A coincidence detection mechanism regulates phagosomal sealing and couples it with phosphoinositide conversion from PtdIns(4,5)P2 enrichment on unsealed phagosomes to PtdIns3P enrichment on fully sealed phagosomes.
Abstract
Phagocytosis requires phosphoinositides (PIs) as both signaling molecules and localization cues. How PIs coordinate to control phagosomal sealing and the accompanying switch of organelle identity is unclear. In this study, we followed dynamic changes in PIs during apoptotic cell clearance in Caenorhabditis elegans. We found that phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2) and phosphatidylinositol-3-phosphate (PtdIns3P), which accumulate transiently on unsealed and fully sealed phagosomes, respectively, are both involved in phagosome closure. We identified PtdIns3P phosphatase MTM-1 as an effector of PtdIns(4,5)P2 to promote phagosomal sealing. MTM-1 coordinates with the class II PI3 kinase PIKI-1 to control PtdIns3P levels on unsealed phagosomes. The SNX9 family protein LST-4 is required for sealing, and its association with unsealed phagosomes is regulated by PtdIns(4,5)P2, PIKI-1, and MTM-1. Loss of LST-4 or its retention on phagosomes disrupts sealing and suppresses PtdIns3P accumulation, indicating close coupling of the two events. Our findings support a coincidence detection mechanism by which phagosomal sealing is regulated and coupled with conversion from PtdIns(4,5)P2 enrichment on unsealed phagosomes to PtdIns3P enrichment on fully sealed phagosomes.
Introduction
Phosphoinositides (PIs) are concentrated on different organelles to direct signaling and membrane trafficking events. For example, phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2) is enriched in the inner leaflet of plasma membranes and is involved in almost all cellular events occurring at the cell surface, whereas phosphatidylinositol-3-phosphate (PtdIns3P) is concentrated on early endosomes and is a key determinant of endosome maturation (Di Paolo and De Camilli, 2006; Bohdanowicz and Grinstein, 2013). The differential distribution of PIs is determined by PI kinases and phosphatases that catalyze local PI synthesis and turnover. PIs mediate signaling or trafficking events by affecting activities of integral membrane proteins or recruiting cytosolic effectors to specific membrane compartments (Di Paolo and De Camilli, 2006; Balla, 2013). PI–protein interactions mediated by electrostatic interaction or by protein modules are of low affinity, and stable protein–membrane association requires additional lipid, protein, or geometry determinants within the membrane (Carlton and Cullen, 2005; Di Paolo and De Camilli, 2006). For instance, EEA1 binds both PtdIns3P and phosphatidylserine through distinct sites in the FYVE domain, suggesting that it can be enriched on membranes containing both lipids (Kutateladze et al., 2004; Carlton and Cullen, 2005). EEA1 also binds RAB-5 and is recruited to early endosomes through both lipid and RAB-5 binding (Lawe et al., 2000). Moreover, SNX1 contains both a PtdIns3P-binding PX domain and a curvature-sensing BAR (Bin–Amphiphysin–Rvs) domain and localizes to high-curvature membrane tubules emanating from endosomes, indicating that membrane geometry can also be a localization cue (Carlton et al., 2004; Carlton and Cullen, 2005). Thus, multiple components of a given membrane serve as the coincidence detection code to define organelle identity and direct membrane trafficking by controlling the spatial and temporal recruitment and release of effectors. Nascent transport vesicles must change identity through interconversion of PIs before fusing with the next membrane. How PI conversion is controlled and coordinated with fusion and/or fission events remains poorly understood.
Eukaryotic cells engulf and eliminate large particles like microorganisms and apoptotic cells through phagocytosis, which involves many typical features of vesicle biogenesis and maturation (Flannagan et al., 2012). In Caenorhabditis elegans hermaphrodites, 131 somatic cells and about half of the germ cells die through apoptosis, and the resulting cell corpses are phagocytosed and cleared by neighboring cells (Pinto and Hengartner, 2012). Apoptotic cells are recognized and engulfed through evolutionarily conserved pathways, leading to cytoskeleton reorganization and formation of a membrane-bound vesicle, namely the phagosome (Wang et al., 2003; Pinto and Hengartner, 2012; Chen et al., 2013). Maturation of apoptotic cell–containing phagosomes, which in many ways parallels endosome progression and maturation of phagosomes enclosing foreign bodies, involves sequential interactions with early endosomes, late endosomes, and lysosomes to yield phagolysosomes where cell corpses are degraded (Kinchen and Ravichandran, 2008; Pinto and Hengartner, 2012). Rab GTPases (RAB-5, -7, and -14 and UNC-108/RAB-2) and their regulatory and effector proteins act sequentially to regulate phagosome maturation (Kinchen and Ravichandran, 2008; Pinto and Hengartner, 2012). PtdIns3P, a marker of mammalian early phagosomes (Vieira et al., 2001; Flannagan et al., 2012; Bohdanowicz and Grinstein, 2013), accumulates transiently on apoptotic cell–containing phagosomes at an early maturation stage controlled by the class II PI3 kinase PIKI-1 and the class III PI3 kinase VPS-34 (Li et al., 2009; Lu et al., 2012; Cheng et al., 2013). Loss of MTM-1, a myotubularin phosphatase that hydrolyzes PtdIns3P, results in accelerated internalization but impaired degradation of cell corpses (Zou et al., 2009; Neukomm et al., 2011; Lu et al., 2012). However, how cell corpse degradation is regulated by MTM-1 remains unclear.
PtdIns(4,5)P2 plays a key role in forming phagosomes that enclose invading microorganisms or opsonized particles (Flannagan et al., 2012; Bohdanowicz and Grinstein, 2013). PtdIns(4,5)P2 is present constitutively in the inner leaflet of plasma membranes, enriches transiently in advancing pseudopods, and decreases rapidly when phagosomes seal (Botelho et al., 2000; Scott et al., 2005). The dynamic change in PtdIns(4,5)P2 levels during phagocytosis is thought to contribute to the localized increase in actin polymerization required for pseudopod extension and the subsequent actin disassembly needed for completion of phagocytosis (Scott et al., 2005; Sarantis et al., 2012). PtdIns(4,5)P2 and PtdIns3P are key markers of unsealed and fully sealed phagosomes, respectively, but how their conversion is controlled to couple with phagosome sealing is unknown. In fact, very little is known about phagosome scission. In clathrin-mediated endocytosis, the large GTPase dynamin and its partner protein sorting nexin 9 (SNX9) are important for vesicle scission (Ramachandran, 2011). SNX9 contains an SH3 domain that interacts with dynamin, a PI-binding PX domain, and a BAR domain that modulates and stabilizes membrane shape (Lundmark and Carlsson, 2009). It contributes to vesicle scission by recruiting and stimulating the GTPase activity of dynamin, as well as coordinating actin polymerization (Lundmark and Carlsson, 2004, 2009; Soulet et al., 2005). In C. elegans, apoptotic cell removal requires both DYN-1, the C. elegans dynamin, and LST-4, an SNX9 family protein, which are thought to function early in phagosome maturation (Yu et al., 2006; Kinchen et al., 2008; Almendinger et al., 2011; Chen et al., 2013). LST-4 interacts with DYN-1 and promotes phagosomal association of the latter (Lu et al., 2010; Chen et al., 2013). However, it is unclear whether LST-4 and DYN-1 regulate early maturation events or are involved in the sealing process.
Here, we identify a coincidence detection mechanism that regulates phagosomal sealing through LST-4 and couples sealing with the switch of membrane identity from PtdIns(4,5)P2-enriched unsealed phagosomes to PtdIns3P-enriched fully sealed phagosomes.
Results
PtdIns(4,5)P2 and PtdIns3P accumulate sequentially on apoptotic cell–containing phagosomes
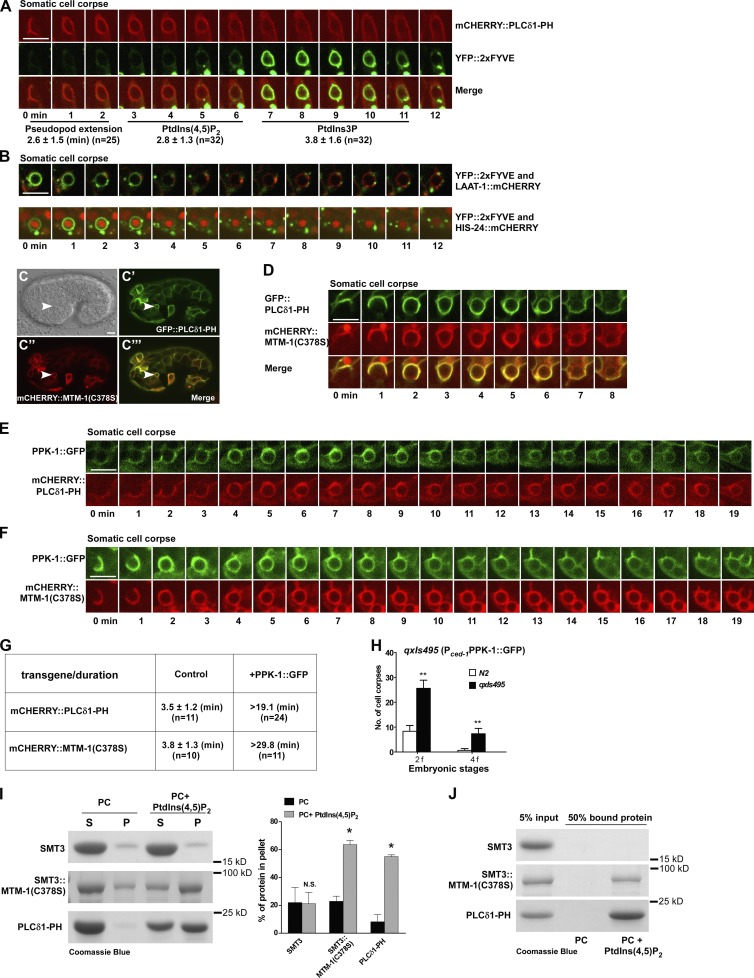
To examine how PIs change during apoptotic cell clearance in C. elegans, we expressed genetically coded PI sensors, including PLCδ1-PH (PtdIns(4,5)P2), 2xFYVE (PtdIns3P), TAPP1 (PtdIns(3,4)P2), BTK1-PH (PtdIns(3,4,5)P3), and AKT1-PH (PtdIns(3,4)P2 and PtdIns(3,4,5)P3), which do not affect apoptotic cell clearance (Fig. S1, A–Q; Dowler et al., 2000; Lemmon, 2008). In embryos, PLCδ1-PH and 2xFYVE mainly labeled plasma membranes and intracellular vesicles, respectively, which is consistent with enrichment of PtdIns(4,5)P2 and PtdIns3P on cell membranes and endosomes (Fig. S1, A–B’). We followed cell corpse clearance by four-dimensional microscopy analyses. PLCδ1-PH labeled extending pseudopods, which gradually surrounded cell corpses, a process that took ∼2.6 min (Fig. 1 A). PLCδ1-PH stayed on phagosomes for 2.8 min, and its release was followed immediately by enrichment of 2xFYVE that persisted for ∼3.8 min (Fig. 1 A and Video 1). No overlapping appearance of PLCδ1-PH and 2xFYVE was observed, suggesting that PtdIns(4,5)P2 and PtdIns3P accumulate transiently on apoptotic cell–containing phagosomes at distinct stages. Later, 2xFYVE-negative phagosomes fused with lysosomes, marked by the lysosomal membrane protein LAAT-1 (Liu et al., 2012), to form phagolysosomes where apoptotic cells are degraded (Fig. 1 B and Video 2). The sequential and nonoverlapping appearance of these reporters divides the phagosome formation and maturation process into different stages, including PLCδ1-PH/PtdIns(4,5)P2-positive, FYVE/PtdIns3P-positive, FYVE/PtdIns3P-negative, and phagolysosomal (LAAT-1 positive) stages. In gla-3 RNAi worms, which have increased germ cell apoptosis but normal cell corpse clearance (Kritikou et al., 2006), PLCδ1-PH and 2xFYVE were observed on phagosomes enclosing germ cell corpses (Fig. S1, F and G). In contrast, no obvious phagosomal association was seen in either embryos or germline when TAPP1, BTK1-PH, or AKT1-PH was expressed (Fig. S1, C–E’ and H–O). This suggests that PtdIns(3,4)P2 and PtdIns(3,4,5)P3 do not accumulate significantly on apoptotic cell–containing phagosomes. Consistent with this, AGE-1 and DAF-18, which produce and hydrolyze PtdIns(3,4,5)P3, respectively, or R01H10.7, an INPP4A homologue that dephosphorylates PtdIns(3,4)P2, are all dispensable for apoptotic cell clearance (Fig. S1, R–X).
Figure 1.
