Abstract
The role of grape berry skin as a protective barrier against damage by physical injuries and pathogen attacks requires a metabolism able to sustain biosynthetic activities such as those relating to secondary compounds (i.e., flavonoids). In order to draw the attention on these biochemical processes, a proteomic and metabolomic comparative analysis was performed among Riesling Italico, Pinot Gris, Pinot Noir, and Croatina cultivars, which are known to accumulate anthocyanins to a different extent. The application of multivariate statistics on the dataset pointed out that the cultivars were distinguishable from each other and the order in which they were grouped mainly reflected their relative anthocyanin contents. Sorting the spots according to their significance 100 proteins were characterized by LC-ESI-MS/MS. Through GC-MS, performed in Selected Ion Monitoring (SIM) mode, 57 primary metabolites were analyzed and the differences in abundance of 16 of them resulted statistically significant to ANOVA test. Considering the functional distribution, the identified proteins were involved in many physiological processes such as stress, defense, carbon metabolism, energy conversion and secondary metabolism. The trends of some metabolites were related to those of the protein data. Taken together, the results permitted to highlight the relationships between the secondary compound pathways and the main metabolism (e.g., glycolysis and TCA cycle). Moreover, the trend of accumulation of many proteins involved in stress responses, reinforced the idea that they could play a role in the cultivar specific developmental plan.
Keywords: anthocyanins, exocarp grape berry, metabolomics, proteomics, stress response, Vitis vinifera
Introduction
Grape berry of the perennial and deciduous woody vines of the genus Vitis is one of the economically most important fruit crop in the world. As recounted in the 2012 report of Food and Agriculture Organization (FAO, http://faostat.fao.org/site/567/DesktopDefault.aspx?PageID=567#ancor), 69.7694 hectares are dedicated to the cultivation of grapevine (Vitis vinifera L), with an estimation of a production of about 67 million tons per year.
During recent years, there was a burst of genomic information about the development and ripening in grape berries. After the first pioneer gene-specific molecular approaches, the appearance of large collections of grapevine ESTs (Ablett et al., 2000) and the construction of grapevine nucleotide microarrays (Terrier et al., 2005; Waters et al., 2005) opened new horizons in the study of grape berry ripening (Grimplet et al., 2007; Zamboni et al., 2010; Fortes et al., 2011). Moreover, the work of grape genome sequencing by Jaillon et al. (2007) has contributed to provide the necessary genomic information (on 2th July 2015, 94,556 protein sequences were available in the NCBI database, http://www.ncbi.nlm.nih.gov/sites/entrezz). These studies paved the way for investigating berry proteome (Deytieux et al., 2007; Giribaldi et al., 2007; Negri et al., 2008a,b, 2011; Grimplet et al., 2009; Giribaldi and Giuffrida, 2010; Zamboni et al., 2010; Martínez-Esteso et al., 2011; Niu et al., 2013; Fraige et al., 2015).
Grape is a non-climacteric fruit formed by three major tissues: exocarp (i.e., skin), mesocarp (pulp) and endocarp which surrounds seeds. Exocarp represents a physical barrier between the external environment and the inner tissues, protecting them by physical damage and pathogen attack (Grimplet et al., 2007). It is metabolically active during all developing phases being the site of the synthesis of exclusive compounds, such as aroma and some phenolic classes. Aromas arise from volatile molecules, such as terpenes, norisoprenoids and thiols that are usually stored as amino acid and sugar conjugates in the vacuole (Lund and Bohlman, 2006). Among phenolic compounds synthetized in the exocarp cells there are anthocyanins. Their color promotes seed dispersal thanks to the high contrast between background foliage and fruits (Burns and Dalen, 2002). Moreover, these compounds are involved in the protection from UV light exposure (Solovchenko and Schmitz-Eiberger, 2003). The biosynthesis of these compounds begins at véraison and continues throughout the ripening phase. The levels of anthocyanins are influenced by many factors, such as genetic background, pedo-climatic conditions and vineyard management while their profiles are relatively stable for each variety (Mattivi et al., 2006; Castellarin et al., 2012; Zheng et al., 2013). The studies on the regulation of anthyocyanin pathway revealed that the synthesis of these compounds requires the UDP-glucose flavonoid glycosyl transferase (UFGT) expression. Northern blot analysis conducted on the exocarp of a range of white and red cultivars showed, in fact, that the transcript of this enzyme was detectable only in colored grapes (Boss et al., 1996a,b). Moreover, by the isolation of several myb-related genes from berries, it was shown that the lack of expression of VvmybA1 in white cultivars results from the insertion of a retrotransposon in its promoter region absent in red cultivars, thus suggesting that the expression of this transcription factor may be the trigger of color set in grape (Kobayashi et al., 2004). Nevertheless, Ageorges et al. (2006) found that in addition to UFGT, at least three other isogenes related to the anthocyanin pathway, such as chalcone synthase 3 (CHS3) and the downstream elements glutathione S-transferase (GST) and caffeoyl methyl transferase (CaoMT) can be clearly associated to color in grape berries.
In the last years, studies performed on separate tissues pointed out several peculiar traits in gene expression, according to their functional role. As showed by the comparison among the main mature berry tissues of cultivar Cabernet Sauvignon, through the use of Affymetrix GeneChip® technology, more than 28% of the genes with significant differential expression showed differences in a particular tissue (Grimplet et al., 2007). The most expressed transcripts in the exocarp were those relating to secondary metabolism, amino acid and lipid metabolism. Moreover, some transcripts that related to some enzymes involved in carbon metabolism were over-represented in this tissue as well as some pathogenesis-related proteins were more abundant in the exocarp at the maturity (Deytieux et al., 2007; Grimplet et al., 2007). A further proteomic investigation performed on exocarp of cultivar Barbera showed that during the final ripening stage an increase in abundance of enzymes involved in primary metabolism, such as the glycolytic pathway, occurred (Negri et al., 2008b).
As expected, some -omic studies revealed that the environmental conditions, such as water availability or sunlight exclusion, deeply affected the metabolism of skin tissue (Grimplet et al., 2007, 2009; Niu et al., 2013; Zheng et al., 2013). Furthermore, a study, in which metabolite and transcript profiling of berry skin of Cabernet Sauvignon and Shiraz were compared, found that there were peculiar metabolic differences between the two cultivars (Degu et al., 2014).
The aim of the present work was to investigate possible relationships among the main biochemical pathways characterizing grape berry ripening and anthocyanin accumulation. For this purpose, four grape cultivars with increasing anthocyanin contents [Riesling Italico (synonymous: Welschriesling) (R), Pinot Gris (PG), Pinot Noir (PN), and Croatina (C)] were compared at the mature berry stage. To mitigate the seasonal effects, the study was conducted analyzing samples harvested in two different years and was performed combining a proteomic analysis (i.e., 2-DE gels/ LC-ESI-MS/MS) with a metabolomic one (i.e., GC-MS). Using multivariate statistical analysis (i.e., FS-LDA) on spot volume dataset, it was possible to discriminate the differences among the four cultivars and to sort the matches according to their discriminating power.
Through the integration of proteomic and metabolomic analyses, this work provided new insights on the ripening process in the skin and on the grape heterogeneity. The results clearly showed that the process of grape ripening in the skin differs among cultivars in some central metabolic traits. These variations appeared to be linked to the different trends of accumulation of secondary metabolites, but they appeared also related to the ripening plan.
Materials and methods
Plant material
Exocarps were harvested during the 2005 and 2006 seasons from Vitis vinifera L. cv. R, PG, PN, and C RS38 berries (full-ripe berries according to modified E–L system, Coombe, 1995). The plants were grown at the Experimental Station of the “Ente Regionale per i Servizi all'Agricoltura e alle Foreste” (ERSAF) of Regione Lombardia (Montebello della Battaglia, PV, Italy).
About 50 g of isolated skins of at least five different randomly chosen plants of one cultivar were detached immediately by squishing the berries in order to remove the seeds and the bulk of the mesocarp. By pressing and smearing the inner part of the skin on two layers of cheesecloth the residual pulp was completely taken away. This operation was repeated three times and each pool, representing a biological replicate (4 cultivars × 2 years × 3 replicas = 24 pools), was frozen in liquid nitrogen, ground and stored at −80°C until use. From each powder pool both proteomic (5 g/extraction) and metabolomic (150 g/extraction) technical replicates were obtained.
Evaluation of anthocyanin contents
Anthocyanins were extracted and measured as previously described by Fumagalli et al. (2006) and Negri et al. (2008b), respectively.
Protein extraction and two-dimensional polyacrylamide gel electrophoresis analysis
Protein fraction was extracted using the method previously described by Negri et al. (2008b) with two modifications. In detail: (i) the cold acetone powder was firstly resuspended in phenol and then incubated for 30 min at 4°C, afterwards an equal volume of extraction buffer was added to proceed with repartition step (Hurkman and Tanaka, 1986); (ii) proteins were resuspended in the IEF pH 4–7 buffer (GE Healthcare).
Protein concentration was determined by 2-D Quant Kit (GE Healthcare). Five-hundred μg of protein sample was used for each 2-DE analysis that was performed using the pH 4–7, 24 cm IPG strips (GE Healthcare) as previously described in Negri et al. (2008b).
The gels were stained according to colloidal Coomassie Brilliant Blue G-250 (cCBB) procedure, (Neuhoff et al., 1988). The gels were then scanned by an Epson Expression 1680 Pro Scanner. For each of the 6 biological replicates of a sample (3 per year), two 2-DE gels were obtained (n = 12).
Gel and proteomic data analysis
Gels were analyzed with ImageMaster 2-D Platinum Software v. 6.0 (GE Healthcare) matching the gels using the landmark-assisted procedure with one of the 48 gels as reference. Since the relevant number of gels and the possibility of spreading the matching errors, an incremental checking method was set up. Briefly, a sub-reference gel was chosen for the group of maps relating to the skins of the same cultivar and year. The spots of the sub-reference gel were ordered according to their decreasing %Vol and only the matching of the 1000 largest spots were checked. The procedure was repeated for all the 8 distinct samples taking into account only their relative 6 gels. In the second step, the analysis comprised the 12 maps of a single cultivar obtained from the skins of both years. Also in this case, only the matches of the 1000 largest spots of the sub-reference gels were evaluated. Similarly, the gels of the different pairs of cultivars were compared. Only in the final step the check was performed considering all the 48 maps. In addition to the cutting off spots of really low abundance, matches grouping less than 3 spots in a sample were discarded. Through this procedure it was possible to eliminate a relevant part of noise in the dataset and to rescue significant matches that are often lost during automatic matching.
The molecular weight and pI of the spots were estimated as previously described by Negri et al. (2008b).
The differences among the four cultivars were assessed analyzing the %Vol dataset through the application of Principal Component Analysis (PCA) and Forward Stepwise Linear Discriminant Analysis (FS-LDA) on the first 10 PCA scores as previously described by Negri et al. (2011).
Significant differences relative to identified proteins were analyzed through the one-way hierarchical clustering methodology using the software PermutMatrix (Caraux and Pinloche, 2005; Meunier et al., 2007). The 2-DE data were converted into a binary matrix replacing the missing values by zero. The row by row normalization of data was performed using the classical zero-mean and unit-standard deviation technique. Pearson's distance and Ward's algorithm were used for the analysis.
Protein in-gel digestion and LC-ESI-MS/MS analysis
Spots excised from gels stained with cCBB were digested as described by Prinsi et al. (2011).
