The transcription factor TFEB is activated in a Parkin- and Atg5-dependent manner during mitophagy, and MiT/TFE transcription factor family members are required for the efficient clearance of damaged mitochondria.
Abstract
The kinase PINK1 and ubiquitin ligase Parkin can regulate the selective elimination of damaged mitochondria through autophagy (mitophagy). Because of the demand on lysosomal function by mitophagy, we investigated a role for the transcription factor EB (TFEB), a master regulator of lysosomal biogenesis, in this process. We show that during mitophagy TFEB translocates to the nucleus and displays transcriptional activity in a PINK1- and Parkin-dependent manner. MITF and TFE3, homologues of TFEB belonging to the same microphthalmia/transcription factor E (MiT/TFE) family, are similarly regulated during mitophagy. Unlike TFEB translocation after starvation-induced mammalian target of rapamycin complex 1 inhibition, Parkin-mediated TFEB relocalization required Atg9A and Atg5 activity. However, constitutively active Rag guanosine triphosphatases prevented TFEB translocation during mitophagy, suggesting cross talk between these two MiT/TFE activation pathways. Analysis of clustered regularly interspaced short palindromic repeats–generated TFEB/MITF/TFE3/TFEC single, double, and triple knockout cell lines revealed that these proteins partly facilitate Parkin-mediated mitochondrial clearance. These results illuminate a pathway leading to MiT/TFE transcription factor activation, distinct from starvation-induced autophagy, which occurs during mitophagy.
Introduction
Loss-of-function mutations in the mitochondrial kinase PINK1 and cytosolic E3 ligase Parkin cause early onset familial Parkinson’s disease (PD; Kitada et al., 1998; Lücking et al., 2000; Valente et al., 2004). Studies in Drosophila melanogaster and mammals suggest that PINK1 functions upstream of Parkin (Clark et al., 2006; Park et al., 2006; Yang et al., 2006) in a quality control pathway that selectively eliminates damaged mitochondria via autophagy (Narendra et al., 2008). PINK1 is maintained at low levels through rapid cleavage by mitochondrial proteases and proteasomal degradation by the N-end rule pathway (Jin et al., 2010; Meissner et al., 2011; Yamano and Youle, 2013). Upon loss of membrane potential, full-length PINK1 accumulates on the outer mitochondrial membrane and recruits Parkin (Matsuda et al., 2010; Narendra et al., 2010; Vives-Bauza et al., 2010), initiating ubiquitination and proteasomal degradation of several outer membrane proteins and autophagosome-dependent lysosomal degradation of the damaged organelle (Narendra et al., 2008; Matsuda et al., 2010; Tanaka et al., 2010; Yoshii et al., 2011; Chan et al., 2011; Sarraf et al., 2013).
Our understanding of the downstream steps of Parkin-mediated mitophagy is limited. Widespread ubiquitination of outer membrane proteins by Parkin initiates assembly of the autophagosomal isolation membrane by recruiting the unc-51–like autophagy activating kinase 1/2 complex (ULK1/2–Atg13–FIP200–Atg101), transmembrane autophagy protein Atg9A-containing structures, and the class III phosphatidylinositol 3-kinase complex (Beclin-1–Atg14(L)–Vps15–Vps34) proximal to mitochondria (Itakura et al., 2012). Two ubiquitin ligase-type reactions that conjugate Atg12 to Atg5 and microtubule-associated protein light chain 3 (LC3) to the lipid phosphatidylethanolamine are further needed to elongate isolation membranes and complete the autophagosome (Klionsky and Schulman, 2014). Subsequently, the autophagosome fuses with the lysosome to degrade and recycle the damaged material.
Mitophagy requires lysosomal function; yet the role of lysosomal biogenesis in mitophagy has not been addressed. Coordinated expression of lysosomal and autophagic genes has recently been shown to be regulated by transcription factor EB (TFEB; Sardiello et al., 2009; Palmieri et al., 2011; Settembre et al., 2011). TFEB is a member of the microphthalmia/transcription factor E (MiT/TFE) subfamily of basic helix-loop-helix leucine zipper transcription factors that bind to the E-box core sequence (CANNTG; Hemesath et al., 1994; Aksan and Goding, 1998). TFEB binds specifically to a 10-bp motif (GTCACGTGAC) found in the promoter regions of many genes encoding lysosomal and autophagic proteins, including several involved in lysosomal acidification and degradation of cytoplasmic substrates, autophagosome formation, and autophagosome–lysosome fusion (Sardiello et al., 2009; Settembre et al., 2011; Palmieri et al., 2011). Activation of TFEB thereby positively regulates both autophagosomes and lysosomes, enabling a coordinated and efficient response to increased degradative needs.
Mammalian target of rapamycin complex 1 (mTORC1) is a major regulator of TFEB transcriptional activity (Martina et al., 2012; Settembre et al., 2012; Roczniak-Ferguson et al., 2012). Under nutrient-rich conditions, TFEB undergoes cycles of transient association with lysosomes where it binds the heterodimeric Rag GTPases and interacts with active mTORC1 (Martina and Puertollano, 2013). mTORC1 phosphorylates TFEB on several residues, including serine 211 (S211), creating a binding site for the cytosolic chaperone 14-3-3 that sequesters TFEB inactive in the cytosol (Martina et al., 2012; Roczniak-Ferguson et al., 2012; Settembre et al., 2012). Conversely, the Rag GTPases assume an inactive conformation upon starvation, causing mTORC1 inactivation and the dissociation of both mTORC1 and TFEB from lysosomes (Sancak et al., 2008; Martina and Puertollano, 2013). This prevents TFEB phosphorylation by mTORC1 and its interaction with 14-3-3, permitting TFEB translocation to the nucleus where it induces transcription of target genes.
The dynamic regulation of lysosomal biogenesis during starvation-induced autophagy raises the question of whether lysosomal biogenesis is regulated during other types of cellular stress that lead to autophagy. Here, we show that TFEB is activated in a Parkin- and Atg5-dependent manner during mitophagy and that MiT/TFE transcription factor family members are required for the efficient clearance of damaged mitochondria. Our work also provides new insights into a previously uncharacterized mechanism of MiT/TFE transcription factor regulation downstream of Atg5.
Results
TFEB is activated during mitophagy in a PINK1- and Parkin-dependent manner
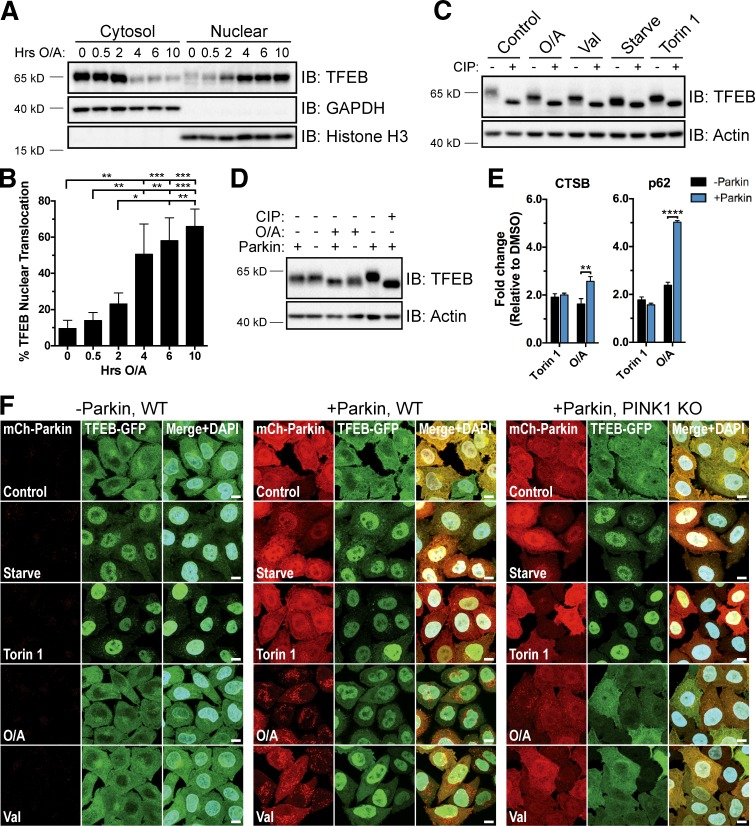
To investigate whether TFEB is activated during mitophagy, we treated HeLa cells stably expressing YFP-Parkin for up to 10 h with both the ATP synthase inhibitor oligomycin and the complex III inhibitor antimycin A (oligomycin/antimycin A [O/A]; Fig. 1, A and B) or valinomycin (Fig. S1, A and B) to induce mitophagy. Separation of cytosolic and nuclear components by subcellular fractionation revealed a dramatic change in the relative abundance of endogenous TFEB in these compartments over time. This progressive relocalization of TFEB resulted in more than 65% of total TFEB in the nuclear fraction by 10 h of O/A treatment and 80% upon treatment with valinomycin. TFEB localization was unaffected by the addition of a pan-caspase inhibitor (QVD; Fig. S1 D) that was included with O/A treatments to prolong cell viability. Furthermore, O/A treatment caused translocation of both endogenous TFEB and TFEB-GFP in BE(2)-M17 cells (Fig. S1 C), a neuroblastoma cell line containing endogenous Parkin (Hasson et al., 2015).
Figure 1.
TFEB translocates to the nucleus during mitophagy in a Parkin- and PINK1-dependent manner. (A) YFP-Parkin HeLa cells were treated with O/A for up to 10 h, fractionated, and immunoblotted. (B) Quantification of data in A. Endogenous TFEB expression was normalized to GAPDH (cytosol) or histone H3 (nuclear) and nuclear TFEB expressed as a percentage of total TFEB. Data are means ± SD (n = 3). (C) mCherry-Parkin HeLa cells were left untreated (Control), starved (2 h), or treated with torin 1 (2 h), O/A (6 h), or valinomycin (Val; 6 h). CIP treatment of cell lysates was performed before immunoblotting. (D) WT and mCherry-Parkin HeLa cells were treated with DMSO or O/A (6 h), lysed, and immunoblotted. A CIP-treated control was included as a reference for total TFEB dephosphorylation. (E) WT and mCherry-Parkin HeLa cells were treated with DMSO, torin 1, or O/A for 18 h and analyzed by quantitative PCR for TFEB target gene expression. Data are means ± SD (n = 3). (F) WT and PINK1 KO HeLa cells stably expressing TFEB-GFP with or without mCherry-Parkin were treated as in C. Fixed cells were stained with DAPI and analyzed by immunofluorescence. Bars, 10 µm. See Fig. S1 F for quantification. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Consistent with previous studies of starvation-induced TFEB activation (Martina et al., 2012; Roczniak-Ferguson et al., 2012; Settembre et al., 2012), we observed a faster migrating endogenous TFEB band in cells deprived of amino acids or treated with the mTOR catalytic inhibitor torin 1 compared with untreated cells (Fig. 1 C). Additional treatment with calf intestinal phosphatase (CIP) further reduced the apparent size of the TFEB band in all samples to the same molecular mass (MM). That the TFEB band in starved and torin 1–treated samples further decreased in size upon CIP treatment indicates that TFEB remains partially phosphorylated under these conditions. This intermediate phosphorylated form of TFEB also appeared in O/A- and valinomycin-treated samples (Fig. 1 C), consistent with the redistribution of TFEB to the nucleus (Fig. 1, A and B; and Fig. S1, A and B) and stimulation of TFEB transcriptional activity toward known lysosome and autophagy target genes cathepsin B and p62 (Fig. 1 E; Sardiello et al., 2009; Settembre et al., 2011). The migration pattern of TFEB was unchanged by a combined O/A + torin 1 treatment or QVD (Fig. S1 E).
