Abstract
Antibiotic resistant bacteria are ubiquitous in the natural environment. The introduction of effluent derived antibiotic resistance genes (ARGs) into aquatic environments is of concern in the spreading of genetic risk. This study showed the prevalence of sulfonamide and tetracycline resistance genes, sul1, sul2, sul3, and tet(M), in the total bacterial assemblage and colony forming bacterial assemblage in river and estuarine water and sewage treatment plants (STP) in South Africa. There was no correlation between antibiotic concentrations and ARGs, suggesting the targeted ARGs are spread in a wide area without connection to selection pressure. Among sul genes, sul1 and sul2 were major genes in the total (over 10-2 copies/16S) and colony forming bacteria assemblages (∼10-1 copies/16S). In urban waters, the sul3 gene was mostly not detectable in total and culturable assemblages, suggesting sul3 is not abundant. tet(M) was found in natural assemblages with 10-3 copies/16S level in STP, but was not detected in colony forming bacteria, suggesting the non-culturable (yet-to-be cultured) bacterial community in urban surface waters and STP effluent possess the tet(M) gene. Sulfamethoxazole (SMX) resistant (SMXr) and oxytetracycline (OTC) resistant (OTCr) bacterial communities in urban waters possessed not only sul1 and sul2 but also sul3 and tet(M) genes. These genes are widely distributed in SMXr and OTCr bacteria. In conclusion, urban river and estuarine water and STP effluent in the Durban area were highly contaminated with ARGs, and the yet-to-be cultured bacterial community may act as a non-visible ARG reservoir in certain situations.
Keywords: antibiotic resistance, sul, tet(M), yet-to-be cultured, South Africa, sewage treatment plant
Introduction
Antibiotic resistance genes (ARGs) are found not only in the clinical but also the natural environment, which can eventually produce antibiotic resistant bacteria (ARB). Antibiotics and ARB are released to the environment from hospitals, livestock facilities, and sewage treatment plants (STP) (Pruden et al., 2013). Although antibiotics are decomposed and diluted in the aquatic environment water, even at low concentrations they may act as signaling molecules in microbes (Fajardo and Martinez, 2008). Selection of ARG mutation by very low concentrations of antibiotics is reported (Gullberg et al., 2011). It is, therefore, critical to understand the fate of released antibiotics, ARB and ARGs in the environment, and whether residual ARGs in the environment pose a risk to humans. The aim of this study was to assess the status of antibiotics and ARGs in anthropogenically impacted surface waters in one area of South Africa.
The status of antibiotic use and STP operation differs between countries. Consequently, the status of antibiotic contamination and presence of ARBs and ARGs in aquatic ecosystems must be established on a case by case basis. In previous monitoring we showed the status of antibiotic contamination (Shimizu et al., 2013) and ARGs (Suzuki et al., 2013) in numerous Asian countries. In many tropical Asian countries an integrated system of agriculture is followed, which includes animal husbandry, aquaculture, and crop farming (Suzuki and Hoa, 2012). In this system the major antibiotic used for animals is sulfonamides. Tetracyclines are also used in aquaculture. STPs receive wastewater and excreta from humans and livestock facilities, which intimates the mixing of waters containing various antibiotics, ARB and ARGs. The main purpose of conventional STPs is to prevent the spread of infectious diseases and reduce solid and nutrient loads from excreta entering surface waters, not to decompose pharmaceuticals and genes. Although advanced disinfection technologies can greatly reduce the danger of waterborne diseases (United States Environmental Protection and Agency, 2004), antibiotics and ARGs are not completely decomposed in the STP process and are released into the environment (Rizzo et al., 2013; Berkner et al., 2014).
The populations and economies of African countries are developing. Although South Africa has a relatively well developed economy by African standards, many STPs are not functioning efficiently and are overloaded and has been identified as a serious cause for concern (Snyman et al., 2006; Water Research and Commission, 2006). Furthermore, most South African cities are characterized by large informal settlements where sanitation facilities are poor and in some cases essentially non-existent, with pit latrines and mobile toilets usually the only form of sanitation. This might result in the introduction of antibiotics, ARB and ARGs into the aquatic environment. Omulo et al. (2015) reviewed many articles on ARB research from Eastern Africa, which mainly studied on human and animal bacteria. Environmental ARB needs to be studied further.