PtdIns(4,5)P2 regulates the phagosomal dynamics of MTM-1. (A) Time-lapse images of a cell corpse in a wild-type embryo expressing mCHERRY::PLCδ1-PH and YFP::2xFYVE. “0 min” represents the time point when PLCδ1-PH was first detected around the cell corpse. The time period until the cell corpse was surrounded by PLCδ1-PH was defined as the pseudopod extension stage. Quantification of the pseudopod extension time and phagosomal duration of PLCδ1-PH and 2xFYVE is shown beneath the images. (B) Time-lapse images of cell corpses in wild-type embryos coexpressing YFP::2xFYVE and LAAT-1::mCHERRY or HIS-24::mCHERRY. “0 min” represents the time point when the 2xFYVE ring appeared. (C–C’’’) DIC and fluorescent images of a wild-type embryo coexpressing GFP::PLCδ1-PH and mCHERRY::MTM-1(C378S). Arrowheads indicate a cell corpse surrounded by both PLCδ1-PH and MTM-1(C378S). (D) Time-lapse images of a cell corpse in a wild-type embryo coexpressing GFP::PLCδ1-PH and mCHERRY::MTM-1(C378S). “0 min” represents the time point when PLCδ1-PH and MTM-1(C378S) were first detected around the cell corpse. (E and F) Time-lapse images of cell corpses in wild-type embryos coexpressing PPK-1::GFP and mCHERRY::PLCδ1-PH (E) or mCHERRY::MTM-1(C378S) (F). “0 min” represents the time point when PPK-1 and PLCδ1-PH (E) or MTM-1(C378S) (F) were first seen around the cell corpse. (G) Quantification of the duration of mCHERRY::PLCδ1-PH and mCHERRY::MTM-1(C378S) on phagosomes with or without PPK-1 expression. (H) Cell corpses were quantified at twofold (2f) and fourfold (4f) embryonic stages in wild type without (N2) or with PPK-1 expression (qxIs495). At least 15 animals were scored in each strain. Data at each embryonic stage were compared with unpaired t tests. **, P < 0.0001. (I) MTM-1(C378S) bound to PC liposomes containing 4% PtdIns(4,5)P2 is more abundant in pellets (P) than in supernatants (S). Three independent experiments were performed. Unpaired t tests were performed. *, P < 0.05. (J) MTM-1(C378S) bound to PC liposomes containing 10% PtdIns(4,5)P2 is recovered in the top fraction in a liposome flotation assay. (A and G) “n” indicates the number of quantified phagosomes. (A, B, and D–F) Images in 20–25 z series were captured. Representative images are shown. Data are shown as mean ± SD. Bars, 5 µm.
PtdIns(4,5)P2 regulates the phagosomal dynamics of the PtdIns3P phosphatase MTM-1
The absence of 2xFYVE from PtdIns(4,5)P2-positive extending pseudopods and phagosomes indicates that PtdIns3P does not accumulate at this stage, which may be because of a lack of PtdIns3P production or PtdIns3P hydrolysis. To test this, we examined the dynamics of C. elegans myotubularin MTM-1, a plasma membrane–localizing PtdIns3P phosphatase involved in apoptotic cell clearance (Fig. S2, A–C; Zou et al., 2009; Neukomm et al., 2011). As overexpression of wild-type MTM-1 causes persistent cell corpses (Zou et al., 2009), we used MTM-1(C378S), a catalytically inactive MTM-1 that does not affect cell corpse removal (Fig. S2, D and E). MTM-1(C378S) colocalized very well with PLCδ1-PH on plasma membranes and apoptotic cell–containing phagosomes (Fig. 1, C–C’’’). It displayed identical dynamics to PLCδ1-PH on extending pseudopods and phagosomes (Fig. 1 D, Fig. S2 F, and Video 3). The simultaneous appearance and release of MTM-1 and PLCδ1-PH prompted us to investigate whether MTM-1 dynamics on phagosomes are regulated by PtdIns(4,5)P2 levels. We expressed PPK-1, the sole C. elegans PtdIns4P-5-kinase, and found that it displayed identical phagosomal dynamics to PLCδ1-PH (Fig. 1 E). PPK-1 expression significantly extended phagosomal duration of PLCδ1-PH, suggesting that PPK-1 produces PtdIns(4,5)P2 on forming phagosomes (Fig. 1, E and G). Similarly, MTM-1(C378S) appeared simultaneously with PPK-1 and persisted for significantly longer on phagosomes when PPK-1 was coexpressed (Fig. 1, F and G). Inactivation of OCRL-1, the inositol 5-phosphatase that hydrolyzes PtdIns(4,5)P2, caused prolonged MTM-1(C378S) duration on phagosomes like PPK-1 expression (see Fig. 4, F and G). These data suggest that MTM-1 dynamics on phagosomes are regulated by PtdIns(4,5)P2 levels. Overexpression of PPK-1 or inactivation of ocrl-1 affected apoptotic cell clearance, causing accumulation of cell corpses, indicating the importance of maintaining PtdIns(4,5)P2 and MTM-1 dynamics in this process (Fig. 1 H; Fig. 2, P–X; and Fig. S4 G). MTM-1(C378S) bound efficiently to phosphocholine (PC) liposomes containing PtdIns(4,5)P2 but not to other PIs or to PC-only liposomes in liposome sedimentation and/or flotation assays (Fig. 1, I and J; and Fig. S2 G). Thus, MTM-1 specifically binds to, and is regulated by, PtdIns(4,5)P2 to maintain a dynamic association with apoptotic cell–containing phagosomes.
Figure 4.
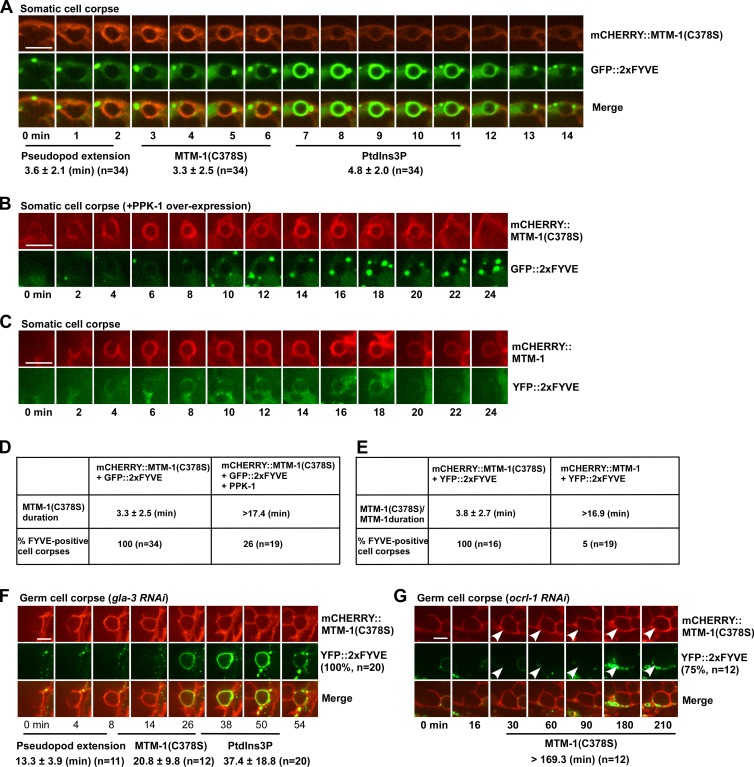
MTM-1 regulates PtdIns3P accumulation on phagosomes. (A) Time-lapse images of a cell corpse in a wild-type embryo expressing both mCHERRY::MTM-1(C378S) and GFP::2xFYVE. “0 min” represents the time point when MTM-1(C378S) was first detected around the cell corpse. The time period until the cell corpse was surrounded by MTM-1(C378S) was defined as the pseudopod extension stage. The pseudopod extension time and duration of MTM-1(C378S) and 2xFYVE were quantified. (B and C) Time-lapse images of cell corpses in wild-type embryos coexpressing mCHERRY::MTM-1(C378S), GFP::2xFYVE, and PPK-1 (B) or mCHERRY::MTM-1 and YFP::2xFYVE (C). (D and E) MTM-1(C378S) duration and 2xFYVE labeling with or without PPK-1 expression are quantified in D. MTM-1(C378S) and MTM-1 duration and 2xFYVE labeling are quantified in E. (F and G) Time-lapse images of germ cell corpses in gla-3RNAi (F) or ocrl-1RNAi (G) animals coexpressing mCHERRY::MTM-1(C378S) and YFP::2xFYVE. The pseudopod extension time and duration of MTM-1(C378S) and 2xFYVE are shown beneath the images. In ocrl-1RNAi animals, most phagosomes (75%, arrowheads) were not labeled by 2xFYVE during the time-lapse analysis, whereas all phagosomes followed in gla-3RNAi were positive for 2xFYVE. (A and D–G) “n” indicates the number of quantified phagosomes. (A–C, F, and G) Images in 20–25 z series were captured. Representative images are shown. Data are shown as mean ± SD. Bars, 5 µm.
Figure 2.
Apoptotic cell–containing phagosomes are arrested at the PtdIns(4,5)P2-positive stage in mtm-1 and ocrl-1 RNAi worms. DIC and fluorescent images of the gonads in gla-3RNAi, mtm-1RNAi, or ocrl-1RNAi animals expressing GFP::PLCδ1-PH (A–B’, Q, and Q’), GFP::RAB-5 (E–F’, S, and S’), GFP::RAB-7 (G–H’, T, and T’), NUC-1::nCHERRY (I–J’, U, and U’), LST-4::GFP (K–L’, V, and V’), and DYN-1::GFP (M–N’, W, and W’) or stained by LysoSensor green (C–D’, R, and R’). White arrowheads and boxes indicate germ cell corpses labeled by phagosomal markers, and yellow ones designate unlabeled corpses. Quantification is shown in O and X. (P) Germ cell corpses were quantified in controlRNAi and ocrl-1RNAi worms at 48 h after L4/adult molt. (O, P, and X) At least 15 animals were scored in each strain, and data are shown as mean ± SD. Recruitment of each phagosomal marker in mtm-1(RNAi) or ocrl-1(RNAi) was individually compared with that in the control group by unpaired t tests. **, P < 0.0001. Bars, 10 µm.
Phagosomes are arrested at the PtdIns(4,5)P2-positive stage in mtm-1(lf) mutants
We found that MTM-1 was enriched on PtdIns(4,5)P2-positive extending pseudopods and phagosomes and was released before the appearance of 2xFYVE, RAB-5, and RAB-7, indicating that MTM-1 acts very early during phagosome formation and/or maturation (Fig. 1 D and Fig. S2, H and I). We isolated a recessive mutation of mtm-1, qx322, which contained a C to T transition that resulted in an early stop codon after Lys99. qx322 worms contained significantly more germ cell corpses than the wild type (Fig. S2 J). This phenotype is similar to inactivation of mtm-1 by RNAi as previously reported (Neukomm et al., 2011) or gk890934, which contains two missense mutations in mtm-1 (Fig. S2 J). Expression of wild-type, but not catalytically inactive, MTM-1 significantly reduced germ cell corpses in qx322 and gk890934, indicating that phosphatase activity is important for MTM-1 function in apoptotic cell clearance (Fig. S2 K). Expression of human myotubularin MTM1, which localized to plasma membranes and phagosomal surfaces like worm MTM-1, rescued the cell corpse phenotype of mtm-1(qx322) (Fig. S2, L–N’), suggesting that human MTM1 can substitute for worm MTM-1 in removing germ cell corpses. The persistent cell corpses in mtm-1RNAi animals were all surrounded by Myri-GFP, a plasma membrane reporter, as in gla-3RNAi worms, suggesting that cell corpse recognition and initiation of engulfment are not disrupted (Fig. S2, O–Q). However, mtm-1(lf) phagosomes were not stained by LysoSensor green, indicating defects in phagosomal acidification (Fig. 2, C–D’ and O). Moreover, phagosomal association of LST-4, DYN-1, RAB-5, and RAB-7, which are recruited at early or late maturation stages, were all disrupted, and the lysosome reporter NUC-1 was also absent from phagosomes in mtm-1RNAi animals (Fig. 2, E–O; Guo et al., 2010). In contrast, phagosomal labeling by PLCδ1-PH was significantly increased in mtm-1(lf) worms (Fig. 2, A–B’ and O). These data indicate that apoptotic cell–containing phagosomes are arrested very early at the PtdIns(4,5)P2-positive stage in mtm-1(lf) worms. This is consistent with association of MTM-1 with PtdIns(4,5)P2-positive phagosomes and strongly supports the hypothesis that MTM-1 acts at the PtdIns(4,5)P2-positive stage to regulate phagocytosis of apoptotic cells.