To investigate whether TFEB nuclear translocation upon O/A or valinomycin treatment requires PINK1 and Parkin, we analyzed TFEB localization in wild-type (WT) HeLa cells, which express little to no endogenous Parkin (Denison et al., 2003; Pawlyk et al., 2003), as well as in WT and PINK1 knockout (KO) HeLa cells stably expressing fluorescently tagged Parkin. PINK1 KO cells were generated by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 gene editing (Table S1) and inhibition of Parkin mitochondrial translocation and mitophagy flux was confirmed by microscopy (Fig. 1 F) and FACS (Fig. S5 G), respectively. Upon starvation or torin 1 treatment, all three cell lines exhibited a similar degree of TFEB nuclear accumulation assessed by cell fractionation (Fig. S1, G and H) and confocal imaging (Figs. 1 F and S1 F). However, valinomycin and O/A induced TFEB nuclear translocation only in cells expressing both ectopic Parkin and endogenous PINK1 (Fig. 1 F and Fig. S1, F–H). Immunostaining of endogenous TFEB confirmed these findings (Fig. S3, A and B) and higher concentrations of O/A were not able to promote TFEB nuclear translocation independently of Parkin (Fig. S1, I and J). A greater increase in cathepsin B and p62 mRNA levels also occurred in +Parkin cells relative to –Parkin cells upon O/A, but not torin 1, treatment (Fig. 1 E). Furthermore, O/A treatment of –Parkin cells produced a TFEB band that migrated between that of untreated cells and torin 1–treated cells, which was absent in O/A-treated +Parkin cells (Figs. 1 D and S1 E). These data show that although partial O/A-induced dephosphorylation occurs, Parkin activity is necessary for additional dephosphorylation events correlating with TFEB translocation and activation.
Effects of Parkin activation on mTORC1 activity and TFEB association with 14-3-3 proteins
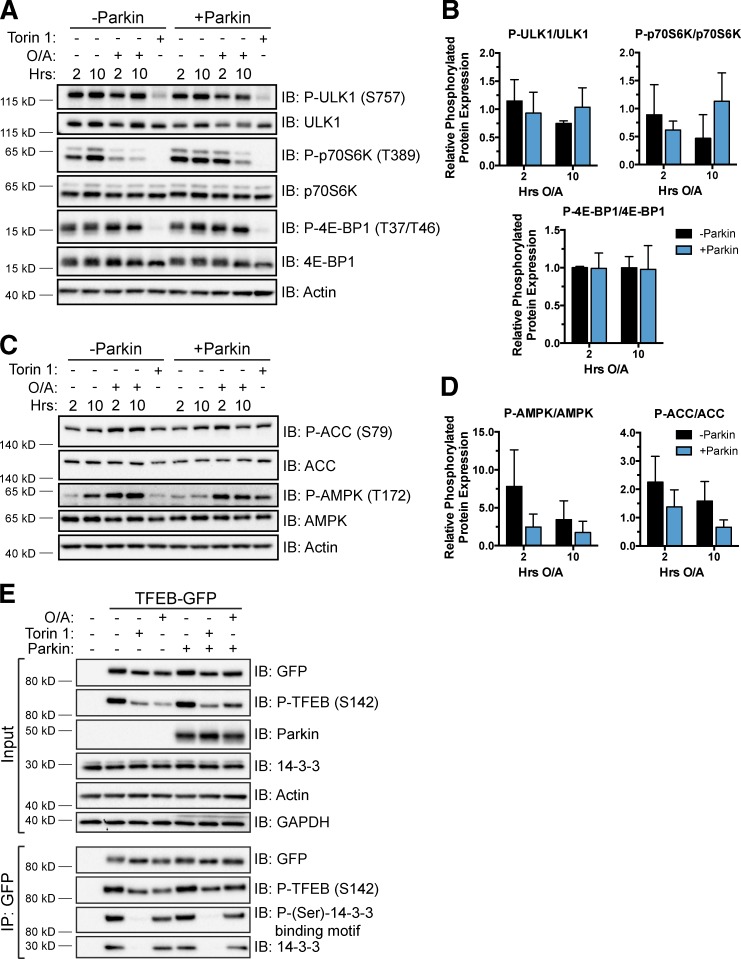
We explored the role of Parkin in modulating mTORC1 activity during mitophagy by examining the phosphorylation levels of three well-established mTORC1 substrates: eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1), p70 S6 kinase (p70S6K), and ULK1 (Brown et al., 1995; Brunn et al., 1997; Burnett et al., 1998; Kim et al., 2011). Whereas torin 1 all but eliminated phosphorylation of these proteins, O/A did not affect the phosphorylation of 4E-BP1 (Fig. 2, A and B). Furthermore, upon O/A treatment, similar decreases in phosphorylation of ULK1 and p70S6K were observed in cells both with and without Parkin, in contrast to O/A-induced TFEB translocation that requires Parkin expression.
Figure 2.
Analysis of Parkin-dependent effects on mTORC1 activity and TFEB association with 14-3-3 proteins. (A and C) WT and YFP-Parkin HeLa cells were treated with DMSO, O/A, or torin 1 as indicated, lysed, and immunoblotted. (B and D) Quantification of data in A and C, respectively. Protein levels were normalized to actin and the ratio of phosphorylated to total protein in treated samples is expressed relative to DMSO controls. Data are means ± SD (n = 3); no differences observed were statistically significant. (E) HeLa cells stably expressing TFEB-GFP were transfected with control or untagged Parkin DNA and treated the next day with DMSO (6 h), torin 1 (2 h), or O/A (6 h). Cells were lysed and TFEB-GFP was immunoprecipitated with anti-GFP beads. Cell lysates (Input) and immunoprecipitated proteins were immunoblotted. Images are representative of n = 2 experiments.
Additionally, we investigated the activity of starvation-responsive AMP-activated protein kinase (AMPK), which can inhibit mTORC1 via phosphorylation of the mTORC1 subunit raptor and upstream inhibitor tuberous sclerosis 2 (Inoki et al., 2003; Gwinn et al., 2008). Various cellular stresses stimulate AMPK activity through phosphorylation of threonine 172 (Hawley et al., 1996; Mihaylova and Shaw, 2011). We found that O/A treatment stimulated phosphorylation of AMPK and its substrate acetyl-CoA carboxylase (Ha et al., 1994) in both WT and YFP-Parkin HeLa cells by 2 h (Fig. 2, C and D), in agreement with previous studies showing that O/A increases the AMP/ATP ratio (Witters et al., 1991; Marsin et al., 2000; Hawley et al., 2002). Thus, AMPK activity is unaffected by the presence or absence of Parkin and is unlikely to account for Parkin-dependent TFEB activation.
To corroborate these findings, we assessed the phosphorylation status of TFEB residues S142 and S211, established to be critical regulators of TFEB translocation and transcriptional activity through the actions of mTORC1 and extracellular signal-regulated kinase 2 (Settembre et al., 2011, 2012; Martina et al., 2012; Roczniak-Ferguson et al., 2012). Immunoblotting of phospho-S142 and indirect analysis of phospho-S211 via coimmunoprecipitation of 14-3-3 bound to TFEB-GFP revealed a pronounced decrease in the former but little to no reduction in the latter in cells lacking endogenous Parkin (Fig. 2 E). No further changes in S142 phosphorylation or 14-3-3 binding were observed in the presence of Parkin. Inhibition of mTORC1 with torin 1 served as a positive control, causing decreased phospho-S142 and total loss of bound 14-3-3. The O/A-dependent decrease in phospho-S142 corresponds with the shift in MM of TFEB caused by O/A alone (Fig. 1 D) and may be the result of the O/A-dependent effects on both AMPK and mTORC1 activity, which likely reflect a loss of mitochondrial function that is independent of mitophagy. Thus, phosphorylation changes at S142 and S211 alone are not sufficient to explain Parkin-induced TFEB translocation. These results indicate that a different critical residue may be targeted for dephosphorylation in a Parkin-dependent manner or that additional mechanisms beyond what has been established may contribute to the regulation of TFEB activity.
Rag GTPases function independently and downstream of Parkin to regulate TFEB subcellular localization
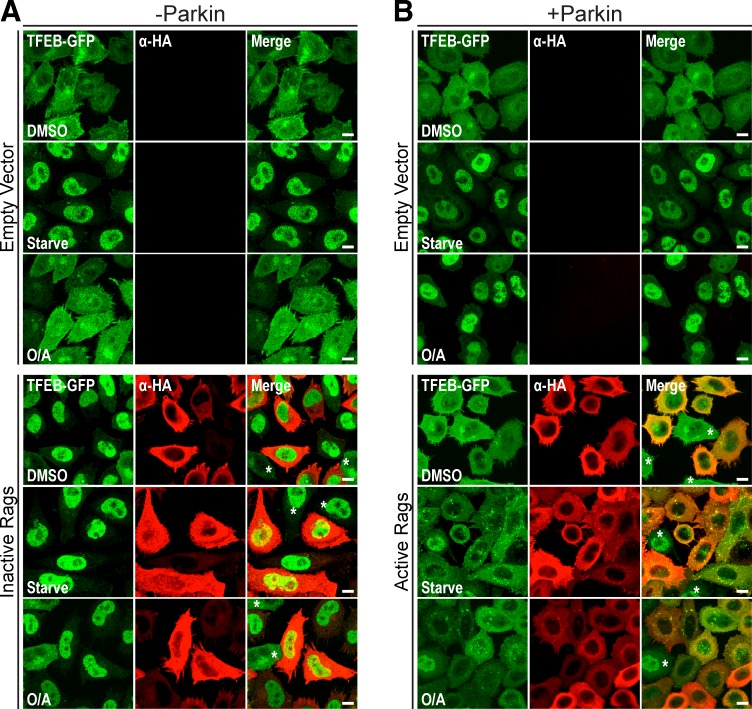
Rag GTPases play a central regulatory role for MiT/TFE transcription factors during starvation (Settembre et al., 2012; Martina and Puertollano, 2013; Martina et al., 2014b). To examine their contribution to mitophagy-induced TFEB activation, we expressed a dominant-negative (RagBGDP/RagDGTP) or constitutively active (RagBGTP/RagDGDP; Sancak et al., 2008) heterodimer in WT and mCherry-Parkin HeLa cells, respectively. Inactive Rags triggered robust TFEB nuclear translocation in control, starved, and O/A-treated WT cells, independently of Parkin-mediated mitophagy (Fig. 3 A). In contrast, active Rags completely inhibited starvation and Parkin-mediated TFEB translocation (Fig. 3 B). These data suggest that Parkin functions upstream of the Rag complex and that, although Parkin does not appear to directly modulate mTORC1 activity, Parkin activity intersects the mTORC1 pathway on lysosomes to mediate TFEB activation.
Figure 3.
Parkin acts upstream of Rag GTPases to regulate TFEB subcellular localization. (A and B) HeLa cells stably expressing TFEB-GFP with (B) or without (A) mCherry-Parkin were transfected with empty vector or active or inactive RagB/D heterodimer DNA (detected with αHA antibody). The indicated cells were starved (2 h) or treated with DMSO or O/A (6 h) before fixation and immunofluorescence analysis. Images are representative of n = 2 experiments. 75–100% of cells observed (mean of 175 cells per condition) exhibited the given phenotype. Asterisks indicate αHA-negative cells. Bars, 10 µm.