The LC-ESI-MS/MS experiments were conducted using a Surveyor (MS pump Plus) HPLC system directly connected to the ESI source of a Finnigan LCQ DECA XP MAX ion trap mass spectrometer (ThermoFisher Scientific Inc). Chromatography separations were performed by using an Inerstil WP300 C18 column (200 μm I.D × 150 mm length, 5 μm particle size) and a gradient from 5% to 80% solvent B [solvent A: 0.1% (v/v) formic acid; solvent B: ACN containing 0.1% (v/v) formic acid] as described by Niessen and co-workers (Niessen et al., 2006), with a flow of 2 μl min−1. ESI was performed in positive ionization mode with the following parameters: (i) spray voltage: 2.5 kV (ii) capillary temperature: 220°C. Data were collected in the full-scan and data dependent MS/MS mode with collision energy of 35% and a dynamic exclusion window of 3 min.
Protein identification was performed by Spectrum Mill MS Proteomics Workbench (Rev B.04.00.127; Agilent Technologies). Cysteine carbamidomethylation and methionine oxidation were set as fixed and variable modifications, respectively, accepting two missed cleavages per peptide. The search was conducted against the subset of Vitis protein sequences (ID 3603; June 2015, 94558 entries) downloaded from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and concatenated with the reverse one. The threshold used for peptide identification was Spectrum Mill score >10, Score Peak Intensity ≥70%, mass tolerance of ±2 Da for parent ion and ±1 Da fragment ions, and Database Fwd-Rev Score ≥2. Physical properties of the proteins were predicted by in silico tools at ExPASy (http://web.expasy.org/compute_pi/).
The identified proteins were sorted in metabolic functional classes according to the MapMan BIN ontology. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (Vizcaíno et al., 2014) via the PRIDE partner repository with the dataset identifier PXD002539.
Metabolite extraction and derivatization
In order to extract the metabolites of the polar fraction, the protocol by Lisec et al. (2006) was used with some modifications. One hundred-fifty milligrams (150 mg) of the frozen powder relative to the 6 biological replicates (3 per year) were resuspended in 1.4 ml of −20°C cooled methanol and 60 μl of 2 mg ml−1 ribitol were added as internal standard. The samples were incubated at 70°C for 15 min in continuous agitation (1200 rpm) with a thermo mixer and subsequently centrifuged for 10 min at 11,000 g at room temperature. After recovering the surnatant, 750 μl of chloroform and 1.5 ml of distilled water were added. The samples were vortexed and then centrifuged for 15 min at 2200 g at 4°C in order to separate the phases with different polarity. Aliquots of 150 μl of the water/methanol supernatant were then transferred to a clean eppendorf tube and dried on a Speedvac (RVC 2–18 CDplus, CHRIST) without heating for 16 h. The dried residues were redissolved in 40 μl of methoxyamination reagent (20 mg ml−1 of metoxyamine hydrochloride in pyridine) at 30°C and 700 rpm for 2 h and derivatized in 60 μl of N-methyl-N-(trimethylsilyl)-trifluoroacetamide (MSTFA) in the same conditions for 6 h. A retention time standard (10 μl), obtained diluting a saturated C7–C40 Alkane Mixture (Supelco, 1000 μg/mL for each component) 1:20 in MSTFA, was added to each sample. Before further analysis, the samples were transferred in glass vials. For each biological replicate, the analysis was repeated twice.
The employed standard substances were dissolved in distilled water or methanol at the concentration of 10 mg ml−1 and 1 μl was dried in vacuum and derivatized as described above to get spectral information.
GC-MS analysis and data processing
Metabolite quantification was conducted using the instrument GC-MSD comprising the gas chromatograph 7890 and the single-quadrupole spectrometer 5975 (Agilent Technologies). The employed method was the one defined by Golm Metabolome Database (http://csbdb.mpimp-golm.mpg.de/cgi-bin/madb2ml.cgi?org=msri&c=ml&o=ht&typ=met&inp=m%5B2%5D) and here briefly summarized. One microliter of sample was injected at 230°C in splitless mode through the autosampler CTC PAL (CTC Analytics AG). The analysis was conducted using a 30 m × 0.25 mm ID × 0.25 μm film thickness DB-5 column (Agilent Technologies). Helium BIP™ (Sapio) was used as carrier gas with a constant flux of 1 ml min−1. The oven ramp was so set up: 1 min at 70°C, 6 min ramp to 76°C, 45 min ramp to 350°C, 1 min at 350°C, 10 min at 330°C. The spectra and the retention times of the standard substances were acquired in a m/z range between 40 and 600. Metabolite analysis was performed in SIM mode following a maximum of 7 ions per time-subgroup and setting a dwell time of 20 ms. The MS source and quad were maintained at 230°C and 150°C, respectively, using the ionization for electronic impact at −70 eV.
Spectral integration was carried out through the software MetaQuant 1.3 (Bunk et al., 2006).
Integrated peaks were normalized by the peak area of the ion with m/z = 219 of ribitol. Data were Box-Cox transformed and the differences were evaluated though ANOVA with p≤0.05 using the software STATISTICA v. 7.1 (StatSoft Inc., Tulsa, OK, USA).
Results
Anthocyanin contents
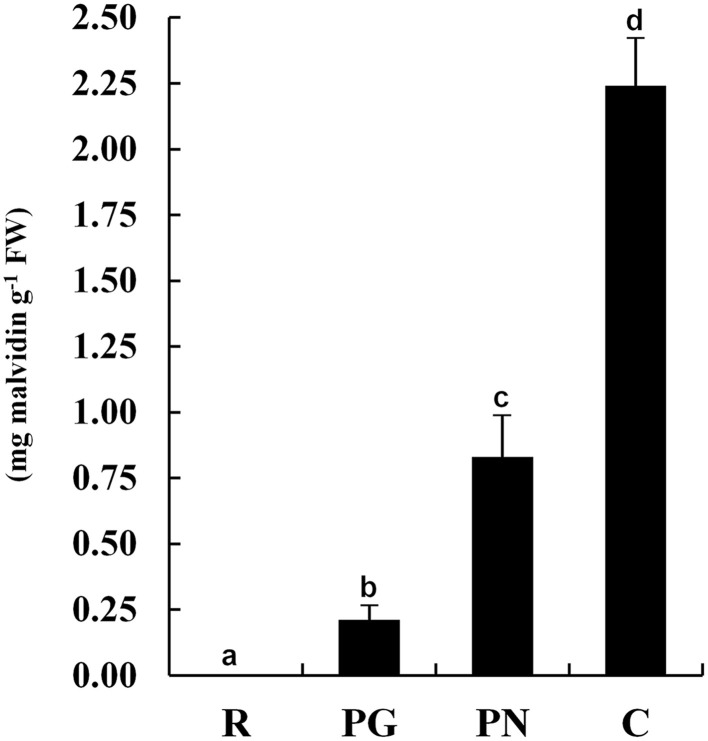
The anthocyanin content of the exocarp berry of four cultivars (Figure 1) moved from the undetectable levels of R to the high levels of C (2.25 mg g−1) passing through the pale PG (0.21 mg g−1) and PN (0.83 mg g−1).
Figure 1.
Anthocyanin contents in grape berry exocarp of Riesling Italico, Pinot Gris, Pinot Noir, and Croatina. R, Riesling Italico; PG, Pinot Gris; PN, Pinot Noir; C, Croatina. Data are the means ± ES, n = 6. Samples indicated with the different letters significantly differ according to Tukey's test (p < 0.01).
2-DE analysis
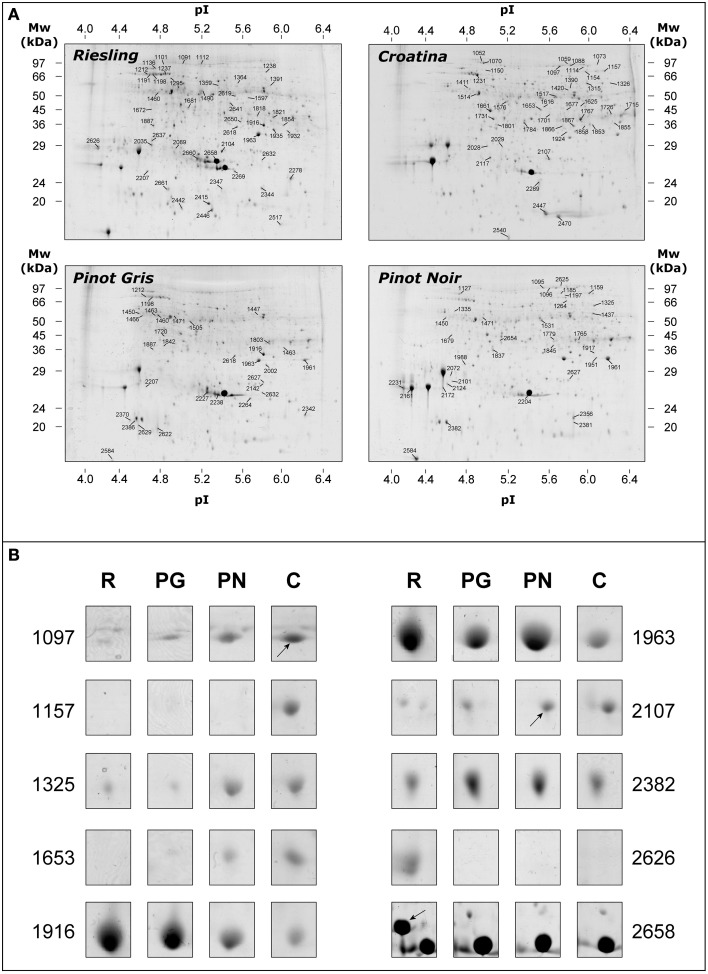
The proteomic analysis was achieved comparing 2-DE gels relating to exocarp of the four cultivars harvested in 2005 and 2006 vintages (n = 12). The number of the detected spots were comparable in the four samples and resulted to be of about 1300 spots per gels. Figure 2A shows the representative 2-DE maps of total protein fraction from exocarp berries of R, PG, PN, and C. Although the gel analysis pointed out a similar pattern, some differences in spot abundance were detected among the four cultivars. After automatic matching and filtering, the correspondence among the spots was assessed by manual checking. We thus focused our attention on the resulting 732 matches. In Figure 2B, some spots that resulted to be present with different abundance in the analyzed genotypes were reported.
Figure 2.
Representative 2-DE maps of total protein fraction from grape berry exocarp of Riesling Italico, Pinot Gris, Pinot Noir, and Croatina. (A) whole maps of the four experimental cultivars. (B) the image of 10 spots that resulted to be significantly different in abundance among the cultivars. Proteins (500 μg) were separated at pH 4–7, followed by 12.5% SDS-PAGE and visualized by cCBB-staining.