Loss of mtm-1 affects phagosomal sealing
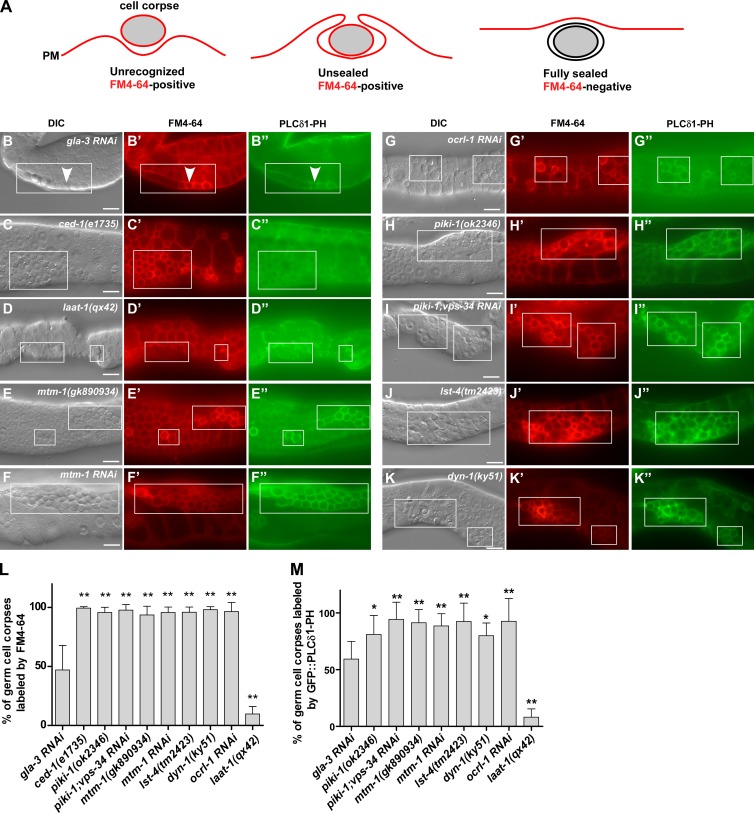
As PtdIns(4,5)P2 associates with unsealed but not sealed phagosomes in mammalian cells (Botelho et al., 2000; Scott et al., 2005; Sarantis et al., 2012), we investigated whether loss of mtm-1 affects phagosomal sealing, causing accumulation of PtdIns(4,5)P2-enriched unsealed phagosomes. We used FM4-64 staining, which was used previously to determine sealing of bacterium-containing phagosomes (Sarantis et al., 2012), and found that it stained the boundary but not the cytosol or nuclei of living germ cells, consistent with its inability to penetrate membranes (Fig. 3, A–B’). We examined two mutants, ced-1(lf), in which germ cell corpses are not recognized and engulfed (Zhou et al., 2001), and laat-1(lf), in which corpses are engulfed but not degraded in phagolysosomes (Liu et al., 2012). FM4-64 labeled corpses in ced-1(lf) but not laat-1(lf), indicating that fully internalized cell corpses are not accessible to FM4-64 (Fig. 3, C–D’ and L). About 50% of cell corpses were stained in gla-3RNAi worms, consistent with continued cell corpse clearance (Fig. 3, B, B’, and L). We found that FM4-64–positive, but not FM4-64–negative, cell corpses were stained by Annexin V, a phosphatidylserine-binding protein that selectively labels apoptotic cells, confirming that FM4-64 labels cell corpses that were not fully internalized (Fig. S3, A–C’’’, H, and I). FM4-64 staining correlated well with PLCδ1-PH labeling in gla-3RNAi and laat-1(qx42) worms, supporting the idea that PtdIns(4,5)P2 associates with unsealed but not fully separated phagosomes (Fig. 3, B–B’’, D–D’’, L, and M). The persistent cell corpses in ced-1(lf) were not labeled by PLCδ1-PH, confirming defects in cell corpse recognition (Fig. 3, C–C’’). Phagosomes in mtm-1(gk890934) and mtm-1RNAi worms were all labeled by FM4-64, Annexin V, and PLCδ1-PH, indicating defects in phagosomal sealing (Fig. 3, E–F’’, L, and M; and Fig. S3, D–D’’’, H, and I). Inactivation of OCRL-1 causes persistent cell corpses, which are labeled by FM4-64, Annexin V, and PLCδ1-PH, indicating that PtdIns(4,5)P2 hydrolysis is required for sealing of apoptotic cell–containing phagosomes (Fig. 2, P–Q’; Fig. 3, G–G’’, L, and M; and Fig. S3, E–E’’’, H, and I). Consistent with the sealing defect, ocrl-1(lf) phagosomes were not acidified, and the recruitment of maturation factors (RAB-5 and RAB-7) and lysosomes (NUC-1) was blocked (Fig. 2, R–U’ and X).
Figure 3.
MTM-1, OCRL-1, PIKI-1, and LST-4 are required for phagosomal sealing. (A) Cartoon illustrating the examination of phagosomal sealing by FM4-64 staining. An unrecognized cell corpse (left) or unsealed phagosome (middle) can be stained by the membrane-impermeable dye FM4-64 (red), whereas a fully separated phagosome (right) is not accessible to FM4-64. (B–K’’) DIC and fluorescent images of the gonads in the indicated strains expressing GFP::PLCδ1-PH and stained by FM4-64. Germ cell corpses are boxed, and arrowheads point to one cell corpse that is labeled by both FM4-64 and GFP::PLCδ1-PH. (L and M) The percentage of cell corpses labeled by FM4-64 (L) and GFP::PLCδ1-PH (M) was quantified in the indicated strains. At least 10 animals were scored in each strain. Data are shown as mean ± SD. The gla-3 RNAi dataset was compared with all the other datasets by a one-way ANOVA with Tukey’s post-test. PM, plasma membrane. *, P < 0.05; **, P < 0.0001. Bars, 10 µm.
MTM-1 and PIKI-1 coordinate to control PtdIns3P levels on PtdIns(4,5)P2-positive phagosomes
To investigate how MTM-1 regulates phagosomal sealing, we followed the phagosomal dynamics of MTM-1(C378S) and 2xFYVE simultaneously. MTM-1(C378S) and 2xFYVE appeared sequentially on phagosomes, with MTM-1 release coinciding with 2xFYVE appearance (Fig. 4, A and F; Video 4; and Video 5). The coincidence of MTM-1 release and 2xFYVE appearance suggests that MTM-1 may hydrolyze PtdIns3P to repress its accumulation on PtdIns(4,5)P2-positive phagosomes. To test this, we first determined whether increased MTM-1 on phagosomes affects 2xFYVE appearance. Overexpression of PPK-1 extended the PtdIns(4,5)P2-positive stage, causing prolonged phagosomal association of MTM-1(C378S) (Fig. 1, E–G). Similarly, RNAi inactivation of the inositol 5-phosphatase OCRL-1 led to sustained MTM-1(C378S) duration on phagosomes (Fig. 4 G). In both cases, the appearance of 2xFYVE on phagosomes was severely impaired, suggesting that sustained MTM-1 suppresses PtdIns3P accumulation (Fig. 4, B, D, and G). Consistent with this, expression of wild-type MTM-1 reduced phagosomal labeling by 2xFYVE (Fig. 4, C and E).
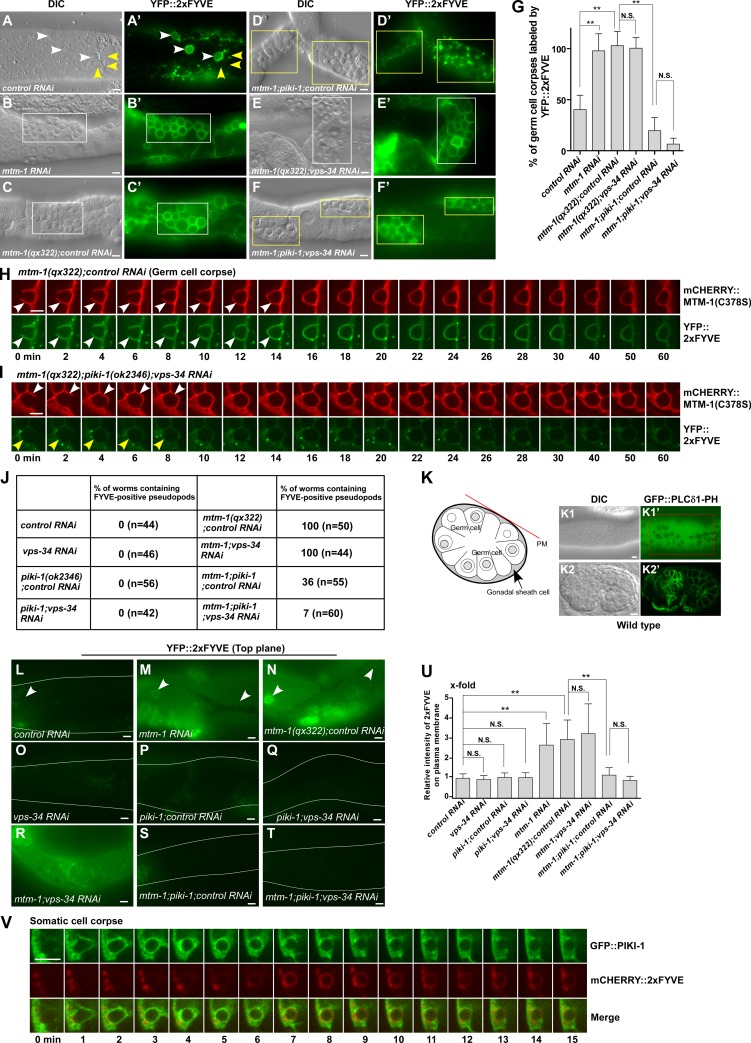
As sustained MTM-1 abrogates 2xFYVE appearance on phagosomes, we next examined whether loss of mtm-1 increases PtdIn3P accumulation. In mtm-1(qx322) or mtm-1RNAi worms, almost all germ cell corpses were labeled by YFP::2xFYVE, whereas <50% of them were YFP positive in wild type (Fig. 5, A–C’ and G). Moreover, extending pseudopods, which are positive for PLCδ1-PH and MTM-1(C378S) but not 2xFYVE in wild type, were labeled by YFP::2xFYVE in mtm-1(lf) (Fig. 5, H and J; Fig. S3, J–K’’; and Video 6). C. elegans germ cell corpses are removed by gonadal sheath cells, which form a single layer covering germline components (Fig. 5 K). In wild-type sheath cells, 2xFYVE formed small puncta in the cytosol but was absent from the cell surface (Fig. 5 L). In mtm-1(qx322), however, YFP::2xFYVE appeared on the surface of sheath cells, similar to PLCδ1-PH, which labels plasma membranes (Fig. 5, K–K2’, M, and N). This suggests that PtdIns3P accumulates on the cell surface in mtm-1(lf) sheath cells. Endosomal and phagosomal labeling by YFP::2xFYVE were also seen in mtm-1(qx322) when images were taken at the top focal plane because of the extremely thin cytosol of the sheath cell layer (Fig. 5, M and N, arrowheads). The cell surface and increased phagosomal labeling of 2xFYVE in mtm-1(qx322) can be suppressed by expressing wild-type but not catalytically inactive MTM-1, indicating that increased PtdIns3P accumulation is caused by loss of MTM-1 function (Fig. S3, L–P). Together, these data indicate that loss of MTM-1 leads to increased PtdIns3P accumulation, causing the appearance of 2xFYVE on cell surfaces, extending pseudopods, and phagosomes.
Figure 5.
MTM-1 and PIKI-1 coordinate to regulate PtdIns3P levels during phagosome formation. (A–G) DIC and fluorescent images of the gonads in the indicated strains expressing YFP::2xFYVE. White arrowheads and boxes indicate germ cell corpses labeled by YFP::2xFYVE, and yellow ones designate unlabeled corpses. Quantifications are shown in G. At least 15 animals were scored in each strain. (H and I) Time-lapse images of germ cell corpses in mtm-1(qx322);controlRNAi (H) or mtm-1(qx322);piki-1(ok2346);vps-34RNAi (I) animals coexpressing mCHERRY::MTM-1(C378S) and YFP::2xFYVE. White and yellow arrowheads point to labeled and nonlabeled extending pseudopods, respectively. (J) The percentage of worms containing 2xFYVE-positive pseudopods in the gonads was quantified in the indicated strains. At least 40 animals were examined in each strain. (K–K2’) A cross section of a pair of gonadal sheath cells is shown in K. The red line indicates the plasma membrane (PM) of the sheath cell, and the arrow points to the sheath cell cytosol separated by germ cell nuclei. DIC and fluorescent images of a gonad (K1 and K1’) and an embryo (K2 and K2’) expressing GFP::PLCδ1-PH. The red box in K1’ indicates the surface of a gonadal sheath cell, designated by the red line in the cross section (K). (L–T) Fluorescent images of gonadal sheath cells in the indicated strains expressing YFP::2xFYVE were captured at the top focal plane to show labeling on the cell surface. The curves indicate outlines of the gonads, and arrowheads point to 2xFYVE-positive endosomes or phagosomes beneath the cell surface. (U) Quantification of the relative intensity of 2xFYVE labeling on the cell surface in the strains shown in L–T. Three different regions were chosen in each animal, and ≥10 animals were quantified in each strain. (V) Time-lapse images of a cell corpse in a wild-type embryo coexpressing GFP::PIKI-1 and mCHERRY::2xFYVE. “0 min” represents the time point when PIKI-1 was first detected around the cell corpse. (G and U) Data are shown as mean ± SD. A one-way ANOVA with Tukey’s post-test was performed. (H, I, and V) Images in 20–25 z series were captured. Representative images are shown. **, P < 0.0001. Bars, 5 µm.
