology (Hu et al., 2009; Anwar et al., 2012). Second, ER–ER fusion during yeast mating may use a different set of protein players and molecular mechanisms than ER fusion that generates and maintains the unique ER structure during the normal cell cycle. This is extremely likely because the expression of nearly 200 genes changes significantly during mating in S. cerevisiae (Roberts et al., 2000; de Godoy et al., 2008). Finally, when ER fusion is monitored based on intermixing between different ER-localized fluorescent proteins during mating, it is nearly impossible to distinguish between the incorporation of newly synthesized fluorescent proteins and the redistribution of preexisting fluorescent proteins between fusing ER membranes (Anwar et al., 2012). Thus, the in vitro assay for Sey1p-mediated ER fusion described here is a valuable complement to in vivo ER fusion assays.
ER microsome preparations are generally contaminated by other organelles and vesicles; therefore, reconstitution of luciferase activity in our in vitro fusion reactions containing isolated microsomes could conceivably be achieved not only by homotypic ER fusion but also by other events, such as vesicle-mediated transport of a luciferase fragment to compartments containing the other fragment. For instance, because luciferase fragments contain an ER-retrieval signal, fusion between ER membranes containing one fragment and cis-Golgi–derived COPI-coated vesicles that carry the other fragment could result in reconstitution of functional luciferase. However, in vitro reconstitution of vesicle-mediated transport generally requires cytosol (Baker et al., 1988; Spang and Schekman, 1998) or purified cytosolic proteins (Barlowe, 1997), such as coat proteins. Furthermore, Ypt1p is required for retrograde Golgi-ER transport (Kamena et al., 2008), but not for our in vitro fusion reactions (Fig. 3, A and B). Finally, recent studies have suggested that Sey1p plays a role in nuclear fusion during yeast cell mating (Chen et al., 2012; Rogers et al., 2013). Deletion of KAR5, a gene required for homotypic nuclear fusion during mating (Kurihara et al., 1994; Beh et al., 1997; Erdman et al., 1998), did not markedly influence Sey1p-dependent fusion in vitro (Fig. S4), suggesting that nuclear fusion does not significantly contribute to the luciferase activity of our in vitro assay. Therefore, luciferase activity detected in our assay results primarily from Sey1p-dependent ER membrane fusion events.
Although SNAREs mediate atlastin-independent ER fusion as recent studies suggested (Anwar et al., 2012; Rogers et al., 2013), this does not necessarily exclude the possibility that atlastin-mediated ER fusion involves the function of SNAREs for efficient fusion. Moreover, our estimation of the abundance of Sey1p in the ER membrane indicates that Sey1p alone is not sufficient to drive ER fusion at its physiological concentration. Thus, it is possible that although Sey1p can initiate ER fusion, Sey1p needs to recruit SNAREs to fusion sites for efficient fusion. If Sey1p has a limited fusogenic capability as our data indicate, what would be its major function in ER fusion? Our data show that Sey1p is absolutely required for ER fusion and seems to function early, before Sec22p. Also, Sey1p-mediated ER fusion does not require the function of Rab GTPases, which can function as a tether by themselves or by recruiting proteins involved in membrane tethering. One feasible function of Sey1p is as a tether for ER fusion. ER tubules are dispersed throughout the cytoplasm and thus make physical contact with various organelles and the plasma membrane; therefore, homotypic ER fusion must be tightly regulated to avoid nonspecific membrane fusion events with the contacted organelles. To achieve this extremely high specificity, the ER may use a specialized tether, distinct from Rab GTPases, for its homotypic fusion. Consistent with this, a recent study showed that the tethering activity of Drosophila atlastin can be uncoupled from its fusogenic activity (Saini et al., 2014).
Our in vitro ER fusion assay may not reflect the in vivo situation because ER fusion in this assay occurs in the absence of cytosol. Thus, the strict Sey1p dependency of our ER fusion assay is possibly caused by the absence of cytosolic factors, which could otherwise play a critical role in ER fusion in vivo. One such factor is the Dsl1p complex comprising the subunits Tip20p, Dsl1p, and Dsl3p, which was recently reported to function in Sey1p-independent ER fusion (Rogers et al., 2014). Because the Dsl1p complex functions as a tether for ER SNARE-mediated membrane fusion events (Kraynack et al., 2005; Ren et al., 2009; Diefenbacher et al., 2011), the tethering activity of Sey1p could become aberrantly dominant over its fusogenic activity in the absence of the Dsl1p complex. Although we cannot completely exclude this possibility, the strict Sey1p dependency was clearly unaffected by the addition of purified yeast cytosol (Fig. S5). Furthermore, subcellular fractionation studies revealed that two subunits of the Dsl1p complex, Dsl1p and Dsl3p, are found exclusively on the membrane, but not in the cytosol (Andag et al., 2001; Kraynack et al., 2005), suggesting that the Dsl1p complex is also present largely on the ER membrane. Although no cytosolic factors have been identified to date that regulate Sey1p-dependent ER fusion, purified yeast cytosol appears to have inhibitory and stimulatory activities for ER fusion (Fig. S5). Therefore, our in vitro ER fusion assay may provide a facile tool for identifying cytosolic factors that regulate Sey1p-dependent ER fusion.
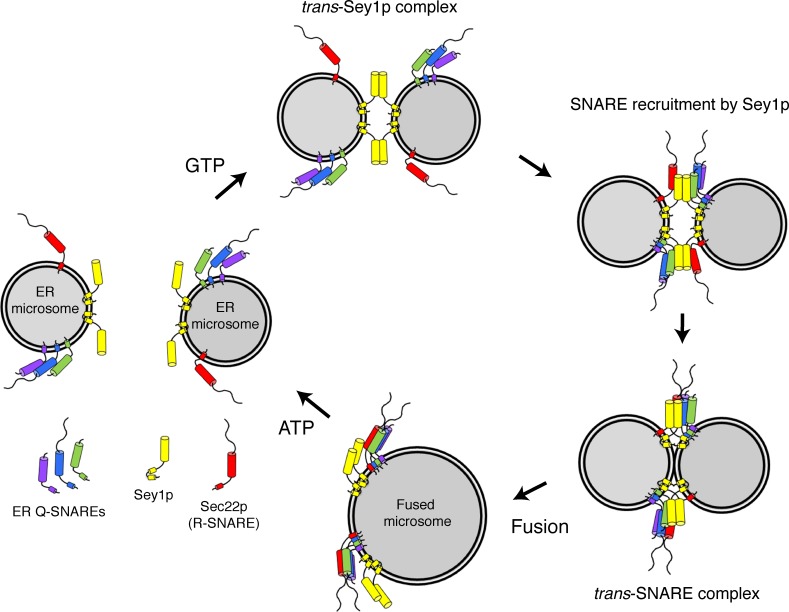
Although further studies are required to elucidate the molecular mechanism by which the yeast atlastin Sey1p and ER-associated SNAREs work together to mediate homotypic ER fusion, we propose a working model (Fig. 10) based on the data presented in this study: GTP-dependent dimerization of Sey1p in trans mediates the initial contact between two fusing ER membranes. In the meantime, Sey1p interacts with ER-associated SNARE molecules and recruits them to the contact site between the two fusing ER membranes. A conformational change in Sey1p induced by GTP hydrolysis brings the two membranes into close proximity, which directly causes some degree of ER membrane fusion or allows ER SNARE proteins between the apposed membranes to form trans-SNARE complexes around trans-Sey1p complexes. These trans-SNARE complexes, in turn, facilitate and complete ER membrane fusion.