It is well known that the majority of bacteria in aquatic environments are non-culturable or yet-to-be cultured bacteria (Bloomfield et al., 1988; Amann et al., 1995; Takami et al., 2009). In recent monitoring in the Philippines we showed that the total bacterial community in seawater possessed minor sulfonamide resistance gene sul3, which was not detected in colony forming bacteria (Suzuki et al., 2013). This suggests that the abundant non-culturable or yet-to-be cultured bacteria in aquatic environments are a reservoir of ARGs, but these are not detectable by culture methods. The sul3 gene was detected in human and non-human isolates of Salmonella in Portuguese waters, although sul3 was a minor contributor compared to sul1 and sul2 genes (Antunes et al., 2005). In Denmark, Escherichia coli isolated from pork and pigs possessed sul3, but this gene was not found in human isolates (Hammerum et al., 2006). In Germany, sul3 was not found in E. coli of human isolates, but was found in cattle, pig, and poultry isolates (Guerra et al., 2003). These studies suggest that sul3 is spreading widely amongst animals but not amongst humans, possibly due to the use of sulfonamide for animal husbandry but not in humans in developed countries, and that sul3 is transferred by a different gene cassette to sul1 and sul2 (Antunes et al., 2005). Although recent advances in metagenomics can detect total resistome, quantitative estimation of ARGs in the microbial community is not yet possible. An understanding of the reservoir of culturable- and non-culturable bacteria in the environment might thus be useful in assessing whether environmental ARGs are posing a risk.
The aim of this study was to assess the abundance of sul1, sul2, sul3, and tet(M) genes in total- and colony forming-bacterial assemblages in surface waters and STP effluent in the eThekwini area of South Africa. As far as we are aware, no information in this context is available for this area, or indeed for other areas in South Africa. Sulfonamides and tetracyclines have a long use as human and animal therapeutic agents and animal growth promoters. Sulfonamide resistance occurs mainly by mutation of the dihydropteroate synthase (DHPS) gene, although other mechanisms are known (Radstrom and Swedberg, 1988; Huang et al., 2004). As to tetracycline resistance, 45 tet genes are known at this time (Roberts et al., 2012). Among the tet genes, tet(M), a ribosomal protection protein gene is suspected of having the broadest host range (Roberts et al., 2012) and its origin is reported to be ancient (Kobayashi et al., 2007). Additionally, tet(M) shows high genetic diversity (Rizzotti et al., 2009) and wide distribution in the natural environment (D’Costa et al., 2011). Therefore, we focused on the sul genes and tet(M) as monitoring targets. We hypothesized that effluent from inefficient STPs or wastewater derived from poor sanitary conditions should contain high concentrations of ARGs from human bacteria. The comparison of ARGs using culture-dependent and independent methods should, therefore, provide an understanding on whether bacterial communities of natural or human origin are the major reservoir of ARGs in aquatic ecosystems.
Materials and Methods
Sampling of Water
Samples were collected with an ethanol rinsed stainless steel bucket between September 3 and 5 in 2012, in the eThekwini Metropolitan Municipality area in the province of KwaZulu-Natal, on the subtropical northeast cost of South Africa (Figure 1). Characteristics of the sampling sites are summarized in Supplementary Table S1. The municipality has a population size of about 3,400,000 (Statistics South Africa, http://www.statssa.gov.za/?page_id=1021&id=ethekwini-municipality). The city of Durban and a number of smaller towns fall in the municipal area. Rainfall in the eThekwini area is seasonal, falling predominantly in summer. Although it was not raining at the times that samples were collected, about 31 mm of rain was recorded at rain monitoring gage in Durban during the sampling period. Because samples were collected at a single point in time we recognize this study does not provide an understanding on the temporal variability of ARBs and ARGs in surface waters and STP effluents in the study area.