TFEB translocation during mitophagy requires Atg5 and Atg9A
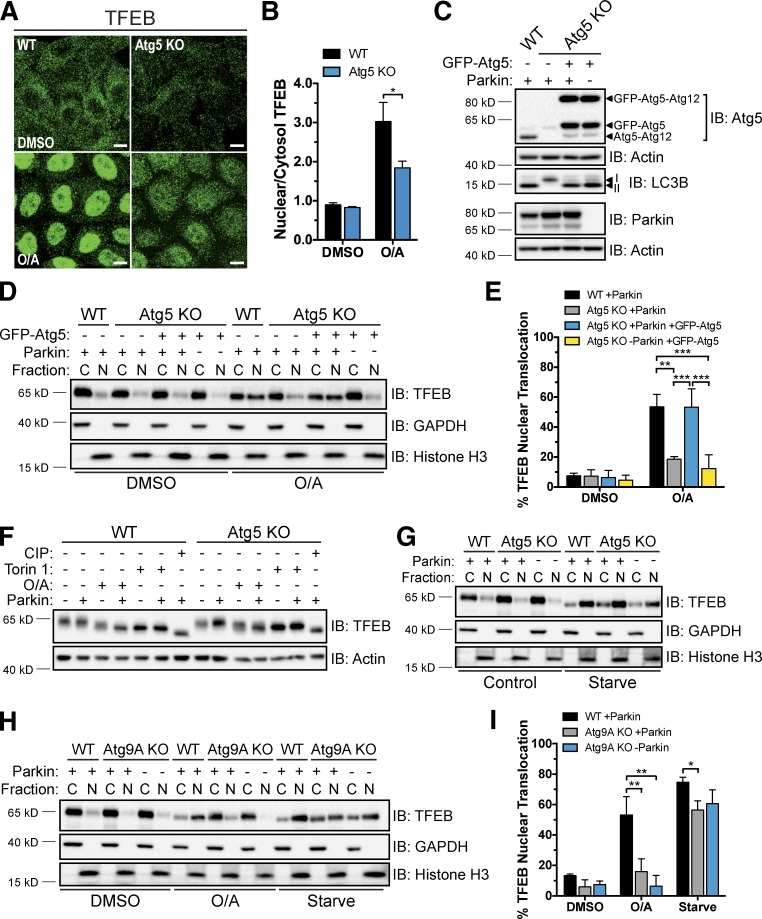
Next, we examined if Parkin-mediated TFEB activation depends on core components of autophagosome formation. Atg5 forms a conjugate with Atg12 and, in complex with Atg16L, facilitates the lipidation and incorporation of LC3 into the forming autophagosome membrane during autophagy (Hanada et al., 2007; Fujita et al., 2008). Loss of Atg5 causes defects in both general autophagy and mitophagy (Mizushima et al., 2001; Narendra et al., 2008). We generated Atg5 KO cells that were unable to produce lipidated LC3 (LC3B-II; Fig. S2 A) by CRISPR/Cas9 gene editing (Table S1). Although normal PINK1 accumulation and efficient mitochondrial recruitment and ligase activity of Parkin were observed in Atg5 KO +Parkin cells upon O/A treatment (Fig. S2, C and D), immunostaining (Fig. 4, A and B) and fractionation experiments (Fig. 4, D and E) revealed that O/A-induced TFEB nuclear localization was inhibited. Whereas TFEB translocation to the nucleus was largely blocked, dephosphorylation of TFEB still occurred, again dissociating the two processes (Fig. 4 F). As in WT cells, increased doses of O/A were not sufficient to induce TFEB nuclear accumulation independently of Parkin or Atg5 (Fig. S2 E), and any decreases in TFEB phospho-S142 and 14-3-3 binding observed were O/A dependent and were not associated with Parkin-mediated TFEB translocation (Fig. S2 F). Stable expression of WT GFP-Atg5, but not a conjugate-defective K130R mutant, rescued LC3 lipidation (Figs. 4 C and S2 G) and TFEB translocation in Atg5 KO +Parkin cells treated with O/A (Fig. 4, D and E; and Fig. S2, H and I). However, loss of Atg5 did not inhibit starvation-stimulated TFEB relocalization (Fig. 4 G), similar to what was observed in mouse embryonic fibroblasts (Zhou et al., 2013). These findings show that unlike during nutrient deprivation, Atg5 activity is required for Parkin-mediated TFEB nuclear accumulation, consistent with Parkin functioning independently of mTORC1 (Fig. 2).
Figure 4.
Atg5 and Atg9A are required for Parkin-mediated TFEB translocation. (A) WT and Atg5 KO cells stably expressing mCherry-Parkin were treated with DMSO or O/A (6 h), fixed, immunostained for TFEB, and analyzed by immunofluorescence. Bars, 10 µm. (B) Quantification of endogenous TFEB nuclear localization in A. The nuclear/cytosol ratio for each condition was calculated from mean fluorescence intensity/volume measurements made for each compartment across a field (four to seven) of cells (40–60 cells/field). Data are means ± SD (n = 3). (C) Untreated WT and Atg5 KO cells stably expressing mCherry-Parkin and GFP-Atg5 as indicated were lysed and immunoblotted. (D) Cells from C were treated with DMSO or O/A (6 h), lysed, fractionated, and immunoblotted. (E) Quantification of data in D. Endogenous TFEB expression was normalized to GAPDH (cytosol) or histone H3 (nuclear) and nuclear TFEB was expressed as a percentage of total TFEB. Data are means ± SD (n = 3). (F) WT and Atg5 KO HeLa cells stably expressing mCherry-Parkin as indicated were treated with DMSO (6 h), torin 1 (2 h), CIP (1 h), and O/A (6 h) as indicated, lysed, and immunoblotted. Images are representative of n = 3 experiments. (G) WT and Atg5 KO cells expressing mCherry-Parkin as indicated were starved (2 h) or left untreated, lysed, fractionated, and immunoblotted. Images are representative of n = 3 experiments. (H) WT and Atg9A KO HeLa cells expressing mCherry-Parkin as indicated were starved (2 h), or treated with DMSO (Ctrl) or O/A for 6 h. Cell lysates were processed as in D. (I) Quantification of endogenous TFEB nuclear localization in H, performed as in E. Data are means ± SD (n = 3). C, cytosol; N, nuclear. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To distinguish whether the block in TFEB translocation observed in the absence of Atg5 was caused by the inhibition of autophagy or by the specific loss of LC3–phosphatidylethanolamine conjugation, we examined TFEB localization in the absence of Atg9A, an autophagy protein required for the emergence of the early autophagosomal isolation membrane (phagophore; Kishi-Itakura et al., 2014). Atg9A KO HeLa cells were generated using CRISPR/Cas9 (Fig. S2 B and Table S1) and, as in the Atg5 KO cells, exhibited normal PINK1 accumulation and Parkin translocation and ligase activity upon O/A treatment (Fig. S2, C and D). However, O/A-induced TFEB nuclear accumulation was prevented in these Atg9A KO cells regardless of Parkin expression and to the same degree as in Atg5 KO cells, whereas TFEB relocalization still occurred upon starvation (Fig. 4, H and I). Because WT levels of LC3B-II were detected in these Atg9A KO cells (Fig. S2 B), these results suggest that lipidated LC3 is not the critical signaling factor required for TFEB translocation during mitophagy.
Autophagosome–lysosome fusion is not required for Parkin-mediated TFEB activation
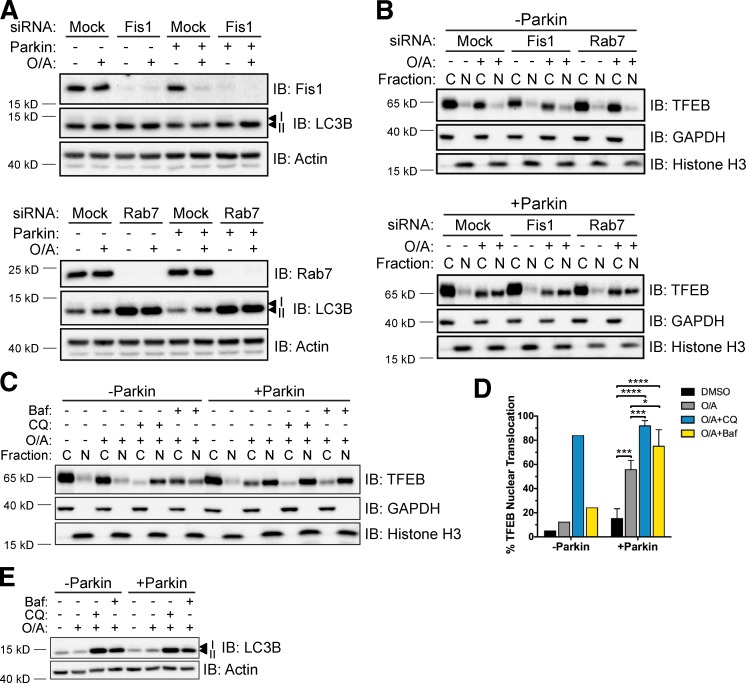
To investigate at which stage of the mitophagy process downstream of Atg9A and Atg5 TFEB is activated, we impaired autophagosome elongation by targeting the small GTPase Rab7 and its upstream effector Fis1 with siRNA. Recent work established a role for Rab7 in autophagosome elongation downstream of Parkin activation (Yamano et al., 2014). The aberrant accumulation of LC3 that occurs when Rab7 activity is not counterbalanced by TBC1D15 suggests a potential link between Rab7 and Atg5. However, although efficient knockdown of Fis1 and Rab7 was attained (Fig. 5 A), this did not influence the Parkin-dependent relocalization of TFEB in cells treated with O/A (Fig. 5 B).
Figure 5.
Fis1, Rab7, and autophagosome–lysosome fusion are dispensable for TFEB nuclear accumulation during mitophagy. (A) WT and mCherry-Parkin HeLa cells were transfected with control (Mock), Fis1, or Rab7 siRNA for 48 h before treatment with DMSO or O/A for 6 h, lysis, and immunoblotting. (B) WT (top) or mCherry-Parkin (bottom) HeLa cells were transfected and treated with DMSO or O/A as in A. Cell lysates were fractionated and localization of endogenous TFEB was analyzed by immunoblotting. Images in A and B are representative of n = 2 experiments. (C) WT and mCherry-Parkin HeLa cells were treated with O/A, CQ, and bafilomycin A1 (Baf) for 6 h as indicated, lysed, fractionated, and immunoblotted. (D) Quantification of data in C. Endogenous TFEB expression was normalized to GAPDH (cytosol) or histone H3 (nuclear) and nuclear TFEB was expressed as a percentage of total TFEB. Data are the mean of n = 2 experiments for −Parkin; means ± SD (n = 3) for +Parkin. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. (E) WT and mCherry-Parkin HeLa cells were treated as in C, lysed, and immunoblotted. C, cytosol; N, nuclear.
Downstream of phagophore elongation, we examined the role of autophagosome–lysosome fusion in Parkin-mediated TFEB activation by treating cells with O/A in combination with chloroquine (CQ) or bafilomycin A1. As expected, disruption of lysosomal function and autophagosome–lysosome fusion with these treatments, respectively, resulted in accumulated LC3B-II (Fig. 5 E). Consistent with previous studies (Roczniak-Ferguson et al., 2012; Settembre et al., 2012), robust TFEB nuclear translocation occurred in response to O/A+CQ regardless of Parkin expression (Fig. 5, C and D), an effect likely a result of mTORC1 inhibition by CQ demonstrated earlier (Settembre et al., 2012). Unlike CQ, bafilomycin A1 treatment only slightly augmented TFEB nuclear abundance in the absence of Parkin (Fig. 5, C and D). This enhancement was also observed in +Parkin cells after mitophagy induction, indicating that autophagosome–lysosome fusion is not required for Parkin-induced TFEB translocation. Thus, either autophagosome maturation, which requires Atg9A and Atg5, or some yet-uncharacterized upstream interaction involving both of these core autophagy proteins is necessary for TFEB activation.