Figure 10.
Working model for the involvement of SNAREs in Sey1p-mediated homotypic ER fusion. See text in Discussion.
Materials and methods
Yeast strains and strain construction
Yeast strains used in this study are listed in Table 1. Microsomes from BJ3505 vam3Δ or its derivatives were used for the in vitro ER fusion assay. Strains expressing ssZIP-Gluc1-HDEL or ssZIP-Gluc2-HDEL in their ER lumen were generated by transforming BJ3505 vam3Δ with pYJ406-KAR2-SS-ZIP-Gluc1-HDEL or pYJ406-KAR2-SS-ZIP-Gluc2-HDEL, respectively. The resulting strains, BJ-Gluc1 and BJ-Gluc2, were used to generate the deletion strains sey1Δ, sec22Δ, rtn1Δ, and yop1Δ using the KanMX4 gene, which was PCR-amplified from the plasmid vector pRS400 (Brachmann et al., 1998). The deletion of each endogenous gene was confirmed by genomic PCR using specific primers. BJ-Gluc1 sey1Δ and BJ-Gluc2 sey1Δ were transformed with BamHI-linearized pRS408-SEY1(K50A), generating BJ-Gluc1 (sey1-K50A) and BJ-Gluc2 (sey1-K50A). RSY250 and RSY275 (Kaiser and Schekman, 1990; gifts from C. Barlowe, Dartmouth College, Hanover, NH) were transformed with BsaBI-linearized pYJ406-KAR2-SS-ZIP-Gluc1-HDEL or pYJ406-KAR2-SS-ZIP-Gluc2-HDEL, generating tester strains for the in vitro ER fusion assay. BY4742 (Winzeler et al., 1999) and its derivatives were transformed with linearized pYJ406-KAR2-SS-EGFP-HDEL and used for ER morphology analysis. BJ3505 and DKY6281 were used for the in vitro vacuole fusion assay.
Table 1. Yeast strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| BJ3505 | MATa ura3-52 trp1-Δ101 his3- Δ200 lys2-801 gal2 (gal3) can1 prb1-Δ1.6R pep4Δ::HIS3 | Jones, 2002 |
| DKY6281 | MATα ura3-52 leu2-3,112 trp1-Δ901 his3-Δ200 lys2-801 suc2-Δ9 pho8Δ::TRP1 | Haas et al., 1994 |
| BJ-GLuc1 | BJ3505 vam3Δ::TRP1 containing pYJ406-ssZIP-GLuc1-HDEL | This study |
| BJ-GLuc2 | BJ3505 vam3Δ::TRP1 containing pYJ406-ssZIP-GLuc2-HDEL | This study |
| BJ-GLuc1 sey1Δ | BJ-GLuc1 with sey1Δ::KanMX4 | This study |
| BJ-GLuc2 sey1Δ | BJ-GLuc2 with sey1Δ::KanMX4 | This study |
| BJ-GLuc1 sec22Δ | BJ-GLuc1 with sec22Δ::KanMX4 | This study |
| BJ-GLuc2 sec22Δ | BJ-GLuc2 with sec22Δ::KanMX4 | This study |
| BJ-GLuc1 rtn1Δ | BJ3505 vam3Δ GLuc1 with rtn1Δ::KanMX4 | This study |
| BJ-GLuc2 rtn1Δ | BJ3505 vam3Δ GLuc2 with rtn1Δ::KanMX4 | This study |
| BJ-GLuc1 yop1Δ | BJ3505 vam3Δ GLuc1 with yop1Δ::KanMX4 | This study |
| BJ-GLuc2 yop1Δ | BJ3505 vam3Δ GLuc2 with yop1Δ::KanMX4 | This study |
| BJ-GLuc1 kar5Δ | BJ3505 vam3Δ GLuc1 with kar5Δ::KanMX4 | This study |
| BJ-GLuc2 kar5Δ | BJ3505 vam3Δ GLuc2 with kar5Δ::KanMX4 | This study |
| BJ-GLuc1 sec22Δ SEY1-OE | BJ-GLuc1 sec22Δ containing pRS408-pTDH3-SEY1 | This study |
| BJ-GLuc2 sec22Δ SEY1-OE | BJ-GLuc2 sec22Δ containing pRS408-pTDH3-SEY1 | This study |
| BJ-GLuc1 Sey1-K50A | BJ-GLuc1 sey1Δ containing pRS408-SEY1-K50A | This study |
| BJ-GLuc2 Sey1-K50A | BJ-GLuc2 sey1Δ containing pRS408-SEY1-K50A | This study |
| RSY250 | MATα ura3-52 | Kaiser and Schekman, 1990 |
| RSY275 | MATα ura3-52 his4-619 s20-1 | Kaiser and Schekman, 1990 |
| RSY250 GLuc1 | RSY250 containing pYJ406-ssZIP-GLuc1-HDEL | This study |
| RSY250 GLuc2 | RSY250 containing pYJ406-ssZIP-GLuc2-HDEL | This study |
| RSY275 GLuc1 | RSY275 containing pYJ406-ssZIP-GLuc1-HDEL | This study |
| RSY275 GLuc2 | RSY275 containing pYJ406-ssZIP-GLuc2-HDEL | This study |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Winzeler et al., 1999 |
| BY-EGFP-HDEL | BY4742 with pYJ406-ssEGFP-HDEL | This study |
| BY-EGFP-HDEL sey1Δ | BY4742 EGFP-HDEL with sey1Δ::KanMX4 | This study |
| BY-EGFP-HDEL rtn1Δ | BY4742 EGFP-HDEL with rtn1Δ::KanMX4 | This study |
| BY-EGFP-HDEL sec22Δ | BY4742 EGFP-HDEL with sec22Δ::KanMX4 | This study |
| BY-EGFP-HDEL sey1Δ rtn1Δ | BY4742 rtn1Δ EGFP-HDEL with sey1Δ::LEU2 | This study |
| BY-EGFP-HDEL sec22Δ rtn1Δ | BY4742 sec22Δ EGFP-HDEL with rtn1Δ::LEU2 | This study |
| BY-EGFP-HDEL sec22Δ sey1Δ | BY4742 sec22Δ EGFP-HDEL with sey1Δ::LEU2 | This study |
DNA constructs and plasmids
Plasmids used in this study are listed in Table S1. pYJ406-ssZIP-Gluc1-HDEL and pYJ406-ssZIP-Gluc2-HDEL were generated by inserting a DNA fragment encoding the signal sequence (the first 45 aa) of Kar2p and the GCN4 leucine zipper-coding sequence (ZIP) fused to either Gluc1-HDEL or ZIP-Gluc2-HDEL (PCR-amplified from pcDNA3.1-Zip-hGluc1 and pcDNA3.1-Zip-hGluc2 [Remy and Michnick, 2006], respectively) into pYJ406 (Jun and Wickner, 2007). pGST-ZIP was generated by inserting a DNA fragment encoding ZIP into pGST-Parallel1 (Sheffield et al., 1999). The SEY1 gene was PCR-amplified from purified yeast genomic DNA and inserted into pRS408 (plasmid #11255; Addgene), which was a gift from F. Cross (Rockefeller University, New York, NY), generating pRS408-SEY1. pRS408-SEY1-K50A was generated by site-directed mutagenesis using pRS408-SEY1 and confirmed by sequencing. For overexpression of Sey1p, pRS408-pTDH3-SEY1 was generated by replacing the SEY1 promoter of pRS408-SEY1 with the TDH3 promoter (PCR-amplified from pYM-N14; Janke et al., 2004). pYJ406-ssEGFP-KDEL was generated by replacing the DNA fragment encoding the ZIP-Gluc1-HDEL of pYJ406-ssZIP-Gluc1-HDEL with a DNA fragment encoding EGFP-HDEL that was PCR-amplified from pEGFP-N3 (Takara Bio Inc.). DNA fragments encoding the CD of Sey1p, the CD of Sec22p, and the CD of Ufe1p were PCR-amplified from yeast genomic DNA and inserted into pGST-Parallel1, pHIS-Parallel1, and pMBP-Parallel1 (Sheffield et al., 1999), generating pGST-SEY1-CD (1–681 aa), pHIS-SEC22-CD (1–188 aa), and pMBP-UFE1-CD (1–326 aa), respectively. DNA fragments encoding Gdi1p and Gyp1p-46 (248–637) were PCR-amplified from yeast genomic DNA and subcloned into pHIS-Parallel1, generating pHIS-GDI1 and pHIS-GYP1-46, respectively.