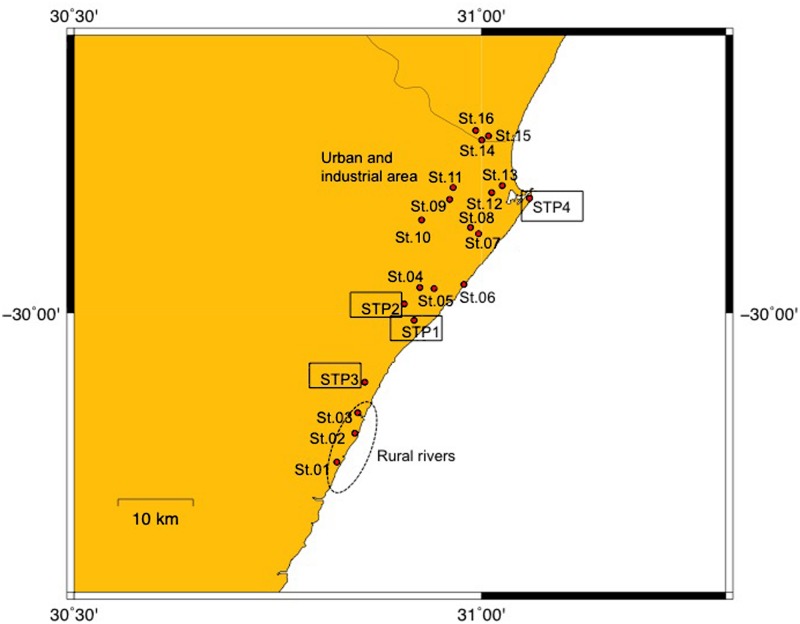
FIGURE 1.
Map of sampling sites. Sites 01–03 are rural river sites, Sites 04–16 are urban river sites, and STP1-4 are sewage treatment plants (STPs).
Surface water samples were taken at three sites (Sites 01–03) in the estuarine parts of rivers situated in rural locations. Thirteen sites (Sites 04–16) were sampled in the riverine and estuarine parts of rivers with urbanized and industrialized catchments in the greater Durban area, and effluent was collected from four STPs (STP1–4). The water and effluent was filtered through 50 μm mesh plankton net to remove large debris, and stored on ice for a few hours until analysis. Further detail on the condition of surface waters at the river and estuarine sites is provided in Segura et al. (2015). Water samples indicate present status of contamination (Takasu et al., 2011).
Antibiotic Concentration
Sulfonamides and tetracyclines were analyzed using a liquid chromatograph (Accela, Thermo Scientific) equipped with a tandem mass spectrometer (LC-MS/MS; Quantum Access, Thermo Scientific) after extraction using a solid-phase cartridge (Oasis HLB resin, Waters). The analytical process was the same as that provided in Segura et al. (2015).
Bacterial Count
Total bacterial cell number was counted by DAPI staining according to Sato-Takabe et al. (2015). Total viable count and sulfamethoxazole resistant (SMXr) and oxytetracycline resistant (OTCr) bacterial numbers were enumerated on nutrient agar plates (LB plus 1.5% agar) incubated at 30°C for 24 h. To estimate SMXr and OTCr bacteria, 60 μg/mL of each drug was supplemented to the medium (Hoa et al., 2011). All plate counts were performed in duplicate.