Nuclear translocation of MITF and TFE3 occurs during mitophagy in a Parkin- and Atg5-dependent manner
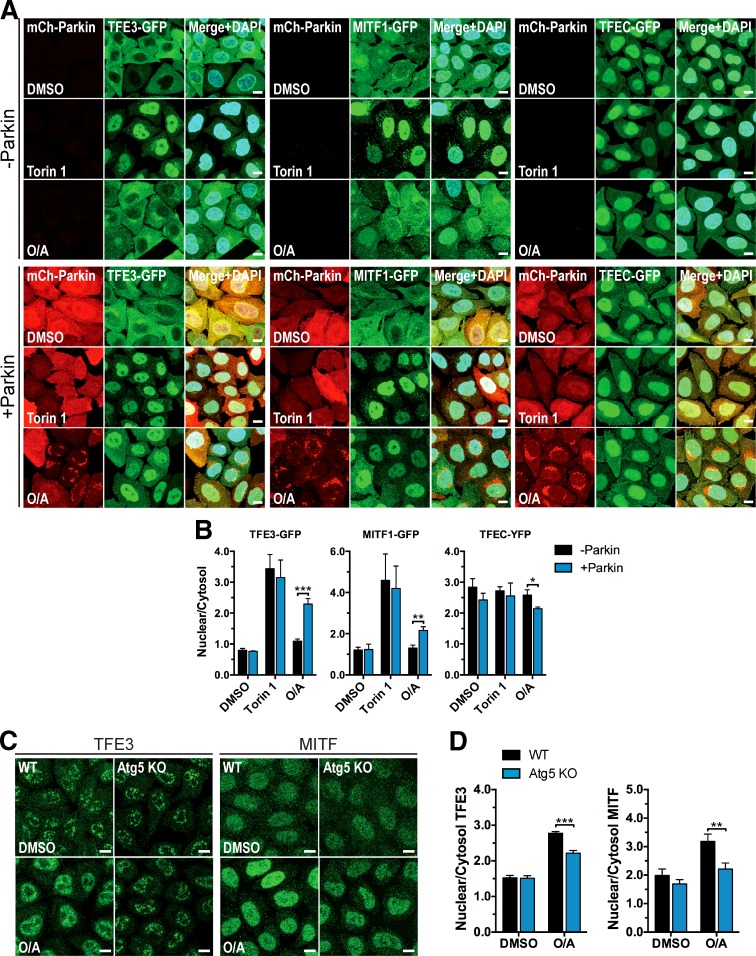
MITF, TFE3, and TFEC comprise the other members of the MiT/TFE subfamily of transcription factors to which TFEB belongs. Homo- and heterodimerization between these four proteins yield their capacity to bind specific DNA sequences via a conserved basic motif (Hemesath et al., 1994). The ability of MiT/TFE family members to cooperatively regulate transcription suggests their concurrent expression and/or activity under certain circumstances. Indeed, recent studies showed MITF and TFE3 to be regulated by Rag GTPases and mTOR in a fashion similar to that of TFEB during starvation, and the transcriptional repertoire of both overlaps with that of TFEB (Martina and Puertollano, 2013; Martina et al., 2014b). Therefore, we sought to determine whether these homologues share the same mechanism of regulation during mitophagy. Microscopy analysis revealed that O/A treatment significantly increases the nuclear intensity of GFP-tagged TFE3 and MITF1 isoform (MITF-A) in the presence of Parkin, whereas torin 1 induces translocation even in the absence of Parkin (Fig. 6, A and B). TFEC-YFP, however, exhibits substantial nuclear abundance in DMSO-treated cells that remained constant across all treatments, except for a small decrease in +Parkin cells upon O/A treatment. The findings for ectopic TFE3 and MITF1 were corroborated by immunostaining of endogenous TFE3 and MITF (Fig. S3, A and B) and subcellular fractionation of endogenous TFE3, TFE3-GFP, and MITF1-GFP (Fig. S3, C–E) using the same cells and treatments. In addition, relocalization of endogenous TFE3 and MITF to the nucleus was significantly inhibited in Atg5 KO cells (Fig. 6, C and D), similar to TFEB. These data reveal a common mechanism for Parkin- and Atg5-dependent regulation of several MiT/TFE family members and suggest that more than one member may be important for mitophagy.
Figure 6.
Parkin- and Atg5-dependent regulation of TFEB homologue subcellular localization. (A) WT HeLa cells stably expressing mCherry-Parkin, TFE3-GFP, MITF1-GFP, and TFEC-YFP as indicated were treated with DMSO (6 h), torin 1 (2 h), or O/A (6 h). Fixed cells were analyzed by immunofluorescence. (B) Quantification of ectopic TFE3, MITF1, and TFEC nuclear localization in A. The nuclear/cytosol ratio for each condition was calculated from mean fluorescence intensity/volume measurements made for each compartment across a field (four to seven) of cells (40–60 cells/field). Data are means ± SD (n = 3). (C) WT and Atg5 KO cells stably expressing mCherry-Parkin treated with DMSO or O/A (6 h) were fixed, immunostained for TFE3 or MITF, and analyzed by immunofluorescence. (D) Quantification of endogenous TFE3 and MITF nuclear localization in C. Analysis was performed as in B (40–60 cells/field, 4 fields/condition, n = 3 experiments). Data are means ± SD. For all graphs: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Bars, 10 µm.
TFEB is not essential for Parkin-mediated mitophagy
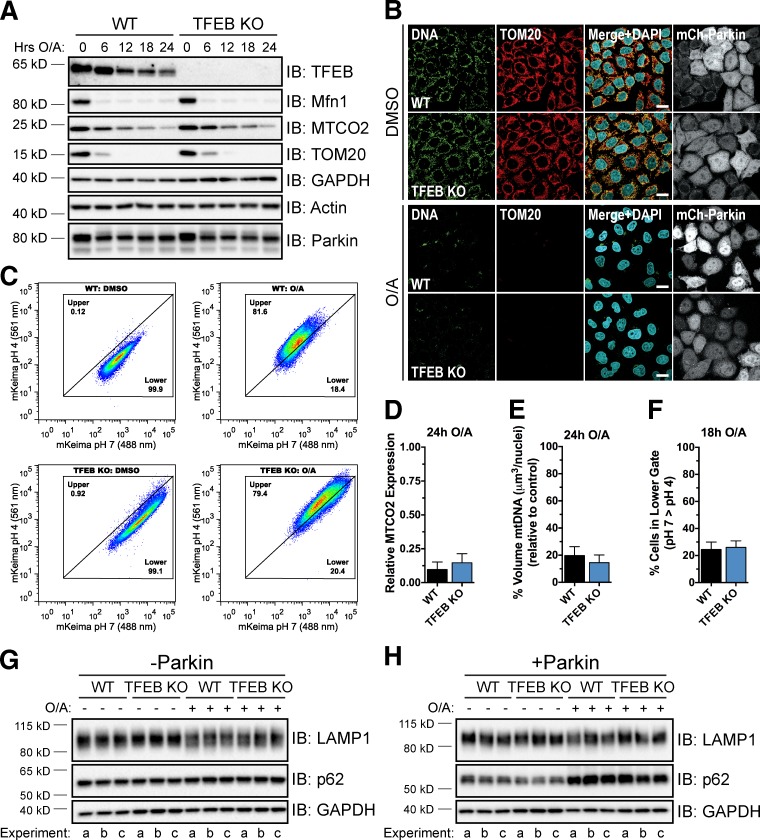
To determine whether TFEB is required for Parkin-mediated mitophagy, we used CRISPR/Cas9 technology to knock out TFEB in HeLa cells by targeting exon 4, confirming the loss of endogenous TFEB expression by PCR screening, DNA sequencing, and immunoblotting (Fig. S4, A–D; and Table S1). To initially assess mitophagy in these cells, we monitored the degradation of the outer mitochondrial membrane proteins mitofusin 1 (Mfn1) and translocase of outer membrane 20 (TOM20) and the inner membrane protein mitochondrial-encoded subunit 2 of cytochrome c oxidase (MTCO2) by immunoblotting. Complete degradation of Mfn1 and TOM20 was observed in both WT and TFEB KO cells expressing mCherry-Parkin (Fig. 7 A), reflecting some level of mitophagy-independent proteasomal degradation (Tanaka et al., 2010; Yoshii et al., 2011). MTCO2 expression also decreased by ∼90% in both cell lines by 24 h of O/A treatment (Fig. 7, A and D), indicating normal mitophagy in TFEB KO cells. Similarly, immunostaining of mitochondrial nucleoids with anti-DNA antibody revealed no major difference in clearance of mitochondrial DNA between WT and TFEB KO cells after 24-h O/A treatment (Fig. 7, B and E).
Figure 7.
TFEB KO cells do not exhibit a defect in mitophagy. (A) WT and TFEB KO cells stably expressing mCherry-Parkin were treated with DMSO or O/A for up to 24 h, lysed, and immunoblotted. (B) Cells from A were treated with DMSO or O/A for 24 h. Fixed cells were immunostained for DNA and TOM20 and analyzed by immunofluorescence. Bars, 20 µm. (C) WT and TFEB KO cells stably expressing YFP-Parkin and mt-mKeima were treated with DMSO or O/A for 18 h and subjected to FACS analysis. Plots are representative of n = 2 experiments. (D) Quantification of MTCO2 levels in A. MTCO2 expression was normalized to actin and expressed relative to 0-h treatment. Data are means ± SD (n = 3). (E) Quantification of anti-DNA stain in B. Z stacks of five to seven fields per condition (20–30 cells/field) were analyzed per experiment (n = 3). Data are means ± SD. (F) Proportion of cells in the lower gate in C. Data are means ± range (n = 2 experiments). (G and H) WT and TFEB KO cells lacking endogenous Parkin (G) or stably expressing mCherry-Parkin (H) were treated with DMSO or O/A for 18 h, lysed, and immunoblotted. Three independent experiments (a–c) are shown.
The lack of an obvious mitophagy phenotype assessed by immunoblotting and microscopy in TFEB KO cells led us to use FACS analysis in combination with mitochondria-targeted Keima (mt-mKeima), a coral-derived acid-stable fluorescent protein that exhibits a bimodal excitation spectrum (Katayama et al., 2011), to more sensitively quantify mitochondrial clearance. A shift from a low to a high ratio of excitation at 561/488 nm with this probe signals its movement into a more acidic environment. We observed a significantly higher 561/488 nm (pH 4/pH 7) ratio signal in WT cells (Fig. 7, C and F), but not in PINK1 KO control cells (Fig. S5 G), after 18 h of O/A treatment relative to DMSO treatment, indicating delivery of depolarized mitochondria into lysosomes. This same shift to an acidic environment was observed in TFEB KO cells (Fig. 7, C and F), confirming that these cells do not exhibit a detectable defect in Parkin-mediated clearance of damaged mitochondria. Consistent with these findings, protein levels for TFEB targets lysosomal-associated membrane protein (LAMP) 1 and p62 were similar between WT and TFEB KO cells (Fig. 7, G and H) and O/A treatment did not affect LAMP1 expression in either cell line, though p62 levels increased in a Parkin-dependent manner (Fig. 7, G and H), suggesting MiT/TFE homologues may be compensating for TFEB loss. Thus, although TFEB is activated during Parkin-mediated mitophagy, it appears to be dispensable for mitophagy under the experimental conditions in the present study. It remains a distinct possibility, however, that TFEB is important for the clearance of damaged mitochondria in other cell lines or under certain physiological conditions.
Loss of multiple MiT/TFE transcription factors causes defects in lysosomes and Parkin-mediated mitophagy
Because of high homology, all MiT/TFE family members bind to E-box DNA recognition sequences and may regulate some of the same genes (Hemesath et al., 1994; Aksan and Goding, 1998). Therefore, we investigated whether TFEB loss may be compensated for by MITF, TFE3, or TFEC. We used the CRISPR/Cas9 system to excise an entire frame-shifting exon from within the basic helix-loop-helix domain of the three remaining homologues (Fig. S4 A) and confirmed the successful generation of several MiT/TFE single KO, double KO (DKO), and triple KO (TKO) cell lines by PCR, immunoblotting, and DNA sequencing (Fig. S4, B–D; and Table S1). Complete degradation of TOM20 and Mfn1 occurred in all cell lines expressing Parkin upon prolonged O/A treatment. Additionally, a similar level of MTCO2 degradation relative to WT cells was observed for all KO, DKO, and TKO cell lines except those lacking TFE3, which showed a slight block in MTCO2 loss that was statistically significant for TFE3 KO cells (Fig. S5, A–D).
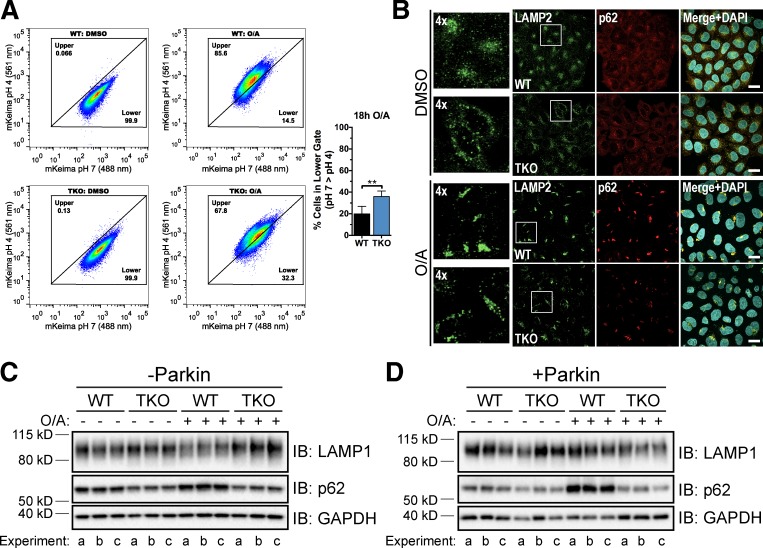
As TFEB, MITF, and TFE3 are all regulated in a similar manner during mitophagy, we expanded our analysis of mitophagy in the TFEB/MITF/TFE3 TKO cell line (subsequently referred to as TKO). By confocal microscopy, there was no difference in mitochondrial DNA clearance between WT and TKO cells after 18-h O/A treatment (Fig. S5, E and F) or in LAMP1 protein levels in all conditions tested (Fig. 8, C and D). The more quantitative mt-mKeima assay, however, revealed a substantially reduced shift to a higher 561/488 nm (pH 4/pH 7) ratio signal upon 18-h O/A treatment in the TKO cells compared with WT cells (Fig. 8 A), indicating a considerable impairment of mitophagy. Consistent with these results, TKO cells displayed a significant defect in the Parkin-dependent up-regulation of p62 by O/A (Fig. 8, C and D), validating that this p62 induction, also seen for p62 mRNA by quantitative PCR (Fig. 1 E), reflects MiT/TFE transcriptional activity. Immunostaining experiments confirmed the defect in p62 expression after Parkin activation and also revealed that lysosome morphology in the TKO cells appeared to be more disorganized than in WT cells (Fig. 8 B). Together, these data support the hypothesis that TFE3 and/or MITF compensate for TFEB loss and establish a role for MiT/TFE transcription factors in Parkin-mediated clearance of damaged mitochondria.