Microsome preparation
Microsomes were prepared as described previously (Baker et al., 1990), with modifications. In brief, yeast cells were grown in YPD to an OD600 of 2–3 and collected by centrifugation at 1,000 g for 5 min at 25°C. Cells were resuspended in 100 mM Tris-HCl, pH 9.4, containing 10 mM DTT and incubated at 30°C for 10 min. Cells were collected and resuspended to an OD600 of 25–40 in spheroplasting buffer (0.1% yeast extract, 0.2% bacteriological peptone, 0.2% glucose, 50 mM KPO4, and 0.6 M sorbitol, pH 7.5). Lyticase (0.4 mg/ml) was added, and the suspension was incubated at 30°C for 35 min. Spheroplasts were collected by centrifugation through a cushion of 0.8 M sucrose, 1.5% Ficoll, and 20 mM Hepes, pH 7.4; resuspended to 100 OD600 U/ml in lysis buffer (20 mM Hepes, pH 7.4, 100 mM sorbitol, 50 mM potassium acetate, 2 mM EDTA, and 1 mM DTT); and homogenized with a Teflon-glass homogenizer (20 strokes). After the lysate was centrifuged at 3,000 g (10 min, 4°C), the resulting supernatant was collected and centrifuged at 12,000 g (10 min, 4°C). The resulting microsomal pellet was resuspended in 900 µl of lysis buffer and loaded onto a 3.0-ml sucrose step gradient (1.5 ml each of 1.5 M sucrose and 1.2 M sucrose prepared in lysis buffer) and centrifuged at 100,000 g (1 h, 4°C). Microsomes were collected from the 1.2 M/1.5 M interface and washed with PS buffer (10 mM Pipes-KOH, pH 6.8, and 200 mM sorbitol) containing 125 mM KCl. After centrifugation at 12,000 g (10 min, 4°C), the pellet was resuspended in PS buffer. Alternatively, vacuolar contamination in microsomal preparations was minimized by preparing microsomes using a previously described protocol for the preparation of vacuoles (Bankaitis et al., 1986), with modifications. In brief, spheroplasts were carefully resuspended in cold 15% Ficoll solution (15% Ficoll, 10 mM Pipes-KOH, pH 6.8, and 200 mM sorbitol), and 300 µl of a 10-mg/ml DEAE-dextran solution prepared in PS-buffered 15% Ficoll was added. The suspension was incubated on ice for 2 min and then at 30°C for 3 min. Microsomes were isolated by flotation through a discontinuous Ficoll step gradient, which was accomplished by transferring the suspension (5 ml) to an SW40 tube (Beckman Coulter), overlaying it with 2 ml of buffered 8% Ficoll, 4 ml of buffered 4% Ficoll, and 2 ml of PS buffer, and centrifuging at 110,000 g (90 min, 4°C). Microsomes were collected from the 4%/8% interface. Microsomes prepared using either of these methods produced similar results in in vitro ER fusion assays.
In vitro ER membrane fusion assay
The standard ER fusion reaction (50 µl) contained 5 µg of Gluc1 microsomes, 5 µg of Gluc2 microsomes, reaction buffer (10 mM Pipes-KOH, pH 6.8, 125 mM KCl, 5 mM MgCl2, and 200 mM sorbitol), an energy-regenerating system (1 mM MgCl2, 1 mg/ml creatine kinase, and 29 mM creatine phosphate), 264 nM Pbi2p (IB2), 10 µM coenzyme A, 1 mM ATP and/or 1 mM GTP (Roche), and 100 µM GST-ZIP. Fusion mixtures were incubated at 27°C. After 90 min, 30 µl of the reaction mixture was mixed with 30 µl of coelenterazine (40 µM; Promega) and transferred to a 96-well white plate for measurement of luminescence using a luminometer (Centro XS3 LB 960; Berthold Technologies).