Quantitative Analysis of Antibiotic Resistance Genes (ARGs)
The sulfonamide resistance genes, sul1, sul2, and sul3, and tetracycline resistance gene, tet(M), were quantified by quantitative PCR (qPCR) from total assemblage using total DNA trapped on 0.2 μm pore filter. For the culturable bacterial assemblage, all colonies on agar plates were mixed and used for qPCR. DNA extraction from the filter and mixtures of colonies was previously reported (Suzuki et al., 2013). DNA from filters and colonies were obtained from triplicate biological samples. qPCR was performed using a CFX 96 Real-Time system (BioRad, Laboratories, Hercules, CA, USA) to detect an increase of double-stranded DNA with an increase in fluorescence according to Suzuki et al. (2013). PCR amplifications were performed in a 20 μl reaction volume containing 1 X Sso Fast EvaGreen Supermix (Bio-Rad), 500 nM of each primer and 1 μl of sample DNA. qPCR was performed using previously designed primers; bacterial 16S rRNA genes (Suzuki et al., 2000), sul1 (Heuer and Smalla, 2007), sul2 (Heuer et al., 2008), sul3 (Pei et al., 2006), and tet(M) (Tamminen et al., 2011). Serial 1:10 dilutions of plasmids constructed from the pGEM-T Easy vector (Promega, Madison, WI, USA) and 16S rRNA gene from E. coli K12, sul1 from plasmid R388, sul2 from plasmid RSF1010, sul3 from plasmid pUVP4401 (Heuer and Smalla, 2007), and tet(M) from pFD310 fragments (Smith et al., 1992) were used as standards for quantification. The qPCR program consisted of an initial denaturation of 30 s at 95°C and 40 cycles of 5 s at 95°C and 10 s at 50°C for 16S rRNA gene and 10 s at 51°C for sul1 and sul2 and 20 s at 60°C for sul3, and 20 s at 57°C for tet(M), respectively. Melting curves for the amplicons were measured by raising the temperature slowly from 60°C and 65°C to 95°C for 16S rRNA gene, sul1, sul2, sul3, tet(M), and sul3, respectively, while monitoring fluorescence. Each sample was measured in triplicate. The copy numbers of sul1, sul2, sul3, and tet(M) were normalized by dividing by the 16S rRNA gene copy number at the respective time points to take into account any temporal variation in bacterial cell numbers. Unit of the copy number is described as copies/16S in the text. The results were analyzed using a Big Dye terminator kit on a 3130 ABI Prism sequencer (Applied Biosystems, Foster City, CA, USA). PCR products were sequenced to confirm they were not non-specific products.
Results and Discussion
Drug Contamination
The distribution of antibiotic concentrations in surface waters and STP effluent showed that SMX was a major contaminant along with trimethoprim, which is a combination drug. The SMX concentrations were: rural surface waters - 48.2 ± 71.2 ng/L (n = 3), urban surface waters - 2561 ± 51.3 ng/L (n = 13), STP effluent - 3612 ± 1733.4 ng/L (n = 4). High SMX concentrations in urban surface waters and STP effluent indicate its frequent use in human chemotherapy. It is also reported that SMX is frequently used in African countries to control bacteria and protozoan infections in HIV patients (Zachariah et al., 2007). Recently report in Ghana, Mozambique, Kenya, and South Africa showed that the SMX is the highest concentration among selected 18 antibiotics in all countries (Segura et al., 2015). Data from STP in the present study showed high concentration compared to these. Tetracyclines were mostly not detectable in surface waters and STP effluent (maximum 18 ng/L, and mostly below detection limit). At one STP (STP4), however, 291 ng/L of OTC was detected, indicating real time use of the drug. The results suggest that SMX is used frequently in the Durban area. The concentration over 1000 ng/L was similar to a pig farm in Vietnam (Hoa et al., 2011; Shimizu et al., 2013), and double that of STP effluent in Michigan, U. S. (Gao et al., 2012b). Erythromycin (1194 ng/L) was also present in STP4 effluent, but was not particularly prevalent in surface water samples, suggesting the antibiotics originated from human medicines. The high contamination of surface waters and STP effluents by antibiotics suggests that ARGs in hospitals are also likely entering the environment (Pruden, 2014).
