Abstract
Bacteria switch between two distinct life styles – planktonic (free living) and biofilm forming – in keeping with their ever-changing environment. Such switch involves sophisticated signaling and tight regulation, which provides a fascinating portal for studying gene function and orchestrated protein interactions. In this work, we investigated the molecular mechanism underlying biofilm formation in Shewanella oneidensis MR-1, an environmentally important model bacterium renowned for respiratory diversities, and uncovered a gene cluster coding for seven proteins involved in this process. The three key proteins, BpfA, BpfG, and BpfD, were studied in detail for the first time. BpfA directly participates in biofilm formation as extracellular “glue” BpfG is not only indispensable for BpfA export during biofilm forming but also functions to turn BpfA into active form for biofilm dispersing. BpfD regulates biofilm development by interacting with both BpfA and BpfG, likely in response to signal molecule c-di-GMP. In addition, we found that 1:1 stoichiometry between BpfD and BpfG is critical for biofilm formation. Furthermore, we demonstrated that a biofilm over-producing phenotype can be induced by C116S mutation but not loss of BpfG.
Keywords: biofilm, bpfA bpfG, bpfD, regulation mechanism, S. oneidensis
Introduction
Biofilm is a type of surface-attached structure composed of microbial cells embedded in their self-produced extracellular polymeric substances (EPS), mainly exopolysaccharides, proteins, and extracellular DNA (Flemming and Wingender, 2010). It has been recognized as the principle life style for microbes in nature (O’Toole, 2003; Hall-Stoodley et al., 2004). Although planktonic cells are advantageous to look for favorable niches, biofilm allows cells to remain and thrive in such places. Thus, switching between planktonic and biofilm-forming modes represents a major life style change for microbes, and has been shown to be a tightly regulated process (O’Toole, 2003; Hall-Stoodley et al., 2004). Many regulatory cascades controlling transition of microbial life-styles studied to date involve regulatory factors to mediate transcription and translation of proteins for biosynthesis of EPS, including sigma factors, transcriptional factors, several nucleotide messengers, and sRNAs (Karatan and Watnick, 2009; Fazli et al., 2014).
However, there are exceptions. In Pseudomonas fluorescens Pf0–1, a protein network is reported to regulate biofilm development through sophisticated signaling and protein interactions rather than mediating EPS production (Newell et al., 2011). This system consists of multiple proteins encoded by genes in a lap cluster (Figure 1A). Of these Lap proteins, LapA, LapG, and LapD are critical to the process of biofilm formation. LapA, a Bap/RTX hybrid cell surface protein, is exported by a type I secretion system (TISS) encoded by three genes (tolC–Pfl0135–hlyD) immediately downstream of lapA and serves as a cell surface attached adhesin responsible for “gluing” cells together (El-Kirat-Chatel et al., 2014). LapG is a periplasmic proteinase, which cuts LapA off the outer membrane (Boyd et al., 2014). As a result, compared to the wild-type mutants lacking LapG are more robust in forming biofilm and overproduction of LapG promotes biofilm dispersion (Boyd et al., 2012). LapD is a transmembrane protein regulating LapG activity in response to signal molecule c-di-GMP (Newell et al., 2009). The environmental cue for the modification of LapA is conditions unfavorable for biofilm formation, such as low inorganic phosphate concentrations (Newell et al., 2011).
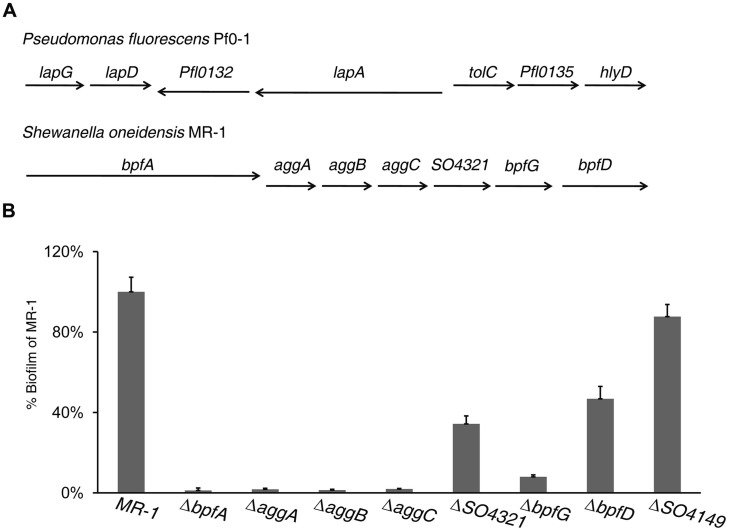
FIGURE 1.
Identification of a cluster of genes involved in biofilm formation in Shewanella oneidensis MR-1. (A) Arrangement of genes in the lapA operon of Pseudomonas fluorescens Pf0–1 and their homologous in MR-1. The type I secretion system (TISS) is formed by ORF tolC–Pfl0135–hlyD in Pf0–1 and aggA–C (SO4318–SO4320) in MR-1. The ORFs SO4321, SO4322 (bpfG), and SO4323 (bpfD) encode an OmpA-like protein, a TISS associated periplasmic transglutaminase-like cysteine proteinase and a bifunctional diguanylate cyclase/phosphodiesterase protein, respectively. Pfl0132 codes for a protein with no impact on biofilm formation. (B) Relative biofilm biomass of mutants lacking genes indicated. Biofilm formation was determined using the standard plate assay, and normalized to the value of MR-1 (wild-type) to yield the relative biofilm for comparison across different experiments. Error bar represents SE of three experiments.
Shewanella oneidensis MR-1 is a Gram-negative facultative anaerobe, the best studied representative of the genus Shewanella (Venkateswaran et al., 1999). Following its isolation (Myers and Nealson, 1988), the bacterium was soon found to be able to use a wide range of solid electron acceptors (EA), a subset of which include minerals containing heavy metal ions such as Fe(III), Cr(VI), and U(VI), to name a few (Fredrickson et al., 2008). The feature renders MR-1 an appealing agent for bioremediation, developing microbial fuel cells (MFC) and synthesizing metal nanomaterials (Logan and Regan, 2006; Marshall et al., 2006; Pirbadian et al., 2014).
Previous studies of biofilm formation in MR-1 have identified BpfA (Bpf stands for Biofilm promoting factor) and AggA as essential proteins for the process (De Windt et al., 2006; Liang et al., 2010, 2012; Theunissen et al., 2010). Sequence comparison reveals that BpfA and AggA are analogous to P. fluorescens LapA and TolC, respectively (Boyd et al., 2014). Like LapA and TolC of P. fluorescens, BpfA and AggA are encoded by two genes next to each other, albeit oppositely oriented (Figure 1A). Additionally, AggA is the outer-membrane component of the accompanying TISS system (AggA–AggB–AggC), resembling TolC–Pfl0135–HlyD (Theunissen et al., 2009, 2010). Despite these similarities, the BpfA-mediated biofilm formation carries novel features as the low inorganic phosphate concentrations could not differentiate the bpfA mutant from the wild-type. Therefore, this work aims to further explore the BpfA-mediated biofilm formation in MR-1. Here, we first identified BpfA, BpfG, and BpfD to be the important players in biofilm development, through bioinformatic, mutational, and molecular analyses. Further investigation uncovered interactions between these proteins and their effects on biofilm formation and dispersion. Although these three proteins constitute a protein network vaguely resembling the one in P. fluorescens, mechanisms underlying roles played by each component differed significantly.