Protein and antibody preparation
His6-Gdi1p, His6-Gyp1-46p, and GST-ZIP were prepared as described previously (Jun et al., 2006; Ko et al., 2014). In brief, these recombinant proteins were produced in Escherichia coli Rosette 2 (λDE3; Merck Millipore) in LB medium containing ampicillin by induction with 1 mM IPTG for 12–20 h at 20°C. Cells were harvested, resuspended in sonication buffer (PBS, 2 mM DTT, 1 mM PMSF, and a protease inhibitor cocktail) containing 20 mM imidazole for His6-Gdi1p and His6-Gyp1-46p or 2 mM EDTA for GST-ZIP and lysed by sonication. After centrifugation at 23,000 g for 30 min at 4°C, the supernatant was further clarified by filtration using a 0.45 mm filter. For His6-Gdi1p and His6-Gyp1-46p, the resulting supernatant was incubated with 2.5 ml of Ni-NTA resin (QIAGEN) at 4°C for 12 h with nutation. After washing with PBS containing 20 mM imidazole, proteins were eluted with PBS containing 250 mM imidazole and dialyzed against PS buffer containing 125 mM KCl. For GST-ZIP, the filtered supernatant was incubated with 1 ml glutathione beads (QIAGEN) at 4°C for 8 h with nutation. After washing with PBS, GST-ZIP was eluted with PBS containing 10 mM reduced glutathione (Thermo Fisher Scientific) and dialyzed against PS buffer containing 125 mM KCl. GST-Sey1p-CD was produced in E. coli Rosette 2 (λDE3) in LB medium containing ampicillin by induction with 0.5 mM IPTG for 9 h at 20°C. Cells were harvested, resuspended in sonication buffer (25 mM Hepes-KOH, pH 7.4, 200 mM KCl, 2 mM DTT, 2 mM EDTA, 5% glycerol, and a protease inhibitor cocktail), and lysed by sonication. After centrifugation at 40,000 rpm (70Ti; Beckman Coulter) for 45 min at 4°C, the supernatant was mixed with 1 ml of glutathione beads and incubated at 4°C for 12 h with nutation. After washing the beads with wash buffer (25 mM Hepes-KOH, pH 7.4, 100 mM KCl, 5% glycerol, 2 mM DTT, and 1 mM EDTA), GST-Sey1p-CD was eluted with wash buffer containing 10 mM reduced glutathione and dialyzed against PS buffer containing 125 mM KCl. For antibody production, GST was cleaved from GST-Sey1p-CD using TEV protease. His6-Sec22p-CD and MBP-Ufe1p-CD were similarly produced in E. coli Rosette 2 (λDE3) as previously described (Jun et al., 2006). In brief, His6-Sec22p-CD was produced in E. coli Rosette 2 (λDE3) in LB medium by induction with 0.5 mM IPTG for 20 h at 16°C. Cells were harvested, resuspended in sonication buffer (PBS, 2 mM DTT, 0.5 mM PMSF, 20 mM imidazole, and a protease inhibitor cocktail), and lysed by sonication. After centrifugation, the supernatant was further clarified by filtration using a 0.45 µm filter and mixed with 1 ml of Ni-NTA resin and incubated at 4°C for 12 h with nutation. After washing with PBS containing 20 mM imidazole, His6-Sec22p-CD was eluted with PBS containing 250 mM imidazole and dialyzed against PS buffer containing 125 mM KCl. MBP-Ufe1p-CD was produced in E. coli Rosette 2 (λDE3) by induction with 0.5 mM IPTG for 12 h at 20°C. Cells were harvested, resuspended in sonication buffer (25 mM Hepes-KOH, pH 7.4, 200 mM NaCl, 0.5 mM DTT, 1 mM EDTA, 5% glycerol, and a protease inhibitor cocktail), and lysed by sonication. After centrifugation, the supernatant was further clarified by filtration using a 0.45-µm filter, mixed with 3 ml of amylose resin (New England BioLabs, Inc.), and incubated at 4°C for 4 h with nutation. After washing, MBP-Ufe1p-CD was eluted with sonication buffer containing 10 mM maltose. Anti-Sey1p, anti-Sec22p, and anti-Ufe1p rabbit polyclonal antibodies were raised against cleaved Sey1p-CD, his6-Sec22p-CD, and MBP-Ufe1p-CD, respectively (AbFrontier). These antibodies were affinity-purified using their antigens bound to Sulfolink resin (Thermo Fisher Scientific) and dialyzed against PS buffer containing 125 mM KCl. Concentrations used in the in vitro ER fusion assay (unless otherwise noted) were 88 nM affinity-purified anti-Sey1p antibody and 0.85 µM affinity-purified anti-Sec22p antibody. Affinity-purified anti-Sec18p and anti-Sec17p rabbit antibodies were gifts from W. Wickner (Dartmouth Medical School, Hanover, NH), and anti-Yet3p (raised against GST-Yet3p) and anti-Kar2p (raised against a TrpE-Kar2p fusion protein) rabbit sera were provided by C. Barlowe (Dartmouth Medical School). Anti-Ypt1p goat antibodies (Santa Cruz Biotechnology, Inc.) and anti-Gaussia luciferase rabbit antibodies (New England BioLabs, Inc.) were purchased.
Extraction and measurement of microsomal lipids
Microsomal lipids were extracted using a modification of the Bligh-Dyer method (Bligh and Dyer, 1959), as described previously (Ko et al., 2014). In brief, chloroform (100 µl) and methanol supplemented with 0.1 M HCl (200 µl) were added to 37 µg of microsomes, as measured by their protein content, in 80 µl of PS buffer, mixed thoroughly by vortexing, and incubated at RT for 1 h. After PS buffer (100 µl) and chloroform (100 µl) were added, the sample was vortexed thoroughly and centrifuged at 14,000 g at RT for 30 s. The organic layer was transferred to a 13 × 100-mm bound-bottom glass tube. Chloroform (200 µl) was added to the remaining aqueous layer, the mixture was vortexed and centrifuged at 14,000 g at RT for 30 s, and the organic layer was removed and added to the organic layer from the first extraction. PS buffer (360 µl) and methanol-HCl (400 µl) were added to the combined organic layers. This mixture was vortexed thoroughly and centrifuged at RT for 30 s, and the aqueous layer was removed by aspiration. The organic layer was then dried in a Sorvall Speed-Vac SC100 (Thermo Fisher Scientific) and used for estimation of microsomal lipid levels. Microsomal lipid levels were measured using a lipid phosphorous assay. 10 µl of 2% ammonium molybdate was added to extracted vacuolar lipids and to standards (0, 5, 10, 25, 50, 75, 100, and 125 µl of a 1 mM K2HPO4 solution). Samples were dried completely in Speed-Vac. After the addition of perchloric acid (300 µl of a 70% solution), samples were incubated at 180°C for 30 min, and then cooled to RT. Ammonium molybdate (1.5 ml of a 0.4% solution) and ascorbic acid (225 µl of a 10% solution) were added. The mixture was incubated at 90°C for 10 min, and then cooled to RT. Absorbance at 820 nm was measured, and phospholipid concentration was estimated by comparing microsomal lipid samples to K2HPO4 standards. To obtain the final lipid concentrations, measured phospholipid concentrations were multiplied by 1.26 to correct for a reported ergosterol/phospholipid molar ratio of 0.26 in microsomal lipids (Zinser et al., 1991), as described previously (Ko et al., 2014).
Measurement of the microsomal Sey1p level
To estimate the microsomal Sey1p level, BJ-Gluc1 and BJ-Gluc2 microsomes were analyzed by SDS-PAGE and immunoblotting for Sey1p. Protein levels were estimated by comparing band intensities from microsomal samples to those from standards of purified recombinant GST-Sey1p-CD.
Yeast vacuole preparation and in vitro vacuole fusion assay
Vacuoles were isolated as described previously (Haas, 1995). In brief, yeast cells were grown in YPD to an OD600 of 0.8–1.2 and collected by centrifugation at 1,000 g for 5 min at 25°C. Cells were resuspended in wash buffer (100 mM Tris-HCl, pH 9.4, 10 mM DTT) and incubated at 30°C for 10 min. Cells were collected and resuspended to an OD600 of 25–40 in spheroplasting buffer. Lyticase (0.4 mg/ml) was added, and the suspension was incubated at 30°C for 35 min. Spheroplasts were collected by centrifugation and carefully resuspended in cold 15% Ficoll solution, and 300 µl of a 10-mg/ml DEAE-dextran solution was added. The suspension was incubated on ice for 2 min and then at 30°C for 3 min. Vacuoles were isolated by flotation through a discontinuous Ficoll step gradient, which was accomplished by transferring the suspension to an SW40 tube, overlaying it with 2 ml of buffered 8% Ficoll, 4 ml of buffered 4% Ficoll, and 2 ml of PS buffer, and centrifuging at 110,000 g (90 min, 4°C). Vacuoles were collected from the 0%/4% interface. Standard in vitro vacuole fusion reactions (30 ml) contained 3 µg of BJ3505 vacuoles and 3 µg of DKY6281 vacuoles, reaction buffer (125 mM KCl, 5 mM MgCl2, 10 mM Pipes-KOH, pH 6.8, and 200 mM sorbitol), 264 nM purified Pbi2p, 10 µM CoA, 1 mM ATP, 1 mg/ml creatine kinase, and 29 mM creatine phosphate. After 90 min of incubation at 27°C, fusion was measured by assaying alkaline phosphatase.