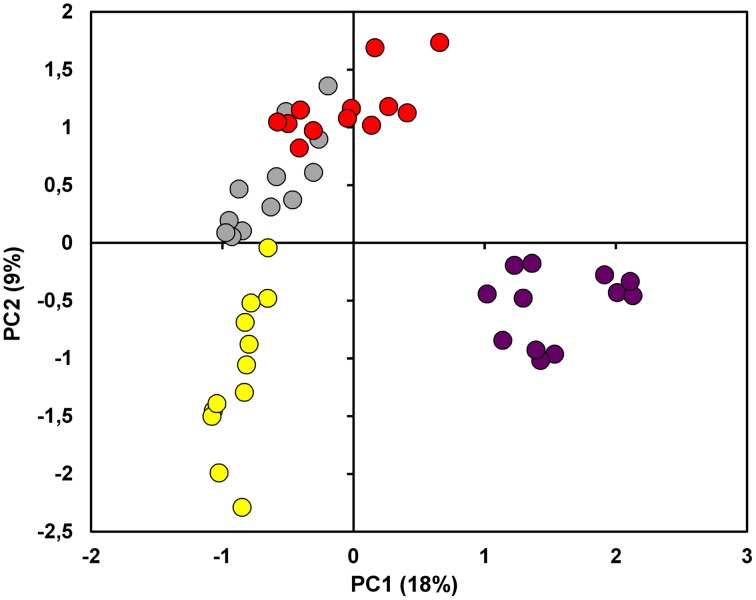
Through PCA it was possible to provide a first description of the relationship among the proteomes. PC1 accounted for the 18% of explained variance and showed a tendency to move apart the different cultivars. PC2 (9% of explained variance), on the other hand, put PG and PN at positive values while R and C were placed in the 3rd and 4th quadrant, respectively (Figure 3 and Supplementary Table S1). Overall, the score plot of the PCA clearly isolated C from the other 3 cultivars, while it showed the great affinity between PG and PN.
Figure 3.
Principal Component Analysis (PCA). The score plot showed in the figure was performed on the overall dataset considering the first two PCs. The samples under investigation are coded by a twelve-circle symbol colored in yellow, gray, red and violet for Riesling Italico, Pinot Gris, Pinot noir, and Croatina, respectively.
In order to distinguish the cultivars and to isolate the spots that were responsible of the observed differences, a linear discriminant analysis (LDA) was applied on the first 10 PCs calculated. The analysis was conducted by means of the forward stepwise (FS) algorithm (Fto_Enter = 13) that selected only 3 PCs (PC1, PC2, PC3) for the classification.
The evaluation of matrix classification showed that the 12 gels of each cultivar were correctly attributed to the class of their belonging (Table 1). Principal components and spot volumes are linked by a linear combination: this evidence permits to calculate each discrimination model according to the spots present on the gels. After the calculation of the 4 discrimination models, it was possible to order the spots in terms of their coefficients that is according of the weight by which they contribute in classifying the samples (Table 2). For each of the four cultivars the 30 most variables showing largest positive and negative coefficients were considered for identification. Since some of the selected spots overlapped among the four discrimination models the potential number of analyzed spots (n = 240) decrease to 144.
Table 1.
Classification matrix of Forward Stepwise—Linear Discriminant Analysis (FS-LDA).
| Percent | Riesling | Pinot gris | Pinot noir | Croatina | |
|---|---|---|---|---|---|
| Riesling | 100.0 | 12 | 0 | 0 | 0 |
| Pinot gris | 100.0 | 0 | 12 | 0 | 0 |
| Pinot noir | 100.0 | 0 | 0 | 12 | 0 |
| Croatina | 100.0 | 0 | 0 | 0 | 12 |
| Total | 100.0 | 12 | 12 | 12 | 12 |
The analyses was performed on the principal components (PCs) calculated, allowing the discrimination of the classes of samples and sorting the variables according to their relevance.
Table 2.
Matrix of the classification functions of Forward Stepwise—Linear Discriminant Analysis (FS-LDA).
| Riesling | Pinot gris | Pinot noir | Croatina | |
|---|---|---|---|---|
| “PC1” | −38,5467 | −11,8858 | 4,32349 | 46,1091 |
| “PC2” | −20,5383 | 5,1528 | 13,11723 | 2,2683 |
| “PC3” | −19,0547 | −1,0812 | 5,84695 | 14,2889 |
| Constant | −37,2914 | −6,1952 | −9,12430 | −38,4861 |
Squared Mahalanobis distances from group centroids were obtained from PC1, PC2, and PC3.
Protein identification and functional distribution
One hundred spots were identified by LC-ESI-MS/MS with a high degree of confidence among the group of 144 selected from FS-LDA (Table 3 and Supplementary Table S3). The missed identifications referred both to spots present in lower abundance that were not pickable from the gels (14% of the spots) and to the excised spots for which the analysis gave results under the set level of significance (19% of analyzed spots). Some spots, such as ACO (spots 1095, 1096, 1097, and 1114), NADP-ME (spots 1315 and 1325), ENO (spots 1505, 1531, and 2619), ENO-1 (spots 1471 and 1517), IFL-5 (spots 1924, 1935, and 2618), MET, (spots 1154, 1157, and 1159), VHA-B2-X1 (spots 1411 and 1450), PPO (spots 1821 and 2386) and many spots classified in stress group (HSP70-2, Thaumatin-like protein, TLP, PRP-10, and PRP-4) were identified as the same protein, indicating the presence of different forms with peculiar pI and/or Mr. Nevertheless, different isoforms of some proteins were found (i.e., ENO and IFR).
Table 3.
List of spots identified by LC-ESI-MS/MS.
| Spot ID | Accession number | Species | Protein description | Abbreviation | Mra / pI a | Mrb / pI b | Cov (%)c |
|---|---|---|---|---|---|---|---|
| PHOTOSYNTHESIS (FIGURE 5A) | |||||||
| 1238 | XP_002266494.2 | Vitis vinifera | Transketolase, chloroplastic | TK | 68.8 / 5.9 | 78.8/ 6.6 | 10 |
| 1988 | XP_002274796.1 | Vitis vinifera | Oxygen-evolving enhancer protein 1, chloroplastic | OEE-1 | 30.4 / 4.8 | 35.3 / 6.1 | 19 |
| C-COMPOUND/CARBOHYDRATE/ENERGY METABOLISM (FIGURE 5B) | |||||||
| 1095 | XP_002278138.1 | Vitis vinifera | Aconitate hydratase, cytoplasmic | ACO | 95.1 / 5.6 | 110.1 / 6.7 | 12 |
| 1096 | XP_002278138.1 | Vitis vinifera | Aconitate hydratase, cytoplasmic | ACO | 95.1 / 5.7 | 110.1 / 6.7 | 17 |
| 1097 | XP_002278138.1 | Vitis vinifera | Aconitate hydratase, cytoplasmic | ACO | 94.3 / 5.7 | 110.1 / 6.7 | 26 |
| 1114 | XP_002263337.1 | Vitis vinifera | Aconitate hydratase 1 | ACO-1 | 91.5 / 5.9 | 98.2 / 6.0 | 8 |
| 1197 | XP_002270157.1 | Vitis vinifera | NADH dehydrogenase [ubiquinone] iron-sulfur protein1, mitochondrial | NDUFS-1 | 74.2 / 5.8 | 80.9 / 6.5 | 14 |
| 1315 | P51615.1 | Vitis vinifera | NADP-dependent malic enzyme (NADP-ME) | NADP-ME | 62.2 / 6.0 | 65.2 / 6.1 | 18 |
| 1325 | P51615.1 | Vitis vinifera | NADP-dependent malic enzyme (NADP-ME) | NADP-ME | 61.8 / 6.1 | 65.2 / 6.1 | 27 |
| 1326 | XP_002284729.1 | Vitis vinifera | Phosphoglucomutase, cytoplasmic | PGMc | 61.8 / 6.2 | 63.6 / 5.8 | 15 |
| 1437 | XP_002263180.1 | Vitis vinifera | Dihydrolipoyl dehydrogenase, mitochondrial | DLD | 55.0 / 6.1 | 52.9 / 6.7 | 29 |
| 1460 | XP_002283951.1 | Vitis vinifera | ATP synthase subunit beta, mitochondrial | ATPase-β | 53.5 / 4.8 | 59.6 / 5.8 | 32 |
| 1471 | XP_002283632.1 | Vitis vinifera | Enolase 1 | ENO-1 | 53.5 / 5.0 | 47.9 / 5.7 | 18 |
| 1505 | XP_002267091.2 | Vitis vinifera | Enolase | ENO | 52.1 / 5.1 | 55.8 / 8.4 | 36 |
| 1517 | XP_002283632.1 | Vitis vinifera | Enolase 1 | ENO-1 | 51.5 / 5.7 | 47.9 / 7.0 | 15 |
| 1531 | XP_002267091.2 | Vitis vinifera | Enolase | ENO | 50.9 / 5.6 | 55.8 / 8.4 | 37 |
| 1576 | XP_002280514.1 | Vitis vinifera | ATP-citrate synthase alpha chain protein 2 | ACLA-2 | 48.6 / 5.2 | 46.4 / 5.3 | 9 |
| 1715 | XP_002278444.1 | Vitis vinifera | Formate dehydrogenase, mitochondrial | FDH | 40.1 / 6.3 | 42.0 / 6.9 | 6 |
| 1726 | XP_002278444.1 | Vitis vinifera | Formate dehydrogenase, mitochondrial | FDH | 39.4 / 6.2 | 42.0 / 6.9 | 10 |
| 1767 | XP_002263950.1 | Vitis vinifera | Phosphoglycerate kinase, cytosolic | PGK | 36.6 / 5.9 | 42.4 / 6.3 | 52 |
| 1779 | XP_002283381.1 | Vitis vinifera | Fructose-bisphosphate aldolase cytoplasmic isozyme | FbPA | 37.2 / 5.9 | 38.6 / 8.0 | 7 |
| 1951 | XP_002277446.1 | Vitis vinifera | Glucan endo-1,3-beta-glucosidase | Glu-β-EnGluco | 31.6 / 6.1 | 36.7 / 8.4 | 25 |
| 1961 | XP_002277446.1 | Vitis vinifera | Glucan endo-1,3-beta-glucosidase | Glu-β-EnGluco | 31.2 / 6.2 | 36.7 / 8.4 | 45 |
| 1963 | XP_002277446.1 | Vitis vinifera | Glucan endo-1,3-beta-glucosidase | Glu-β-EnGluco | 31.2 / 5.8 | 36.7 / 8.4 | 25 |
| 2619 | XP_002267091.2 | Vitis vinifera | Enolase | ENO | 50.5 / 5.6 | 55.8 / 5.4 | 28 |
| CELL WALL (FIGURE 5C) | |||||||
| 1765 | XP_002263490.1 | Vitis vinifera | UDP-arabinopyranose mutase 1 | UDP-Arab-M1 | 36.5 / 5.7 | 40.8/ 6.2 | 12 |
| 2002 | BAB78506.1 | Vitis labrusca x Vitis vinifera | Xyloglucan endo-transglycosylase | XTHs | 30.1 / 5.9 | 32.7 / 5.5 | 7 |
| NITROGEN METABOLISM/AMINO ACID METABOLISM (FIGURE 5D) | |||||||
| 1154 | XP_002276438.1 | Vitis vinifera | 5-methyltetrahydropteroyltriglutamate–homocysteine methyltransferase | MET | 80.8 / 6.0 | 85.0/ 6.1 | 9 |
| 1157 | XP_002276438.1 | Vitis vinifera | 5-methyltetrahydropteroyltriglutamate–homocysteine methyltransferase | MET | 79.8 / 6.1 | 85.0/ 6.1 | 21 |
| 1159 | XP_002276438.1 | Vitis vinifera | 5-methyltetrahydropteroyltriglutamate–homocysteine methyltransferase | MET | 80.1 / 6.0 | 85.0 / 6.1 | 12 |
| 1625 | XP_003635049.1 | Vitis vinifera | Cysteine desulfurase 1, mitochondrial-like | CyD-1 | 46.2 / 5.9 | 49.9 / 6.7 | 17 |