Materials and Methods
Strains and Growth Conditions
Shewanella oneidensis strains and Escherichia coli strains were cultured in LB medium at 30°C and 37°C, respectively. In detecting biofilm formation in high Pi and low Pi medium, the composition of medium is: low-phosphate (50 mM Tris-HCl, 0.2% Tryptone, lactate, 0.6 mM MgSO4); high-phosphate (add 1 mM K2HPO4 into low-phosphate medium). When needed, chemicals were added at the following concentrations: 2, 6-diaminopimelic acid, 0.3 mM; ampicillin, 50 μg/ml; gentamycin, 15 μg/ml; kanamycin, 50 μg/ml; and tetracycline, 12.5 μg/ml; chloramphenicol, 25 μg/ml; streptomycin, 12.5 μg/ml.
Mutant Construction and Complementation
MR-1 in-frame deletion strains were constructed by the att-based Fusion PCR method (Jin et al., 2013). In brief, two fragments flanking the target gene were generated by PCR, with primers containing attB and gene specific sequences, and then were fused by a second round of PCR. The fusion fragments were introduced into plasmid pHGM01 by site-specific recombination using the BP Clonase (Invitrogen) according to the manufacturer’s instruction. The resulting mutagenesis vectors were transformed into E. coli WM3064, and the verified ones were transferred into proper MR-1 strains via conjugation. Integration of the mutated constructs into the chromosome were selected by resistance to gentamicin and confirmed by PCR. Verified trans-conjugants were grown in LB broth in the absence of NaCl and plated on LB supplemented with 10% sucrose. Gentamicin-sensitive and sucrose-resistant colonies were screened by PCR for deletion of the target gene. All mutations were verified by sequencing the mutated regions.
Plasmids pHG102 (Wu et al., 2011) and pHGE-Ptac were used in genetic complementation of mutants. Genes of interest were amplified and inserted into MCS of pHG102 under the control of MR-1 arcA promoter, which is constitutively active (Gao et al., 2010) or pHGE-Ptac, which is under IPTG induction (Luo et al., 2013). The resulting complementation vectors were transferred into the corresponding mutation strain via conjugation and their presence were confirmed by plasmid purification and restriction enzyme digestion.
pHGE-Ptac containing SO4322 (bpfG) or SO4323 (bpfD) were used as the templates for site-directed mutagenesis with a QuikChange II XL site-directed mutagenesis kit (Stratagene) as described previously (Sun et al., 2013). Mutated PCR products were generated, subsequently digested by DpnI, and transformed into E. coli WM3064. After sequencing verification, the resulting plasmid was transferred into the MR-1 strains by conjugation.
To construct bpfG mutants lacking one of GGDEF, EAL, and HAMP domains, location of each domain was defined based on GenBank annotation and sequence alignment to characterized homologous proteins. The HAMP, GGDEF, and EAL domains are composed of residues from 174 to 221, from 238 to 383, and from 410 to 639, respectively. Mutants expressing truncated proteins (lacking one of the three domains) were constructed by the att-based Fusion PCR method described above.
Biofilm Formation Assay and Quantification
Biofilm assays were carried out as described before (O’Toole and Kolter, 1998; Yuan et al., 2013), with some modifications. Briefly, an aliquot (20 μl) of an overnight culture grown in LB was transferred into 2 ml fresh LB medium contained in each well of a 24-well plate and grown with shaking at 250 rpm at 30°C for specified hours (or 8 h, if not specified). Biofilm quantification was conducted as following: Biofilm formed on the plate walls was stained using 0.25% crystal violet for 15 min at room temperature, rinsed with water and air-dried. Absorbed crystal violet was dissolved using 30% (v/v) acetic acid and transferred to a fresh flat bottom microtiter plate and the absorption of the solution at 540 nm was determined by microplate spectrophotometer. The wild-type was included in every plate to serve as an “internal standard” to control for batch-to-batch variation. Biofilm of all other strains were normalized to the value of the wild-type to yield the relative biofilm for comparison across different experiments. Readings from no less than three experiments per strain were used to calculate the average and SE.
Promoter Activity Assay
Promoters for bpfA and the SO4318–SO4323 operon were first predicted with bioformatics analysis. To construct the PbpfA-lacZ reporter, ∼400 bp DNA fragments upstream of the bpfA operon was amplified by PCR cloned into pTP327 (Shi et al., 2013). After verification by DNA sequencing, the reporter plasmid was transferred into each MR-1 strain by conjugation. Cell culture of targeted density was harvested by centrifugation. Cell pellets were washed once with PBS buffer, resuspended to an optical density of OD600 ≈ 1.0 and lysed. β-galactosidase activity assay was performed using an assay kit (Beyotime, China) as described previously (Wu et al., 2011).
BpfA Extraction
Bacterial cultures were grown overnight in LB, and then sub-cultured into 40 ml of fresh LB at a 1:100 dilution and grown shaking at 250 rpm. After 6 h of incubation, cells were vortexed and harvested by centrifugation (12,000 g, 5 min). All resulting supernatant was passed through a 0.22 μM filter (Millipore, Billerica, MA, USA) to remove residual cells, and then concentrated in 100,000 MWCO filter column (Millipore, Billerica, MA, USA). The final volume in concentrator was the supernatant fraction. For membrane-anchored BpfA, cells were harvested as above; the pellet was washed once in 30 mM Tris-HCl (pH 8.1) and repelleted. Cell pellet were then resuspended in 20 ml 30 mM Tris-HCl-20% sucrose buffer, followed by the addition of 200 μl of 20 mg/ml lysozyme-0.1 mM EDTA (pH 7.3) and incubated on ice for 30 min. Following lysozyme treatment, 20 ml of 3 mM ETDA (pH 7.3) was added and the resulting extract was sonicated for 40 cycles (3 s sonication, 4 s interval). A 40 ml fraction of the extract was then centrifuged at 16,000 g for 60 min., the resulting pellet was resuspended in 500 μl of SDS sample buffer to yield the membrane fraction. Sample was boiled at 100°C for 5 min prior to SDS-PAGE display. Finally, 40 μl samples were displayed on 6 M urea-5% SDS gels.