The class III and II PI3 kinases VPS-34 and PIKI-1 coordinate to regulate PtdIns3P generation and accumulation on phagosomes (Fig. S3, Q–U; and Table S1; Lu et al., 2012; Cheng et al., 2013). We found that loss of piki-1 but not vps-34 significantly suppressed 2xFYVE labeling on extending pseudopods and phagosomes in mtm-1(qx322) mutants, and 2xFYVE labeling on pseudopods was further reduced in mtm-1;piki-1;vps-34RNAi worms (Fig. 5, D–J). Moreover, piki-1 mutation reduced cell surface labeling by 2xFYVE in mtm-1(qx322) sheath cells (Fig. 5, S and U). Like MTM-1 and PLCδ1-PH, GFP::PIKI-1 appeared on extending pseudopods and nascent phagosomes, and its release coincided with enrichment of 2xFYVE (Fig. 5 V). These data indicate that PIKI-1 functions at the same stage as MTM-1 to control PtdIns3P levels on plasma membranes, extending pseudopods, and nascent phagosomes. RNAi of vps-34 further reduced FYVE labeling on mtm-1;piki-1 pseudopods (Fig. 5 J), suggesting that VPS-34 may contribute to PtdIns3P production at this stage.
As in mtm-1(lf), persistent phagosomes in piki-1 or piki-1;vps-34RNAi worms were labeled by both FM4-64 and PLCδ1-PH, indicating that phagosomal sealing is defective (Fig. 3, H–I’’, L, and M; and Fig. S3, F–F’’’, H, and I). Thus, MTM-1 and PIKI-1, which control PtdIns3P levels at the PtdIns(4,5)P2-positive stage, are both required for phagosomal sealing. Consistent with this, germ cell corpse numbers were similar in mtm-1(lf) and piki-1;vps-34RNAi and slightly increased in mtm-1;piki-1;vps-34RNAi triple mutants (Table S1).
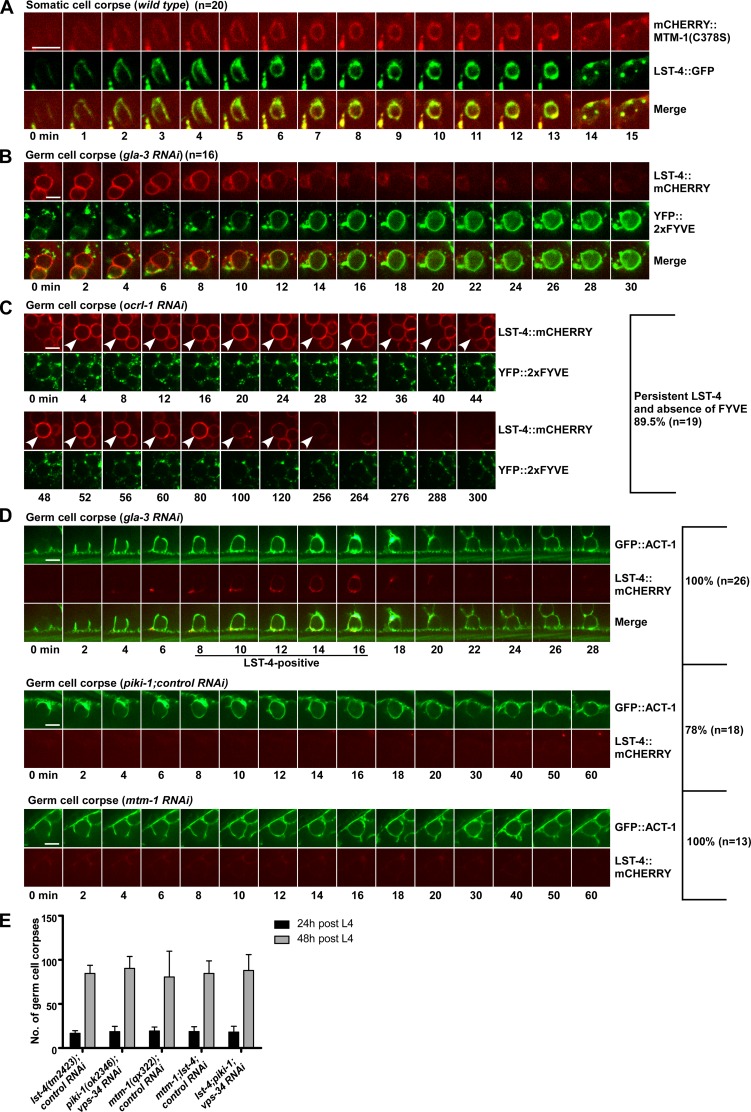
The SH3-PX-BAR protein LST-4 acts at the PtdIns(4,5)P2-positive stage to regulate phagosomal sealing
Scission of endocytic vesicles requires the large GTPase dynamin and its partner SNX9. In C. elegans, the SNX9 family protein LST-4 acts through DYN-1, the worm dynamin, to regulate the early stage of phagosome maturation (Almendinger et al., 2011; Chen et al., 2013). In lst-4 mutants, phagosomes are arrested at the early stage like in mtm-1(lf) (Chen et al., 2013). We found that GFP::LST-4 appeared on extending pseudopods and nascent phagosomes simultaneously with MTM-1(C378S), and its release coincided with, or preceded the enrichment of, 2xFYVE on phagosomes (Fig. 6, A and B; Fig. S4 A; and Video 7 and Video 8). The germ cell corpses in lst-4(lf) and dyn-1(lf) mutants were all labeled by FM4-64 and PLCδ1-PH, suggesting that phagosomal sealing is defective (Fig. 3, J–M; and Fig. S3, G–G’’’, H, and I). lst-4(lf) mutants accumulated similar numbers of germ cell corpses as in mtm-1(qx322) or piki-1(ok2356);vps-34RNAi worms, and this phenotype was unchanged in mtm-1;lst-4 double or lst-4;piki-1;vps-34RNAi triple mutants (Fig. 6 E and Table S1). These data indicate that LST-4 acts at the PtdIns(4,5)P2-positive stage in the same pathway with MTM-1 and PIKI-1 to regulate phagosomal sealing.
Figure 6.
LST-4 acts at the PtdIns(4,5)P2-positive stage, and its dynamics are regulated by PtdIns(4,5)P2, PIKI-1, and MTM-1. (A) Time-lapse images of a cell corpse in a wild-type embryo coexpressing mCHERRY::MTM-1(C378S) and LST-4::GFP. (B and C) Time-lapse images of germ cell corpses in a gla-3RNAi (B) or an ocrl-1RNAi (C) animal coexpressing LST-4::mCHERRY and YFP::2xFYVE. (D) Time-lapse images of germ cell corpses in the indicated strains coexpressing GFP::ACT-1 and LST-4::mCHERRY. GFP::ACT-1 indicates internalization of apoptotic cells. (C and D) The percentage of phagosomes with the representative pattern is shown at the right. “n” indicates the number of phagosomes that were followed and quantified. Arrowheads in C point to the LST-4::mCHERRY-positive phagosome in the ocrl-1RNAi animal. (A–D) Images in 20–25 z series were captured. Representative images are shown. (E) Germ cell corpses were quantified in the indicated strains at 24 and 48 h after L4/adult molt. At least 15 animals were scored in each strain at each stage. Data are shown as mean ± SD. A two-way ANOVA was performed. No significant differences were found (genotype variation, P = 0.7157). Bars, 5 µm.
Phagosomal association of LST-4 is regulated by PtdIns(4,5)P2, PIKI-1, and MTM-1
We next investigated whether LST-4 is regulated by PtdIns(4,5)P2, PtdIns3P, or both. Overexpression of PPK-1 or inactivation of OCRL-1, which causes increased PtdIns(4,5)P2 on phagosomes, led to extended LST-4 duration accompanied by defective 2xFYVE appearance (Fig. 2, V–X; Fig. 6 C; and Fig. S4, B–F). This suggests that the phagosomal dynamics of LST-4 are regulated by PtdIns(4,5)P2 levels. As previously reported (Lu et al., 2012), loss of piki-1, which affects PtdIns3P generation, abolished phagosomal association of LST-4, suggesting that PtdIns3P is important for recruiting LST-4 to phagosomes (Fig. 6 D). Surprisingly, loss of mtm-1, which led to increased PtdIns3P accumulation on plasma membranes and forming phagosomes, also abrogated LST-4 recruitment (Fig. 2, K–O; Fig. 6 D; and Fig. S4 H). These data suggest that phagosomal association of LST-4 may require precisely controlled PtdIns3P levels or that MTM-1 may recruit LST-4 through a PtdIns3P-independent mechanism, or that both mechanisms are involved.
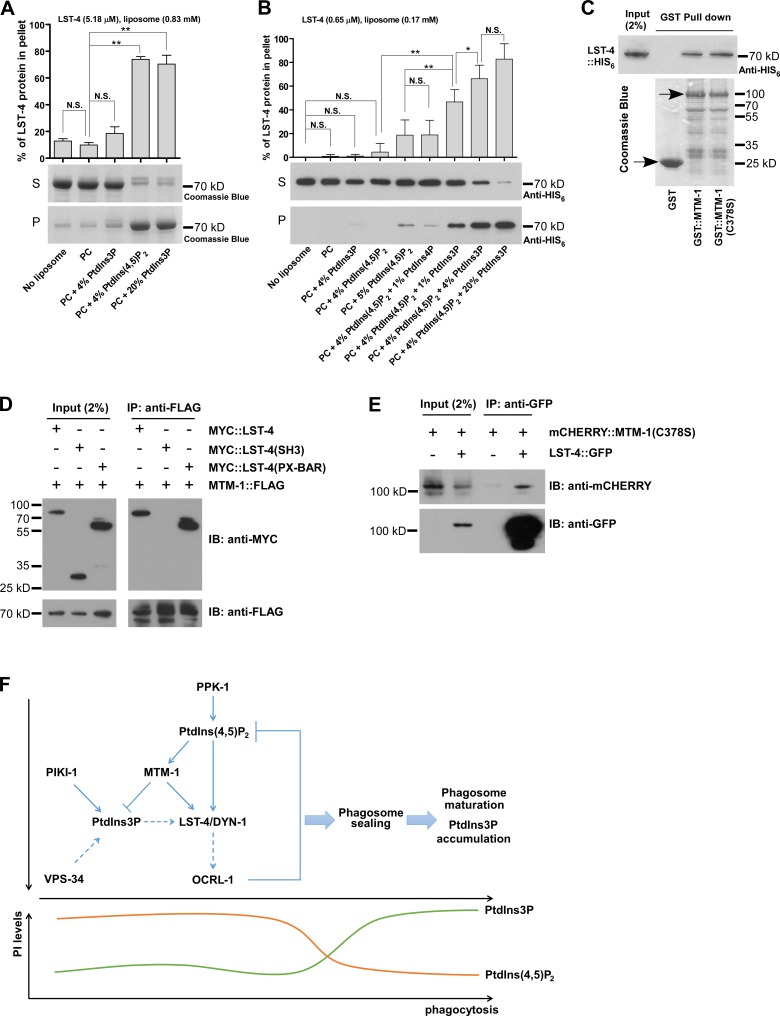
We attempted to test these hypotheses by examining the lipid-binding activity of LST-4 in vitro. LST-4 bound efficiently to PC liposomes containing 4% PtdIns(4,5)P2, but not to PC liposomes containing 4% PtdIns3P (Fig. 7 A). When PtdIns3P was incorporated into PC liposomes at 20%, LST-4 binding became obvious (Fig. 7 A). The PI-binding property of LST-4 is similar to SNX9 (Lundmark and Carlsson, 2003; Yarar et al., 2008) and is consistent with it associating with PtdIns(4,5)P2- but not PtdIn3P-enriched phagosomes in vivo. We performed liposome sedimentation assays using limited amounts of LST-4 protein and a lower concentration of liposomes, a condition under which only minimal binding of LST-4 to PtdIns(4,5)P2 liposomes was detected (Fig. 7 B). Addition of PtdIns3P but not PtdIns4P significantly increased binding of LST-4 to PtdIns(4,5)P2 liposomes, suggesting that PtdIns3P may enhance LST-4 binding to PtdIns(4,5)P2 (Fig. 7 B and Fig. S5 A). LST-4::GFP was diffuse in the cytoplasm of wild-type sheath cells but appeared on plasma membranes in mtm-1(lf) worms, which accumulate both PtdIns(4,5)P2 and PtdIns3P on their cell membranes (Fig. 5, K–N; and Fig. S5, B–D). The plasma membrane localization of LST-4 decreased in mtm-1(lf);piki-1;vps-34RNAi triple mutants, suggesting that PtdIns3P facilitates LST-4 binding to PtdIns(4,5)P2-containing membranes (Fig. S5 E). His-tagged LST-4 was successfully pulled down by GST–MTM-1 and GST–MTM-1(C378S) but not GST alone, suggesting that MTM-1 interacts with LST-4 and the interaction is independent of MTM-1 phosphatase activity (Fig. 7 C). The PX-BAR region, but not the SH3 domain of LST-4, was coprecipitated with FLAG-tagged MTM-1, indicating that MTM-1 interacts with LST-4 through the PX-BAR domain (Fig. 7 D). GFP::LST-4 coprecipitated with mCHERRY::MTM-1(C378S) in worms expressing both proteins, indicating that LST-4 associates with MTM-1 in C. elegans (Fig. 7 E).