| 1720 | CAC39216.1 | Vitis vinifera | Glutamine synthetase | GS | 39.8 / 4.9 | 39.0 / 5.4 | 13 |
| 1731 | CAC39216.1 | Vitis vinifera | Glutamine synthetase | GS | 38.6 /5.1 | 39.0 / 5.4 | 34 |
| SECONDARY METABOLISM (FIGURE 5E) | |||||||
| 1616 | BAB41020.1 | Vitis vinifera | UDP-glucose:flavonoid 3-O-glucosyltransferase | UFGT | 46.7 / 5.6 | 50.1 / 6.2 | 6 |
| 1653 | ABM67590.1 | Vitis vinifera | Anthocyanidin synthase | ANS | 45.0 / 5.5 | 40.2 / 5.6 | 28 |
| 1818 | 2P3X | Vitis vinifera | Polyphenol oxidase | PPO | 35.2 / 5.8 | 38.4 / 5.5e | 5 |
| 1821 | AAB41022.1 | Vitis vinifera | Polyphenol oxidase | PPO | 35.2 / 5.9 | 67.4 / 6.4 | 7 |
| 1916 | CAI56335.1 | Vitis vinifera | Isoflavone reductase-like protein 6 | IFL-6 | 32.7 / 5.9 | 33.9 / 6.0 | 43 |
| 1924 | CAI56334.1 | Vitis vinifera | Isoflavone reductase-like protein 5 | IFL-5 | 32.4 / 5.8 | 33.9 / 5.8 | 35 |
| 1935 | CAI56334.1 | Vitis vinifera | Isoflavone reductase-like protein 5 | IFL-5 | 32.0 / 6.0 | 33.9 / 5.8 | 24 |
| 2117 | P51117.1 | Vitis vinifera | Chalcone-flavonone isomerase 1 | CHI-1 | 27.0 / 5.0 | 25.1 / 5.3 | 22 |
| 2382 | S52629 | Vitis vinifera | Catechol oxidase (EC 1.10.3.1) precursor - grape | Cat-OX | 21.8 / 4.7 | 67.3 / 6.3 | 9 |
| 2386 | AAB41022.1 | Vitis vinifera | Polyphenol oxidase | PPO | 21.7 / 4.6 | 67.4 / 6.4 | 7 |
| 2618 | CAI56334.1 | Vitis vinifera | Isoflavone reductase-like protein 5 | IFL-5 | 33.1 / 5.6 | 33.9 / 5.8 | 29 |
| HORMONE METABOLISM (FIGURE 5F) | |||||||
| 1391 | AAX48772 | Vitis vinifera | 9,10[9',10']carotenoid cleavage dioxygenase | CCD | 58.1 / 5.9 | 61.1 / 6.0 | 9 |
| 1853 | CAN68994.1 | Vitis vinifera | Aldo-keto reductase 4d (auxin regulated) | Ald-Keto-Red-4 | 34.5 / 6.0 | 37.5 / 6.5 | 7 |
| 1854 | CAN68994.1 | Vitis vinifera | Aldo-keto reductase 4d (auxin regulated) | Ald-Keto-Red-4 | 34.5 / 6.1 | 37.5 / 6.5 | 7 |
| 1855 | CAN68994.1 | Vitis vinifera | Aldo-keto reductase 4d (auxin regulated) | Ald-Keto-Red-4 | 34.5 / 6.2 | 37.5 / 6.1 | 6 |
| 2227 | XP_002280658.1 | Vitis vinifera | Stem-specific protein TSJT1 (auxin regulated) | TSJT-1 | 25.6 / 5.3 | 25.2 / 5.7 | 22 |
| 2029 | XP_002283483.1 | Vitis vinifera | Stem-specific protein TSJT1 (auxin regulated) | TSJT-1 | 28.9 / 5.1 | 27.2 / 5.5 | 22 |
| REDOX (FIGURE 5G) | |||||||
| 1784 | XP_002282603.1 | Vitis vinifera | Probable protein disulfide-isomerase A6 | PDI-A6-1 | 36.1 / 5.4 | 39.3 / 5.6 | 23 |
| 2107 | XP_002284767.1 | Vitis vinifera | L-ascorbate peroxidase 2, cytosolicc | AscPOX-2 | 27.2 / 5.6 | 27.6 / 5.7 | 54 |
| 2269 | AKG51696.1 | Vitis vinifera | Superoxide dismutase | SOD | 25.0 / 5.6 | 25.3 / 6.8 | 17 |
| PROTEIN (FIGURE 5H) | |||||||
| 1070 | XP_002282146.1 | Vitis vinifera | Cell division control protein 48 homolog | CDC48 | 100.2 / 5.0 | 89.6 / 5.1 | 24 |
| 1088 | XP_002266780.1 | Vitis vinifera | elongation factor 2 | EF-2 | 95.6 / 5.8 | 94.0 / 5.8 | 9 |
| 1150 | XP_002273246.1 | Vitis vinifera | Probable Xaa-Pro aminopeptidase P | AMPP | 82.0 / 5.0 | 71.7 / 5.2 | 19 |
| 1390 | XP_002283510.1 | Vitis vinifera | T-complex protein 1 subunit zeta | TCP-1 | 58.0 / 5.8 | 59.1 / 6.0 | 20 |
| 1447 | XP_002284370.1 | Vitis vinifera | Probable mitochondrial-processing peptidase subunit beta | MPP-β | 54.9 / 5.8 | 58.5 / 6.4 | 13 |
| 1463 | XP_002283310.1 | Vitis vinifera | Mitochondrial-processing peptidase subunit alpha | MPP-α | 53.7 / 4.8 | 54.5 / 5.7 | 14% |
| 1490 | XP_002276114.1 | Vitis vinifera | Leucine aminopeptidase 1 | LAP-1 | 52.8 / 5.2 | 60.7 / 6.7 | 11 |
| 1597 | XP_002283140.2 | Vitis vinifera | Aminoacylase-1 isoform X1 | AmCyl-X1 | 47.6 / 5.7 | 54.1 / 5.6 | 5 |
| 1679 | XP_002276099.1 | Vitis vinifera | DNA damage-inducible protein 1 | DDI-1 | 43.1 / 4.7 | 45.1 / 5.0 | 11 |
| 1803 | XP_002278975.1 | Vitis vinifera | 26S proteasome non-ATPase regulatory subunit 7 homolog A | PSMD-7 | 35.6 / 5.9 | 34.8 / 6.0 | 25 |
| 1917 | XP_002284566.1 | Vitis vinifera | 26S proteasome non-ATPase regulatory subunit 14 | PSMD-14 | 32.7 / 6.1 | 34.5/ 6.3 | 11 |
| CELL ORGANIZATION/SIGNAL (FIGURE 5I) | |||||||
| 1514 | XP_002285721.1 | Vitis vinifera | Tubulin alpha-3 chain | TUB-3 | 51.5 / 4.9 | 49.7 / 4.9 | 15 |
| 1661 | XP_002282516.1 | Vitis vinifera | Actin-7 | ACT-7 | 44.0 / 5.0 | 41.7 / 5.3 | 37 |
| TRANSPORT (FIGURE 5L) | |||||||
| 1411 | XP_010664138.1 | Vitis vinifera | V-type proton ATPase subunit B 2 isoform X1 | VHA-B2-X1 | 56.0 / 4.8 | 56.2 / 5.0 | 6 |
| 1450 | XP_010664138.1 | Vitis vinifera | V-type proton ATPase subunit B 2 isoform X1 | VHA-B2-X1 | 54.5 / 4.6 | 56.2 / 5.0 | 8 |
| 2627 | XP_002270168.1 | Vitis vinifera | V-type proton ATPase subunit E1 | VHA-E1 | 27.0 / 5.9 | 26.0 / 6.2 | 6 |
| OTHER FUNCTIONS (FIGURE 5M) | |||||||
| 1677 | XP_010649306.1 | Vitis vinifera | Protein YLS2-like | YLS2 | 43.3 / 5.8 | 42.0 / 5.9 | 9 |
| 1801 | XP_002284459.1 | Vitis vinifera | 11S globulin subunit beta | 11S-Globulin | 35.6 / 5.1 | 38.3 / 5.4 | 16 |
| 1866 | XP_002270155.1 | Vitis vinifera | Glutelin type-A 3 | Glutelin-A3 | 34.3 / 5.7 | 38.5 / 5.6 | 9 |
| 1867 | XP_002270155.1 | Vitis vinifera | Glutelin type-A 3 | Glutelin-A3 | 34.2 / 5.8 | 38.5 / 5.6 | 21 |
| 1887 | ABC86739.1 | Vitis pseudoreticulata | Cyclase | Cyclase | 33.5 / 4.8 | 29.8 / 5.5 | 16 |
| 2028 | XP_002267609.1 | Vitis vinifera | Remorin | Remorin | 28.9 / 5.0 | 21.8 / 5.3 | 14 |
| 2104 | XP_010654260.1 | Vitis vinifera | Uncharacterized protein At5g02240 | Unc-1 | 27.2 / 5.4 | 36.1 / 9.4 | 15 |
| 2264 | XP_002283286.1 | Vitis vinifera | NAD(P)H dehydrogenase (quinone) FQR1 | FQR-1 | 25.0 / 5.7 | 21.7 / 5.8 | 46 |
| 2289 | NP_001268052.1 | Vitis vinifera | Ripening-related protein grip22 precursor | Grip22 | 24.1 / 5.5 | 22.9 / 4.8 | 6 |
| 2517 | XP_002271352.1 | Vitis vinifera | Nucleoside diphosphate kinase B | NdPK-B | 16.6 / 6.0 | 16.3 / 6.8 | 32 |
| 2637 | NP_001268052.1 | Vitis vinifera | Ripening-related protein grip22 precursor | Grip22 | 29.7 / 4.7 | 22.9 / 4.8 | 17 |
| STRESS (FIGURE 5N) | |||||||
| 1136 | XP_002273244.1 | Vitis vinifera | Heat shock cognate protein 80-like | HSP80 | 84.5 / 4.8 | 80.8 / 5.0 | 24 |
| 1198 | XP_002283532.2 | Vitis vinifera | Heat shock cognate 70 kDa protein 2 | HSP70-2 | 72.9 / 4.8 | 71.2 / 5.2 | 28 |
| 1231 | XP_002283532.2 | Vitis vinifera | Heat shock cognate 70 kDa protein 2 | HSP70-2 | 66.9 / 4.9 | 71.2 / 5.2 | 45 |
| 1237 | XP_002283532.2 | Vitis vinifera | Heat shock cognate 70 kDa protein 2 | HSP70-2 | 67.5 / 4.9 | 71.2 / 5.2 | 28 |
| 2124 | XP_010657212.1 | Vitis vinifera | Carboxymethylenebutenolidase homolog | Carboxy-Ase | 27.0 / 4.7 | 26.3 / 5.0 | 20 |
| 2161 | 4L5H_A | Vitis vinifera | Thaumatin-like protein | TLP | 26.5 / 4.5 | 21.3 / 4.8 | 14 |
| 2172 | 4L5H_A | Vitis vinifera | Thaumatin-like protein | TLP | 26.4 / 4.6 | 21.3 / 4.8 | 28 |
| 2204 | AAZ93634.1 | Vitis vinifera | MSA | MSA | 25.7 / 5.4 | 16.7 / 5.7 | 14 |
| 2231 | CAB85637.1 | Vitis vinifera | Putative thaumatin-like protein | TLP | 25.7 / 4.2 | 24.0 / 4.9 | 9 |
| 2278 | ABB02395.1 | Vitis vinifera | Temperature-induced lipocalin | TInLi | 24.6 / 6.1 | 21.5 / 6.6 | 13 |
| 2342 | XP_002281506.1 | Vitis vinifera | Class I heat shock protein | HSP-I | 22.8 / 6.2 | 16.3/ 6.9 | 18 |
| 2344 | XP_010657906.1 | Vitis vinifera | 22.0 kDa class IV heat shock protein-like | HSP-IV | 22.8 / 5.8 | 21.3/ 5.9 | 30 |
| 2356 | CAC16165.1 | Vitis vinifera | Pathogenesis-related protein 10 | PRP-10 | 22.5 / 5.9 | 17.1 / 6.0 | 23 |
| 2381 | CAC16165.1 | Vitis vinifera | Pathogenesis-related protein 10 | PRP-10 | 22.0 / 5.9 | 17.1 / 6.0 | 23 |
| 2415 | XP_002281285.1 | Vitis vinifera | 18.2 kDa class I heat shock protein | HSP-I | 19.9 / 5.3 | 17.1 / 5.8 | 9 |
| 2446 | XP_003631809.1 | Vitis vinifera | 17.3 kDa class II heat shock protein-like | HSP-s | 18.7 / 5.3 | 17.5 / 5.7 | 27 |
| 2540 | ADG35965.1 | Vitis hybrid cultivar | Pathogenesis-related protein 4 | PRP-4 | 15.9 / 5.2 | 15.2 / 5.5 | 14 |
| 2584 | ADG35965.1 | Vitis hybrid cultivar | Pathogenesis-related protein 4 | PRP-4 | 14.7 / 4.4 | 15.2 / 5.5 | 24 |
| 2626 | AAB65776.1 | Vitis vinifera | Class IV endochitinase | EnChi-4 | 29.0 / 4.2 | 27.2 / 5.4 | 32 |
| 2658 | AAZ93634.1 | Vitis vinifera | MSA | MSA | 26.7 / 5.4 | 16.7 / 5.7 | 31 |
Proteins were divided according to their functions.