Bacterial Two-Hybrid Assay
A bacterial two-hybrid (B2H) system (BacterioMatch II Two-Hybrid System Vector Kit) was used to investigate protein–protein interactions in vivo in E. coli cells as described previously (Wu et al., 2011). Briefly, plasmid constructs were created by cloning the bait (DNAs for BpfG, BpfGC116S, BpfD, BpfDperi) and target (DNAs for BpfD and BpfANterm) into the pBT and pTRG vectors and verified by sequencing. BpfDperi is the periplasmic domain (∼100 a.a.) of BpfD and BpfANterm is the N terminus (∼200 a.a.) of BpfA. BpfANterm was used since cloning of the entire BpfA was hindered by its large size and repetitive nature. As for BpfDperi, the rationale was that the transmembrane regions of full-length BpfD might be a problem and lead to the negative result between BpfG and BpfD in the B2H assay, and since BpfG locates in the periplasm, the periplasmic region of BpfD was used to probe BpfD and BpfG interaction. Moreover, because the N-terminal cytosolic domain of BpfD has only 39 a.a. (including signal peptide), we did not construct a fractional BpfD containing only this domain to probe its interaction with BpfA. We did not construct a BpfD The resulting plasmids were used to co-transform BacterioMatch II Validation Reporter competent cells on M9 salt agar plates containing 25 mg/ml chloramphenicol and 12.5 mg/ml tetracycline with or without 3-amino-1,2,4-triazole (3-AT). Plasmid pair of pBT-L/pTRG-G was used as the positive control, and Plasmid pair of pBT/pTRG was used as the negative control. The plates were incubated for 24 h at 37°C. If colonies are not apparent, transfer the plates to room temperature and continue to incubate the plates in a dark location (to preserve the tetracycline) for an additional 16 h. The second incubation may allow the growth of cells containing toxic proteins or weak interactors. Then, the positive interactions were confirmed by streaking colonies onto plates containing both 3-AT and streptomycin (12.5 mg/ml).
Bioinformatics Analyses
Promoters were predicted with the program Neural Network (Reese, 2001). Membrane protein structure and domain functionality were predicted with TopPred and SMART, respectively (Claros and von Heijne, 1994; Letunic et al., 2015).
Results
The SO4317–SO4323 Gene Cluster is involved in MR-1 Biofilm Formation
As shown in Table 1 and Figure 1A, an in silico analysis revealed that two genes downstream of bpfA (SO4317), namely SO4322 and SO4323, resemble P. fluorescens lapG and lapD genes in terms of sequence, predicted function, and gene arrangement in the seven-gene cluster SO4317–SO4323. To be consistent, we named SO4322 and SO4323 as bpfG and bpfD, respectively. To examine roles of these genes within the cluster in biofilm formation, we created in-frame deletion mutants for each gene. In agreement with previous reports (De Windt et al., 2006; Liang et al., 2010; Theunissen et al., 2010), BpfA and the TISS system encoded by aggABC genes (SO4318–20) were found to be crucial for biofilm formation in MR-1. Additionally, mutants missing bpfG or bpfD gene were also significantly defective in biofilm formation, indicating that both of the genes are required for the process (Figure 1B). Moreover, we found that SO4321 (annotated to encode an OmpA family protein), the gene located between the TISS system coding genes and bpfG, had a role in biofilm formation as well (Figure 1B). Interestingly, MR-1 has another potential LapA homolog, SO4149, which may participate in Ca2+-mediated cell-cell adhesion because it contains a Cadherin repeat-like domain (Cao et al., 2005). However, impact of the SO4149 loss on biofilm formation was rather minor (Figure 1B). Given the essentiality of LapA to biofilm formation, a combination of the distinct phenotypes resulting from losses of BpfA and SO4149 and the syntenic similarity between lapA and bpfA concludes that BpfA is the counterpart of P. fluorescens lapA.
Table 1.
BlastP results of Pseudomonas fluorescens LapA, LapG, and LapD against Shewanella oneidensis.
| P. fluorescens | S. oneidensis | Annotation for S. oneidensis proteins | E-value |
|---|---|---|---|
| LapA | BpfA (SO4317) | Biofilm-promoting protein BpfA | 2e–32 |
| SO4149 | Secreted VCBS domain protein | 4e–41 | |
| LapG | BpfG (SO4322) | Type I secretion system (TISS) associated periplasmic transglutaminase-like cysteine proteinase | 3e–41 |
| LapD | BpfD (SO4323) | Bifunctional diguanylate cyclase/phosphodiesterase LasD-like protein | 3e–52 |
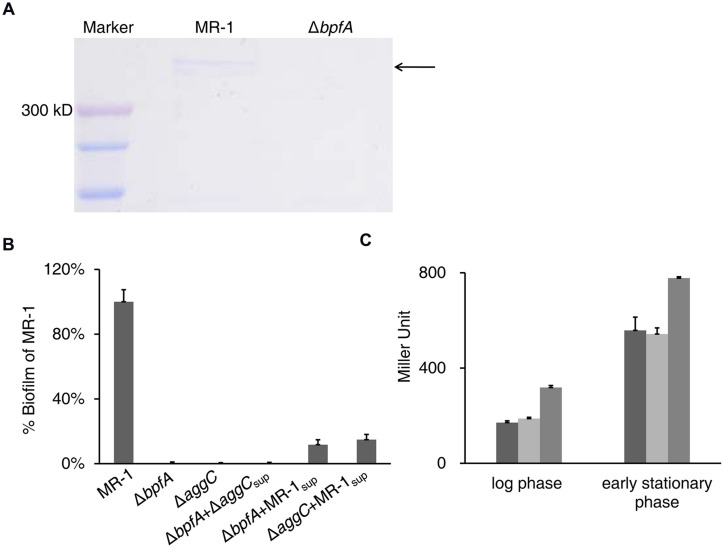
BpfA possesses a Type I secretion C-terminal target domain per InterPro prediction and is believed to be exported by the TISS system coded by aggABC (De Windt et al., 2006; Theunissen et al., 2009). We found that BpfA protein can be collected from the supernatant of vigorously vortexed liquid cultures (Figure 2A), and such supernatant could partially rescue the biofilm defect of the BpfA mutant (Figure 2B). This observation supports the notion that BpfA is an outer-membrane adhesion. Moreover, BpfA promoter activity displayed a significant rise as cells enter the stationary phase, in good accordance with the expected timeline for biofilm development (Figure 2C).
FIGURE 2.
BpfA is an extracellular adhesin required for biofilm formation. (A) BpfA from culture supernatants of the indicated strains visualized by SDS-PAGE. Arrow indicates the band corresponding to BpfA protein. (B) Culture supernatant of wild-type could partially rescue the biofilm defect of ΔbpfA and ΔaggC. Supernatant prepared from wild-type or ΔaggC cells were added to the wells in a polystyrene plate containing LB-cultures of ΔbpfA or ΔaggC. Error bar represents SE of three experiments. (C) Promoter activity of bpfA in strains MR-1 (dark gray), ΔbpfG (light gray), or ΔbpfD (gray). The samples were collected in late log phase (OD600 ≈ 0.6) and early stationary phase (OD600 ≈ 1.0) for comparison. Error bar represents SE of three experiments.