Preparation of reconstituted Sey1p liposomes
Reconstitution of proteoliposomes bearing purified recombinant Sey1p was performed as described previously with modifications (Orso et al., 2009; Anwar et al., 2012). All nonfluorescent lipids were purchased from Avanti Polar Lipids, Inc., except for ergosterol (Sigma-Aldrich). The fluorescent lipids N-(7-nitro-2,1,3-benzoxadiazole-4-yl)-PE (NBD-PE), N-(lissamine rhodamine B sulfonyl)-PE (Rh-PE), and dansyl-PE were purchased from Molecular Probes. ER-mimicking lipid mixes for Sey1p liposomes contained 1-palmitoyl-2-oleoyl-PC (POPC; 44% or 46% [mol/mol] for donor and acceptor Sey1p proteoliposomes, respectively), POPE (20%), Soy-PI (10%), POPS/DOPS (8.0%), POPA (3.0%), ergosterol (10%), cardiolipin (1.0%), diacylglycerol (1.0%), and fluorescent lipids (1.5% each of NBD-PE/Rh-PE or 1.0% of dansyl-PE for donor and acceptor liposomes, respectively). These lipids in chloroform were mixed and dried under a stream of N2 gas. The resulting dried lipid films were dissolved in RB150 (20 mM Hepes-NaOH, pH 7.4, 150 mM NaCl, and 10% [vol/vol] glycerol) containing 1 mM EDTA and incubated at 37°C for 30 min. Large unilamellar vesicles (LUVs) were formed by five freeze-thaw cycles in liquid N2 and water at 37°C. To form uniform-sized unilamellar liposomes, LUVs were extruded 11 times through a polycarbonate filter (Avanti Polar Lipids, Inc.) with a pore size of 100 nm. Sey1p reconstitution was performed by detergent-assisted insertions as described previously (Rigaud and Lévy, 2003). In brief, purified Sey1p in 0.1% Triton X-100 was mixed with preformed liposomes at the indicated protein-to-lipid ratio and an effective detergent-to-lipid ratio (Reff) below 0.64, and incubated at 4°C for 2 h. Reff is defined by the equation Reff = (Dtotal – Dwater)/[total lipid], in which Dtotal is the total detergent concentration and Dwater is the monomeric detergent concentration (0.18 mM for Triton X-100). After proteins and lipids were allowed to mix for 1 h at 4°C, detergent was removed by incubating with BioBeads SM-2 adsorbent beads (Bio-Rad Laboratories) at 4°C for 3 h with stirring. After another incubation with fresh beads at 4°C for 16 h with continuous stirring, proteoliposomes were brought to 2 mM lipid in RB150 containing 1 mM EDTA, and small aliquots were frozen in liquid nitrogen and stored at –80°C.
In vitro Sey1p proteoliposome fusion assay
Liposome fusion assays were based on previously described methods (Orso et al., 2009; Anwar et al., 2012), except that donor and acceptor proteoliposomes were mixed at a molar ratio of 1:2.5. In brief, labeled donor proteoliposomes (200 µM of total lipid) were mixed with unlabeled acceptor proteoliposomes (500 µM of total lipid) in RB150. The reaction mixture was transferred to a black polystyrene 384-well plate and incubated at 30°C for 10 min. The fusion reaction was started by adding 1 mM GTP and 2 mM Mg2+ (final). NBD fluorescence was measured at 30-s intervals. After 30 min, 90 mM β-octylglucoside (final) was added to determine total fluorescence in the sample. Fusion is expressed as the percentage of total fluorescence.
Coimmunoprecipitation
Microsomes were incubated in ER fusion reaction buffer (10 mM Pipes-KOH, pH 6.8, 125 mM KCl, 5 mM MgCl2, and 200 mM sorbitol) containing an ATP/GTP-regenerating system at 27°C. After 60 min, membranes were pelleted by centrifugation at 11,000 g (10 min, 4°C). The supernatant was removed, and the sedimented membranes were resuspended in ice-cold solubilization buffer (PBS containing 0.5% Triton X-100, 1 mM PMSF, and 10 µM leupeptin) and incubated on ice for 20 min. Detergent-insoluble material was removed by centrifugation at 11,000 g (10 min, 4°C). The resulting post-centrifugation supernatants were precleared by incubation with protein A Sepharose (GE Healthcare) at 4°C for 1 h. Affinity-purified anti-Sec22p antibodies, affinity-purified anti-Ufe1p antibodies, or preimmune IgG were added to the precleared supernatants and incubated at 4°C on a nutator for 4 h. Protein A Sepharose was then added and further incubated for 1 h. Beads were collected by centrifugation at 3,000 g (1 min, RT) and washed three times with ice-cold solubilization buffer. Bound proteins were eluted with SDS sample buffer for SDS-PAGE analysis followed by immunoblotting using anti-Sec22p, anti-Sey1p, and anti-Ufe1p antibodies. For immunoprecipitation of Sec22p in the absence or presence of anti-Sec18p antibodies, affinity-purified anti-Sec22p antibody was conjugated to Dynabeads (Life Technologies) according to the manufacturer’s instructions, and the beads were added to detergent-solubilized microsomes. After incubation at 4°C on a nutator for 4 h, beads were collected using magnetic separation and washed three times with ice-cold solubilization buffer. Bound proteins were eluted by boiling in SDS sample buffer and analyzed by SDS-PAGE and immunoblotting using anti-Sec22p, anti-Sey1p, and anti-Ufe1p antibodies. In vivo cross-linking of Sey1p to Sec22p was performed as described previously (Siniossoglou and Pelham, 2001), with modifications. In brief, 500 mg of spheroplasts from strain BJ3505 were resuspended in a cross-linking buffer (25 mM potassium phosphate, pH 7.4, and 200 mM sorbitol) containing protease inhibitors and 4 mM DSP (Thermo Fisher Scientific), and incubated for 30 min at 4°C. DSP was then quenched for 5 min by the addition of 150 mM Tris-HCl, pH 7.4. After centrifugation at 11,000 g (10 min, 4°C), pelleted cells were resuspended in PBS containing 0.5% Triton X-100, 1 mM PMSF, and 10 µM leupeptin, and lysed in the presence of glass beads by vortexing. Detergent-insoluble material was removed by centrifugation at 11,000 g (10 min, 4°C). The resulting post-centrifugation supernatants were used for immunoprecipitation using anti-Sec22p antibodies or preimmune IgG. Cross-links were cleaved using β-mercaptoethanol in SDS sample buffer, and proteins cross-linked with Sec22p were analyzed by SDS-PAGE followed by immunoblotting using anti-Sec22p, anti-Sey1p, and anti-Yet3p antibodies.