Figure 7.
LST-4 interacts with PtdIns(4,5)P2, PtdIns3P, and MTM-1. (A and B) Binding of His-tagged LST-4 to the indicated liposomes was detected by Coomassie blue staining (A) or Western blotting (B). The lipid–protein complex is recovered in the pellet (P) but not the supernatant (S). The final concentration of LST-4 and liposomes used in each experiment is indicated. Three (A) and six (B) independent experiments were performed. Data (shown as mean ± SD) were compared by one-way ANOVA with Tukey’s post-test. *, P < 0.05; **, P < 0.0001. (C) His-tagged LST-4 is pulled down by GST::MTM-1::FLAG and GST::MTM-1(C378S)::FLAG but not GST::FLAG. Purified GST fusion proteins stained by Coomassie blue (arrows) are shown underneath. (D) Full-length LST-4 and the PX-BAR region, but not the SH3 domain, are coprecipitated from HEK293T cells by MTM-1. (E) mCHERRY::MTM-1(C378S) is coprecipitated by LST-4::GFP in lysates prepared from mix-staged worms coexpressing both fusion proteins. (F) Proposed model of coregulation of phagosomal sealing and PI conversion by the PtdIns(4,5)P2–PtdIns3P–MTM-1–LST-4 module in apoptotic cell clearance. Dashed arrows indicate regulations that need further investigation. IB, immunoblotting; IP, immunoprecipitation.
Discussion
Sealing of apoptotic cell–containing phagosomes involves both PtdIns(4,5)P2 and PtdIns3P
PtdIns(4,5)P2 and PtdIns3P are key determinants of phagosome formation and maturation, respectively (Bohdanowicz and Grinstein, 2013). Here, we found that both are involved in closure of apoptotic cell–containing phagosomes. PtdIns(4,5)P2 associates with extending pseudopods and unsealed but not sealed phagosomes. Inhibition of PtdIns(4,5)P2 hydrolysis by ocrl-1 RNAi or its increased generation by PPK-1 overexpression causes defective clearance of apoptotic cells and accumulation of unsealed phagosomes. Thus, like mammalian phagocytosis, sealing of apoptotic cell–containing phagosomes coincides with, and requires timely depletion of, PtdIns(4,5)P2. The inositol 5-phosphatase OCRL-1, which contributes to PtdIns(4,5)P2 elimination in mammalian phagocytosis (Bohdanowicz et al., 2012), is important for this process. Loss of ocrl-1 leads to prolonged association of MTM-1, LST-4, and DYN-1 with unsealed phagosomes, suggesting that defective sealing may be caused by sequestration of sealing factors by PtdIns(4,5)P2-enriched membranes. Our finding is consistent with a recent study that shows OCRL is required for dissociation of SNX9 from endocytic vesicles during uncoating (Nández et al., 2014).
Our data indicate that PtdIns3P is produced on plasma membranes and forming phagosomes where PtdIns(4,5)P2 is enriched, but its accumulation is suppressed by the PtdIns(4,5)P2 effector MTM-1. Loss of MTM-1 causes PtdIns3P accumulation on plasma membranes, extending pseudopods, and nascent phagosomes and numerous unsealed PtdIns(4,5)P2-positive phagosomes. Inactivation of the PI3 kinases PIKI-1 and VPS-34 completely suppresses PtdIns3P accumulation caused by mtm-1 mutation but also leads to defects in phagosomal sealing, indicating that PtdIns3P must be tightly controlled in this process. Consistent with this, the 2xFYVE reporter is only visible on plasma or unsealed phagosomal membranes in mtm-1(lf) but not wild-type worms. The timely depletion of PtdIns(4,5)P2 by OCRL-1 may release MTM-1, LST-4, and DYN-1, which completes the scission process and allows subsequent accumulation of PtdIns3P on fully sealed phagosomes (Fig. 7 F). In this way, phagosomal sealing and the switch of membrane identity from unsealed (PtdIns(4,5)P2 enriched) to sealed (PtdIns3P enriched) phagosomes are coregulated and precisely coupled. More work is needed to determine how OCRL-1 is regulated when apoptotic cell–containing phagosomes are being sealed.
The PtdIns(4,5)P2 effector MTM-1 coordinates with PIKI-1 to control PtdIns3P levels on unsealed phagosomes
Previously, we and others reported that MTM-1 localizes to plasma membranes and negatively regulates cell corpse engulfment through the CED-5/12/10 module (Zou et al., 2009; Neukomm et al., 2011). Here, we found that MTM-1 associates with forming phagosomes to promote sealing. MTM-1 binds specifically to PtdIns(4,5)P2, and its membrane association is regulated by the latter. Thus, MTM-1 acts as an effector of PtdIns(4,5)P2 to regulate both initiation and completion of engulfment. The acceleration in the initiation step but impairment in the sealing of phagosomes caused by loss of mtm-1 suggest that PtdIns3P turnover may have distinct effects on apoptotic cell clearance at different stages, a phenomenon that is also observed in autophagy regulation (Vergne et al., 2009; Taguchi-Atarashi et al., 2010; Cebollero et al., 2012; Wu et al., 2014).
Our data indicate that PIKI-1, the class II PI3 kinase, acts with MTM-1 to control a subpool of PtdIns3P on plasma membranes and unsealed phagosomes for cell corpse removal. The class III PI3 kinase VPS-34 may play a major role in producing PtdIns3P on sealed phagosomes, as both MTM-1 and PIKI-1 are released at this stage. Human class II PI3 kinase C2α is reported to control clathrin-mediated endocytosis by generating PtdIns(3,4)P2 (Posor et al., 2013). In our study, however, 2xFYVE but not TAPP1, a PtdIns(3,4)P2 reporter, is readily seen on phagosomes and abrogated by loss of PIKI-1. Loss or overexpression of R01H10.7, the C. elegans homologue of INPP4A, which converts PtdIns(3,4)P2 to PtdIns3P, has no effect on cell corpse removal. Moreover, TAPP1 was not observed on phagosomes in r01h10.7(lf) or r01h10.7(lf);mtm-1RNAi worms (Fig. S1, Y and Z). Thus, PtdIns(3,4)P2 is probably not involved in apoptotic cell clearance either directly or as a precursor of PtdIns3P.
The PI3 kinases PIKI-1 and VPS-34 were reported to regulate phagosome maturation by acting sequentially to control PtdIns3P oscillation on phagosomes, a process antagonized by MTM-1 (Lu et al., 2012). By examining >100 embryonic cell corpses, however, we found that PtdIns3P oscillation is not readily observed on cell corpses other than the three designated corpses (C1, C2, and C3) on which two waves of 2xFYVE were previously observed and characterized. Thus, PtdIns3P oscillation may associate with clearance of specific corpses like C1, C2, and C3. MTM-1(C378S) and 2xFYVE appear sequentially on the C3 phagosome, and MTM-1 release coincides with 2xFYVE enrichment (Fig. S5 H and Video 9), confirming that MTM-1 acts before PtdIns3P accumulation on phagosomes. MTM-1 remains absent when 2xFYVE disappears and reappears on the C3 phagosome, indicating that MTM-1 is probably not involved in regulating PtdIns3P oscillation on the C3 phagosome (Fig. S5 H and Video 9).
The copresence of PtdIns(4,5)P2, PtdIns3P, and MTM-1 recruits LST-4 to unsealed phagosomes
LST-4 was reported as a PtdIns3P effector that promotes phagosome maturation (Lu et al., 2010). However, we found that LST-4 associates with phagosomes at the PtdIns(4,5)P2-positive stage and is released before PtdIns3P accumulates. This is consistent with the lipid-binding property of LST-4 and two studies showing that LST-4 acts earlier than RAB-5 in cell corpse removal (Almendinger et al., 2011; Chen et al., 2013). We observed a large number of unsealed phagosomes in worms lacking LST-4 or DYN-1, indicating essential roles of these proteins in the sealing process. Thus, LST-4 may act with DYN-1 to control scission of apoptotic cell–containing phagosomes as in endocytosis.
Our data strongly support the hypothesis that PtdIns(4,5)P2, PtdIns3P, and MTM-1 serve as a coincidence detection code on unsealed phagosomes to recruit LST-4 for the sealing of apoptotic cell–containing phagosomes (Fig. 7 F). LST-4 binds efficiently to PtdIns(4,5)P2 liposomes in vitro and associates with PtdIns(4,5)P2- but not PtdIns3P-enriched phagosomes in vivo, indicating an essential role of PtdIns(4,5)P2 in membrane recruitment of LST-4. Loss of the PtdIns3P-producing kinase PIKI-1 or the LST-4–interacting PtdIns3P phosphatase MTM-1 disrupts phagosomal association of LST-4, causing accumulation of unsealed phagosomes. This indicates that PtdIns3P is important for LST-4 recruitment and its levels may be tightly controlled during sealing. Expression of MTM-1(C378S), which interacts with LST-4 but lacks catalytic activity, failed to rescue the defective phagosomal association of LST-4 in mtm-1(lf);piki-1(lf) double mutants (Fig. S5, F–G’’’), further supporting the role of PtdIns3P in LST-4 recruitment to phagosomes. Addition of PtdIns3P, which by itself displays limited affinity to LST-4, significantly enhances LST-4 binding to PtdIns(4,5)P2 liposomes in vitro, suggesting that PtdIns3P may facilitate or stabilize the binding of LST-4 with PtdIns(4,5)P2. Our results are consistent with a recent study that shows SNX9 is selectively recruited by coincidence of PtdIns(4,5)P2 and PtdIns3P on curved liposomal membranes (Gallop et al., 2013). Binding of SNX9 to liposomes is significantly increased by the combination of PtdIns(4,5)P2 and PtdIns3P compared with PtdIns(4,5)P2 alone and PtdIns3P alone (Gallop et al., 2013). We found that LST-4 interacts with MTM-1 through the lipid-binding PX-BAR domain but not the protein–protein-interacting SH3 domain, raising the possibility that binding of LST-4 to MTM-1 and lipids may be mutually regulated. However, more work is needed to uncover how PtdIns3P and/or MTM-1 may modulate LST-4 binding to PtdIns(4,5)P2 liposomes or phagosomes.
The PtdIns(4,5)P2–PtdIns3P–MTM-1–LST-4 regulatory module may be used in mammals
When expressed in worms, human MTM1 localizes to plasma membranes and efficiently rescues the persistent cell corpse phenotype of mtm-1(lf), suggesting evolutionarily conserved roles of myotubularin in this process. Mutations in MTM1, BIN1 (which encodes an N-BAR and SH3 domain–containing protein), and DNM2 lead to X-linked, autosomal recessive, and autosomal dominant centronuclear myopathies, respectively, and cause similar T tubule defects in skeletal muscle, suggesting that they may act in a common pathway important for muscle function (Cowling et al., 2012). MTM1 interacts with BIN1 in skeletal muscle and enhances BIN1-mediated membrane tubulation dependent on both binding and phosphatase activity (Royer et al., 2013). A PI-binding motif, which is present only in the muscle isoform of BIN1, facilitates PtdIns(4,5)P2 binding and promotes membrane tubulation (Lee et al., 2002). Thus, the PtdIns(4,5)P2–PtdIns3P–MTM-1–BAR regulatory module may play a conserved role to maintain muscle function and could be relevant to pathological alterations in centronuclear myopathies.