The dual-band of BpfA in SDS-PAGE (Figure 2A) was intriguing, although not unusual for Bap family proteins (Cucarella et al., 2001). By excising and subjecting these bands to LC-MS/MS analysis, it was concluded that both bands arose from BpfA, and no perceivable post-translational modification was detected (Supplemental Tables S2 and S3). Efforts to clone bpfA, unfortunately, were hindered by its large size and repetitive nature. One interesting observation was that BpfA appears to be larger than the size deduced from the annotated genome sequence, both in terms of PCR product and SDS-PAGE result, which corresponded to each other well (Supplemental Figure S1). This is likely due to the sequencing error well known to be associated with genome regions rich in repeat sequences (Lesk, 2012). By resequencing, we confirmed 2038 bp from the translation initiation codon and 4167 bp ending at the stop codon. The remaining sequence between these two fragments is ∼2.6 kb longer than that reported in the genome sequence (2183 bp; Heidelberg et al., 2002). This fragment of ∼4.8 kb is composed of up to 16 300-bp repeats, based on sequence features of LapA (Boyd et al., 2014).
BpfG is a Bifunctional Protein Necessary for BpfA Secretion and Cleavage
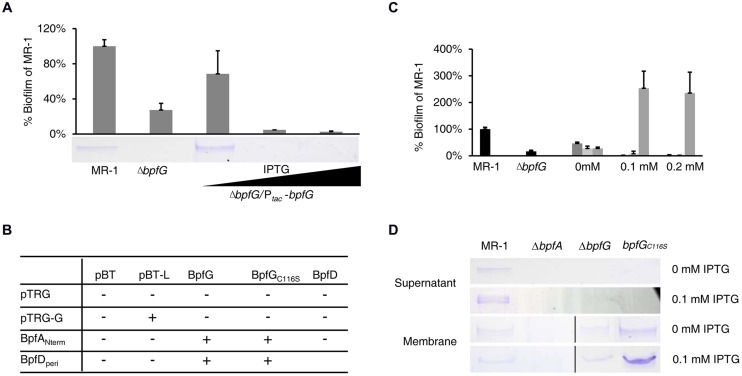
As mentioned in the introduction, P. fluorescens lapG mutant (devoid of the periplasmic cysteine proteinase) displayed a biofilm-overproducing phenotype (Boyd et al., 2012). In contrast, deletion of bpfG led to a defect in MR-1 biofilm formation (Figure 1B), indicating a distinct role of BpfG compared with P. fluorescens LapG. Further analysis of the supernatant of vortexed ΔbpfG cultures revealed that BpfA was missing (Figure 3A), suggesting that BpfG influenced biofilm formation by modulating BpfA. Given that BpfG is annotated as a TISS associated periplasmic transglutaminase-like cysteine proteinase and located in the periplasm (Heidelberg et al., 2002; Brown et al., 2010), one possible scheme would be that BpfG is involved in certain process critical for BpfA excretion and function.
FIGURE 3.
BpfG is a periplasmic cysteine proteinase required for BpfA export and cleavage. (A) The biphasic impact of BpfG on biofilm. ΔbpfG mutant was complemented with a plasmid carrying a Ptac-bpfG construct, treated with different concentrations of IPTG (the black triangle underneath indicates the increase of IPTG level. 0, 0.1 mM, 0.2 mM from left to right, same in all figures unless specified otherwise). Error bar represents SE of three experiments. BpfA in the supernatant fraction were shown by protein electrophoresis. Arrow indicates the BpfA protein band. (B) Bacterial two-hybrid assay results of indicated proteins. “+” indicates positive results while “–” indicates that no interaction was detected in this assay. BpfANterm is the N terminus (∼200 a.a.) of BpfA, and BpfDperi is the periplasmic domain (∼100 a.a.) of BpfD, see Section “Materials and Methods” for more explanations. (C) Comparison of wild-type BpfG (dark gray) with BpfGC15S (light gray) and BpfGC116S (gray) in their ability to complement ΔbpfD for biofilm formation. The wild-type and ΔbpfD were provided for reference. Ptac recombinant plasmids were treated with different concentrations of IPTG. Error bar represents SE of three experiments. BpfGC18S and BpfGC28S displayed similar phenotype as BpfGC15S and were omitted in the figure for simplicity. (D) BpfA from the extracellular and membrane preparations of indicated strains were visualized by protein electrophoresis. Gels separated by a black line were from samples derived from the same experiment and processed in parallel.
Interestingly, complementation of ΔbpfG with an IPTG-inducible Ptac plasmid revealed a biphasic effect of the protein on biofilm formation and BpfA (Figure 3A). More specifically, complementation in terms of biofilm formation and BpfA excretion was partially successful when no IPTG was added (the promoter was slightly leaky (Shi et al., 2014; Fu et al., 2015; Supplemental Figure S2), whereas IPTG at 0.1 mM or more abolished such effect, indicating that either missing or over-production of BpfG had negative impact on biofilm formation. By using the B2H assay, we detected direct interaction between BpfA and BpfG in vivo (Figure 3B). This finding, along with the fact that LapG modifies LapA in P. fluorescens, prompted us to hypothesize that BpfG might also be involved in BpfA cleavage, causing defect in biofilm formation when either in absence or over-abundance.
To provide evidence to support this hypothesis, we examined effects of the four cysteine residues of BpfG on biofilm formation, some of which are likely essential to the proteinase activity of BpfG. Plasmids expressing BpfG variants carrying C15S, C18S, C28S, and C116S mutations were individually introduced into the ΔbpfG strain and biofilm formation of resulting strains was examined. As shown in Figure 3C, phenotype of BpfGC15S, BpfGC18S, and BpfGC28S were comparable to that of wild-type BpfG. However, overexpression of the BpfGC116S mutant resulted in biofilm over-production. To our surprise, the amount of extracellular BpfA isolated in the supernatant of this mutant cultures was reduced to levels below the detection limit (Figure 3D), a scenario that was quite puzzling considering the established importance of BpfA in biofilm formation. Since BpfA most likely functioned as a cell-surface-attached extracellular adhesin, and the C116S mutation of BpfG did not block its interaction with BpfA (Figure 3B), we hypothesized that BpfGC116S might change the nature of the association between BpfA and the cell membrane. SDS-PAGE analysis confirmed that BpfA isolated from the membrane fraction of the strain producing BpfGC116S was more firmly associated with the cell membrane (Figure 3D). These results suggest that the 116th Cys residue is responsible for the observed negative impact of over-produced BpfG on biofilm formation, possibly by disrupting BpfA attachment to cell surface and even leading to degradation of unbounded BpfA. Taken together, these observations manifest that BpfG is a periplasmic cysteine proteinase that plays a role in both BpfA excretion and cleavage and the 116th residue is most likely to be the catalytic residue responsible for releasing BpfA from the membrane.