Experimental molecular weight (kDa) or isoelectric point.
Theoretical molecular weight (kDa) or isoelectric point.
Amino acid coverage (%).
Information obtained by blastp (protein-protein BLAST) algorithm.
Partial sequence.
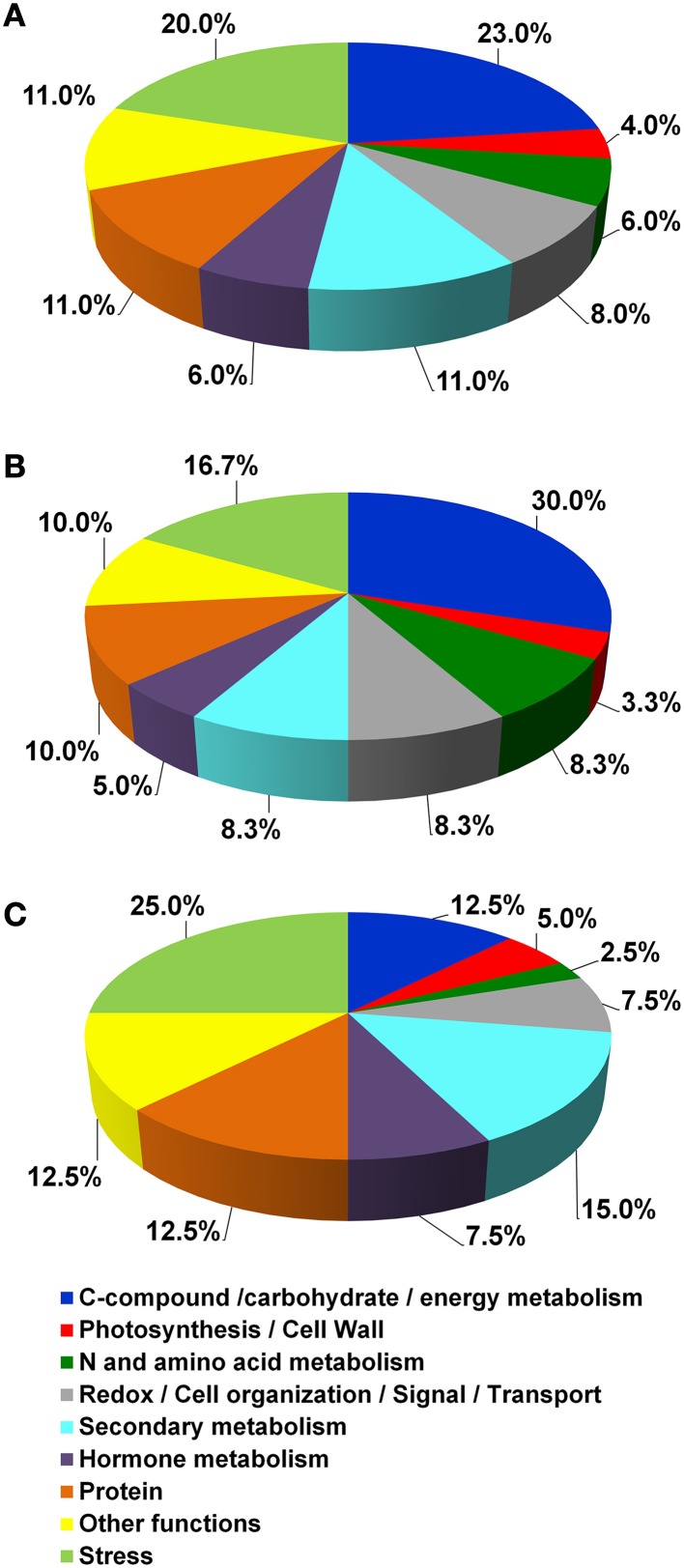
According to their function, the identified proteins were grouped in 12 main classes (Table 3 and Figure 4A). In detail, they were involved in Photosynthesis/Cell Wall (4%), C-compound/carbohydrate/energy metabolism (23%), N and amino acid metabolism (6%), Redox/Cell organization/Signal/Transport (8%), Secondary metabolism (11%), Hormone metabolism (6%), Protein (11%), Other functions (11%) and Stress (20%).
Figure 4.
Functional distribution of the identified proteins. (A) functional distribution of all the identified proteins reported in Table 3. (B,C) functional distribution of proteins that showed greater abundance in Croatina and in Riesling Italico, respectively.
The quantitative differences among the four genotypes are showed in the Supplementary Figure S1, in which spot volume percentages were reported for all identified proteins. As a whole, the results showed that the greater differences occurred between R and C genotypes. In this view, we created two functional distribution pie charts in which R and C genotypes were compared. Figures 4B,C showed the proteins having higher abundance in C and in R, respectively. It is interesting to observe that in C the proteins with higher abundance belonged to the C-compound/carbohydrate/energy metabolism, N and amino acid metabolism functional classes, while in R Secondary metabolism and Stress ones prevailed.
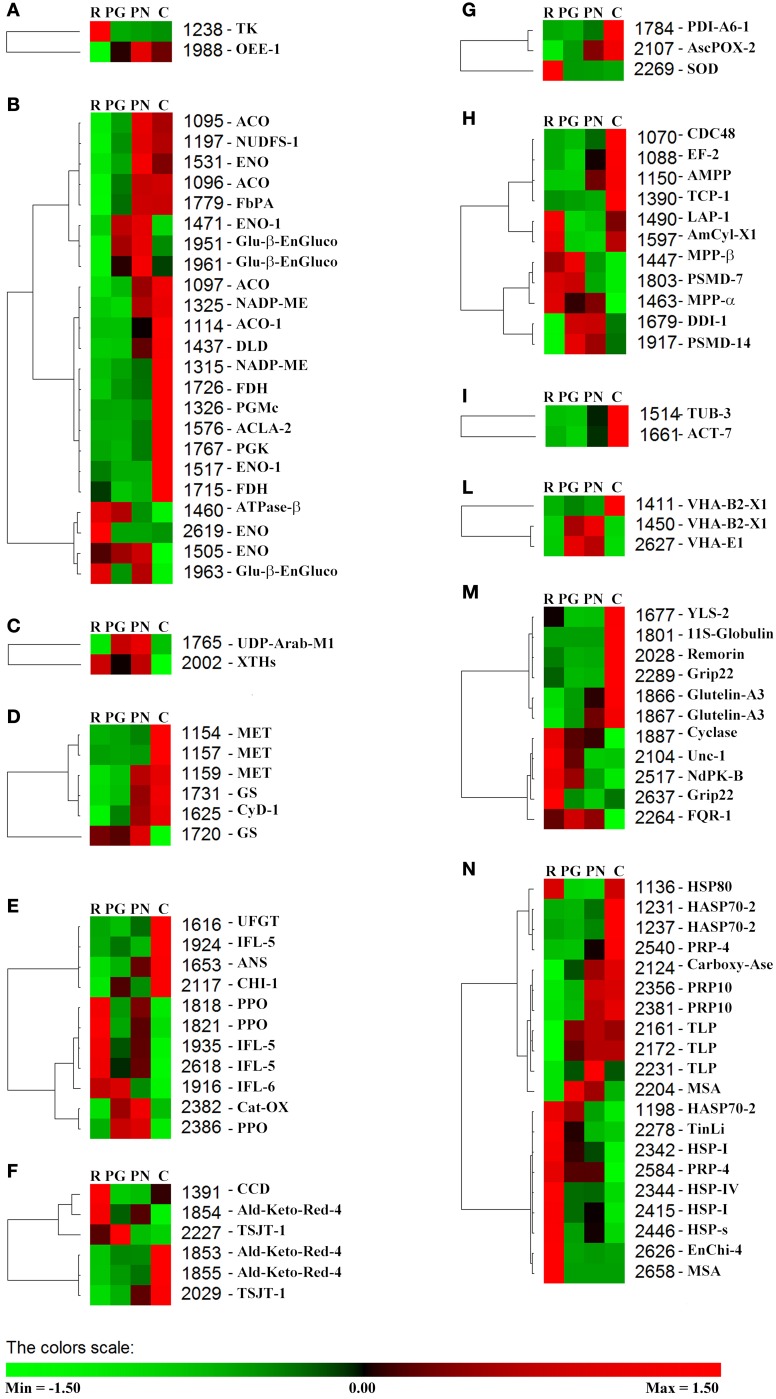
Using PermutMatrix software, a hierarchical clustering of the different functional groups was created to depict in detail the differences in protein abundances as well as to appreciate the gap among the proteomes of the four cultivars (Figure 5). The most striking evidence was that many proteins showed a direct or inverse relation with anthocyanin content. R and C showed the most divergent proteomes, while PG and PN looked more similar.
Figure 5.
Clustering analysis of the spots grouped according to their functional class. Two-way hierarchical clustering analysis of proteins that resulted to be significantly different in their relative spot volumes and identified by LC-ESI-MS/MS (Table 3) was performed with PermutMatrix. Pearson's distance and Ward's algorithm were used for the analysis. Each colored cell represents the average of the relative spot value, according to the color scale at the bottom of the figure. (A) Phothosynthesis. (B) C-compound/carbohydrate/energy metabolism. (C) Cell wall. (D) Nitrogen metabolism and amino acid metabolism. (E) Secondary metabolism. (F) Hormone metabolism. (G) Redox. (H) Protein. (I) Cell organization/Signal. (L) Transport. (M) Other functions. (N) Stress.