Materials and methods
C. elegans strains
Strains of C. elegans were cultured at 20°C using standard protocols (Brenner, 1974). The N2 Bristol strain was used as the wild-type strain. The following strains were used in this work: linkage group (LG) I, mtm-1(gk890934), mtm-1(qx322), and ced-1(e1735); LG II, laat-1(qx42), age-1(hx546), and age-1(mg44); LG III, R01H10.7(ok489); LG IV, daf-18(ok480) and lst-4(tm2423); LG X, piki-1(ok2346) and dyn-1(ky51). age-1(mg44) was maintained as mIn1/age-1, and the homozygous progenies were examined. dyn-1(ky51) was maintained at 20°C, and L4 larvae were moved to 25°C for 36 h before germ cell corpses were examined.
mtm-1(qx322) was isolated from a genetic screen by defective clearance of residual bodies. qx322 contains a C to T transition that results in an early stop codon after Lys99 and is probably a null mutation of mtm-1. mtm-1(gk890934), which was provided by the Caenorhabditis Genetics Center, contains the amino acid substitutions V188M and R229K. qx322 and gk890934 were out-crossed with the wild-type N2 strain six and four times, respectively, before further analysis.
The reporter strains used in this study are listed below. Transgenic animals carrying extrachromosomal arrays (qxEx) were generated using standard microinjection methods, and genome-integrated arrays (qxIs) were obtained by γ irradiation to achieve stable expression from arrays with low copy numbers (Evans, 2006).
The reporter strains used in this study include qxIs513(Pced-1mCHERRY::PLCδ1-PH), qxIs352(Pced-1LAAT-1::mCHERRY), qxIs476, qxIs518(Pced-1GFP::PLCδ1-PH), qxIs477, qxIs478(Pced-1mCHERRY::MTM-1(C378S)), qxIs495(Pced-1PPK-1::GFP), qxIs408(Pced-1GFP::RAB-5), qxIs66(Pced-1GFP::RAB-7), qxIs68(Pced-1nCHERRY::RAB-7), qxIs257(Pced-1NUC-1::nCHERRY), qxIs139(Pced-1DYN-1::GFP), qxIs482(Pced-1mCHERRY::MTM-1), qxIs405(Pced-1GFP::ACT-1), qxIs535(Pced-1TAPP1::GFP), qxIs539(Pced-1BTK1-PH::GFP), qxIs517(Pced-1AKT1-PH::nCHERRY), qxIs248(Pced-1GFP::MTM-1), qxIs390(Pced-1myriGFP), qxEx4539(Pced-1mCHERRY::MTM-1(C378S) + Pced-1GFP::PLCδ1-PH), qxEx4423(Pced-1mCHERRY::MTM-1(C378S) + Pced-1GFP::2xFYVE), qxEx5254(Pced-1PPK-1 + Pced-1mCHERRY::MTM-1(C378S) + Pced-1GFP::2xFYVE), qxEx5346(Pced-1GFP::PIKI-1 + Pced-1mCHERRY::2xFYVE), qxEx5344(Pced-1LST-4::GFP + Pced-1mCHERRY::2xFYVE), qxEx5341(Pced-1PPK-1 + Pced-1LST-4::GFP + Pced-1mCHERRY::2xFYVE), qxEx5825(Pced-1R01H10.7::GFP), qxEx5564(Pced-1DAF-18::GFP), qxEx5828(Pced-1AGE-1::GFP), and qxEx5649(Pced-1GFP::hMTM-1).
We obtained opIs334(Pced-1YFP::2xFYVE) from K.S. Ravichandran (University of Virginia, Charlottesville, VA) and M.O. Hengartner (University of Zurich, Zurich, Switzerland), stIs10226(Phis-72HIS-24::mCHERRY) from Z. Bao (Sloan Kettering Institute, New York, NY), yqIs114(Plst-4LST-4::GFP), yqIs111(Pced-1LST-4::GFP), yqIs119(Plst-4LST-4::mCHERRY), and yqIs96(PhspLST-4::GFP) from C. Yang (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China), and lst-4(tm2423) from S. Mitani (Tokyo Women's Medical University, Tokyo, Japan).
Quantification of cell corpses and phagosome maturation
The number of somatic cell corpses in the head region of living embryos and the number of germ cell corpses in one gonad arm at various adult ages (24 or 48 h after L4/adult molt) were scored as described previously (Gumienny et al., 1999; Wang et al., 2002). The cell corpses were identified by their raised-button–like morphology using Nomarski optics. At least 15 animals were scored at each stage in each strain. To study apoptotic cell clearance in C. elegans germlines, gla-3 RNAi was performed to increase germ cell apoptosis without affecting cell corpse removal (Kritikou et al., 2006), thus allowing examination of more phagosomes. To induce gene expression by heat-shock treatment, L4 larvae were picked to fresh nematode growth medium plates and treated at 33°C every 12 h. The number of germ cell corpses was scored at 48 h after L4/adult molt. To examine phagosome maturation, differential interference contrast (DIC) and fluorescent images were captured using a microscope (Axioimager A1; Carl Zeiss) as described previously (Lu et al., 2008). The percentage of germ cell corpses labeled by various phagosomal markers was quantified in adult animals (36 or 48 h after L4/adult molt) by dividing the total number of cell corpses by the number of labeled ones. Corpses were examined at multiple focal planes. At least 15 animals were scored in each strain.
Microscopy and imaging analysis
DIC and fluorescent images were captured with a microscope (Axioimager A1) equipped with epifluorescence (filter set 13 for GFP [excitation bandpass (BP) 470/20, beam splitter FT 495, emission BP 503–530] and filter set 20 for Cherry [excitation BP 546/12, beam splitter FT 560, emission BP 575–640]) and a monochrome digital camera (AxioCam; Carl Zeiss). Images were processed and viewed using Axiovision Rel. 4.7 software (Carl Zeiss). A 100× objective (Plan-Neofluar; NA 1.30) was used with Immersol 518F oil (Carl Zeiss). For confocal images, an inverted confocal microscope (LSM 5 Pascal; Carl Zeiss) with 488 (emission filter BP 503–530) and 543 (emission filter BP 560–615) lasers was used, and images were processed and viewed using LSM Image Browser software (Carl Zeiss). All images were taken at 20°C.
Spinning-disk time-lapse recording
Embryos at the precomma or comma stage, or adult worms at 24 or 36 h after L4/adult molt, were mounted on agar pads in M9 buffer without (embryos) or with 5-mM levamisole (adult worms; levamisole prevents animals from moving but does not affect the germline or the gonad). Fluorescent images were captured using a 100× objective (CFI Plan Apochromat Lambda; NA 1.45; Nikon) with immersion oil (type NF) on an inverted fluorescence microscope (Eclipse Ti-E; Nikon) with a spinning-disk confocal scanner unit (UltraView; PerkinElmer) with 488 (emission filter 525 [W50]) and 561 (dual-band emission filter 445 [W60] and 615 [W70]) lasers. Images in 20–25 z series (1.0 µm/section) were captured every 1 or 2 min for 1–2 h (embryos) or 2–6 h (adult worms) at 20°C and were viewed and analyzed using Volocity software (PerkinElmer). The numbers of phagosomes that were followed and quantified are indicated in the figures and figure legends. In general, at least 10 phagosomes from three independent animals were followed and quantified in each strain.
FM4-64, Annexin V, and LysoSensor staining
Adult worms (36 or 48 h after L4/adult molt) were dissected to expose their gonads in 200 µl gonad dissection buffer with or without 2 µl Alexa Fluor 488–conjugated Annexin V (Invitrogen; Wang et al., 2007). 155 µl gonad dissection buffer was removed, and 5 µl FM4-64 (200 µg/ml; Invitrogen) was added to make a final concentration of 20 µg/ml. Staining was performed at 4°C for 5 min before examination by fluorescence microscopy. At least 10 animals were scored in each strain. The fluorescence intensity of Annexin V conjugates varied on different apoptotic cells, which is likely the result of differences in phosphoserine levels on cell corpses that persist for different periods of time. LysoSensor staining was performed as described previously with some modifications (Guo et al., 2010). In brief, adult animals (48 h after L4/adult molt) were dissected in gonad dissection buffer with 1-µM LysoSensor green DND-189 (Invitrogen) and examined by fluorescence microscopy.
Visualization of plasma membrane labeling by 2xFYVE in gonadal sheath cells
Adult worms (48 h after L4/adult molt) were dissected in gonad dissection buffer to expose their gonads. The distal gonadal sheath cells near the gonad loop were imaged at the top focal plane using a microscope (Axioimager A1). Three circular regions (3 µm in radius) along the gonad arm were chosen in each image, and the mean intensity per unit area was measured with Axiovision Rel. 4.7 software. At least 10 worms were imaged under the same conditions and quantified in each strain.
Liposome sedimentation assay
1,2-dioleoyl-sn-glycero-3-PC alone or mixed with 1,2-dioleoyl-sn-glycero-3-phospho-l-serine, PtdIns3P, PtdIns4P, and/or PtdIns(4,5)P2 (Avanti Polar Lipids, Inc.) in the required molar ratio was prepared in chloroform and dried under nitrogen. Lipid films were hydrated in a buffer containing 50-mM Tris-HCl, pH 7.5, and 150-mM NaCl with a 1-mM total lipid concentration. Unilamellar vesicles (100 nm) were generated using a mini extruder (Avanti Polar Lipids, Inc.). For sedimentation assays, 2.5–30 µg proteins were incubated with 4–50 µl (1 mM) liposomes in a 60-µl reaction at room temperature for 10 min before sedimentation at 60,000 rpm for 40 min in a rotor (TLA-100; Beckman Coulter). Both supernatant and pellet were resolved with SDS-PAGE and revealed by Coomassie blue staining or Western blotting. At least three independent experiments were performed for all liposome sedimentation assays.
Liposome flotation assay
1,2-dioleoyl-sn-glycero-3-PC alone or mixed with other phospholipids (1,2-dioleoyl-sn-glycero-3-phospho-l-serine, PtdIns3P, PtdIns4P, PtdIns(3,4)P2, PtdIns(4,5)P2, or PtdIns(3,4,5)P3; Avanti Polar Lipids, Inc.) in a molar ratio of 9:1 was prepared in chloroform and dried under nitrogen. Lipid films were hydrated in TBS buffer containing 50-mM Tris-HCl, pH 7.5, and 150-mM NaCl with a 1-mM total lipid concentration. For flotation assays, 6–18 µg proteins were incubated with 90 µl (1 mM) liposomes in a 100-µl reaction at room temperature for 15 min. A 900-µl 50% (wt/vol) sucrose solution in TBS was added to the protein-liposome mixtures to a final concentration of 45% (wt/vol). A 2.2-ml 30% (wt/vol) sucrose solution in TBS was layered on the samples, followed by 1 ml TBS layered on top. The samples were then subjected to ultracentrifugation at 45,000 rpm for 5 h in a rotor (SW 60 Ti; Beckman Coulter). The top fraction containing floating liposomes (100 µl) was recovered, and half of it (50 µl) was resolved with SDS-PAGE and revealed by Coomassie blue staining. At least three independent experiments were performed for each assay, and representative results are shown.
RNAi
The bacteria-feeding protocol was used in RNAi experiments. In each experiment, 3–10 L4 larvae (P0) were cultured on the RNAi plate, and F1 progeny at the L4 stage were transferred to fresh RNAi plates and aged for 24, 36, or 48 h before examination of germ cell corpses.
The DNA sequences that are targeted by the double-stranded RNAs in the RNAi experiments are as follows: gla-3 (T02E1, 12,950–13,970 nt), mtm-1 (Y110A7A, 35,061–37,303 nt), ocrl-1 (C16C2, 2,650–4,215 nt), and vps-34 (B0025, 10,243–11,439 nt; Table S3).
Phosphatase assay
The phosphatase activity of MTM-1 was assayed using a Malachite green assay kit (Echelon Biosciences, Inc.) by measuring the liberated free phosphate after the phosphatase reaction. Experiments were performed according to the manufacturer’s instructions with some modifications. In brief, 500 ng His-tagged MTM-1 or MTM-1(C378S) was incubated with 1,000 pmol of substrate (DiC8 PtdIns3P, DiC8 PtdIns(3,5)P2, DiC8 PtdIns(4,5)P2, DiC8 PtdIns(3,4)P2, and DiC8 PtdIns(3,4,5)P3; Echelon Biosciences, Inc.) in a 25-µl reaction containing 50-mM Tris-HCl, pH 7.5, 150-mM NaCl, 2-mM CaCl2, 2-mM DTT, and 5% glycerol overnight at 20°C. The substrate-only controls were prepared in the same way to measure the background phosphate. Phosphate standards were prepared by diluting the phosphate standard solution provided in the kit according to the instructions. 100 µl Malachite green solution was added to each reaction, control, and phosphate standard for 20 min at room temperature, and absorbance was measured at 620 nm. Averages of triplicate samples were used to draw the phosphate standard curve and determine the free phosphate in each reaction and control.