Microscopy
The ER was visualized by expressing EGFP-HDEL in yeast cells. Live yeast cells were imaged in growth medium at room temperature using a 100×/NA 1.40 oil immersion objective lens (UPlan-SApochromat) of a laser-scanning confocal microscope (FV1000; Olympus). Images were obtained using Olympus Fluoview software (FV10-ASV 3.1) and prepared with Photoshop (Adobe).
Statistical analysis
ANOVA with Tukey’s test for multiple comparisons and the Student’s t test were performed using SIGMAPLOT 12 (Systat Software). The normal distribution of data points was checked using the Shapiro-Wilk method. All data presented showed a normal distribution and equal variance. Only statistical significance is marked.
Trypsin digestion assay
Sey1p-proteoliposomes (150 µg/ml) were treated with trypsin (6 µg/ml) in the absence or presence of 1% Triton X-100 at 30°C. After 30 min, the mixtures were separated by SDS-PAGE, and protein bands were visualized by Coomassie brilliant blue staining. The ratio of the intensities of the full-length Sey1p band before and after trypsin digestion was analyzed by densitometry. To examine the GTP hydrolysis-induced dimerization capability of Sey1p proteins reconstituted into preformed liposomes, Sey1p-proteoliposomes (150 µg/ml) were incubated in the presence of 2 mM GDP and/or AlF4 (2 mM AlCl3 and 20 mM NaF) for 20 min at 30°C. The mixtures were then further incubated with or without trypsin (2 µg/ml) for 20 min at 30°C, and analyzed by SDS-PAGE and Coomassie brilliant blue staining.
Yeast cytosol preparation
Yeast cytosol was prepared as described previously (Barlowe, 1997), with modifications. In brief, strain BJ3505 was grown to mid-log phase in 4 liters of YPD medium. Cells were harvested, washed once with PS buffer containing 150 mM KOAc and 5 mM MgOAc, resuspended in the same buffer containing 1 mM PMSF and 10 µM leupeptin, and quick-frozen by drop-wise addition to liquid nitrogen. The frozen cells were mixed with liquid nitrogen in a Waring blender for 10 min to prepare a cell lysate. This lysate was thawed on ice and centrifuged at 8,000 g for 10 min at 4°C. The resulting supernatant was centrifuged at 200,000 g (45,000 rpm in a Type 70 Ti rotor; Beckman Coulter) for 90 min at 4°C. The clarified portion of this supernatant fraction was either frozen in liquid nitrogen for storage at –80°C or used as cytosol for the in vitro ER fusion assay.
Online supplemental material
Fig. S1 shows that both PCA fragments are required for fusion-dependent luciferase activity and the reaction kinetics of ATP/GTP-driven ER fusion and GTP-driven ER fusion reactions. Fig. S2 indicates that ER-shaping proteins are not essential for ER membrane fusion per se. Fig. S3 shows that a majority of Sey1p molecules are incorporated into proteoliposomes in the correct orientation. Fig. S4 indicates that nuclear fusion contributes little to the luciferase signal of in vitro ER fusion reactions. Fig. S5 shows that the strict Sey1p dependency of the in vitro ER fusion reaction is unaffected by the addition of cytosol. Table S1 lists the plasmids used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201501043/DC1.
Supplementary Material
Acknowledgments
We thank Drs. Charles Barlowe and William Wickner for reagents. We also thank Dr. Joji Mima (Osaka University) for suggestions on proteoliposome preparation and the lipid-mixing assay.
This work was supported by a grant from the Integrative Aging Research Center at GIST and by the National Research Foundation (NRF; grant NRF-2014R1A2A1A11051229), funded by the Korea government (MSIP). This work was also supported by a grant from Cell Dynamics Research Center, NRF (2007-0056157).
The authors declare no competing financial interests.
Author contributions: M. Lee, Y.-J. Ko, Y. Moon, and Y. Jun designed the research; M. Lee, Y.-J. Ko, Y. Moon, M. Han, K.J. Ko, H.-W. Kim, S.H. Lee, and Y. Jun performed the research; M. Lee, Y.-J. Ko, Y. Moon, K.J. Ko, S.H. Lee, and Y. Jun analyzed data; and M. Lee, Y.-J. Ko, K.J. Ko, and Y. Jun wrote the paper.
Footnotes
Abbreviation used in this paper:
- CD
- cytosolic domain
- DSP
- dithiobis-succinimidyl-propionate
- Gluc
- Gaussia princeps luciferase
- PCA
- protein fragment complementation assay
- ZIP
- GCN4 leucine zipper-coding sequence
References
- Albert S., Will E., and Gallwitz D.. 1999. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J. 18:5216–5225. 10.1093/emboj/18.19.5216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andag U., Neumann T., and Schmitt H.D.. 2001. The coatomer-interacting protein Dsl1p is required for Golgi-to-endoplasmic reticulum retrieval in yeast. J. Biol. Chem. 276:39150–39160. 10.1074/jbc.M105833200 [DOI] [PubMed] [Google Scholar]
- Anwar K., Klemm R.W., Condon A., Severin K.N., Zhang M., Ghirlando R., Hu J., Rapoport T.A., and Prinz W.A.. 2012. The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae. J. Cell Biol. 197:209–217. 10.1083/jcb.201111115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A., Desai A., and Oegema K.. 2007. A role for Rab5 in structuring the endoplasmic reticulum. J. Cell Biol. 178:43–56. 10.1083/jcb.200701139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D., Hicke L., Rexach M., Schleyer M., and Schekman R.. 1988. Reconstitution of SEC gene product-dependent intercompartmental protein transport. Cell. 54:335–344. 10.1016/0092-8674(88)90196-1 [DOI] [PubMed] [Google Scholar]
- Baker D., Wuestehube L., Schekman R., Botstein D., and Segev N.. 1990. GTP-binding Ypt1 protein and Ca2+ function independently in a cell-free protein transport reaction. Proc. Natl. Acad. Sci. USA. 87:355–359. 10.1073/pnas.87.1.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis V.A., Johnson L.M., and Emr S.D.. 1986. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc. Natl. Acad. Sci. USA. 83:9075–9079. 10.1073/pnas.83.23.9075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C. 1997. Coupled ER to Golgi transport reconstituted with purified cytosolic proteins. J. Cell Biol. 139:1097–1108. 10.1083/jcb.139.5.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beh C.T., Brizzio V., and Rose M.D.. 1997. KAR5 encodes a novel pheromone-inducible protein required for homotypic nuclear fusion. J. Cell Biol. 139:1063–1076. 10.1083/jcb.139.5.1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian X., Klemm R.W., Liu T.Y., Zhang M., Sun S., Sui X., Liu X., Rapoport T.A., and Hu J.. 2011. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc. Natl. Acad. Sci. USA. 108:3976–3981. 10.1073/pnas.1101643108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh E.G., and Dyer W.J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917. 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., and Boeke J.D.. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 14:115–132. [DOI] [PubMed] [Google Scholar]