Figure 8.
Concurrent deletion of TFEB, MITF, and TFE3 results in lysosomal and mitophagy defects. (A, left) WT and TFEB/MITF/TFE3 TKO (TKO) cells stably expressing YFP-Parkin and mt-mKeima were treated with DMSO or O/A for 18 h and subjected to FACS analysis. Plots are representative of n = 4 experiments. (A, right) Proportion of cells in the lower gate in left panels after 18-h O/A treatment. Data are means ± SD. **, P < 0.01. (B) WT and TKO cells stably expressing mCherry-Parkin were treated with DMSO or O/A for 18 h, fixed, immunostained for p62 and LAMP2, and analyzed by immunofluorescence. Left-most panels are enlarged versions of boxed regions in LAMP2 panels. Images are representative of n = 3 experiments where z stacks of five fields per condition (40–50 cells/field) per experiment were collected. Bars, 10 µm. (C and D) WT and TKO cells lacking endogenous Parkin (C) or stably expressing mCherry-Parkin (D) were treated with DMSO or O/A for 18 h, lysed, and immunoblotted. Three independent experiments (a–c) are shown.
Discussion
The recent characterization of MiT/TFE transcription factors as activators of lysosomal and autophagic gene expression during starvation-induced autophagy has revealed that cells dynamically alter their transcriptome according to nutrient status (Martina et al., 2014a). Our study expands the role of MiT/TFE family members to another type of cellular stress response by showing their activation during Parkin-mediated mitophagy. After treatment with mitochondrial depolarizing compounds to activate PINK1 and Parkin, TFEB, TFE3, and MITF relocalize from the cytosol to the nucleus. Loss of either PINK1 or Parkin prevents such nuclear translocation and their subsequent transcriptional activity.
In the current model of TFEB activation upon nutrient depletion, regulation of TFEB phosphorylation at residues S142 and S211 by mTORC1 plays a critical role, particularly as dephosphorylation of S211 is predicted to expose a nuclear import sequence that is otherwise occluded by 14-3-3 binding (Martina et al., 2012; Roczniak-Ferguson et al., 2012; Settembre et al., 2012). As occurs upon mTOR inhibition (Settembre et al., 2011, 2012), Parkin-mediated TFEB nuclear accumulation corresponds with TFEB dephosphorylation. We observed several distinct steps of TFEB MM shift. Mitochondrial depolarization with O/A causes a decrease in TFEB MM in +Parkin cells that is slightly greater than the shift seen in −Parkin cells and equivalent to the lower TFEB band seen with torin 1. Combined O/A + torin 1 treatment does not cause an additional MM shift in TFEB possibly because these treatments affect the same residues or an unstable dephosphorylated TFEB species is produced and degraded under these conditions. Phosphatase treatment induces a further shift downward, indicating that phosphorylation of some residues might stabilize activated TFEB (Peña-Llopis et al., 2011; Ferron et al., 2013).
In agreement with previous studies of mitochondrial stress reducing mTORC1 activity (Dennis et al., 2001; Kwon et al., 2011; Kim et al., 2013), we detected decreases in ULK1 and p70S6K phosphorylation upon O/A-induced mitochondrial depolarization. However, these changes were not Parkin dependent and thus did not correlate with TFEB activation. We also observed an O/A-dependent reduction in TFEB phosphorylation at S142, which could be attributed to the reduced mTORC1 activity. These data suggest that O/A treatment alone inhibits mTOR activity toward select substrates including TFEB, but this is not sufficient for TFEB nuclear translocation in the conditions tested. However, mTORC1 inhibition may be necessary as constitutively active Rag GTPases prevented Parkin-mediated TFEB translocation.
Furthermore, we found, counterintuitively, that TFEB translocation in WT +Parkin cells treated with O/A does not correspond with a loss of TFEB-bound 14-3-3. Although mostly cytosolic 14-3-3 proteins are also found bound to substrates in the nucleus (Brunet et al., 2002; Obsilová et al., 2008), so perhaps a nuclear TFEB–14-3-3 complex exists after mitophagy induction. This hypothesis implies that 14-3-3 may regulate TFEB subcellular localization by one or more mechanisms in addition to masking a nuclear import signal. One such mechanism may be stabilization of either an inactive or active TFEB conformation depending on external signals. Alternatively, because 14-3-3 is active as homo- and heterodimers in most cases, one 14-3-3 monomer on TFEB might stimulate interactions with other 14-3-3–bound proteins important for the regulation of TFEB localization and activity (Mackintosh, 2004). Parkin activation could thus alter such interactions without decreasing the amount of 14-3-3 bound to TFEB. These possibilities, in addition to the roles of differential 14-3-3 posttranslational modifications, isoform expression, and binding specificity (Aitken, 2011), require further investigation.
Downstream of Parkin activation, we demonstrate that Atg5 is required for TFEB, TFE3, and MITF nuclear translocation. In contrast, loss of Atg5 does not impair TFEB nuclear accumulation upon nutrient deprivation, similar to previous work performed in Atg5 KO mouse embryonic fibroblasts that showed starvation-induced TFEB translocation to the nucleus to be uninhibited, although subsequent activation of lysosomal function was suppressed (Zhou et al., 2013). These findings provide compelling evidence that MiT/TFE transcription factors are differentially regulated during starvation and mitophagy.
Atg9A is also necessary for Parkin-mediated translocation of TFEB. This requirement for another core autophagy protein that acts at a different stage of autophagosome biogenesis suggests that the common denominator behind the inhibition of TFEB translocation in Atg5 and Atg9A KO cells may be a defect in autophagosome formation. However, depletion of either Fis1 or Rab7, factors involved in phagophore elongation during mitophagy, did not affect O/A-induced TFEB nuclear accumulation in Parkin-expressing cells. We also did not observe any diminution of Parkin-mediated TFEB translocation after impairment of autophagosome–lysosome fusion with bafilomycin A1. Kishi-Itakura et al. (2014) recently expanded upon the specific functions of Atg9A and Atg5, reporting that Atg9A is required for the emergence of the isolation membrane and that isolation membranes could elongate in the absence of Atg5 but mature autophagosomes that are fully closed could not form without Atg5. TFEB may thus be activated during autophagosome maturation, which requires both Atg9A and Atg5, or at an earlier stage of the autophagy process. A key question remains concerning the process that intermediates between Parkin, Atg9A/Atg5, and TFEB. To this end, Medina et al. (2015) recently reported TFEB as a substrate of the phosphatase calcineurin, which is activated by calcium signaling mediated by the lysosomal calcium channel MCOLN1 upon starvation. Therefore, a future avenue of research may explore the possibility of a similar mechanism of phosphatase stimulation in the context of Parkin-mediated TFEB activation.
We also expand the understanding of events occurring downstream of Parkin activation by showing that cells lacking MiT/TFE transcription factors exhibit abnormal lysosome morphology and impaired degradation of damaged mitochondria. Although TFEB, MITF, and TFE3 share a common mechanism of regulation, analysis of MiT/TFE family member KO cell lines suggests that TFE3 is the predominant mediator of lysosomal and autophagic induction during prolonged mitochondrial stress in HeLa cells. However, this may vary by cell type because of differential expression of these homologues (Kuiper et al., 2004; Martina et al., 2014a). Quantitative analysis of mitophagy showed TFEB/MITF/TFE3 TKO cells, but not TFEB KO cells, to be moderately defective in mitophagy. We also observed a loss of induction of MiT/TFE target p62 protein expression after O/A treatment in the TFEB/MITF/TFE3 TKO cells, but not in TFEB KO cells. These results are consistent with previous studies suggesting that MiT/TFE family members share redundant functions (Steingrimsson et al., 2002; Martina et al., 2014a). They also suggest that other transcription factors may be involved in autophagy and lysosomal gene expression during mitophagy such as ATFS-1, which up-regulates autophagy genes in response to mitochondrial stress independently of TFEB in Caenorhabditis elegans (Guo et al., 2014). Further investigation is needed to fully characterize the transcriptional network specifically activated upon perturbation of mitochondrial function.
Quality control of mitochondria appears important in maintaining cellular health, as excessive mitochondrial dysfunction is implicated in numerous metabolic and neurodegenerative diseases, including PD (DiMauro and Schon, 2008). Increasing evidence indicates that lysosomal dysfunction may also contribute to the pathogenesis of such diseases. Notably, mutations in the lysosomal enzyme glucocerebrosidase, which cause the lysosomal storage disorder (LSD) Gaucher disease, are a risk factor for PD (Sidransky et al., 2009). Overexpression of TFEB has been used to promote clearance of toxic storage materials and rescue cellular and protein homeostasis in several animal and cell models of LSDs and neurodegenerative disorders, including Gaucher disease and Tay-Sachs disease (Song et al., 2013); Pompe disease (Medina et al., 2011; Spampanato et al., 2013); multiple sulfatase deficiency, Batten disease, and mucopolysaccharidosis type IIIA (Medina et al., 2011); Alzheimer’s disease (Polito et al., 2014); Huntington disease (Tsunemi et al., 2012); and PD (Decressac et al., 2013). Overall, this study identifies a novel way in which TFEB and its homologues can be activated by the PINK1–Parkin pathway and may open new avenues for targeting therapeutics to treat diseases such as PD and LSDs.
Materials and methods
Cell culture and treatments
All experiments were performed in HeLa cells or neuroblastoma BE(2)-M17 cells cultured at 37°C in high glucose DMEM or Opti-MEM (Gibco), respectively, supplemented with 100 µM MEM nonessential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), 2 mM GlutaMAX (Gibco), and 10% FBS (BenchMark; Gemini Bio Products) under a humidified atmosphere of 5% CO2. For starvation experiments, cells were washed twice with HBSS and incubated for 2–3 h in starvation buffer (140 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 5 mM glucose, 20 mM Hepes, and 1% BSA, pH 7.4; Axe et al., 2008). For drug treatment experiments, cells were incubated in medium containing one or more of the following compounds: 250 nM torin 1 (2 h; Tocris Bioscience), 100 µM CQ (6 h; Sigma-Aldrich), 400 nM bafilomycin A1 (6 h; Sigma-Aldrich), 10 µM valinomycin (Sigma-Aldrich; see figures for treatment times), a mixture of 10 µg/ml oligomycin (EMD Millipore) and 5 µg/ml antimycin A (Sigma-Aldrich; O/A; see figures for treatment times), or 10 µM quinolyl-valyl-O-methylaspartyl-[-2,6-difluorophenoxy]-methyl ketone (QVD; APExBio). Valinomycin and O/A were always used in combination with QVD, a broad-spectrum caspase inhibitor, except in Fig. S1 (D and E).