BpfD Affects Biofilm Formation by Modulating BpfG Activity
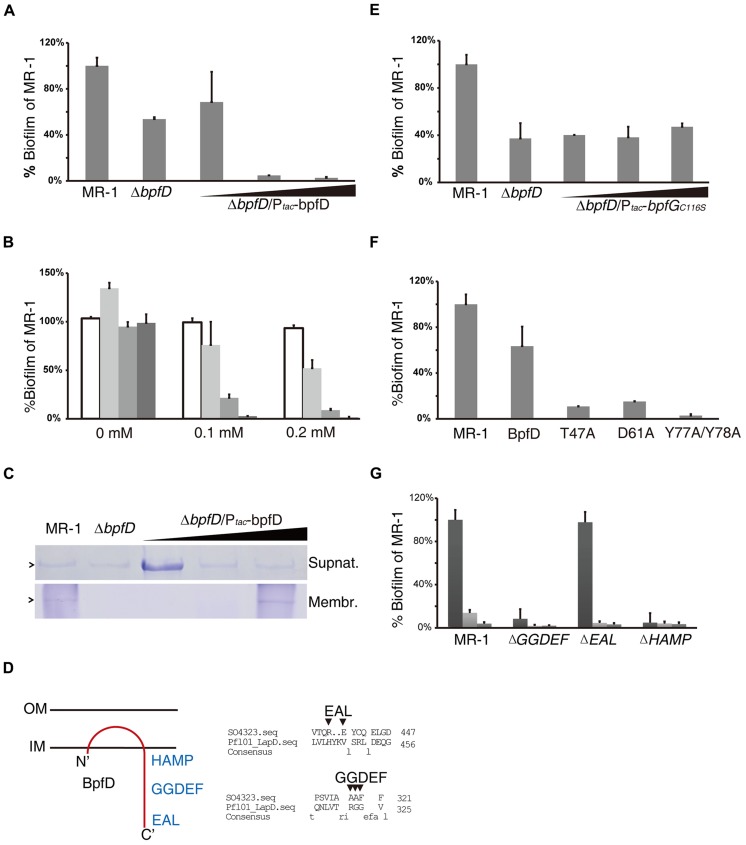
Complementation of ΔbpfD with IPTG-induced bpfD expression displayed a biphasic impact similar to that of BpfG (Figure 4A). However, complementation of the ΔbpfGΔbpfD double mutant with these two genes driven by one promoter was constantly successful (Figure 4B, light gray bars), indicating that a 1:1 stoichiometry between BpfG and BpfD may be crucial for biofilm formation. Furthermore, over-expression of either BpfG or BpfD in MR-1 undermined biofilm formation (Figure 4B); over-expression of GFP, on the other hand, did not have such impact, excluding the possibility of plasmid-related artifact (Figure 4B). One logical explanation for the observation would be that BpfG and BpfD interact with each other stoichiometrically, and their interaction is essential for regulating biofilm development. Moreover, we found that when BpfD in excess amounts of membrane-associated BpfA increased significantly, in addition to the biofilm defect (Figure 4C). Our interpretation of these data is that BpfD probably captures and holds BpfA to the cytoplasmic membrane, while interaction of BpfG and BpfD is required for BpfA release and export.
FIGURE 4.
BpfD is an inner membrane protein regulating biofilm formation by interacting with BpfG and BpfA. (A) The biphasic impact of BpfD. ΔbpfD mutant was complemented with a plasmid carrying a Ptac-bpfD construct, treated with different concentrations of IPTG. Error bar represents SE of three experiments. (B) A biofilm assay examining wild-type strain carrying Ptac-GFP (white), ΔbpfDΔbpfG strain carrying Ptac-bpfDbpfG (light gray), Ptac-bpfD (gray) and Ptac-bpfG (dark gray). All Ptac recombinant plasmids were treated with a range of concentrations of IPTG as indicated. Error bar represents SE of three experiments. (C) BpfA from the extracellular and membrane preparations were visualized by protein electrophoresis for the indicated strains, treated with different concentrations of IPTG. (D) Predicted orientation of BpfD (left), and alignment of BpfD and P. fluorescens LapD showing missing key residues as arrows indicate, numbers indicate amino acid position (right). (E) Over-expression of BpfGC116S in ΔBpfD did not result in biofilm over-production. Ptac-bpfGC116S was treated with IPTG as indicated. (F) Point mutants BpfDT47A, BpfDD61A, and BpfDY 77AY 78A led to biofilm defect. Wild-type and mutant versions of BpfD were all carried individually on a Ptac plasmid and assayed in ΔbpfD background with no IPTG added (the plasmid is leaky). (G) Biofilm formation of the wild-type, BpfD domain mutants ΔGGDEF, ΔEAL, and ΔHAMP for extended incubation (24 h: dark gray, 48 h: light gray, 72 h: gray).
Consistent with this postulation, TopPred (Claros and von Heijne, 1994) predicted that BpfD is likely to be an inner-membrane protein encompassing a periplasmic domain close to the N-terminus, and a C-terminal cytosolic domain. In addition, the cytosolic domain might harbor a diguanylate cyclase GGDEF domain, a phosphodiesterase EAL domain, and a HAMP domain. SMART-based sequence analysis (Letunic et al., 2015) suggested that these domains are most likely degenerated since many key residues are absent (Figure 4D).
The predicted structure of BpfD, function of BpfG, and the resemblance of BpfG–BpfD to the P. fluorescens LapG–LapD system naturally led us to speculate that both systems may regulate biofilm formation through similar mechanisms. More specifically, BpfD may modulate BpfG activity (and thereby biofilm development) upon signals transmitted through its cytosolic domain. Consistent with this possibility, over-expression of BpfGC116S in ΔbpfD mutant did not result in biofilm over-production as much as in ΔbpfG (Figure 4E), suggesting that BpfD probably regulates BpfG-mediated BpfA releasing from the membrane. A positive B2H assay result indicated that such modulation is achieved through direct interaction (Figure 3B). To confirm this, a semi-random point mutation screen was conducted to identify residues within BpfD important for the interaction. As shown in Figure 4F, when residue Thr47, Asp61, Tyr77, and Tyr78 were mutated to alanine (separately), amounts of biofilm reduced greatly. A comparative analysis of predicted tertiary structures for these BpfD variants revealed that none of these mutations altered protein configuration significantly (Supplemental Figure S3). Hence, the observed negative impact of the mutations on biofilm formation may be specific to the interaction between BpfG and BpfD.
To assess the function of the GGDEF, EAL, and HAMP domains of BpfD, mutants lacking each individual domain were constructed and their biofilm forming ability was evaluated. As shown in Figure 4G, the mutant lacking the EAL domain had capability to form biofilm comparable to the wild-type, whereas loss of either GGDEF or HAMP caused a defective phenotype. Additionally, dispersion of biofilm was also slower (judged from the reduction of bar height overtime in Figure 4G) in the latter two mutants as well as in the ΔbpfD strain, indicating that these domains might be important in regulating biofilm dispersion.