- Burri L., Varlamov O., Doege C.A., Hofmann K., Beilharz T., Rothman J.E., Söllner T.H., and Lithgow T.. 2003. A SNARE required for retrograde transport to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 100:9873–9877. 10.1073/pnas.1734000100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes L.J., and Sondermann H.. 2011. Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc. Natl. Acad. Sci. USA. 108:2216–2221. 10.1073/pnas.1012792108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Araç D., Wang T.-M., Gilpin C.J., Zimmerberg J., and Rizo J.. 2006. SNARE-mediated lipid mixing depends on the physical state of the vesicles. Biophys. J. 90:2062–2074. 10.1529/biophysj.105.071415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Novick P., and Ferro-Novick S.. 2012. ER network formation requires a balance of the dynamin-like GTPase Sey1p and the Lunapark family member Lnp1p. Nat. Cell Biol. 14:707–716. 10.1038/ncb2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K.M., and Wickner W.T.. 2007. Trans-SNARE complex assembly and yeast vacuole membrane fusion. Proc. Natl. Acad. Sci. USA. 104:8755–8760. 10.1073/pnas.0702290104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean N., and Pelham H.R.. 1990. Recycling of proteins from the Golgi compartment to the ER in yeast. J. Cell Biol. 111:369–377. 10.1083/jcb.111.2.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Godoy L.M.F., Olsen J.V., Cox J., Nielsen M.L., Hubner N.C., Fröhlich F., Walther T.C., and Mann M.. 2008. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 455:1251–1254. 10.1038/nature07341 [DOI] [PubMed] [Google Scholar]
- Dennison S.M., Bowen M.E., Brunger A.T., and Lentz B.R.. 2006. Neuronal SNAREs do not trigger fusion between synthetic membranes but do promote PEG-mediated membrane fusion. Biophys. J. 90:1661–1675. 10.1529/biophysj.105.069617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbacher M., Thorsteinsdottir H., and Spang A.. 2011. The Dsl1 tethering complex actively participates in soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein receptor (SNARE) complex assembly at the endoplasmic reticulum in Saccharomyces cerevisiae. J. Biol. Chem. 286:25027–25038. 10.1074/jbc.M110.215657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilcher M., Veith B., Chidambaram S., Hartmann E., Schmitt H.D., and Fischer von Mollard G.. 2003. Use1p is a yeast SNARE protein required for retrograde traffic to the ER. EMBO J. 22:3664–3674. 10.1093/emboj/cdg339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L.L., Collins R.N., and Novick P.J.. 1998. Identification of a Sec4p GTPase-activating protein (GAP) as a novel member of a Rab GAP family. J. Biol. Chem. 273:3253–3256. 10.1074/jbc.273.6.3253 [DOI] [PubMed] [Google Scholar]
- English A.R., and Voeltz G.K.. 2012. Rab10 GTPase regulates ER dynamics and morphology. Nat. Cell Biol. 15:169–178. 10.1038/ncb2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman S., Lin L., Malczynski M., and Snyder M.. 1998. Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 140:461–483. 10.1083/jcb.140.3.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., and Voeltz G.K.. 2011. The ER in 3D: a multifunctional dynamic membrane network. Trends Cell Biol. 21:709–717. 10.1016/j.tcb.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett M.D., Zahner J.E., Cheney C.M., and Novick P.J.. 1994. GDI1 encodes a GDP dissociation inhibitor that plays an essential role in the yeast secretory pathway. EMBO J. 13:1718–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondopoulos A., Bastos R.N., Yoshimura S., Anderson R., Carpanini S., Aligianis I., Handley M.T., and Barr F.A.. 2014. Rab18 and a Rab18 GEF complex are required for normal ER structure. J. Cell Biol. 205:707–720. 10.1083/jcb.201403026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham A.K., Sinka R., Torres I.L., Lilley K.S., and Munro S.. 2014. Toward a comprehensive map of the effectors of rab GTPases. Dev. Cell. 31:358–373. 10.1016/j.devcel.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans B.L., Ortiz D., and Novick P.. 2006. Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. USA. 103:11821–11827. 10.1073/pnas.0601617103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A. 1995. A quantitative assay to measure homotypic vacuole fusion in vitro. Methods Cell Sci. 17:283–294. 10.1007/BF00986234 [DOI] [Google Scholar]
- Haas A., Conradt B., and Wickner W.. 1994. G-protein ligands inhibit in vitro reactions of vacuole inheritance. J. Cell Biol. 126:87–97. 10.1083/jcb.126.1.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A., Scheglmann D., Lazar T., Gallwitz D., and Wickner W.. 1995. The GTPase Ypt7p of Saccharomyces cerevisiae is required on both partner vacuoles for the homotypic fusion step of vacuole inheritance. EMBO J. 14:5258–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Shibata Y., Voss C., Shemesh T., Li Z., Coughlin M., Kozlov M.M., Rapoport T.A., and Prinz W.A.. 2008. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 319:1247–1250. 10.1126/science.1153634 [DOI] [PubMed] [Google Scholar]
- Hu J., Shibata Y., Zhu P.-P., Voss C., Rismanchi N., Prinz W.A., Rapoport T.A., and Blackstone C.. 2009. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 138:549–561. 10.1016/j.cell.2009.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M.. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Jones E.W. 2002. Vacuolar proteases and proteolytic artifacts in Saccharomyces cerevisiae. Methods Enzymol. 351:127–150. 10.1016/S0076-6879(02)51844-9 [DOI] [PubMed] [Google Scholar]
- Jun Y., and Wickner W.. 2007. Assays of vacuole fusion resolve the stages of docking, lipid mixing, and content mixing. Proc. Natl. Acad. Sci. USA. 104:13010–13015. 10.1073/pnas.0700970104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun Y., Thorngren N., Starai V.J., Fratti R.A., Collins K., and Wickner W.. 2006. Reversible, cooperative reactions of yeast vacuole docking. EMBO J. 25:5260–5269. 10.1038/sj.emboj.7601413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C.A., and Schekman R.. 1990. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 61:723–733. 10.1016/0092-8674(90)90483-U [DOI] [PubMed] [Google Scholar]
- Kamena F., Diefenbacher M., Kilchert C., Schwarz H., and Spang A.. 2008. Ypt1p is essential for retrograde Golgi-ER transport and for Golgi maintenance in S. cerevisiae. J. Cell Sci. 121:1293–1302. 10.1242/jcs.016998 [DOI] [PubMed] [Google Scholar]