Plasmids and generation of retroviral constructs
The following constructs were obtained from Addgene: plasmid 38119, pEGFP-N1-TFEB, plasmid 38132, pEGFP-N1-MITF-A, and plasmid 38120, pEGFP-N1-TFE3, all contain a CMV promoter within the pEGFP-N1 backbone vector and were gifts from S. Ferguson (Yale University School of Medicine, New Haven, CT; Roczniak-Ferguson et al., 2012); plasmid 38196, pMXs-IP-EGFP-mAtg5, contains an IRES-driven puromycin selection marker within the pMXs-IP-EGFP retrovirus backbone vector and was a gift from N. Mizushima (Tokyo Medical and Dental University, Tokyo, Japan; Hara et al., 2008), and the pMXs-IP-EGFP-mAtg5-K130R mutant was generated by site-directed mutagenesis; plasmid 19302, pRK5-HA-GST-RagB54L, plasmid 19303, pRK5-HA-GST-RagB99L, plasmid 19308, pRK5-HA-GST-RagD77L, and plasmid 19309, pRK5-HA-GST-RagD121L, all contain a CMV promoter within the pRK5-HA backbone vector and were gifts from D. Sabatini (Whitehead Institute for Biomedical Research, Massachusetts Institute of Technology, and Broad Institute, Cambridge, MA; Sancak et al., 2008). The pCMV6-Entry-TFEC-Myc-DDK construct was obtained from OriGene. The viral TFEB-GFP expression vector was generated by PCR amplification of the full-length encoding sequence from pEGFP-N1-TFEB followed by in-frame cloning into HindIII–SalI sites of the retrovirus pBMN-Z vector (Orbigen). The viral MITF-GFP and TFE3-GFP expression vectors were generated by in-frame cloning into HindIII–NotI sites of the pBMN-Z vector. The viral TFEC-GFP expression vector was generated in a two-step process through PCR amplification of the full-length TFEC encoding sequence from pCMV6-Entry-TFEC-Myc-DDK followed by in-frame cloning into XhoI–BamHI sites of the pEYFP-N1 vector (Takara Bio Inc.). The pEYFP-N1-TFEC was then digested with BglII and NotI followed by in-frame cloning into BamHI–NotI sites of the pBMN-Z vector. The mt-mKeima plasmid was a gift from A. Miyawaki (Institute of Physical and Chemical Research Brain Science Institute, Wako, Japan; Katayama et al., 2011) and recloned into retrovirus pCHAC-MCS1-IRES-MCS2 vector (ABP-PVL-BICISIRES; Allele Biotechnology) at XhoI–SalI sites. Construction of pBMN-mCherry-Parkin and pBMN-YFP-Parkin have been previously described (Yamano et al., 2014); both were cloned into BamHI–SalI sites of the pBMN-Z vector. All constructs were confirmed by DNA sequencing.
Generation of stable cell lines
To produce retrovirus, retroviral constructs were transfected into HEK293T cells together with the plasmids encoding Gag-Pol (1.0 µg) and VSV-G (0.5 µg) using Avalanche-Omni Transfection Reagent (EZ Biosystems). Media was replaced at 24 h. Conditioned media containing retroviral particles was collected at 48 h and used to infect HeLa cells to generate stable cell lines. Selection was performed by cell sorting based on fluorescence.
Generation of CRISPR/Cas9-mediated gene KO cell lines
To generate KO cell lines, CRISPR target sites were chosen within an exon (TFEB, Atg5, and Atg9A) or upstream (5′) and downstream (3′) of a frame-shifting exon (MITF, TFEC, and TFE3) found in all predicted splice variants of each gene. Gene-specific guide RNA (gRNA) expression vectors were generated as previously described (Mali et al., 2013). hCas9 (plasmid 41815; Addgene) and gRNA cloning vector (plasmid 41824; Addgene) were gifts from G. Church (Harvard Medical School, Boston, MA; Mali et al., 2013). This gRNA cloning vector contains a U6 promoter and single gRNA scaffold in a pCR-blunt II backbone. For each gene of interest, 19 bp of the selected target sequence adjacent to the protospacer adjacent motif site (NGG) was incorporated into two 60-bp oligonucleotides (forward, 5′-TTTCTTGGCTTTATATATCTTGTGGAAAGGACGAAACACCG(N)19-3′; reverse, 5′-GACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC(N)19C-3′). These two oligos were annealed and extended using Phusion polymerase (Thermo Fisher Scientific) to produce a 100-bp double-stranded DNA fragment, which was then inserted into the AflII site of the gRNA cloning vector using Gibson Assembly (New England Biolabs, Inc.). Cells were cotransfected with CRISPR nuclease (hCas9), gene-specific gRNA, and C1-mCherry (Takara Bio Inc.) vector using Avalanche-Omni Transfection Reagent and sorted based on fluorescence after 48 h into 96-well plates for single colony isolation. Genomic DNA was extracted for analysis with the Quick-gDNA Miniprep kit (Zymo Research). For Atg5, Atg9A, and TFEB, clones were screened by PCR using primers surrounding the CRISPR target site followed by restriction enzyme digestion (SapI, PshAI, and SacI, respectively). For MITF, TFEC, and TFE3, clones were screened by PCR using a forward primer upstream of the 5′ CRISPR target site and a reverse primer downstream of the 3′ CRISPR site. TFEB, Atg5, and Atg9A clones positive for CRISPR digestion (those with PCR products not digested by the restriction enzyme) and MITF, TFEC, and TFE3 clones exhibiting a truncated PCR product were then analyzed by Sanger sequencing (Eurofins MWG Operon) and/or immunoblotting to confirm clones as KOs. For generation of PINK1 KO HeLa cells, two CRISPRs targeting exons 1 and 7 were cotransfected with hCas9 and pEYFP-C1 into HeLa cells. Single colonies were first screened by PCR for the targeted deletion, and then loss of PINK1 expression, Parkin mitochondrial translocation, and mitophagy flux in positive KO clones was confirmed by immunoblotting, microscopy, and FACS, respectively. See Table S1 for CRISPR and screening PCR primer sequences.
Immunostaining and confocal microscopy
Cells were seeded into LabTek 1.5 borosilicate chambered slides (Thermo Fisher Scientific). For Rags experiments, cells were transfected for 16–24 h with 500 ng total of DNA plasmids pRK5 (empty vector), pRK5-HA-GST-RagBQ99L and pRK5-HA-GST-RagDS77L (constitutively active dimer pair), or pRK5-HA-GST-RagBT54L and pRK5-HA-GST-RagDQ121L (dominant-negative dimer pair) using Avalanche-Omni Transfection Reagent according to the manufacturer’s protocol, before treatment. Cells were fixed with 4% paraformaldehyde in PBS for 15 min and blocked with PBS containing 0.1% Triton X-100 and 3% goat serum for 1 h. For immunostaining, cells were incubated with primary antibodies overnight at 4°C and were then incubated with secondary antibodies goat anti–rabbit, anti–mouse, or anti–guinea pig IgG conjugated with Alexa Fluor 488, 594, or 633 (Invitrogen) for 45 min at room temperature. Cells were stained with DAPI (Thermo Fisher Scientific) and imaged in PBS at room temperature using an inverted confocal microscope (LSM510; Carl Zeiss) with a 63× 1.4 NA oil differential interference contrast Plan Apochromat objective using the accompanying LSM software (Carl Zeiss). Z-stack projections were composed using ImageJ v1.47 (National Institutes of Health) and brightness and contrast of images were adjusted in both ImageJ and Photoshop CS6 v13 (Adobe). All images for a given experiment were collected and adjusted in an identical manner.
Immunofluorescence quantification
Volocity v6.3 (PerkinElmer) was used for quantification analysis of immunofluorescence images. For protein nuclear translocation, 6–7 z-sections collected at 1-µm intervals were collected for four to seven fields (40–60 cells/field) per condition per independent experiment. Nuclear intensity of the protein of interest was calculated by subtracting the cytosolic intensity (exclusion of DAPI-stained area) from the total intensity. The corresponding nuclear and cytosolic volumes were then used to calculate a standardized mean intensity/volume measurement for each compartment and the ratio of these was expressed. For mitochondrial DNA staining, three central z-sections collected at 0.6-µm intervals were collected for 5–10 fields (20–40 cells/field) per condition per experiment. The volume of mitochondrial DNA was calculated by subtracting the DAPI-stained volume from the volume stained by an antibody against DNA. The ratio of mitochondrial DNA volume per nuclei was calculated for O/A-treated cells and expressed as a percentage of mitochondrial DNA volume/nuclei in DMSO-treated cells.
Immunoprecipitation
HeLa WT or Atg5 KO cells stably expressing TFEB-GFP were seeded into 10-cm tissue culture dishes at 106 cells/dish. The next day, cells were transfected with 2 µg of either untagged Parkin or pcDNA3.1+ (Takara Bio Inc.) empty vector plasmid DNA using X-tremeGENE HP DNA Transfection Reagent (Roche) according to the manufacturer’s instructions. After 16–24 h of transfection, cells were treated, washed once with ice-cold PBS, and lysed with ice-cold lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, protease, and phosphatase inhibitor cocktails [Roche]). Lysates were passed through a 26.5-gauge needle eight times and incubated on ice for 30 min. The soluble fraction was isolated from the cell lysate by centrifugation at 20,000 rpm for 15 min at 4°C. A small amount of lysate was saved as a whole cell lysate input for immunoblotting and the remaining lysate fractions were then incubated with 20 µl of a 50% slurry of GFP-nAb Magnetic (or Magnetic Agarose) beads (Allele Biotechnology) overnight with rotation at 4°C. Immunoprecipitates were washed four times with lysis buffer lacking Triton X-100. Immunoprecipitated proteins were denatured by the addition of NuPAGE LDS Sample Buffer (Invitrogen) supplemented with NuPAGE Sample Reducing Agent (Invitrogen) and heating for 5–10 min at 70°C.
Cell lysis and subcellular fractionation
Cells were fractionated as previously described (Holden and Horton, 2009) with some modifications. In brief, cells were plated in 6-well dishes and treated the next day. Cells were washed and collected in HBSS, lysed in 400 µl buffer 1 (50 mM Hepes, 150 mM NaCl, and 100 µg/ml digitonin) supplemented with complete protease inhibitor cocktail (Roche) and PhosSTOP phosphatase inhibitor cocktail (Roche), and rotated at 4°C for 10 min. Cell homogenates were centrifuged at 4,000 g for 2 min at 4°C to obtain the cytosolic fraction. The pellet fraction was washed once with cold PBS and then solubilized in 400 µl buffer 3 (50 mM Hepes, 150 mM NaCl, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate) supplemented with 10 U/ml Benzonase (Sigma-Aldrich) for 15 min and shaking at 4°C. Insoluble cell debris was separated from the nuclei-enriched fraction by centrifuging at 13,000 g for 5 min at 4°C. A 1/100 volume of 1.25% sodium deoxycholate was added to the cytosolic samples and then proteins in the cytosolic and pellet supernatant fractions were precipitated by adding 1/5 volume of 72% trichloroacetic acid and incubating on ice for 15 min. Samples were then centrifuged at maximum speed for 25 min at 4°C, washed once with cold acetone, and centrifuged as before. Whole cell lysates and fractionated protein samples were solubilized by boiling in NuPAGE LDS Sample Buffer supplemented with NuPAGE Sample Reducing Agent.
Protein dephosphorylation
For dephosphorylation of proteins, cells were lysed in buffer containing NEB3 (New England Biolabs, Inc.) and 1% Triton X-100 by repeated passage through a 26.5-gauge needle. Half of each sample was digested with CIP (New England Biolabs, Inc.) at 37°C for 1 h. All samples were diluted with 3× sample buffer (180 mM Tris HCl, pH 6.8, 6% SDS, 30% glycerol, 150 mM dithiothreitol, and 0.5 mg/ml bromophenol blue) before immunoblotting.
Immunoblotting
Equal amounts of protein were separated on 8%, 12%, or 4–12% Bis-Tris NuPAGE gels (Invitrogen) in NuPAGE MOPS SDS Running Buffer (Invitrogen) and transferred to polyvinylidene fluoride membrane in tris-glycine transfer buffer (12 mM tris, 96 mM glycine, and 20% methanol). Membranes were blocked in PBS containing 0.1% Tween 20 and 5% nonfat dry milk, incubated with primary antibodies overnight at 4°C, and then incubated with HRP-conjugated secondary antibodies for 1–2 h at room temperature. Bound antibody was visualized with ECL Prime Western Blotting Detection Reagent (GE Healthcare) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific). Images were acquired with a ChemiDoc XRS+ Imaging System and densitometric analysis was performed using Image Lab software (Bio-Rad Laboratories). Band intensities were normalized to GAPDH, actin, or histone H3 loading controls. For subcellular fractionated samples, relative TFEB expression was normalized to GAPDH for cytosol fractions and to histone H3 for nuclear fractions and nuclear TFEB was expressed as a percentage of total TFEB.