Bacterial Numbers
The counts of bacteria in different surface water and STP effluent samples are shown in Table 1, as enumerated by DAPI count (total number), plate count (colony forming number), and SMXr and OTCr bacterial counts. Total cell number was almost the same in the rural and urban surface waters, with 106 cells/ml, but an order of magnitude higher in STP effluents. The colony forming number was two orders of magnitude lower than the total cell number. The contribution of culturable bacteria to the total cell number was 1.0–1.5% in surface waters and 6.8% in STP effluents, a statistically significant difference (p < 0.05, t-test). The culturable bacterial contribution to the total cell number in freshwater is reported to be approximately 0.25% (Amann et al., 1995), indicating that the number of culturable bacteria was higher in surface waters and STP effluents in Durban. Dominance rate of viable number was higher in urban surface waters and STP effluent compared to rural surface waters, suggesting contamination of culturable bacteria is derived from human sources. Iweriebor et al. (2015) reported in South Africa that resistance rate of Enterococcus from hospital and STP effluents was 67–100%. The contribution of ARB in STP effluent was higher than in surface waters in our study (SMXr, p < 0.05 and OTCr, p < 0.01). Culturable bacteria in STP effluent should include enteric bacteria, which form colonies on agar plates with a contribution of 15% (Langendijk et al., 1995) compared to 0.1% in seawater (Amann et al., 1995; Fuhrman and Hagström, 2008). Abundances of SMXr and OTCr bacteria were not positively correlated to antibiotic concentrations. It is reported that drug concentrations and occurrence of ARB are not correlated to fluoroquinolones in environment (Takasu et al., 2011). Although the reason why sulfonamide- and tetracycline-resistance are frequently found in non-contaminated environments is not known, the heavy use of sulfonamides and tetracyclines in the 20th century could be one of the reasons for the selection of SMXr - and OTCr -genes in bacterial communities. Sediment stores sul and tet genes for a long time in non-contaminated areas (Tamminen et al., 2011; Muziasari et al., 2014), whereas water samples indicate present status. The abundance of ARB in surface waters suggests their continuous input into the environment. The ARGs for these drugs should be distributed in various environmental bacteria around the world. There are factors other than antibiotics, such as metals (Knapp et al., 2011), that may select for ARB and ARGs in natural bacterial assemblages.
Table 1.
Bacterial number in three categorized sites.
| Site | Total cell count (cells/ml) | Colony count (CFU/ml) (% of total) | Sulfamethoxazole resistant (SMXr; CFU/ml) (% of colony count) | Oxytetracycline resistant (OTCr; CFU/ml) (% of colony count) |
|---|---|---|---|---|
| Rural river (n = 3) | (1.1 ± 0.47) × 106 | (1.1 ± 1.4) × 104 (1.0%) | (1.8 ± 3.0) × 103(16.8%) | (5.0 ± 7.0) × 102(4.7%) |
| Urban and industrial river (n = 13) | (2.7 ± 2.7) × 106 | (4.1 ± 4.8) × 104(1.5) | (8.1 ± 7.7) × 103 (20.0) | (4.8 ± 6.9) × 103(11.9) |
| Sewage treatment plant (STP) (n = 4) | (1.0 ± 1.0) × 107 | (7.0 ± 11) × 105(6.8) | (2.1 ± 3.0) × 105 (30.4) | (4.8 ± 5.1) × 104(6.9) |
The sul and tet(M) Genes in Total- and Culturable-Assemblages
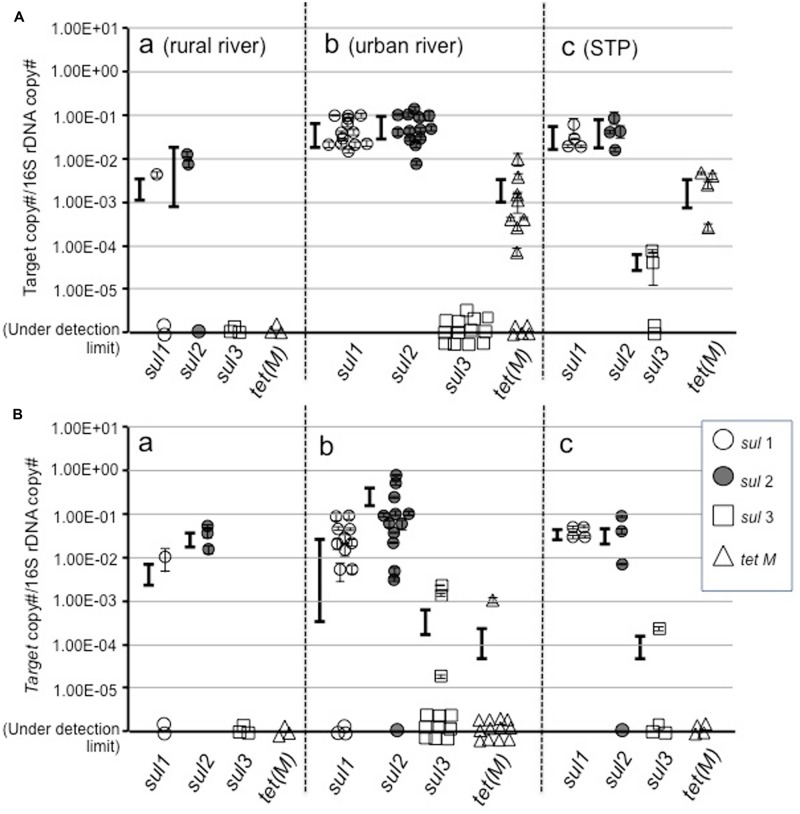
Among sul genes, sul1 and sul2 were detected at a similar copy number in total assemblages in all categories of water (Figure 2A), and also in colony forming bacteria (Figure 2B). This indicates that sul1 and sul2 are ubiquitous in bacterial communities, including yet-to-be cultured and culturable bacteria in aquatic environments in the Durban area. At most urban river and estuarine and STP sites, sul1 and sul2 were present at copy numbers of 10-2–10-1/16S. These values are higher than at rural sites in the Durban area, and in the Philippines (Suzuki et al., 2013) and in Finnish sediment (Muziasari et al., 2014), but are comparable to values reported for suspended solids in lagoon waters (McKinney et al., 2010).