- Ko Y.-J., Lee M., Kang K., Song W.K., and Jun Y.. 2014. In vitro assay using engineered yeast vacuoles for neuronal SNARE-mediated membrane fusion. Proc. Natl. Acad. Sci. USA. 111:7677–7682. 10.1073/pnas.1400036111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraynack B.A., Chan A., Rosenthal E., Essid M., Umansky B., Waters M.G., and Schmitt H.D.. 2005. Dsl1p, Tip20p, and the novel Dsl3(Sec39) protein are required for the stability of the Q/t-SNARE complex at the endoplasmic reticulum in yeast. Mol. Biol. Cell. 16:3963–3977. 10.1091/mbc.E05-01-0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara L.J., Beh C.T., Latterich M., Schekman R., and Rose M.D.. 1994. Nuclear congression and membrane fusion: two distinct events in the yeast karyogamy pathway. J. Cell Biol. 126:911–923. 10.1083/jcb.126.4.911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latterich M., and Schekman R.. 1994. The karyogamy gene KAR2 and novel proteins are required for ER-membrane fusion. Cell. 78:87–98. 10.1016/0092-8674(94)90575-4 [DOI] [PubMed] [Google Scholar]
- Lewis M.J., Rayner J.C., and Pelham H.R.. 1997. A novel SNARE complex implicated in vesicle fusion with the endoplasmic reticulum. EMBO J. 16:3017–3024. 10.1093/emboj/16.11.3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., and Barlowe C.. 2002. Analysis of Sec22p in endoplasmic reticulum/Golgi transport reveals cellular redundancy in SNARE protein function. Mol. Biol. Cell. 13:3314–3324. 10.1091/mbc.E02-04-0204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkus P., Graham L.A., Stevens T.H., and Schekman R.. 2004. Role of Vma21p in assembly and transport of the yeast vacuolar ATPase. Mol. Biol. Cell. 15:5075–5091. 10.1091/mbc.E04-06-0514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., Wickner W., and Haas A.. 1996. Sec18p (NSF)-driven release of Sec17p (α-SNAP) can precede docking and fusion of yeast vacuoles. Cell. 85:83–94. 10.1016/S0092-8674(00)81084-3 [DOI] [PubMed] [Google Scholar]
- McBride H.M., Rybin V., Murphy C., Giner A., Teasdale R., and Zerial M.. 1999. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 98:377–386. 10.1016/S0092-8674(00)81966-2 [DOI] [PubMed] [Google Scholar]
- Nichols B.J., Ungermann C., Pelham H.R., Wickner W.T., and Haas A.. 1997. Homotypic vacuolar fusion mediated by t- and v-SNAREs. Nature. 387:199–202. 10.1038/387199a0 [DOI] [PubMed] [Google Scholar]
- Orso G., Pendin D., Liu S., Tosetto J., Moss T.J., Faust J.E., Micaroni M., Egorova A., Martinuzzi A., McNew J.A., and Daga A.. 2009. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 460:978–983. 10.1038/nature08280 [DOI] [PubMed] [Google Scholar]
- Patel S.K., Indig F.E., Olivieri N., Levine N.D., and Latterich M.. 1998. Organelle membrane fusion: a novel function for the syntaxin homolog Ufe1p in ER membrane fusion. Cell. 92:611–620. 10.1016/S0092-8674(00)81129-0 [DOI] [PubMed] [Google Scholar]
- Remy I., and Michnick S.W.. 2006. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods. 3:977–979. 10.1038/nmeth979 [DOI] [PubMed] [Google Scholar]
- Ren Y., Yip C.K., Tripathi A., Huie D., Jeffrey P.D., Walz T., and Hughson F.M.. 2009. A structure-based mechanism for vesicle capture by the multisubunit tethering complex Dsl1. Cell. 139:1119–1129. 10.1016/j.cell.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaud J.-L., and Lévy D.. 2003. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 372:65–86. 10.1016/S0076-6879(03)72004-7 [DOI] [PubMed] [Google Scholar]
- Rismanchi N., Soderblom C., Stadler J., Zhu P.-P., and Blackstone C.. 2008. Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum. Mol. Genet. 17:1591–1604. 10.1093/hmg/ddn046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C.J., Nelson B., Marton M.J., Stoughton R., Meyer M.R., Bennett H.A., He Y.D., Dai H., Walker W.L., Hughes T.R., et al. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 287:873–880. 10.1126/science.287.5454.873 [DOI] [PubMed] [Google Scholar]
- Rogers J.V., Arlow T., Inkellis E.R., Koo T.S., and Rose M.D.. 2013. ER-associated SNAREs and Sey1p mediate nuclear fusion at two distinct steps during yeast mating. Mol. Biol. Cell. 24:3896–3908. 10.1091/mbc.E13-08-0441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J.V., McMahon C., Baryshnikova A., Hughson F.M., and Rose M.D.. 2014. ER-associated retrograde SNAREs and the Dsl1 complex mediate an alternative, Sey1p-independent homotypic ER fusion pathway. Mol. Biol. Cell. 25:3401–3412. 10.1091/mbc.E14-07-1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini S.G., Liu C., Zhang P., and Lee T.H.. 2014. Membrane tethering by the atlastin GTPase depends on GTP hydrolysis but not on forming the cross-over configuration. Mol. Biol. Cell. 25:3942–3953. 10.1091/mbc.E14-08-1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield P., Garrard S., and Derewenda Z.. 1999. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15:34–39. 10.1006/prep.1998.1003 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Voeltz G.K., and Rapoport T.A.. 2006. Rough sheets and smooth tubules. Cell. 126:435–439. 10.1016/j.cell.2006.07.019 [DOI] [PubMed] [Google Scholar]
- Shimoni Y., and Schekman R.. 2002. Vesicle budding from endoplasmic reticulum. Methods Enzymol. 351:258–278. 10.1016/S0076-6879(02)51852-8 [DOI] [PubMed] [Google Scholar]
- Siniossoglou S., and Pelham H.R.. 2001. An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes. EMBO J. 20:5991–5998. 10.1093/emboj/20.21.5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A., and Schekman R.. 1998. Reconstitution of retrograde transport from the Golgi to the ER in vitro. J. Cell Biol. 143:589–599. 10.1083/jcb.143.3.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai V.J., Jun Y., and Wickner W.. 2007. Excess vacuolar SNAREs drive lysis and Rab bypass fusion. Proc. Natl. Acad. Sci. USA. 104:13551–13558. 10.1073/pnas.0704741104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorngren N., Collins K.M., Fratti R.A., Wickner W., and Merz A.J.. 2004. A soluble SNARE drives rapid docking, bypassing ATP and Sec17/18p for vacuole fusion. EMBO J. 23:2765–2776. 10.1038/sj.emboj.7600286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M.D., Plutner H., and Balch W.E.. 1997. A Rab GTPase is required for homotypic assembly of the endoplasmic reticulum. J. Biol. Chem. 272:13479–13483. 10.1074/jbc.272.21.13479 [DOI] [PubMed] [Google Scholar]
- Voeltz G.K., Prinz W.A., Shibata Y., Rist J.M., and Rapoport T.A.. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 124:573–586. 10.1016/j.cell.2005.11.047 [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 285:901–906. 10.1126/science.285.5429.901 [DOI] [PubMed] [Google Scholar]
- Zhang M., and Hu J.. 2013. Homotypic fusion of endoplasmic reticulum membranes in plant cells. Front. Plant Sci. 4:514 10.3389/fpls.2013.00514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser E., Sperka-Gottlieb C.D., Fasch E.V., Kohlwein S.D., Paltauf F., and Daum G.. 1991. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 173:2026–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.