Role of c-di-GMP
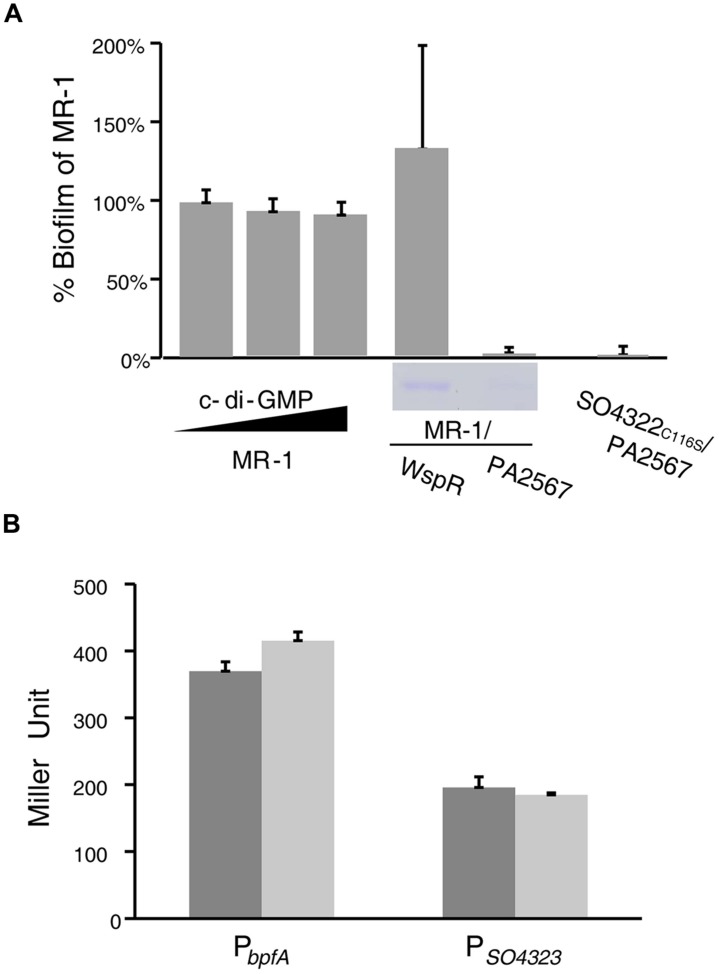
Given the widely reported importance of c-di-GMP in biofilm formation, we examined the impact of this molecule in our system. As shown in Figure 5A, no significant change of biofilm production was observed through increasing c-di-GMP by either adding the purchased pure compound into the medium or over-producing WspR, a known P. aeruginosa diguanylate cyclase in the cell (De et al., 2008). Consistently, over-expression of PA2567, a verified phosphodiesterase from P. aeruginosa (Rao et al., 2009), abolished biofilm formation as well as extracellular presence of BpfA in both wild-type and the biofilm-overproducing BpfGC116S strains.
FIGURE 5.
MR-1 biofilm formation requires c-di-GMP but not sensitive to medium phosphate level. (A) A biofilm assay examining MR-1, MR-1/Ptac-WspR, MR-1/pHG102-PA2567, and BpfGC116S/pHG102-PA2567. MR-1/Ptac-WspR was induced with 0.1 mM IPTG; MR-1 was treated with 0 mM, 60 mM and 120 mM c-di-GMP. BpfA in the supernatant fraction were shown by SDS-PAGE. (B) Promoter activities of bpfA and bpfD in MR-1 (dark) and MR-1/pHG102-PA2567 (light), respectively. The samples were collected in log phase (OD600 = 0.6).
Furthermore, the biofilm defect resulting from the bpfD mutation was not due to transcriptional suppression of the adhesin BpfA as expression of bpfA and bpfG did not change in MR-1 mutants overproducing phosphodiesterase PA2567 (Figure 5B). These data suggest that c-di-GMP is functionally associated with Bpf system but may not exert its impact as a transcriptional/translational cofactor.
Discussion
Regulation of biofilm formation in microorganisms is a topic of both scientific and practical value. From the standpoint of basic scientific research, microbes have to stay tuned with the external environment to seize the cue(s) calling for the life-style switch from free-living to biofilm and make it through. Although how microorganisms make such decision remains largely unknown, it clearly involves orchestrated processes, including modulation of gene expression and interactions between proteins, as well as proteins and small molecules such as c-di-GMP (Karatan and Watnick, 2009). From a practical view point, biofilm has been associated with various problems in human daily life, especially in the food, environment, and biomedical fields (Leaper et al., 2010; Simões et al., 2010), better understanding of biofilm and its formation would help us tackle them.
Thanks to extensive studies on various microbes that have accumulated enormous amount of information, we now have gained considerable understanding of the numerous cellular networks that regulate biofilm formation. However, most of them concern regulation of EPS biosynthesis and other mechanisms are much less understood. In this work, we investigated roles of a protein cascade in biofilm formation in MR-1. We discovered that BpfA–BpfG–BpfD forms an interactive system that governs biofilm formation. Central to this system is BpfA, which is a very large protein although substantially smaller than P. fluorescens LapA. Because of a large number of repeats, the bpfA gene in the released genome sequence is ∼2.6 kb shorter. Each repeat, as illustrated in lapA, is about 300 bp for 100 amino acid residues (Boyd et al., 2014). Also because of these repeats, we failed to clone the full-length bpfA for complementation and other analyses, a situation encountered by O’Toole and Kolter (1998) team on lapA (personal communications). To date, many such proteins have been identified and exclusively function as surface-associated adhesions (Boyd et al., 2014).
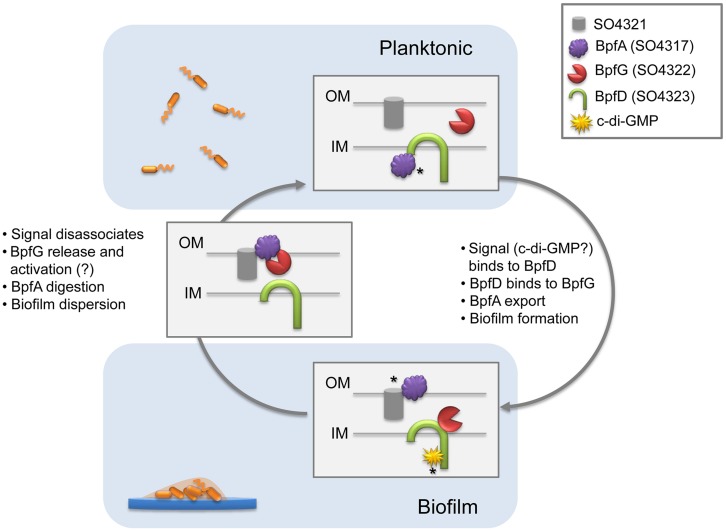
Although this three-membered system partially resembles LapA–LapG–LapD of P. fluorescens (Newell et al., 2011), there are significant differences in certain key features. In the best studied LapA–LapG–LapD system, the transmembrane receptor LapD is activated by high cytoplasmic concentrations of c-di-GMP, which in turn recruits the periplasmic protease LapG, preventing it from cleaving LapA, thereby promoting cell adhesion and subsequent biofilm formation (Newell et al., 2009, 2011; Boyd et al., 2012, 2014; Chatterjee et al., 2014). Our proposed model is as shown in Figure 6: BpfA, exported by the TISS channel formed by AggABC and anchored in the outer membrane, serves as a surface attached adhesin mediating cell–cell or cell–matrix adhesion. Our data suggest that intracellular BpfA molecules are probably held by BpfD before exportation, and free BpfA may not be exported efficiently. On the contrary, bindings of c-di-GMP and of BpfG to the respective cytosolic and the periplasmic domains of BpfD results in rapid BpfA exportation and biofilm formation. This explains the biphasic impact of BpfD on biofilm formation: enrichment of BpfA in the membrane fraction from cells overproducing BpfD and requirement of c-di-GMP and BpfG for biofilm formation. The balancing point is achieved when the stoichiometry between BpfG and BpfD is maintained at 1:1. BpfG may be released from BpfD upon c-di-GMP hydrolyzation or some other cues, which can move to BpfA for cleavage such that BpfA can be released from the outer membrane. When over-produced, BpfG overwhelms the BpfD control, leading to cleavage of BpfA and defect in biofilm formation. The role of the OmpA-like protein coded by SO4321 in the process is not known. But given its location and predicted functions, the protein probably participates in translocation of BpfD and BpfG, as AggABC TISS transports protein from the cytosol to the extracellular space directly. Efforts to test this notion are under way.