Antibodies
The following rabbit-derived antibodies were used in this work: anti–4E-BP1, anti–phospho-4E-BP1-(T37/46) (clones 53H11 and 236B4), anti-p70S6K (clone 49D7), anti–phospho-p70S6K-(T389) (clone 108D2), anti–phospho-ULK1-(S757), anti–phospho-AMPKα-(threonine 172) (clone 40H9), anti–acetyl-CoA carboxylase, anti–phospho-acetyl-CoA carboxylase-(S79), anti-Atg5 (clone D1G9), anti–histone H3 (clone D1H2), anti-LAMP1 (clone D2D11), anti-PINK1 (clone D8G3), anti–pan-14-3-3, anti–phospho-(Ser)-14-3-3 binding motif, and anti-TFEB (initially from E. Sidransky, National Institute of Neurological Disorders and Stroke, Bethesda, MD) from Cell Signaling Technology; anti-Atg9A (clone EPR5973) from Abcam; anti-GAPDH, anti-LC3B, and anti-TFE3 from Sigma-Aldrich; anti-TOM20 and anti-ULK1 from Santa Cruz Biotechnology, Inc.; rabbit anti-Mfn1 (custom-made by Covance; polyclonal antibody was raised against GST-tagged human Mfn1 recombinant protein [residues 667–741]); and rabbit anti–phospho-TFEB-(S142) (a gift from A. Ballabio, Baylor College of Medicine, Houston, TX, and Telethon Institute of Genetics and Medicine, Napoli, Italy; polyclonal antibody was raised against the AGNSAPN{pSer}PMAMLHIC peptide, coupled to KLH [Settembre et al., 2012]). The following mouse-derived antibodies were used in this work: anti-AMPKα1/α2 (clone 34.2) and anti-MTCO2 (clone 12C4F12) from Abcam; anti-actin (clone C4) from EMD Millipore; anti-DNA (clone AC-30-10) from Progen Biotechnik; anti-LAMP2 (clone H4B4) and anti-Parkin (clone PRK8) from Santa Cruz Biotechnology, Inc.; anti-SQSTM1 (p62) for Western blot (clone 2C11) from Abnova; anti-HA (clone 16B12) from Covance; and mouse anti-MITF (a gift from H. Arnheiter, National Institute of Neurological Disorders and Stroke; monoclonal antibody was raised against GST-tagged mouse MITF recombinant protein [residues 296–419]). Additional antibodies used in this work include guinea pig anti-p62 for ICC from Cedarlane; rabbit anti-GFP and goat-derived anti-rabbit, anti-mouse, and anti–guinea pig Alexa Fluor–conjugated secondary antibodies (Alexa Fluor 488, 594, 633) from Invitrogen; and HRP-conjugated sheep anti–mouse and donkey anti–rabbit IgG from GE Healthcare.
Flow cytometry analysis
Cells stably expressing YFP-Parkin and mt-mKeima were analyzed by flow cytometry based on previously described parameters (Katayama et al., 2011). In brief, treated cells were resuspended in sorting buffer (145 mM NaCl, 5 mM KCl, 1.8 mM CaCl2, 0.8 mM MgCl2, 10 mM Hepes, 10 mM glucose, and 0.1% BSA) containing 10 µg/ml DAPI and analyzed on a MoFlo Astrios cell sorter (Beckman Coulter) using Summit software (v6.2.6.16198). Dual-excitation ratiometric pH measurements were made using 488- (pH 7) and 561 (pH 4)-nm lasers with 620/29 nm and 614/20 nm emission filters, respectively. Either 50,000 or 100,000 events were initially collected for each sample and subsequently gated for YFP/mt-mKeima double-positive cells that were DAPI-negative. Data were analyzed using FlowJo (v10; Tree Star).
RNA interference
FlexiTube siRNA duplexes (QIAGEN) were used to achieve efficient knockdown of Fis1 (target sequence: 5′-TACAATGATGACATCCGTAAA-3′) and Rab7 (target sequence: 5′-CACGTAGGCCTTCAACACAAT-3′) genes. In brief, Lipofectamine RNAiMAX Reagent (Life Technologies) was incubated with RNAiDMEM-FBS media (High Glucose DMEM, 100 µM MEM nonessential amino acids [Gibco], 1 mM sodium pyruvate [Gibco], and 2 mM GlutaMAX [Gibco]) at room temperature for 10 min. This mixture was then incubated with either Fis1, Rab7, or AllStars negative control siRNA (QIAGEN) for 25 min before being added to 120,000 cells/well, seeded in an equal volume of RNAiDMEM+2×FBS media (RNAiDMEM-FBS supplemented with 20% FBS [BenchMark]). Treatment and downstream analysis was performed after 48 h.
RNA isolation and relative quantitative PCR
RNA was isolated from cells with the RNAeasy Mini Plus kit (QIAGEN) as recommended by the manufacturer. Purified mRNA (500 ng) was reverse transcribed in a 20-µl reaction to generate cDNA using the iSCRIPT Reverse transcription kit (Bio-Rad Laboratories) according to the manufacturer’s instructions. Relative quantitative PCR was performed in a total reaction volume of 10 µl containing 5 µl of Maxima SYBR green/ROX quantitative PCR Master Mix (Thermo Fisher Scientific), 2 µl cDNA, and 0.2 µM of gene-specific primer mix. Quantification of gene expression was performed in triplicate using a 7900HT Fast real-time PCR system and associated SDS software (Applied Biosystems). The thermal profile of the reaction was the following: 95°C for 10 min and 40 cycles of 95°C for 15 s followed by 60°C for 30 s and 72°C for 30 s. Amplification of the sequence of interest was normalized with a reference endogenous gene (GAPDH). Fold-change values were calculated using the ΔΔCt method and the ratio of drug-treated cells to control-treated cells was expressed. The RQ Manager software (v1.2.2; Applied Biosystems) and Excel (Microsoft Corp.) were used for data analysis. The following gene-specific primers were used: cathepsin B (forward, 5′-AGTGGAGAATGGCACACCCTA-3′; reverse, 5′-AAGAAGCCATTGTCACCCCA-3′), p62 (forward, 5′-AATCAGCTTCTGGTCCATCG-3′; reverse, 5′-TTCTTTTCCCTCCGTGCTC-3′), and GAPDH (forward, 5′-TGCACCACCAACTGCTTAGC-3′; reverse, 5′-GGCATGGACTGTGGTCATGAG-3′).
Statistical analysis
Error bars represent SD or range (for experiments with less than three replicates) as indicated in the figure legends. Data were processed in Excel and statistical analyses were performed using GraphPad Prism v6.0d (GraphPad Software). Statistical analysis of differences between two groups was performed using a two-tailed, unpaired t test and between more than two groups using a one-way analysis of variance test followed by a Tukey’s post-hoc comparison; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Only statistically significant comparisons are indicated in graphs.
Online supplemental material
Fig. S1 provides further validation of mitophagy-induced activation of TFEB and shows treatment control data. Fig. S2 shows Atg5 and Atg9A KO validation and experimental supporting data. Fig. S3 shows Parkin-dependent nuclear accumulation of endogenous and ectopic TFE3 and and MITF. Fig. S4 provides the CRISPR KO strategy for, and confirmation of, the generation of MiT/TFE KO cells. Fig. S5 shows the variable blockage of O/A-induced degradation of mitochondrial proteins or mitochondrial DNA in several MiT/TFE KO cell lines and provides representative FACS plots for mt-mKeima negative control experiments. The CRISPR and screening primers used to generate all KO cell lines are provided in Table S1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201501002/DC1.
Supplementary Material
Acknowledgments
We thank all Youle laboratory members for helpful discussion, S. Smith for assistance, the Neuroscience Light Imaging facility managed by C. Smith, L. Samsel at the National Heart, Lung, and Blood Institute Flow Cytometry Core, and D. Maric at the National Institute of Neurological Disorders and Stroke Flow Cytometry Core.
This work was supported by the National Institute of Neurological Disorders and Stroke intramural program.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AMPK
- AMP-activated protein kinase
- CIP
- calf intestinal phosphatase
- CRISPR
- clustered regularly interspaced short palindromic repeats
- CQ
- chloroquine
- DKO
- double knockout
- KO
- knockout
- LAMP
- lysosomal-associated membrane protein
- LC3
- light chain 3
- LSD
- lysosomal storage disorder
- MiT/TFE
- microphthalmia/transcription factor E
- mTORC1
- mammalian target of rapamycin complex 1
- MM
- molecular mass
- O/A
- oligomycin/antimycin A
- PD
- Parkinson’s disease
- TFEB
- transcription factor EB
- TKO
- triple knockout
- WT
- wild type
References
- Aitken A. 2011. Post-translational modification of 14-3-3 isoforms and regulation of cellular function. Semin. Cell Dev. Biol. 22:673–680. 10.1016/j.semcdb.2011.08.003 [DOI] [PubMed] [Google Scholar]
- Aksan I., and Goding C.R.. 1998. Targeting the microphthalmia basic helix-loop-helix–leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo. Mol. Cell. Biol. 18:6930–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., and Ktistakis N.T.. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701. 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E.J., Beal P.A., Keith C.T., Chen J., Shin T.B., and Schreiber S.L.. 1995. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature. 377:441–446. 10.1038/377441a0 [DOI] [PubMed] [Google Scholar]
- Brunet A., Kanai F., Stehn J., Xu J., Sarbassova D., Frangioni J.V., Dalal S.N., DeCaprio J.A., Greenberg M.E., and Yaffe M.B.. 2002. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 156:817–828. 10.1083/jcb.200112059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunn G.J., Hudson C.C., Sekulić A., Williams J.M., Hosoi H., Houghton P.J., Lawrence J.C. Jr., and Abraham R.T.. 1997. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 277:99–101. 10.1126/science.277.5322.99 [DOI] [PubMed] [Google Scholar]
- Burnett P.E., Barrow R.K., Cohen N.A., Snyder S.H., and Sabatini D.M.. 1998. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. USA. 95:1432–1437. 10.1073/pnas.95.4.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan N.C., Salazar A.M., Pham A.H., Sweredoski M.J., Kolawa N.J., Graham R.L.J., Hess S., and Chan D.C.. 2011. Broad activation of the ubiquitin–proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 20:1726–1737. 10.1093/hmg/ddr048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark I.E., Dodson M.W., Jiang C., Cao J.H., Huh J.R., Seol J.H., Yoo S.J., Hay B.A., and Guo M.. 2006. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 441:1162–1166. 10.1038/nature04779 [DOI] [PubMed] [Google Scholar]
- Decressac M., Mattsson B., Weikop P., Lundblad M., Jakobsson J., and Björklund A.. 2013. TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc. Natl. Acad. Sci. USA. 110:E1817–E1826. 10.1073/pnas.1305623110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison S.R., Wang F., Becker N.A., Schüle B., Kock N., Phillips L.A., Klein C., and Smith D.I.. 2003. Alterations in the common fragile site gene Parkin in ovarian and other cancers. Oncogene. 22:8370–8378. 10.1038/sj.onc.1207072 [DOI] [PubMed] [Google Scholar]
- Dennis P.B., Jaeschke A., Saitoh M., Fowler B., Kozma S.C., and Thomas G.. 2001. Mammalian TOR: a homeostatic ATP sensor. Science. 294:1102–1105. 10.1126/science.1063518 [DOI] [PubMed] [Google Scholar]
- DiMauro S., and Schon E.A.. 2008. Mitochondrial disorders in the nervous system. Annu. Rev. Neurosci. 31:91–123. 10.1146/annurev.neuro.30.051606.094302 [DOI] [PubMed] [Google Scholar]
- Ferron M., Settembre C., Shimazu J., Lacombe J., Kato S., Rawlings D.J., Ballabio A., and Karsenty G.. 2013. A RANKL–PKCβ–TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 27:955–969. 10.1101/gad.213827.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Itoh T., Omori H., Fukuda M., Noda T., and Yoshimori T.. 2008. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell. 19:2092–2100. 10.1091/mbc.E07-12-1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Huang X., Zhang P., Qi L., Liang Q., Zhang X., Huang J., Fang B., Hou W., Han J., and Zhang H.. 2014. Genome-wide screen identifies signaling pathways that regulate autophagy during Caenorhabditis elegans development. EMBO Rep. 15:705–713. 10.1002/embr.201338310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn D.M., Shackelford D.B., Egan D.F., Mihaylova M.M., Mery A., Vasquez D.S., Turk B.E., and Shaw R.J.. 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 30:214–226. 10.1016/j.molcel.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha J., Daniel S., Broyles S.S., and Kim K.H.. 1994. Critical phosphorylation sites for acetyl-CoA carboxylase activity. J. Biol. Chem. 269:22162–22168. [PubMed] [Google Scholar]
- Hanada T., Noda N.N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., and Ohsumi Y.. 2007. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282:37298–37302. 10.1074/jbc.C700195200 [DOI] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J.L., and Mizushima N.. 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181:497–510. 10.1083/jcb.200712064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson S.A., Fogel A.I., Wang C., MacArthur R., Guha R., Heman-Ackah S., Martin S., Youle R.J., and Inglese J.. 2015. Chemogenomic profiling of endogenous PARK2 expression using a genome-edited coincidence reporter. ACS Chem. Biol. 10:1188–1197. 10.1021/cb5010417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley S.A., Davison M., Woods A., Davies S.P., Beri R.K., Carling D., and Hardie D.G.. 1996. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J. Biol. Chem. 271:27879–27887. 10.1074/jbc.271.44.27879 [DOI] [PubMed] [Google Scholar]