GST pull-down assay
GST::FLAG, GST::MTM-1::FLAG, or GST::MTM-1(C378S)::FLAG immobilized on glutathione-agarose beads was incubated with BL21 bacterial lysates containing His-tagged LST-4 for 2 h at 4°C. The beads were washed extensively with PBS, and the bound proteins were resolved with SDS-PAGE and revealed by Western blotting with an antibody against the hexahistidine tag.
Coimmunoprecipitation assay in worms
Mix-staged worms expressing both LST-4::GFP and mCHERRY::MTM-1(C378S), or mCHERRY::MTM-1(C378S) alone, were lysed with a tissue grinder in worm lysis buffer containing 50-mM Tris-HCl, pH 7.5, 150-mM NaCl, 1% Triton X-100, and protease inhibitor cocktail (EDTA free; Roche). The homogenized solutions were kept on ice for 15 min and centrifuged at 14,000 rpm for 20 min at 4°C to remove the debris. 400 µl of precleared solution was incubated with 1 µl GFP antibody (polyclonal antibody raised in rabbits against recombinant GFP; C. Yang laboratory, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China) for 2 h at 4°C, followed by incubation with 20 µl BSA-blocked protein G–Sepharose (Invitrogen) for 2 h. After extensive washing with 50-mM Tris-HCl, pH 7.5, and 250-mM NaCl, the bound proteins were resolved with SDS-PAGE and revealed by Western blotting with an antibody against mCHERRY (mouse, HX1810; Huaxingbio, Inc.).
Coimmunoprecipitation assay in the HEK293T cell line
HEK293T cells were cotransfected with MTM-1::FLAG and MYC::LST-4, MYC::LST-4 (SH3), or MYC::LST-4 (PX-BAR). Cells were harvested 36 h after transfection and lysed in cell lysis buffer containing 50-mM Tris-HCl, pH 7.5, 150-mM NaCl, 1% Triton X-100, and protease inhibitor cocktail. The supernatant was collected and incubated with agarose beads conjugated with M2 anti-FLAG antibody (mouse, A2220; Sigma-Aldrich) at 4°C for 6 h with gentle rocking. Extensive washing with 50-mM Tris-HCl, pH 7.5, and 150-mM NaCl was performed before bound proteins were resolved with SDS-PAGE and revealed by Western blotting with anti-MYC antibody (rabbit, sc-789; Santa Cruz Biotechnology, Inc.).
Statistical analysis
The SD was used as y axis error bars for bar charts plotted from the mean value of the data. Data derived from different genetic backgrounds were compared by Student’s two-way unpaired t test, one-way analysis of variance (ANOVA) followed by Tukey’s post-test, or two-way ANOVA followed by Bonferroni post-test, as indicated in the figure legends. Data were considered statistically different at P < 0.05; *, P < 0.05; **, P < 0.0001 (0.001 in a two-way ANOVA analysis).
Plasmid construction
To generate Pced-1mCHERRY::PLCδ1-PH and Pced-1GFP::PLCδ1-PH, PLCδ1-PH was amplified from a human cDNA library using primers PXL248/PZHW42 and was cloned into pPD49.26-Pced-1mCHERRY1 and pPD49.26-Pced-1GFP1, respectively, through the KpnI site. To construct Pced-1PPK-1 and Pced-1PPK-1::GFP, the full-length cDNA of ppk-1 was amplified from a C. elegans cDNA library (Invitrogen) by PHBW40/PRM5 and PHBW40/PSYC401, respectively, and ligated into pPD49.26-Pced-1 and pPD49.26-Pced-1GFP3 through the KpnI site. Pced-1mCHERRY::MTM-1 and Pced-1mCHERRY::MTM-1(C378S) were constructed by ligating the mtm-1 genomic sequence or the C378S mutant form into pPD49.26-Pced-1mCHERRY1 at the KpnI site. To generate Pced-1TAPP1::GFP, TAPP1 was amplified from a human cDNA library using primers PSYC545/546 and cloned into pPD49.26-Pced-1GFP3 through the KpnI site. BTK1-PH was amplified from a plasmid provided by M.P. Wymann (University of Basel, Basel, Switzerland) using primers PSYC549/550 and ligated into pPD49.26-Pced-1GFP3 through the KpnI site. To generate Pced-1AKT1-PH::nCHERRY, AKT1-PH was amplified from a human cDNA library using primers PPFG370/PXL297 and cloned into pPD49.26-Pvha-6mCHERRY2. Pvha-6 was then replaced by Pced-1 through the BamHI site. The GFP::2xFYVE fragment digested from pPD49.78-GFP::2xFYVE (F. Muller laboratory, University of Fribourg, Fribourg, Switzerland) was ligated into pPD49.26 followed by the insertion of Pced-1 through the BamHI site to obtain Pced-1GFP::2xFYVE. To generate Pced-1mCHERRY::2xFYVE, 2xFYVE was amplified using primers PWZ294/296 and ligated into pPD49.26-Pced-1mCHERRY1 through the KpnI site. Pced-1GFP::PIKI-1 was constructed by ligating the piki-1 cDNA amplified using primers PBL217/218 into pPD49.26-Pced-1GFP1 through the KpnI site. To generate Pced-1R01H10.7::GFP and Pced-1AGE-1::GFP, R01H10.7 genomic DNA amplified using primers PSYC521/522 and age-1 cDNA amplified using primers PSYC501/502 were ligated into pPD49.26-GFP3 through XmaI–NheI sites, followed by the insertion of Pced-1 at the XmaI site. Pced-1DAF-18::GFP was constructed by ligating the daf-18 cDNA amplified using primers PXL353/354 into pPD49.26-Pced-1GFP3 through the KpnI site. To generate Pced-1GFP::hMTM1, hMTM1 cDNA was amplified from pPD49.78-hMTM1 as described previously (Zou et al., 2009) using primers PWZ386/385 and cloned into pPD49.26-Pced-1GFP1 through the KpnI site. To generate PhsphMTM1(C375S), the C375S mutation of hMTM1 was introduced into Pmtm-1hMTM1 by site-directed mutagenesis using primers PWZ445/446, and the C375S fragment was cloned into pPD49.78 and pPD49.83 through the KpnI site.
To generate pET21b-PLCδ1-PH::HIS6, PLCδ1-PH was amplified using primers PXLG1/2 and ligated into pET21b through BamHI–XhoI sites. To generate pET21b-LST-4::HIS6, the c isoform of lst-4 cDNA was amplified using primers PSYC450/452 and cloned into pET21b through NdeI–XhoI sites. To generate pET21b-MTM-1::HIS6 and pET41b-GST::MTM-1::HIS6, mtm-1 cDNA was amplified using primers PWDL108/PWZ706, digested with SpeI and HindIII, and cloned into pET21b and pET41b through NheI–HindIII and SpeI–HindIII sites, respectively. To generate pET21b-MTM-1(C378S)::HIS6, the C378S mutation was introduced into pET41b-GST::MTM-1::HIS6 using primers PQL148/149, and the fragment was ligated into pET21b through NheI–HindIII sites. To generate pET21b-SMT3::HIS6, SMT3 containing a thrombin cleavage site was amplified using primers PSYC609/610 and was cloned into pET21b through NheI–BamHI sites. pET21b-SMT3::MTM-1(C378S) was constructed by ligating the C378S form amplified using primers PSYC602/405 into pET21b-SMT3::HIS6 at HindIII–XhoI sites. To construct pET41b-GST::MTM-1::FLAG and pET41b-GST::MTM-1(C378S)::FLAG, the wild-type and C378S forms of MTM-1 with a Flag tag were amplified using primers PWDL108/PSYC443 and ligated into pET41b through SpeI–KpnI sites. To generate pET41b-GST::FLAG, GST::FLAG was amplified using primers PWK73/70 and cloned into pET41b through NdeI–NotI sites. To generate pCMV-MTM-1::FLAG, mtm-1 cDNA with a Flag tag was amplified using primers PWZ215/PSYC443 and ligated into pCMV-MYC(−) through the KpnI site. To generate pCMV-MYC::LST-4, pCMV-MYC::LST-4(SH3), and pCMV-MYC::LST-4(PX-BAR), the c isoform of full-length lst-4 cDNA, a 516-bp fragment that covers the SH3 domain (SH3, 1–516 bp), and a 1,332-bp fragment that covers the PX-BAR domain (PX-BAR, 367–1,698 bp) of lst-4 cDNA were amplified with PHWD377/375, PHWD377/PSYC645, and PSYC640/PHWD375, respectively, and ligated into pCMV-MYC through the KpnI site. Primers used for plasmid construction are listed in Table S2.
Online supplemental material
Fig. S1 shows that PtdIns(3,4)P2 and PtdIns(3,4,5)P3 do not accumulate significantly on apoptotic cell–-containing phagosomes. Fig. S2 shows that MTM-1 acts at the PtdIns(4,5)P2-positive stage to regulate apoptotic cell clearance. Fig. S3 shows that MTM-1 regulates PtdIns3P levels on plasma membranes, extending pseudopods, and phagosomes. Fig. S4 shows that phagosomal association of LST-4 is regulated by PtdIns(4,5)P2, MTM-1, and PIKI-1. Fig. S5 shows that PtdIns3P enhances LST-4 binding to PtdIns(4,5)P2-containing membranes. Video 1 shows dynamic changes of PtdIns(4,5)P2 and PtdIns3P on apoptotic cell–containing phagosomes. Video 2 shows that PtdIns3P depletion precedes phagolysosome formation. Video 3 shows that MTM-1 and PtdIns(4,5)P2 display identical dynamics on extending pseudopods and forming phagosomes. Videos 4 and 5 show that MTM-1 release coincides with PtdIns3P accumulation on phagosomes containing somatic (Video 4) or germ cell corpses (Video 5). Video 6 shows that loss of MTM-1 causes PtdIns3P accumulation on extending pseudopods and forming phagosomes. Video 7 shows that LST-4 and MTM-1 display identical phagosomal dynamics. Video 8 shows that LST-4 release coincides with PtdIns3P accumulation on phagosomes. Video 9 shows that MTM-1 release coincides with PtdIns3P accumulation on the C3 phagosome. Table S1 shows that LST-4 acts in the same pathway with MTM-1 and PIKI-1 to regulate apoptotic cell clearance. Table S2 lists primers that are used for plasmid construction. Table S3 lists double-stranded RNAs used for RNAi experiments. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201501038/DC1.
Supplementary Material
Acknowledgements
We thank Drs. K. Ravichandran, M.O. Hengartner, Z. Bao, C. Yang, S. Mitani, and M.P. Wymann for strains and plasmids; Dr. Isabel Hanson for editing services; and the imaging core facility at the National Institute of Biological Sciences (Beijing, China) for technical assistance. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research infrastructure programs (grant P40OD010440).