Metabolite analysis
GC-MS analysis permitted the identification and quantification of 56 metabolites (Supplementary Table S2). The choice of performing these analyses through a SIM approach was linked to the fact that, especially in a ripe fruit, the metabolome is dominated by large amount of accumulated sugars. Because of this, the peaks of ions relating to primary metabolites could be reduced or not easily integrated. In order to solve these analytical problems, we thus decided to limit the acquisition to the ions of interest, improving the signal and the quality of ion shape (data not shown). The ions used for the quantification were chosen on the basis of the spectra and the retention times obtained by the isolated injection of standard substances under the same chromatographic and MS conditions. Among the quantified metabolites, there were sugars (16), amino acids (15), organic acids (13), and phenolic acids (5). Moreover, among the identified compounds, 5 were glycolytic intermediates and 7 were intermediates of TCA cycle. Sixteen metabolites resulted significantly different to ANOVA test (p≤0.05) and some of these showed a trend that was related with the anthocyanin one (Supplementary Figure S2).
Discussion
The multivariate statistical analysis permitted to isolate spots useful to discriminate the four cultivar
Through multivariate analyses it was possible to overview the relationship among the proteomes of the 4 cultivars, clearly distinguishing the samples according to the cultivar and sorting the most relevant observed differences in spot abundance.
As expected, the biological dataset resulted to be quite complex since the first 5 PCs accounted only the 43.37% of the explained variance (Supplementary Table S1). In every case, it is interesting to note that in PC1 the cultivars were roughly ordered following their anthocyanin content, moving from R at negative values to C proteomes at positive values. Nevertheless, the placement on PC2, in which there was substantial distinction between Pinot and non-Pinot cultivars, suggested a contribution of genetic factors (Figure 3). At the same time, it was surprising that on the first 2 PCs differences linked to seasonal variation did not emerge. In this view, it must be considered that the technique used in this study permitted to consider only a part of whole proteome and this could be inadequate to find the plasticity of the grapevine berry previously revealed by transcriptomic study (Dal Santo et al., 2013).
The relation between the four proteomes was further investigated through FS-LDA performed on the first 10 PCs calculated. The trick of using PCs instead original variables led to dimensionality reduction and to noise elimination by excluding the less significant ones as performed in Negri et al. (2011). The linear discriminant analysis reflected and reinforced PCA results since, as witnessed by the classification matrix (Table 1), it correctly distinguished the gels according to the cultivar of belonging. Moreover, the squared Mahalanobis distances from group centroids (Table 2) demonstrated that the objects (i.e., the gels) could be ordered following the anthocyanin content of the 4 cultivars: R gels, for instance, are thus really distant from the centroid of C and rather close to the PG one. At the same time, it was interesting to note that although they are quite identical at the genetic level and overlapped in PCA score plot (Figure 3), the gels of the two Pinot cultivars were clearly separated by FS-LDA, showing a clear proximity as inferable by the small values of the relative squared Mahalanobis distances (Table 2).
As described, an interesting number of the variables with the highest or the lowest coefficients of discrimination showed to have a high significance in the models of 3 or even all the 4 cultivars, suggesting that some of the main factors that differentiate them, as for anthocyanin content, could pass through the modulation of some specific proteins.
Proteomic and metabolomic data highlight new peculiar traits of the metabolism operating in grape berry exocarp
Previous proteomic studies revealed that, differently to the mesocarp, in the exocarp tissue of grape berry the glycolysis and the hexose-monophosphate shunt pathways remained active also at full ripe stage (Sarry et al., 2004; Negri et al., 2008b, 2011). This metabolic state was attributed to the demand of C-skeletons and energy to sustain the synthesis of secondary compounds (mainly anthocyanins) and/or other activities linked to defensive mechanisms.
Focusing the attention on primary carbon metabolism (i.e., glycolysis and TCA cycle) and respiration chain, the accumulation trend of the spots referring to PGMc (spot 1326), PGK (spot 1767), ENO-1 and ENO (spot 1517 and 1531), ACLA-2 (spot 1576), DLD (spot 1437), and NDUFS-1 (spot 1197) showed a good association with anthocyanin content also in this study (Figures 1, 2, 5; Table 3; Supplementary Figure S1). According to a greater activation of glycolysis and TCA cycle, some intermediates of these pathways, such as fructose-1,6-phosphate, pyruvate and succinate, showed the same tendency (Supplementary Figure S2).
The greater activation of the carbon metabolism was also suggested by a coherent increase in the red cultivar of aconitate hydratase, an enzyme having a central role in citric acid metabolism (Terol et al., 2010). Three spots identified as ACO showed high similarity with the same accession (XP_002278138.1), so suggesting that the spots could refer to different phosphorylated forms, according to the PTMs previously identified for this enzyme (Bycova et al., 2003; Millar et al., 2005).
The results regarding enolase highlighted the multifunctional role of these glycolytic enzymes. Of the five spots referring to enolase, three revealed to be the same protein (accession XP_002267091.2), while the others shared the greatest similarity with the accessions XP_002283632.1 suggesting the presence of different isoforms (Table 3). Although, the current knowledge on the pattern of Vitis vinifera enolase did not permit further consideration from the classification point of view, by using iPSORT software (http://ipsort.hgc.jp/index.html) we verified the absence of N-terminal signal peptide, suggesting that all identified forms are cytosolic enolase. These data supported the suggestion that also in grape berry exocarp, as previously observed in others plant tissues (Voll et al., 2009), the plastidic isoform could be lacking. As recently emerged, in the absence of a complete glycolysis pathway in the plastids, cytosolic enolase plays a central role to modulate the synthesis of aromatic amino acids and secondary phenyilpropanoid compounds (Voll et al., 2009; Eremina et al., 2015). Moreover, the trend of spots 1517 and 1531 could suggest that they are the forms that modulate shikimate pathway.
On the base of the above conclusion that cytosolic phosphoenolpyruvate is requested for both phenylpropanoid biosynthesis and respiratory pathway, the higher level of pyruvate in red cultivars could appear unexpected (Supplementary Figure S2). In this context, it could be observed that among the spots showing an increase in abundance in red cultivars, two were identified as a NADP-dependent malic enzyme (NADP-ME, spots 1315 and 1325), supporting the idea that this enzyme in the skin tissue could play an important role to sustain pyruvate request. Nevertheless, the level of malic acid was higher in the red cultivars (Supplementary Figure S2), suggesting that a concomitant supply of this organic acid occurred. Previously, Iland and Coombe (1988) showed that during ripening the levels of this organic acid decreased in the mesocarp, whilst it did not change in the exocarp. Moreover, these authors found that the leaching of malate from the mesocarp tissue increased during ripening. Although a direct evidence is not available, an interesting hypothesis is that during ripening malic acid could move from mesocarp to skin, sustaining the carbon demand occurring in this tissue.
Two spots identified as a mitochondrial formate dehydrogenase (spots 1715 and 1726) showed greater abundance in C. Their reciprocal position on the gel appeared ascribable to PTMs, such as a phosphorylation (Bycova et al., 2003; Millar et al., 2005). Although a complete understanding of the functional role of this enzyme is awaited, its accumulation could be interpreted as a further request of reducing power (Plaxton and Podestà, 2015) that occurs in the cultivar with the highest level of anthocyanins.
Previously, a role of glycolytic enzymes, such as aldolase and enolase in the activation of vacuolar H+-ATPase through an association with a subunit VHA-B has been described (Barkla et al., 2009 and references therein). Moreover, an activation of vacuolar proton pumps during grape berry was reported (Terrier et al., 2001; Grimplet et al., 2009; He et al., 2010). In red cultivars an upsurge of both the FbPA (spots 1779) and the subunit VHA-B2-X1 (spots 1411) took place (Figures 5B,L). Although the anthocyanin transport into vacuole in exocarp tissue involves different mechanisms (Coon et al., 2008; Gomez et al., 2009; Sweetman et al., 2009; Francisco et al., 2013), these results suggest that on the whole the transport of these compounds could require an increase of the tonoplastic H+-pump activity.
Some identified spots, such as CHI-1, ANS and UFGT (spots 2117, 1653, and 1616, respectively), were enzymes operating in the anthocyanin synthesis pathway. Their trends were well related to the anthocyanin levels (Figures 1, 5E). These results appear in accordance to previous transcriptional analysis in which the expression of the genes codifying for these proteins were mainly (CHI-1, ANS) or only (UFGT) detected in the red cultivars (Boss et al., 1996b). In this context, the greater abundance in PN or C of two spots that were identified as glutamine synthetase (Spots 1731, 1720) was in agreement with interlinks between anthocyanin metabolism and nitrogen recycling (Singh et al., 1998; Cantón et al., 2005). Moreover, we found that the abundances of three spots corresponding to methionine synthase (spots 1154, 1157, and 1159) related with the anthocyanin contents (Figure 5D). Nevertheless the levels of methionine were not different among the four cultivars, suggesting that a simultaneous transformation of this amino acid occurred in red cultivars (Supplementary Figure S2). Although a more direct evidence is needed, an intriguing hypothesis is that this amino acid could be required for the biosynthesis of ethylene, according to an involvement of this hormone in the anthocyanin production (El-Kereamy et al., 2003; Böttcher and Davies, 2012).
From the proteomic analysis differences in protein metabolism emerged (Figure 5H). In C the Elongation factor 2-like isoform 1 (EF-2, spot 1088), a probable Xaa-Pro aminopeptidase P (AMPP, spot 1150) and the subunit zeta T-Complex protein 1 (TCP-1, spot 1390) resulted more abundant, while some proteins involved in mitochondrial-proteolytic system (MPP-α and MPP-β, spots 1463 and 1447) or in protein degradation (PSMD-7 and PMSD-14, spots 1803 and 1917, respectively) were inversely related to the anthocyanin content. Taken together, these results pointed out that in C prevailed activities involved in protein synthesis, whilst in the other cultivars a more evident protein catabolism seemed to take place. In this view, it is interesting to observe that in C CDC48, TUB-3 and ACT-7 (spots 1070, 1514, and 1661, respectively) resulted most abundant, so sustaining that in this cultivar was operating a more active cellular metabolism (Figures 5H,I).
Many of the identified proteins are involved in the stress-related processes
This work brought to the identification of many proteins that are known to be involved in defense and stress responses. It is interesting to observed that many studies performed on grape berry reported the presence of proteins, such as thaumatin-like and chitinases, even in the absence of pathogen infections (Deytieux et al., 2007; da Silva et al., 2005; Negri et al., 2008b, 2011; Fraige et al., 2015), suggesting that their accumulation could be linked to a preventive defensive plan closely related to the genetic background of the cultivar. Nevertheless, it was suggested that these proteins could also play a multifaceted role in the ripening process (van Hengel et al., 2001, 2002; Kasprzewska, 2003). In our work we find the presence of proteins belonging to the stress class also in healthy fruit as well as we confirm that they resulted to be linked to the genetic background (Figure 5N and Table 3).
Previously, Pilati et al. (2007) described in the red cultivar PN that the start of ripening phase was characterized by an oxidative burst and that this depended on specific changes in gene expression. Recently, these authors found that this process occurs mainly in the skin tissue and reported evidences on possible role of ROS as cellular signal (Pilati et al., 2014). In our work the majority of the proteins classified in the stress group showed an abundance that was both directly or inversely related to the anthocyanin contents. Considering their functions, we found a similar oxidative stress condition, but the data suggested that the analysed cultivars used different strategies in responding to it. This was particular evident comparing C with R. In this last cultivar some proteins that are known to be involved in the oxidative stress responses (i.e., PPO and SOD, spots 1818, 1821, and 2269 respectively) were present in higher abundance (Figures 5E,G). Considering the Mr it was possible to conclude that these PPO spots corresponded to the active form of the enzyme (Dry and Robinson, 1994). According to the possibility that a suffering condition was occurring in the R, five HSPs were more accumulated (spots 1198, 2342, 2344, 2415, 2446). Moreover, the level of ascorbic acid resulted significantly lower in R respect to other cultivars (Supplementary Figure S2). This result could be linked to genetic characteristic as well as to be a symptom of a suffering status.