FIGURE 2.
Abundance of sul1, sul2, sul3, and tet(M) genes in total (A) and culturable bacterial assemblages (B).
Profiles for sul3 and tet(M) were different from sul1 and sul2 between total assemblage and culturable bacteria. The sul3 gene was not detected or was at a very low abundance in the total assemblage at most sites, although two sites showed 10-5–10-4/16S. In the case of the Philippines, sul3 was not detected in natural assemblages or colony forming bacteria in freshwater lakes and rivers, whereas a high copy number was detected in seawater assemblages (Suzuki et al., 2013). The fact that sul3 was not at a high copy number in culturable bacteria (Figure 2B) suggests this gene is not abundant in the Durban area. Gao et al. (2012a) reported similar results in freshwater.
In the case of tet(M), the total assemblage in urban surface waters and STP effluents possessed approximately 10-3/16S, whereas culturable bacteria did not. This suggests the yet-to-be cultured community possesses tet(M). Since the yet-to-be cultured bacteria comprise the major component of the bacterial community, the gene pool of tet(M) in environment should be large. A risk assessment for ARGs amongst this silent majority is required.
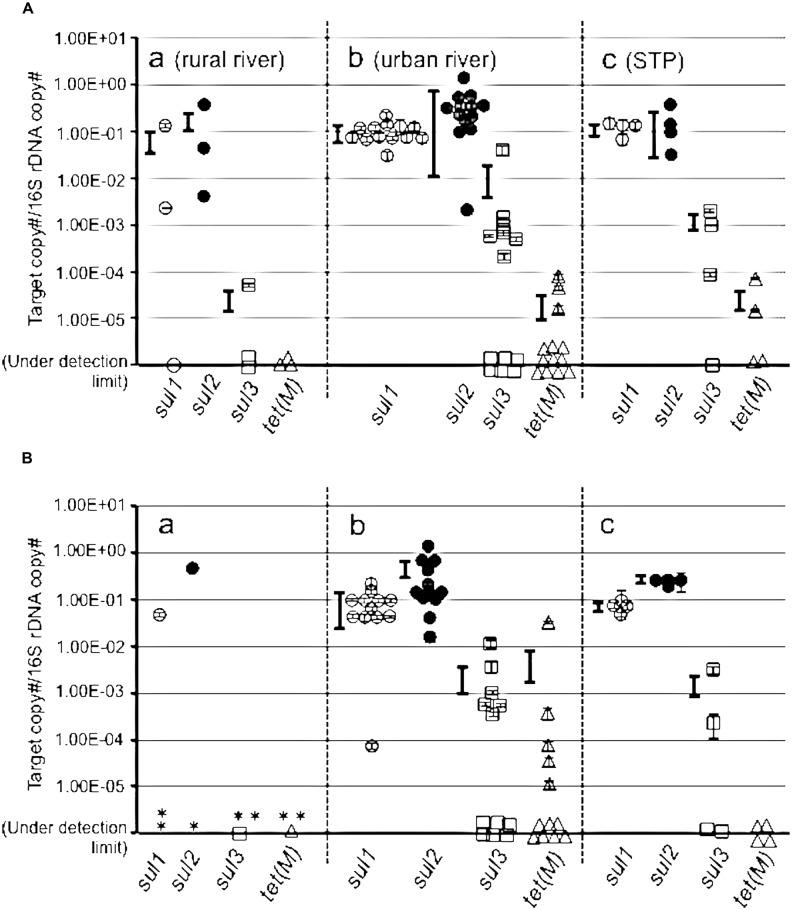
The copy numbers of the targeted ARGs were measured in a pooled colony from SMXr and OTCr bacteria (Figure 3). The sul1 and sul2 were higher than 10-1/16S at urban and STP sites, with sul3 around 10-3∼10-2/16S (Figure 3A). The rural sites also showed high copies of sul1 and sul2, but sul3 was detected at only one site at a low concentration. This indicates that colony forming SMXr bacteria possess sul genes, which were selected on SMX-containing agar plate. The sul genes in the OTCr assemblage also showed a high copy number of sul genes (Figure 3B). It is reported that sul and tet genes are sometimes coded on the same plasmid of aquatic bacteria (Kim et al., 2008; Nonaka et al., 2012), and SMXr and OTCr phenotypes are frequently linked (Hu et al., 2008). The present study supports the findings in terms of gene copy numbers in assemblages by cross checking with SMXr and OTCr bacteria. On the other hand, tet(M) copy number was less than 10-2 in SMXr and OTCr bacteria at most sites, suggesting two possibilities. One is that the selected bacteria by SMX and OTC possess other tet genes than tet(M), and is the other that tet(M) is abundant in total assemblages but not in culturable resistant bacteria.
FIGURE 3.