This work was supported by the National Science Foundation of China (grant 31325015), the National Basic Research Program of China (grant 2013CB910100), and an International Early Career Scientist grant from the Howard Hughes Medical Institute to X. Wang.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ANOVA
- analysis of variance
- BP
- bandpass
- DIC
- differential interference contrast
- PC
- phosphocholine
- PI
- phosphoinositide
- PtdIns3P
- phosphatidylinositol-3-phosphate
- PtdIns(4,5)P2
- phosphatidylinositol-4,5-bisphosphate
References
- Almendinger J., Doukoumetzidis K., Kinchen J.M., Kaech A., Ravichandran K.S., and Hengartner M.O.. 2011. A conserved role for SNX9-family members in the regulation of phagosome maturation during engulfment of apoptotic cells. PLoS ONE. 6:e18325 10.1371/journal.pone.0018325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. 2013. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93:1019–1137. 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohdanowicz M., and Grinstein S.. 2013. Role of phospholipids in endocytosis, phagocytosis, and macropinocytosis. Physiol. Rev. 93:69–106. 10.1152/physrev.00002.2012 [DOI] [PubMed] [Google Scholar]
- Bohdanowicz M., Balkin D.M., De Camilli P., and Grinstein S.. 2012. Recruitment of OCRL and Inpp5B to phagosomes by Rab5 and APPL1 depletes phosphoinositides and attenuates Akt signaling. Mol. Biol. Cell. 23:176–187. 10.1091/mbc.E11-06-0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho R.J., Teruel M., Dierckman R., Anderson R., Wells A., York J.D., Meyer T., and Grinstein S.. 2000. Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J. Cell Biol. 151:1353–1368. 10.1083/jcb.151.7.1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J.G., and Cullen P.J.. 2005. Coincidence detection in phosphoinositide signaling. Trends Cell Biol. 15:540–547. 10.1016/j.tcb.2005.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J., Bujny M., Peter B.J., Oorschot V.M., Rutherford A., Mellor H., Klumperman J., McMahon H.T., and Cullen P.J.. 2004. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high- curvature membranes and 3-phosphoinositides. Curr. Biol. 14:1791–1800. 10.1016/j.cub.2004.09.077 [DOI] [PubMed] [Google Scholar]
- Cebollero E., van der Vaart A., Zhao M., Rieter E., Klionsky D.J., Helms J.B., and Reggiori F.. 2012. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr. Biol. 22:1545–1553. 10.1016/j.cub.2012.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Jian Y., Liu X., Zhang Y., Liang J., Qi X., Du H., Zou W., Chen L., Chai Y., et al. 2013. Clathrin and AP2 are required for phagocytic receptor-mediated apoptotic cell clearance in Caenorhabditis elegans. PLoS Genet. 9:e1003517 10.1371/journal.pgen.1003517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S., Wu Y., Lu Q., Yan J., Zhang H., and Wang X.. 2013. Autophagy genes coordinate with the class II PI/PtdIns 3-kinase PIKI-1 to regulate apoptotic cell clearance in C. elegans. Autophagy. 9:2022–2032. 10.4161/auto.26323 [DOI] [PubMed] [Google Scholar]
- Cowling B.S., Toussaint A., Muller J., and Laporte J.. 2012. Defective membrane remodeling in neuromuscular diseases: insights from animal models. PLoS Genet. 8:e1002595 10.1371/journal.pgen.1002595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G., and De Camilli P.. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443:651–657. 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Dowler S., Currie R.A., Campbell D.G., Deak M., Kular G., Downes C.P., and Alessi D.R.. 2000. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 351:19–31. 10.1042/0264-6021:3510019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T.C., editor. 2006. Transformation and microinjection. The C. elegans Research Community, WormBook. http://dx.doi.org//10.1895/wormbook.1.108.1
- Flannagan R.S., Jaumouillé V., and Grinstein S.. 2012. The cell biology of phagocytosis. Annu. Rev. Pathol. 7:61–98. 10.1146/annurev-pathol-011811-132445 [DOI] [PubMed] [Google Scholar]
- Gallop J.L., Walrant A., Cantley L.C., and Kirschner M.W.. 2013. Phosphoinositides and membrane curvature switch the mode of actin polymerization via selective recruitment of toca-1 and Snx9. Proc. Natl. Acad. Sci. USA. 110:7193–7198. 10.1073/pnas.1305286110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny T.L., Lambie E., Hartwieg E., Horvitz H.R., and Hengartner M.O.. 1999. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 126:1011–1022. [DOI] [PubMed] [Google Scholar]
- Guo P., Hu T., Zhang J., Jiang S., and Wang X.. 2010. Sequential action of Caenorhabditis elegans Rab GTPases regulates phagolysosome formation during apoptotic cell degradation. Proc. Natl. Acad. Sci. USA. 107:18016–18021. 10.1073/pnas.1008946107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen J.M., and Ravichandran K.S.. 2008. Phagosome maturation: going through the acid test. Nat. Rev. Mol. Cell Biol. 9:781–795. 10.1038/nrm2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen J.M., Doukoumetzidis K., Almendinger J., Stergiou L., Tosello-Trampont A., Sifri C.D., Hengartner M.O., and Ravichandran K.S.. 2008. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat. Cell Biol. 10:556–566. 10.1038/ncb1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritikou E.A., Milstein S., Vidalain P.O., Lettre G., Bogan E., Doukoumetzidis K., Gray P., Chappell T.G., Vidal M., and Hengartner M.O.. 2006. C. elegans GLA-3 is a novel component of the MAP kinase MPK-1 signaling pathway required for germ cell survival. Genes Dev. 20:2279–2292. 10.1101/gad.384506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze T.G., Capelluto D.G., Ferguson C.G., Cheever M.L., Kutateladze A.G., Prestwich G.D., and Overduin M.. 2004. Multivalent mechanism of membrane insertion by the FYVE domain. J. Biol. Chem. 279:3050–3057. 10.1074/jbc.M309007200 [DOI] [PubMed] [Google Scholar]
- Lawe D.C., Patki V., Heller-Harrison R., Lambright D., and Corvera S.. 2000. The FYVE domain of early endosome antigen 1 is required for both phosphatidylinositol 3-phosphate and Rab5 binding. Critical role of this dual interaction for endosomal localization. J. Biol. Chem. 275:3699–3705. 10.1074/jbc.275.5.3699 [DOI] [PubMed] [Google Scholar]
- Lee E., Marcucci M., Daniell L., Pypaert M., Weisz O.A., Ochoa G.C., Farsad K., Wenk M.R., and De Camilli P.. 2002. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science. 297:1193–1196. 10.1126/science.1071362 [DOI] [PubMed] [Google Scholar]
- Lemmon M.A. 2008. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9:99–111. 10.1038/nrm2328 [DOI] [PubMed] [Google Scholar]
- Li W., Zou W., Zhao D., Yan J., Zhu Z., Lu J., and Wang X.. 2009. C. elegans Rab GTPase activating protein TBC-2 promotes cell corpse degradation by regulating the small GTPase RAB-5. Development. 136:2445–2455. 10.1242/dev.035949 [DOI] [PubMed] [Google Scholar]
- Liu B., Du H., Rutkowski R., Gartner A., and Wang X.. 2012. LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis. Science. 337:351–354. 10.1126/science.1220281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Zhang Y., Hu T., Guo P., Li W., and Wang X.. 2008. C. elegans Rab GTPase 2 is required for the degradation of apoptotic cells. Development. 135:1069–1080. 10.1242/dev.016063 [DOI] [PubMed] [Google Scholar]
- Lu N., Shen Q., Mahoney T.R., Liu X., and Zhou Z.. 2010. Three sorting nexins drive the degradation of apoptotic cells in response to PtdIns(3)P signaling. Mol. Biol. Cell. 22:354–374. 10.1091/mbc.E10-09-0756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N., Shen Q., Mahoney T.R., Neukomm L.J., Wang Y., and Zhou Z.. 2012. Two PI 3-kinases and one PI 3-phosphatase together establish the cyclic waves of phagosomal PtdIns(3)P critical for the degradation of apoptotic cells. PLoS Biol. 10:e1001245 10.1371/journal.pbio.1001245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundmark R., and Carlsson S.R.. 2003. Sorting nexin 9 participates in clathrin-mediated endocytosis through interactions with the core components. J. Biol. Chem. 278:46772–46781. 10.1074/jbc.M307334200 [DOI] [PubMed] [Google Scholar]
- Lundmark R., and Carlsson S.R.. 2004. Regulated membrane recruitment of dynamin-2 mediated by sorting nexin 9. J. Biol. Chem. 279:42694–42702. 10.1074/jbc.M407430200 [DOI] [PubMed] [Google Scholar]
- Lundmark R., and Carlsson S.R.. 2009. SNX9 – a prelude to vesicle release. J. Cell Sci. 122:5–11. 10.1242/jcs.037135 [DOI] [PubMed] [Google Scholar]
- Nández R., Balkin D.M., Messa M., Liang L., Paradise S., Czapla H., Hein M.Y., Duncan J.S., Mann M., and De Camilli P.. 2014. A role of OCRL in clathrin-coated pit dynamics and uncoating revealed by studies of Lowe syndrome cells. eLife. 3:e02975 10.7554/eLife.02975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukomm L.J., Nicot A.S., Kinchen J.M., Almendinger J., Pinto S.M., Zeng S., Doukoumetzidis K., Tronchère H., Payrastre B., Laporte J.F., and Hengartner M.O.. 2011. The phosphoinositide phosphatase MTM-1 regulates apoptotic cell corpse clearance through CED-5-CED-12 in C. elegans. Development. 138:2003–2014. 10.1242/dev.060012 [DOI] [PubMed] [Google Scholar]
- Pinto S.M., and Hengartner M.O.. 2012. Cleaning up the mess: cell corpse clearance in Caenorhabditis elegans. Curr. Opin. Cell Biol. 24:881–888. 10.1016/j.ceb.2012.11.002 [DOI] [PubMed] [Google Scholar]
- Posor Y., Eichhorn-Gruenig M., Puchkov D., Schöneberg J., Ullrich A., Lampe A., Müller R., Zarbakhsh S., Gulluni F., Hirsch E., et al. 2013. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. 499:233–237. 10.1038/nature12360 [DOI] [PubMed] [Google Scholar]
- Ramachandran R. 2011. Vesicle scission: dynamin. Semin. Cell Dev. Biol. 22:10–17. 10.1016/j.semcdb.2010.09.001 [DOI] [PubMed] [Google Scholar]
- Royer B., Hnia K., Gavriilidis C., Tronchère H., Tosch V., and Laporte J.. 2013. The myotubularin–amphiphysin 2 complex in membrane tubulation and centronuclear myopathies. EMBO Rep. 14:907–915. 10.1038/embor.2013.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarantis H., Balkin D.M., De Camilli P., Isberg R.R., Brumell J.H., and Grinstein S.. 2012. Yersinia entry into host cells requires Rab5-dependent dephosphorylation of PI(4,5)P₂ and membrane scission. Cell Host Microbe. 11:117–128. 10.1016/j.chom.2012.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C.C., Dobson W., Botelho R.J., Coady-Osberg N., Chavrier P., Knecht D.A., Heath C., Stahl P., and Grinstein S.. 2005. Phosphatidylinositol-4,5-bisphosphate hydrolysis directs actin remodeling during phagocytosis. J. Cell Biol. 169:139–149. 10.1083/jcb.200412162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulet F., Yarar D., Leonard M., and Schmid S.L.. 2005. SNX9 regulates dynamin assembly and is required for efficient clathrin-mediated endocytosis. Mol. Biol. Cell. 16:2058–2067. 10.1091/mbc.E04-11-1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi-Atarashi N., Hamasaki M., Matsunaga K., Omori H., Ktistakis N.T., Yoshimori T., and Noda T.. 2010. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic. 11:468–478. 10.1111/j.1600-0854.2010.01034.x [DOI] [PubMed] [Google Scholar]
- Vergne I., Roberts E., Elmaoued R.A., Tosch V., Delgado M.A., Proikas-Cezanne T., Laporte J., and Deretic V.. 2009. Control of autophagy initiation by phosphoinositide 3-phosphatase jumpy. EMBO J. 28:2244–2258. 10.1038/emboj.2009.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira O.V., Botelho R.J., Rameh L., Brachmann S.M., Matsuo T., Davidson H.W., Schreiber A., Backer J.M., Cantley L.C., and Grinstein S.. 2001. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J. Cell Biol. 155:19–26. 10.1083/jcb.200107069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yang C., Chai J., Shi Y., and Xue D.. 2002. Mechanisms of AIF-mediated apoptotic DNA degradation in Caenorhabditis elegans. Science. 298:1587–1592. 10.1126/science.1076194 [DOI] [PubMed] [Google Scholar]
- Wang X., Wu Y.C., Fadok V.A., Lee M.C., Gengyo-Ando K., Cheng L.C., Ledwich D., Hsu P.K., Chen J.Y., Chou B.K., et al. 2003. Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science. 302:1563–1566. 10.1126/science.1087641 [DOI] [PubMed] [Google Scholar]
- Wang X., Wang J., Gengyo-Ando K., Gu L., Sun C.L., Yang C., Shi Y., Kobayashi T., Shi Y., Mitani S., et al. 2007. C. elegans mitochondrial factor WAH-1 promotes phosphatidylserine externalization in apoptotic cells through phospholipid scramblase SCRM-1. Nat. Cell Biol. 9:541–549. 10.1038/ncb1574 [DOI] [PubMed] [Google Scholar]
- Wu Y., Cheng S., Zhao H., Zou W., Yoshina S., Mitani S., Zhang H., and Wang X.. 2014. PI3P phosphatase activity is required for autophagosome maturation and autolysosome formation. EMBO Rep. 15:973–981. 10.15252/embr.201438618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D., Surka M.C., Leonard M.C., and Schmid S.L.. 2008. SNX9 activities are regulated by multiple phosphoinositides through both PX and BAR domains. Traffic. 9:133–146. 10.1111/j.1600-0854.2007.00675.x [DOI] [PubMed] [Google Scholar]
- Yu X., Odera S., Chuang C.H., Lu N., and Zhou Z.. 2006. C. elegans dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev. Cell. 10:743–757. 10.1016/j.devcel.2006.04.007 [DOI] [PubMed] [Google Scholar]
- Zhou Z., Hartwieg E., and Horvitz H.R.. 2001. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 104:43–56. 10.1016/S0092-8674(01)00190-8 [DOI] [PubMed] [Google Scholar]
- Zou W., Lu Q., Zhao D., Li W., Mapes J., Xie Y., and Wang X.. 2009. Caenorhabditis elegans myotubularin MTM-1 negatively regulates the engulfment of apoptotic cells. PLoS Genet. 5:e1000679 10.1371/journal.pgen.1000679 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.