- Hawley S.A., Gadalla A.E., Olsen G.S., and Hardie D.G.. 2002. The antidiabetic drug metformin activates the AMP-activated protein kinase cascade via an adenine nucleotide-independent mechanism. Diabetes. 51:2420–2425. 10.2337/diabetes.51.8.2420 [DOI] [PubMed] [Google Scholar]
- Hemesath T.J., Steingrímsson E., McGill G., Hansen M.J., Vaught J., Hodgkinson C.A., Arnheiter H., Copeland N.G., Jenkins N.A., and Fisher D.E.. 1994. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 8:2770–2780. 10.1101/gad.8.22.2770 [DOI] [PubMed] [Google Scholar]
- Holden P., and Horton W.A.. 2009. Crude subcellular fractionation of cultured mammalian cell lines. BMC Res. Notes. 2:243 10.1186/1756-0500-2-243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K., Zhu T., and Guan K.-L.. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 115:577–590. 10.1016/S0092-8674(03)00929-2 [DOI] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., Koyama-Honda I., and Mizushima N.. 2012. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J. Cell Sci. 125:1488–1499. 10.1242/jcs.094110 [DOI] [PubMed] [Google Scholar]
- Jin S.M., Lazarou M., Wang C., Kane L.A., Narendra D.P., and Youle R.J.. 2010. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 191:933–942. 10.1083/jcb.201008084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama H., Kogure T., Mizushima N., Yoshimori T., and Miyawaki A.. 2011. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem. Biol. 18:1042–1052. 10.1016/j.chembiol.2011.05.013 [DOI] [PubMed] [Google Scholar]
- Kim J., Kundu M., Viollet B., and Guan K.-L.. 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13:132–141. 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.G., Hoffman G.R., Poulogiannis G., Buel G.R., Jang Y.J., Lee K.W., Kim B.Y., Erikson R.L., Cantley L.C., Choo A.Y., and Blenis J.. 2013. Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Mol. Cell. 49:172–185. 10.1016/j.molcel.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi-Itakura C., Koyama-Honda I., Itakura E., and Mizushima N.. 2014. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J. Cell Sci. 127:4089–4102. 10.1242/jcs.156034 [DOI] [PubMed] [Google Scholar]
- Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., and Shimizu N.. 1998. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 392:605–608. 10.1038/33416 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., and Schulman B.A.. 2014. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat. Struct. Mol. Biol. 21:336–345. 10.1038/nsmb.2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper R.P., Schepens M., Thijssen J., Schoenmakers E.F.P.M., and van Kessel A.G.. 2004. Regulation of the MiTF/TFE bHLH-LZ transcription factors through restricted spatial expression and alternative splicing of functional domains. Nucleic Acids Res. 32:2315–2322. 10.1093/nar/gkh571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K.Y., Viollet B., and Yoo O.J.. 2011. CCCP induces autophagy in an AMPK-independent manner. Biochem. Biophys. Res. Commun. 416:343–348. 10.1016/j.bbrc.2011.11.038 [DOI] [PubMed] [Google Scholar]
- Lücking C.B., Dürr A., Bonifati V., Vaughan J., De Michele G., Gasser T., Harhangi B.S., Meco G., Denèfle P., Wood N.W., et al. 2000. Association between early-onset Parkinson’s disease and mutations in the parkin gene. N. Engl. J. Med. 342:1560–1567. 10.1056/NEJM200005253422103 [DOI] [PubMed] [Google Scholar]
- Mackintosh C. 2004. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem. J. 381:329–342. 10.1042/BJ20031332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., and Church G.M.. 2013. RNA-guided human genome engineering via Cas9. Science. 339:823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsin A.S., Bertrand L., Rider M.H., Deprez J., Beauloye C., Vincent M.F., Van den Berghe G., Carling D., and Hue L.. 2000. Phosphorylation and activation of heart PFK-2 by AMPK has a role in the stimulation of glycolysis during ischaemia. Curr. Biol. 10:1247–1255. 10.1016/S0960-9822(00)00742-9 [DOI] [PubMed] [Google Scholar]
- Martina J.A., and Puertollano R.. 2013. Rag GTPases mediate amino acid–dependent recruitment of TFEB and MITF to lysosomes. J. Cell Biol. 200:475–491. 10.1083/jcb.201209135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J.A., Chen Y., Gucek M., and Puertollano R.. 2012. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 8:903–914. 10.4161/auto.19653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J.A., Diab H.I., Li H., and Puertollano R.. 2014a Novel roles for the MiTF/TFE family of transcription factors in organelle biogenesis, nutrient sensing, and energy homeostasis. Cell. Mol. Life Sci. 71:2483–2497. 10.1007/s00018-014-1565-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J.A., Diab H.I., Lishu L., Jeong-A L., Patange S., Raben N., and Puertollano R.. 2014b The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal. 7:ra9 10.1126/scisignal.2004754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., et al. 2010. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189:211–221. 10.1083/jcb.200910140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina D.L., Fraldi A., Bouche V., Annunziata F., Mansueto G., Spampanato C., Puri C., Pignata A., Martina J.A., Sardiello M., et al. 2011. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell. 21:421–430. 10.1016/j.devcel.2011.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina D.L., Di Paola S., Peluso I., Armani A., De Stefani D., Venditti R., Montefusco S., Scotto-Rosato A., Prezioso C., Forrester A., et al. 2015. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 17:288–299. 10.1038/ncb3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner C., Lorenz H., Weihofen A., Selkoe D.J., and Lemberg M.K.. 2011. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J. Neurochem. 117:856–867. 10.1111/j.1471-4159.2011.07253.x [DOI] [PubMed] [Google Scholar]
- Mihaylova M.M., and Shaw R.J.. 2011. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 13:1016–1023. 10.1038/ncb2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., and Yoshimori T.. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152:657–668. 10.1083/jcb.152.4.657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D.F., and Youle R.J.. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183:795–803. 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., and Youle R.J.. 2010. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8:e1000298 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obsilová V., Silhan J., Boura E., Teisinger J., and Obsil T.. 2008. Physiol. Res. 57:S11–S21. [DOI] [PubMed] [Google Scholar]
- Palmieri M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M., and Ballabio A.. 2011. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 20:3852–3866. 10.1093/hmg/ddr306 [DOI] [PubMed] [Google Scholar]
- Park J., Lee S.B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J.-M., and Chung J.. 2006. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 441:1157–1161. 10.1038/nature04788 [DOI] [PubMed] [Google Scholar]
- Pawlyk A.C., Giasson B.I., Sampathu D.M., Perez F.A., Lim K.L., Dawson V.L., Dawson T.M., Palmiter R.D., Trojanowski J.Q., and Lee V.M.. 2003. Novel monoclonal antibodies demonstrate biochemical variation of brain parkin with age. J. Biol. Chem. 278:48120–48128. 10.1074/jbc.M306889200 [DOI] [PubMed] [Google Scholar]
- Peña-Llopis S., Vega-Rubin-de-Celis S., Schwartz J.C., Wolff N.C., Tran T.A.T., Zou L., Xie X.-J., Corey D.R., and Brugarolas J.. 2011. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 30:3242–3258. 10.1038/emboj.2011.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polito V.A., Li H., Martini-Stoica H., Wang B., Yang L., Xu Y., Swartzlander D.B., Palmieri M., di Ronza A., Lee V.M., et al. 2014. Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol. Med. 6:1142–1160. 10.15252/emmm.201303671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roczniak-Ferguson A., Petit C.S., Froehlich F., Qian S., Ky J., Angarola B., Walther T.C., and Ferguson S.M.. 2012. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5:ra42 10.1126/scisignal.2002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., and Sabatini D.M.. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 320:1496–1501. 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., et al. 2009. A gene network regulating lysosomal biogenesis and function. Science. 325:473–477. [DOI] [PubMed] [Google Scholar]
- Sarraf S.A., Raman M., Guarani-Pereira V., Sowa M.E., Huttlin E.L., Gygi S.P., and Harper J.W.. 2013. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 496:372–376. 10.1038/nature12043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science. 332:1429–1433. 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., et al. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31:1095–1108. 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidransky E., Nalls M.A., Aasly J.O., Aharon-Peretz J., Annesi G., Barbosa E.R., Bar-Shira A., Berg D., Bras J., Brice A., et al. 2009. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N. Engl. J. Med. 361:1651–1661. 10.1056/NEJMoa0901281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Wang F., Savini M., Ake A., di Ronza A., Sardiello M., and Segatori L.. 2013. TFEB regulates lysosomal proteostasis. Hum. Mol. Genet. 22:1994–2009. 10.1093/hmg/ddt052 [DOI] [PubMed] [Google Scholar]
- Spampanato C., Feeney E., Li L., Cardone M., Lim J.A., Annunziata F., Zare H., Polishchuk R., Puertollano R., Parenti G., et al. 2013. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol. Med. 5:691–706. 10.1002/emmm.201202176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E., Tessarollo L., Pathak B., Hou L., Arnheiter H., Copeland N.G., and Jenkins N.A.. 2002. Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc. Natl. Acad. Sci. USA. 99:4477–4482. 10.1073/pnas.072071099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A., Cleland M.M., Xu S., Narendra D.P., Suen D.F., Karbowski M., and Youle R.J.. 2010. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 191:1367–1380. 10.1083/jcb.201007013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemi T., Ashe T.D., Morrison B.E., Soriano K.R., Au J., Roque R.A.V., Lazarowski E.R., Damian V.A., Masliah E., and La Spada A.R.. 2012. PGC-1α rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci. Transl. Med. 4:142ra97 10.1126/scitranslmed.3003799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente E.M., Salvi S., Ialongo T., Marongiu R., Elia A.E., Caputo V., Romito L., Albanese A., Dallapiccola B., and Bentivoglio A.R.. 2004. PINK1 mutations are associated with sporadic early-onset parkinsonism. Ann. Neurol. 56:336–341. 10.1002/ana.20256 [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., et al. 2010. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA. 107:378–383. 10.1073/pnas.0911187107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witters L.A., Nordlund A.C., and Marshall L.. 1991. Regulation of intracellular acetyl-CoA carboxylase by ATP depletors mimics the action of the 5′-AMP-activated protein kinase. Biochem. Biophys. Res. Commun. 181:1486–1492. 10.1016/0006-291X(91)92107-U [DOI] [PubMed] [Google Scholar]
- Yamano K., and Youle R.J.. 2013. PINK1 is degraded through the N-end rule pathway. Autophagy. 9:1758–1769. 10.4161/auto.24633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K., Fogel A.I., Wang C., van der Bliek A.M., and Youle R.J.. 2014. Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. eLife. 3:e01612 10.7554/eLife.01612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Gehrke S., Imai Y., Huang Z., Ouyang Y., Wang J.-W., Yang L., Beal M.F., Vogel H., and Lu B.. 2006. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl. Acad. Sci. USA. 103:10793–10798. 10.1073/pnas.0602493103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii S.R., Kishi C., Ishihara N., and Mizushima N.. 2011. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J. Biol. Chem. 286:19630–19640. 10.1074/jbc.M110.209338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Tan S.H., Nicolas V., Bauvy C., Yang N.D., Zhang J., Xue Y., Codogno P., and Shen H.M.. 2013. Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion. Cell Res. 23:508–523. 10.1038/cr.2013.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.