It is interesting to observe that in this cultivar a 9,10[9′,10′]carotenoid cleavage dioxygenase (CCD, spot 1391) was found to be present in the highest amount, an enzyme involved in the carotenoid cleavage to produce apocarotenoid from which important signaling molecules, such as abscisic acid (ABA) and stringolactones derive (Harrison and Bugg, 2014). Recently, Ye et al. (2011) reported interesting evidences on possible role of ABA in the control of catalase gene expression, an enzyme that was described to play a central role in the ROS detoxification also in grape berry skin (Pilati et al., 2014). Although the pH range used in IEF excluded the possibility to detect the catalase, taken together, also these results further sustained the idea that in the white cultivar R biochemical mechanisms to counteract an oxidative stress condition were activated.
This study led to the identification of some forms of isoflavone reductase-like (IFL) protein (spots, 1916, 1924, 1935, and 2618). Three of these (1924, 1935, and 2618) showed high similarity with the same accession (CAI56334.1). On the basis of experimental pI, it could be suggested that the three spots refer to different phosphorylation state, a PTM that is not yet described for this enzyme. Since the stereospecific reduction of isoflavones by isoflavone reductase (IFR) is restricted primarily to legumes, a distinct reductase reaction was postulated for IFL. In fact, this enzyme was related to oxidative stress occurring during the somatic embryogenesis of Vitis vinifera callus (Zhang et al., 2009). Considering that three IFL spots showed to be more abundant in R, our data suggested a similar protective role in grape berry skin.
Differently from the white cultivar R, in the red cultivars many of these responses resulted less active. In this context, it could be underlined that flavonoid biosynthesis produces some compounds, such as quercetin, myrecetin, kaempferol, having high antioxidant activity (Pietta, 2000). The metabolic profiling on berry skin performed on a large number of cultivars, revealed that these compounds are higher in PN and C than in R (Mattivi et al., 2006). Hence, our work revealed that the oxidative burst occurring in grape berry skin ripening stimulates typical antioxidant responses in which the phenolic compounds have a central role. In the red cultivars, in fact, the high levels of phenolic compounds probably compensate for the induction of the other antioxidant mechanisms observed in the white cultivar.
Concluding remarks
This work highlighted new information about the biochemical and physiological events occurring in the skin tissue during grape berry ripening. Considering the functional distribution of the identified proteins and the trends of some metabolites, the results showed how many physiological processes, such as carbon metabolism (e.g., glycolysis and TCA cycle), energy conversion, secondary metabolism and oxidative stress, are involved in the protective role against damage by physical injuries and pathogen attacks. Nevertheless, some biochemical responses appeared requested to counteract oxidative burst, an event that characterizes the ripening step of grape berry skin. In this view, the strategy used strictly depended on flavonoid biosynthesis.
Author contributions
AN contributed to the conception of the experimental design, carried out protein extraction, 2-DE, gel analysis, metabolomic analysis and statistical analysis. BP contributed to the conception of the experimental design, carried out protein characterization by LC-ESI-MS/MS, analyzed the MS data. OF and AS participated to the manuscript revision. LE conceived the study, coordinated the experiments, wrote and edited the manuscript. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Glossary
Abbreviations
- 2-DE
two-dimensional electrophoresis
- ACN
acetonitrile
- cCBB
colloidal Coomassie Brilliant Blue G-250
- C
Croatina
- FS-LDA
Forward Stepwise Linear Discriminant Analysis
- PCA
Principal Component Analysis
- PG
Pinot Gris
- PN
Pinot Noir
- R
Riesling Italico
- SIM
Selected Ion Monitoring.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00603
References
- Ablett E., Seaton G., Scott K., Shelton D., Graham M. W., Baverstock P., et al. (2000). Analysis of grape ESTs: global gene expression patterns in leaf and berry. Plant Sci. 159:87–95. 10.1016/S0168-9452(00)00335-6 [DOI] [PubMed] [Google Scholar]
- Ageorges A., Fernandez L., Vialet S., Merdinoglu D., Terrier N., Romieu C. (2006). Four specific isogenes of the anthocyanin metabolic pathway are systematically co-expressed with the red colour of grape berries. Plant Sci. 170, 372–383. 10.1016/j.plantsci.2005.09.007 [DOI] [Google Scholar]
- Barkla B. J., Vera-Estrella R., Hernández-Coronado M., Pantoja O. (2009). Quantitative proteomics of the tonoplast reveals a role for glycolytic enzymes in salt tolerance. Plant Cell 21, 4044–4058. 10.1105/tpc.109.069211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss P. K., Davies C., Robinson S. P. (1996a). Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv. Shiraz grape berries and the implications for pathway regulation. Plant Physiol. 111, 1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss P. K., Davies C., Robinson S. P. (1996b). Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Mol. Biol. 32, 565–569. 10.1007/BF00019111 [DOI] [PubMed] [Google Scholar]
- Böttcher C., Davies C. (2012). Hormonal control of grape berry development and ripening in The Biochemistry of the Grape Berry, ed Gerós G., Chaves M. M., Delrot S. (Sharjah: Bentham Science Publishers; ), 194–217. [Google Scholar]
- Bunk B., Kucklick M., Jonas R., Mu R., Schobert M., Dieter J., et al. (2006). MetaQuant: a tool for the automatic quantification of GC/MS-based metabolome data. Bioinformatics 22, 2962–2965. 10.1093/bioinformatics/btl526 [DOI] [PubMed] [Google Scholar]
- Burns K. C., Dalen J. L. (2002). Foliage color contrasts and adaptive fruit color variation in a bird-dispersed plant community. Oikos 96, 463–469. 10.1034/j.1600-0706.2002.960308.x [DOI] [Google Scholar]
- Bycova N. V., Egsgaard H., Møller I. A. (2003). Identication of 14 new phosphoproteins involved in important plant mitochondrial processes. FEBS Lett. 540, 141–146. 10.1016/S0014-5793(03)00250-3 [DOI] [PubMed] [Google Scholar]
- Cantón F. R., Suárez M. F., Cánovas F. M. (2005). Molecular aspects of nitrogen mobilization and recycling in trees. Photosyn. Res. 83, 265–278. 10.1007/s11120-004-9366-9 [DOI] [PubMed] [Google Scholar]
- Caraux G., Pinloche S. (2005). PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics 21, 1280–1281. 10.1093/bioinformatics/bti141 [DOI] [PubMed] [Google Scholar]
- Castellarin S. D., Bavaresco L., Falginella L., Gonçalves M. I. V. Z., Di Gaspero G. (2012). Phenolics in grape berry and key antioxidant in The biochemistry of the grape berry, ed Gerós G., Chaves M. M., Delrot S. (Sharjah: Bentham Science Publishers; ), 89–110. [Google Scholar]
- Coombe B. G. (1995). Growth stages of the grapevine: adoption of a system for identifying grapevine growth stages. Aust. J. Grape Wine Res. 1, 104–110. 10.1111/j.1755-0238.1995.tb00086.x [DOI] [Google Scholar]
- Coon S., Curtin C., Bézier A., Franco C., Zhang W. (2008). Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J. Exp. Bot. 59, 3621–3634. 10.1093/jxb/ern217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Santo J., Tornielli G. B., Zenoni S., Fasoli M., Farina L., Anesi A., et al. (2013). The plasticity of the grapevine berry transcriptome. Genome Biol. 14, R54. 10.1186/gb-2013-14-6-r54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva F. G., Iandolino A., Al-Kayal F., Bohlmann M. C., Cushman M. A., Lim H., et al. (2005). Characterizing the grape transcriptome. Analysis of expressed sequence tags from multiple vitis species and development of a compendium of gene expression during berry development. Plant Physiol. 139, 574–559. 10.1104/pp.105.065748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degu A., Hochberg U., Sikron N., Venturini L., Buson G., Ghan R., et al. (2014). Metabolite and transcript profiling of berry skin during fruit development elucidates differential regulation between Cabernet Sauvignon and Shiraz cultivars at branching points in the polyphenol pathway. BMC Plant Biol. 14:188. 10.1186/s12870-014-0188-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deytieux C., Geny L., Lapaillerie D., Claverol S., Bonneu M., Donéche B. (2007). Proteome analysis of grape skins during ripening. J. Exp. Bot. 58, 1851–1862. 10.1093/jxb/erm049 [DOI] [PubMed] [Google Scholar]
- Dry I. B., Robinson S. P. (1994). Molecular cloning and characterization of grape berry polyphenol oxidase. Plant Mol. Biol. 26, 495–502. 10.1007/BF00039560 [DOI] [PubMed] [Google Scholar]
- El-Kereamy A., Chervin C., Roustan J.-P., Cheynier V., Souquet J.-M., Moutounet M., et al. (2003). Exogenous ethylene stimulates the long-term expression of genes related to anthocyanin biosynthesis in grape berries. Physiol. Plant. 119, 175–182. 10.1034/j.1399-3054.2003.00165.x [DOI] [Google Scholar]
- Eremina M., Rozhon W., Yang S., Poppenberger B. (2015). ENO2 activity is required for the development and reproductive success of plants, and is feedback-repressed by AtMBP-1. Plant J. 81, 895–906. 10.1111/tpj.12775 [DOI] [PubMed] [Google Scholar]
- Fortes A. M., Agudelo-Romero P., Silva M. S., Ali K., Sousa L., Maltese F., et al. (2011). Transcript and metabolite analysis in Trincadeira cultivar reveals novel information regarding the dynamics of grape ripening. BMC Plant Biol. 11:149. 10.1186/1471-2229-11-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraige K., González-Fernándezb R., Carrilho E., Jorrín-Novo J. V. (2015). Metabolite and proteome changes during the ripening of Syrah and Cabernet Sauvignon grape varieties cultured in a nontraditional wine region in Brazil. J. Proteomics 113, 206–225. 10.1016/j.jprot.2014.09.021 [DOI] [PubMed] [Google Scholar]
- Francisco R. M., Regalado A., Ageorges A., Burla B. J., Bassin B., Eisenach C., et al. (2013). ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. Plant Cell 25, 1840–1854. 10.1105/tpc.112.102152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli F., Rossoni M., Iriti M., Di Gennaro A., Faoro F., Borroni E., et al. (2006). From field to health: a sample way to increase the nutraceutical content of grape as shown by NO-dependent vascular relaxion. J. Agric. Food Chem. 54, 5344–5349. 10.1021/jf0607157 [DOI] [PubMed] [Google Scholar]
- Giribaldi M., Giuffrida M. G. (2010). Heard it through the grapevine: Proteomic perspective on grape and wine. J. Proteomics 73, 1647–1655. 10.1016/j.jprot.2010.05.002 [DOI] [PubMed] [Google Scholar]