FIGURE 6.
Proposed model for regulatory biofilm formation mechanism in S. oneidensis MR-1. In planktonic cells, BpfA is captured by BpfD to prevent biofilm formation. In biofilm forming stage, c-di-GMP and BpfG bind to BpfD, resulting in BpfA release and export by TISS system (not shown in figure). SO4321 facilitates BpfA localization on the outer membrane, and biofilm forms. c-di-GMP hydrolyzation and disassociation from BpfD leads to BpfG release and possibly activation of its proteinases activity, BpfA is subsequently digested and biofilm disperses. Asterisks in the figure indicate places of pure logical speculation at this point, and invites further investigation.
Given the phylogenic closeness of Shewanella and Pseudomonas (Wu et al., 2008), it is not surprising that the “glue” function of BpfA, proteinase function of BpfG, and transmembrane regulator role of BpfD are shared between our model and the LapA–LapG–LapD network in P. fluorescens. However, the involvement of SO4321 in biofilm formation, the essentiality of BpfG in BpfA export, and the critical 1:1 ratio of BpfG and BpfD are all data-supported novel aspects of the MR-1 Bpf system. In addition, the lack of impact of medium phosphate level on biofilm formation suggests that the upstream regulation is also distinct in Shewanella. It would be interesting to track down how BpfG and BpfD take on their new roles, as well as how the overarching regulatory scheme diverges in these closely related bacteria.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by National Natural Science Foundation of China (31270097, 41476105) to HG; and by National Natural Science Foundation of China (31100600), Fundamental Research Funds for the Central Universities (2014FZA6002), and Ministry of Education Science and Technology Development Center (20120541) to JY.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00790
References
- Boyd C. D., Chatterjee D., Sondermann H., O’toole G. A. (2012). LapG, required for modulating biofilm formation by Pseudomonas fluorescens Pf0-1, is a calcium-dependent protease. J. Bacteriol. 194 4406–4414. 10.1128/JB.00642-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd C. D., Smith T. J., El-Kirat-Chatel S., Newell P. D., Dufrêne Y. F., O’Toole G. A. (2014). Structural features of the Pseudomonas fluorescens biofilm adhesin LapA required for LapG-dependent cleavage, biofilm formation, and cell surface localization. J. Bacteriol. 196 2775–2788. 10.1128/JB.01629-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. N., Romine M. F., Schepmoes A. A., Smith R. D., Lipton M. S. (2010). Mapping the subcellular proteome of Shewanella oneidensis MR-1 using sarkosyl-based fractionation and LC-MS/MS protein identification. J. Proteome Res. 9 4454–4463. 10.1021/pr100215h [DOI] [PubMed] [Google Scholar]
- Cao L., Yan X., Borysenko C. W., Blair H. C., Wu C., Yu L. (2005). CHDL: a cadherin-like domain in Proteobacteria and Cyanobacteria. FEMS Microbiol. Lett. 251 203–209. 10.1016/j.femsle.2005.08.004 [DOI] [PubMed] [Google Scholar]
- Chatterjee D., Cooley R. B., Boyd C. D., Mehl R. A., O’Toole G. A., Sondermann H. (2014). Mechanistic insight into the conserved allosteric regulation of periplasmic proteolysis by the signaling molecule cyclic-di-GMP. Elife 3:e03650 10.7554/eLife.03650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros M. G., von Heijne G. (1994). TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10 685–686. 10.1093/bioinformatics/10.6.685 [DOI] [PubMed] [Google Scholar]
- Cucarella C., Solano C., Valle J., Amorena B., Lasa I., Penades J. R. (2001). Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183 2888–2896. 10.1128/JB.183.9.2888-2896.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De N., Pirruccello M., Krasteva P. V., Bae N., Raghavan R. V., Sondermann H. (2008). Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol. 6:e67 10.1371/journal.pbio.0060067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Windt W., Gao H., Kromer W., Van Damme P., Dick J., Mast J., et al. (2006). AggA is required for aggregation and increased biofilm formation of a hyper-aggregating mutant of Shewanella oneidensis MR-1. Microbiology 152 721–729. 10.1099/mic.0.28204-0 [DOI] [PubMed] [Google Scholar]
- El-Kirat-Chatel S., Beaussart A., Boyd C. D., O’toole G. A., Dufrene Y. F. (2014). Single-cell and single-molecule analysis deciphers the localization, adhesion, and mechanics of the biofilm adhesin LapA. ACS Chem. Biol. 9 485–494. 10.1021/cb400794e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazli M., Almblad H., Rybtke M. L., Givskov M., Eberl L., Tolker-Nielsen T. (2014). Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ. Microbiol. 16 1961–1981. 10.1111/1462-2920.12448 [DOI] [PubMed] [Google Scholar]
- Flemming H.-C., Wingender J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8 623–633. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- Fredrickson J. K., Romine M. F., Beliaev A. S., Auchtung J. M., Driscoll M. E., Gardner T. S., et al. (2008). Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 6 592–603. 10.1038/nrmicro1947 [DOI] [PubMed] [Google Scholar]
- Fu H., Jin M., Wan F., Gao H. (2015). Shewanella oneidensis cytochrome c maturation component CcmI is essential for heme attachment at the non-canonical motif of nitrite reductase NrfA. Mol. Microbiol. 95 410–425. 10.1111/mmi.12865 [DOI] [PubMed] [Google Scholar]
- Gao H., Wang X., Yang Z. K., Chen J., Liang Y., Chen H., et al. (2010). Physiological roles of ArcA, Crp, and EtrA and their interactive control on aerobic and anaerobic respiration in Shewanella oneidensis. PLoS ONE 5:e15295 10.1371/journal.pone.0015295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L., Costerton J. W., Stoodley P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2 95–108. 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- Heidelberg J. F., Paulsen I. T., Nelson K. E., Gaidos E. J., Nelson W. C., Read T. D., et al. (2002). Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20 1118–1123. 10.1038/nbt749 [DOI] [PubMed] [Google Scholar]
- Jin M., Jiang Y., Sun L., Yin J., Fu H., Wu G., et al. (2013). Unique organizational and functional features of the cytochrome c maturation system in Shewanella oneidensis. PLoS ONE 8:e75610 10.1371/journal.pone.0075610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatan E., Watnick P. (2009). Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73 310–347. 10.1128/MMBR.00041-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaper D., Mcbain A. J., Kramer A., Assadian O., Sanchez J. L., Lumio J., et al. (2010). Healthcare associated infection: novel strategies and antimicrobial implants to prevent surgical site infection. Ann. R. Coll. Surg. Engl. 92 453–458. 10.1308/003588410X12699663905276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesk A. M. (2012). “Mapping, sequencing, annotation and databases,” in Introduction to Genomics, 2nd Edn (New York, NY: Oxford university press; ). [Google Scholar]
- Letunic I., Doerks T., Bork P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43 D257–D260. 10.1093/nar/gku949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Gao H., Chen J., Dong Y., Wu L., He Z., et al. (2010). Pellicle formation in Shewanella oneidensis. BMC Microbiol. 10:291 10.1186/1471-2180-10-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Gao H., Guo X., Chen J., Qiu G., He Z., et al. (2012). Transcriptome analysis of pellicle formation of Shewanella oneidensis. Arch. Microbiol. 194 473–482. 10.1007/s00203-011-0782-x [DOI] [PubMed] [Google Scholar]
- Logan B. E., Regan J. M. (2006). Electricity-producing bacterial communities in microbial fuel cells. Trends Microbiol. 14 512–518. 10.1016/j.tim.2006.10.003 [DOI] [PubMed] [Google Scholar]
- Luo Q., Dong Y., Chen H., Gao H. (2013). Mislocalization of Rieske protein PetA predominantly accounts for the aerobic growth defect of tat mutants in Shewanella oneidensis. PLoS ONE 8:e62064 10.1371/journal.pone.0062064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall M. J., Beliaev A. S., Dohnalkova A. C., Kennedy D. W., Shi L., Wang Z., et al. (2006). c-Type cytochrome-dependent formation of U(IV) nanoparticles by Shewanella oneidensis. PLoS Biol. 4:e268 10.1371/journal.pbio.0040268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers C. R., Nealson K. H. (1988). Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240 1319–1321. 10.1126/science.240.4857.1319 [DOI] [PubMed] [Google Scholar]
- Newell P. D., Boyd C. D., Sondermann H., O’toole G. A. (2011). A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol. 9:e1000587 10.1371/journal.pbio.1000587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell P. D., Monds R. D., O’toole G. A. (2009). LapD is a bis-(3’,5’)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc. Natl. Acad. Sci. U.S.A. 106 3461–3466. 10.1073/pnas.0808933106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole G. A. (2003). To build a biofilm. J. Bacteriol. 185 2687–2689. 10.1128/JB.185.9.2687-2689.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole G. A., Kolter R. (1998). Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28 449–461. 10.1046/j.1365-2958.1998.00797.x [DOI] [PubMed] [Google Scholar]
- Pirbadian S., Barchinger S. E., Leung K. M., Byun H. S., Jangir Y., Bouhenni R. A., et al. (2014). Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. Proc. Natl. Acad. Sci. U.S.A. 111 12883–12888. 10.1073/pnas.1410551111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F., Qi Y., Chong H. S., Kotaka M., Li B., Li J., et al. (2009). The functional role of a conserved loop in EAL domain-based cyclic di-GMP-specific phosphodiesterase. J. Bacteriol. 191 4722–4731. 10.1128/JB.00327-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese M. G. (2001). Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 26 51–56. 10.1016/S0097-8485(01)00099-7 [DOI] [PubMed] [Google Scholar]
- Shi M., Gao T., Ju L., Yao Y., Gao H. (2014). Effects of FlrBC on flagellar biosynthesis of Shewanella oneidensis. Mol. Microbiol. 93 1269–1283. 10.1111/mmi.12731 [DOI] [PubMed] [Google Scholar]
- Shi M., Wu L., Xia Y., Chen H., Luo Q., Sun L., et al. (2013). Exoprotein production correlates with morphotype changes of non-motile Shewanella oneidensis mutants. J. Bacteriol. 195 1463–1474. 10.1128/JB.02187-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões M., Simões L. C., Vieira M. J. (2010). A review of current and emergent biofilm control strategies. LWT Food Sci. Technol. 43 573–583. 10.1016/j.lwt.2009.12.008 [DOI] [Google Scholar]
- Sun L., Jin M., Ding W., Yuan J., Kelly J., Gao H. (2013). Post-translational modification of flagellin FlaB in Shewanella oneidensis. J. Bacteriol. 195 2550–2561. 10.1128/JB.00015-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen S., De Smet L., Dansercoer A., Motte B., Coenye T., Van Beeumen J. J., et al. (2010). The 285 kDa Bap/RTX hybrid cell surface protein (SO4317) of Shewanella oneidensis MR-1 is a key mediator of biofilm formation. Res. Microbiol. 161 144–152. 10.1016/j.resmic.2009.12.002 [DOI] [PubMed] [Google Scholar]
- Theunissen S., Vergauwen B., De Smet L., Van Beeumen J., Van Gelder P., Savvides S. N. (2009). The agglutination protein AggA from Shewanella oneidensis MR-1 is a TolC-like protein and forms active channels in vitro. Biochem. Biophys. Res. Commun. 386 380–385. 10.1016/j.bbrc.2009.06.044 [DOI] [PubMed] [Google Scholar]
- Venkateswaran K., Moser D. P., Dollhopf M. E., Lies D. P., Saffarini D. A., MacGregor B. J., et al. (1999). Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49(Pt 2) 705–724. 10.1099/00207713-49-2-705 [DOI] [PubMed] [Google Scholar]
- Wu L., Liu X., Fields M. W., Thompson D. K., Bagwell C. E., Tiedje J. M., et al. (2008). Microarray-based whole-genome hybridization as a tool for determining procaryotic species relatedness. ISME J. 2 642–655. 10.1038/ismej.2008.23 [DOI] [PubMed] [Google Scholar]
- Wu L., Wang J., Tang P., Chen H., Gao H. (2011). Genetic and molecular characterization of flagellar assembly in Shewanella oneidensis. PLoS ONE 6:e21479 10.1371/journal.pone.0021479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Chen Y., Zhou G., Chen H., Gao H. (2013). Investigation of roles of divalent cations in Shewanella oneidensis pellicle formation reveals unique impacts of insoluble iron. Biochim. Biophys. Acta 1830 5248–5257. 10.1016/j.bbagen.2013.07.023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.