- Giribaldi M., Perugini I., Sauvage F. X., Schubert A. (2007). Analysis of protein changes during grape berry ripening by 2-DE and MALDI-TOF. Proteomics 7, 3154–3170. 10.1002/pmic.200600974 [DOI] [PubMed] [Google Scholar]
- Gomez C., Terrier N., Torregrosa L., Vialet S., Fournier-Level A., Verriès C., et al. (2009). Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiol. 150, 402–415. 10.1104/pp.109.135624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimplet J., Deluc L. G., Tillett R. L., Wheatley M. D., Schlauch K. A., Cramer G. R., et al. (2007). Tissue-specific mRNA expression profiling in grape berry tissues. BMC Genomics 8:187. 10.1186/1471-2164-8-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimplet J., Wheatley M. D., Jouira H. B., Deluc L. G., Cramer G. R., Cushman J. C. (2009). Proteomic and selected metabolite analysis of grape berry tissues under well watered and water-deficit stress conditions. Proteomics 9, 2503–2528. 10.1002/pmic.200800158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison P. J., Bugg T. D. H. (2014). Enzymology of the carotenoid cleavage dioxygenases: reaction mechanisms, inhibition and biochemical roles. Arch. Biochem. Biophys. 544, 105–111. 10.1016/j.abb.2013.10.005 [DOI] [PubMed] [Google Scholar]
- He F., Mu L., Yan G.-L., Liang N.-N., Pan Q.-H., Wang J., et al. (2010). Biosynthesis of anthocyanins and their regulation in colored grapes. Molecules 15, 9057–9091. 10.3390/molecules15129057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurkman W. J., Tanaka C. K. (1986). Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol. 81, 802–806. 10.1104/pp.81.3.802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iland P. G., Coombe B. G. (1988). Malate, tartrate, potassium, and sodium in flesh and skin of shiraz grapes during ripening: concentration and compartmentation. Am. J. Enol. Vitic. 39, 71–76. [Google Scholar]
- Jaillon O., Aury J. M., Noel B., Policriti A., Clepet C., Casagrande A., Choisne N., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. 10.1038/nature06148 [DOI] [PubMed] [Google Scholar]
- Kasprzewska A. (2003). Plant chitinases—regulation and function. Cell. Mol. Biol. Lett. 8, 809–824. Available online at: http://www.cmbl.org.pl/vol8_nr3.php# [PubMed] [Google Scholar]
- Kobayashi S., Goto-Yamamoto N., Hirochika H. (2004). Retrotransposon-induced mutations in grape skin color. Science 304, 982. 10.1126/science.1095011 [DOI] [PubMed] [Google Scholar]
- Lisec J., Schauer N., Kopka J., Willmitzer L., Fernie A. R. (2006). Gas chromatography mass spectrometry-based metabo-lite profiling in plants. Nat. Protoc. 1, 387–396. 10.1038/nprot.2006.59 [DOI] [PubMed] [Google Scholar]
- Lund S. T., Bohlman J. (2006). The molecular basis for wine grape quality-a volatile subject. Science 311, 804–805. 10.1126/science.1118962 [DOI] [PubMed] [Google Scholar]
- Martínez-Esteso M. J., Sellés-Marchart S., Lijavetzky D., Pedreño M. A., Bru-Martinez R. (2011). A DIGE-based quantitative proteomic analysis of grape berry flesh development and ripening reveals key events in sugar and organic acid metabolism. J. Exp. Bot. 62, 2521–2569. 10.1093/jxb/erq434 [DOI] [PubMed] [Google Scholar]
- Mattivi F., Guzzon R., Vrhovsek U., Stefanini M., Velasco R. (2006). Metabolite profiling of grape: flavonols and anthicyanins. J. Agric. Food Chem. 54, 7692–7702. 10.1021/jf061538c [DOI] [PubMed] [Google Scholar]
- Meunier B., Dumas E., Piec I., Béchet D., Hébraud M., Hocquette J. F. (2007). Assessment of hierarchical clustering methodologies for proteomic data mining. J. Proteome Res. 26, 358–366. 10.1021/pr060343h [DOI] [PubMed] [Google Scholar]
- Millar A. H., Heazlewood J. L., Kristensen B. K., Braun H.-P., Møller I. M. (2005). The plant mitochondrial proteome. Trends Plant Sci. 10, 36–43. 10.1016/j.tplants.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Negri A. S., Prinsi B., Rossoni M., Failla O., Scienza A., Cocucci M., et al. (2008b). Proteome changes in the skin of the grape cultivar Barbera among different stages of ripening. BMC Genomics 9:378. 10.1186/1471-2164-9-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri A. S., Prinsi B., Scienza A., Morgutti S., Cocucci M. (2008a). Analysis of grape berry cell wall proteome: a comparative evaluation of extraction methods. J. Plant Physiol. 165, 1379–1389. 10.1016/j.jplph.2007.10.011 [DOI] [PubMed] [Google Scholar]
- Negri A. S., Robotti E., Prinsi B., Espen L., Marengo E. (2011). Proteins involved in biotic and abiotic stress responses as the most significant biomarkers in the ripening of Pinot Noir skins. Funct. Integr. Genomics 11, 341–355. 10.1007/s10142-010-0205-0 [DOI] [PubMed] [Google Scholar]
- Neuhoff V., Arold N., Taube D., Ehrhardt W. (1988). Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9, 255–262. 10.1002/elps.1150090603 [DOI] [PubMed] [Google Scholar]
- Niessen S., McLeod I., Yates J. R., III. (2006). HPLC separation of digested proteins and preparation for matrix-assisted laser desorption/ionization analysis. Cold Spring Harb. Protoc. 2006:pdb.prot4663. 10.1101/pdb.prot4663. [DOI] [PubMed] [Google Scholar]
- Niu N., Cao Y., Duan W., Wu B., Li S. (2013). Proteomic analysis of grape berry skin responding to sunlight exclusion. J. Plant Physiol. 170, 748–757. 10.1016/j.jplph.2012.12.020 [DOI] [PubMed] [Google Scholar]
- Pietta P. G. (2000). Flavonoids as antioxidant. J. Nat. Prod. 63, 1035–1042. 10.1021/np9904509 [DOI] [PubMed] [Google Scholar]
- Pilati S., Brazzale D., Guella G., Milli A., Ruberti C., Biasioli F., et al. (2014). The onset of grapevine berry ripening is characterized by ROS accumulation and lipoxygenase-mediated membrane peroxidation in the skin. BMC Plant Biol. 14:87. 10.1186/1471-2229-14-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilati S., Perazzolli M., Malossini A., Cestaro A., Demattè L., Fontana P., et al. (2007). Genome-wide transcriptional analysis of grapevine berry ripening reveals a set of genes similarly modulated during three seasons and the occurrence of an oxidative burst at vèraison. BMC Genomics 8:428. 10.1186/1471-2164-8-428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton W. C., Podestà F. E. (2015). The functional organization and control of plant respiration. CRC. Crit. Rev. Plant Sci. 25, 159–198. 10.1080/07352680600563876 [DOI] [Google Scholar]
- Prinsi B., Negri A. S., Fedeli C., Morgutti S., Negrini N., Cocucci M., et al. (2011). Peach fruit ripening: a proteomic comparative analysis of the mesocarp of two cultivars with different flesh firmness at two ripening stages. Phytochemistry 72, 1251–1262. 10.1016/j.phytochem.2011.01.012 [DOI] [PubMed] [Google Scholar]
- Sarry J. E., Sommerer N., Sauvage F. X., Bergoin A., Rossignol M., Albagnac G., et al. (2004). Grape berry biochemistry revisited upon proteomic analysis of the mesocarp. Proteomics 4, 201–215. 10.1002/pmic.200300499 [DOI] [PubMed] [Google Scholar]
- Singh S., Lewis N. G., Towers N. (1998). Nitrogen recycling during phenylpropanoid metabolism in sweet potato tubers. J. Plant Physiol. 153, 316–323. 10.1016/S0176-1617(98)80157-0 [DOI] [PubMed] [Google Scholar]
- Solovchenko A., Schmitz-Eiberger M. (2003). Significance of skin flavonoids for UV-B-protection in apple fruits. J. Exp. Bot. 54, 1977–1984. 10.1093/jxb/erg199 [DOI] [PubMed] [Google Scholar]
- Sweetman C., Deluc L. G., Cramer G. C., Ford C. M., Soole K. L. (2009). Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry 70, 1329–1344. 10.1016/j.phytochem.2009.08.006 [DOI] [PubMed] [Google Scholar]
- Terol J., Soler G., Talon M., Cercos M. (2010). The aconitate hydratase family from Citrus. BMC Plant Biol. 10:222. 10.1186/1471-2229-10-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier N., Glissant D., Grimplet J., Barrieu F., Abbal P., Couture C., et al. (2005). Isogene specific oligo arrays reveal multifaceted changes in gene expression during grape berry (Vitis vinifera L.) development. Planta 222, 832–847. 10.1007/s00425-005-0017-y [DOI] [PubMed] [Google Scholar]
- Terrier N., Sauvage F.-X., Ageorges A., Romieu C. (2001). Changes in acidity and in proton transport at the tonoplast of grape berries during development. Planta 213, 20–28. 10.1007/s004250000472 [DOI] [PubMed] [Google Scholar]
- van Hengel A. J., Tadesse Z., Immerzeel P., Schols H., van Kammen A., de Vries S. C. (2001). N-Acetylglucosamine and glucosamine containing arabinogalactan proteins control somatic embryogenesis. Plant Physiol. 125, 1880–1890. 10.1104/pp.125.4.1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hengel A. J., van Kammen A., de Vries S. C. (2002). A relationship between seed development, Arabinogalactan proteins (AGPs) and the AGP mediated promotion of somatic embryogenesis. Physiol. Plant. 114, 637–644. 10.1034/j.1399-3054.2002.1140418.x [DOI] [PubMed] [Google Scholar]
- Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., et al. (2014). ProteomeXchange provides globally co-ordinated proteomics data submission and dissemination. Nat. Biotechnol. 30, 223–226. 10.1038/nbt.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voll L. M., Hajirezaei M. R., Czogalla-Peter C., Lein W., Stitt M., Sonnewald U., et al. (2009). Antisense inhibition of enolase strongly limits the metabolism of aromatic amino acids, but has only minor effects on respiration in leaves of transgenic tobacco plants. New Phytol. 184, 607–618. 10.1111/j.1469-8137.2009.02998.x [DOI] [PubMed] [Google Scholar]
- Waters D. L., Holton T. A., Ablett E. M., Lee L. S., Henry R. J. (2005). cDNA microarray analysis of developing grape (Vitis vinifera cv. Shiraz) berry skin. Funct. Integr. Genomics 5, 40–58. 10.1007/s10142-004-0124-z [DOI] [PubMed] [Google Scholar]
- Ye N., Zhu G., Liu L., Li Y., Zhang J. (2011). ABA controls H2O2 accumulation through the induction of OsCATB in rice leaves under water stress. Plant Cell Physiol. 52, 689–698. 10.1093/pcp/pcr028 [DOI] [PubMed] [Google Scholar]
- Zamboni A., Di Carli M., Guzzo F., Stocchero M., Zenoni S., Ferrarini A., et al. (2010). Identification of putative stage-specific grapevine berry biomarkers and omics data integration into networks. Plant Physiol. 154, 1439–1459. 10.1104/pp.110.160275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Mab H., Chen S., Ji M., Perl A., Kovacs L., Chen S. (2009). Stress response proteins' differential expression in embryogenic and non-embryogenic callus of Vitis vinifera L. cv. Cabernet Sauvignon - a proteomic approach. Plant Sci. 177, 103–113. 10.1016/j.plantsci.2009.04.003 [DOI] [Google Scholar]
- Zheng Y., Li J. H., Xin H. P., Wang N., Guan L., Wu B. H., et al. (2013). Anthocyanin profile and gene expression in berry skin of two red Vitis vinifera grape cultivars that are sunlight dependent versus sunlight independent. Aust. J. Grape Wine Res. 19, 238–248. 10.1111/ajgw.1202325158067 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.