Abundance of sul1, sul2, sul3, and tet(M) genes in sulfamethoxazole resistant (SMXr) colonies mix (A) and in oxytetracycline resistant (OTCr) colonies mix (B). (B) Asterisk shows sites where resistant isolate was not obtained. Symbols are the same to Figure 2.
Conclusion
Quantitative PCR and culture methods revealed that sul genes are conveyed by bacterial communities in urban surface waters and STP effluent in the Durban area of South Africa. Additionally, sul3 was detected in the culturable bacteria assemblage. The yet-to-be cultured bacterial community may act as a non-visible reservoir of ARGs in certain situations.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate the assistance of Dr. T. Yokokawa and other members in the field and laboratory. The authors thank Grant-in-Aids from JSPS (90196816, 22241014, 22254001) for funding this study.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00796
References
- Amann R. I., Ludwig W., Schleifer K.-H. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59 143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes P., Machado J., Sousa J. C., Peixe L. (2005). Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob. Agents Chemother. 49 836–839. 10.1128/AAC.49.2.836-839.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkner S., Konradi S., Schönfeld J. (2014). Antibiotic resistance and the environment—there and back again. EMBO Rep. 15 740–744. 10.15252/embr.201438978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield S. F., Gordon S., Stewart A. B., Dodd C. E. R., Booth I. R., Power E. G. M. (1988). The viable but non-culturable phenomenon explained? Microbiology 144 1–3. 10.1099/00221287-144-1-1 [DOI] [PubMed] [Google Scholar]
- D’Costa V. M., King C. E., Kalan L., Morar M., Sung W. W. L., Schwarz C., et al. (2011). Antibiotic resistance is ancient. Nature 477 458–461. [DOI] [PubMed] [Google Scholar]
- Fajardo A., Martinez J. L. (2008). Antibiotics as signals that trigger specific bacterial responses. Curr. Opin. Microbiol. 11 161–167. 10.1016/j.mib.2008.02.006 [DOI] [PubMed] [Google Scholar]
- Fuhrman J. A., Hagström Å. (2008). “Bacterial and archaeal community structure and its patterns,” in Microbial Ecology of the Oceans 2nd Edn ed. Kirchman D. L. (Hoboken, NJ: Wiley Sons; ) 45–90. [Google Scholar]
- Gao P., Mao D., Luo Y., Wang L., Xu B., Xu L. (2012a). Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Wat. Res. 46 2355–2364. 10.1016/j.watres.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Gao P., Munir M., Xagoraraki I. (2012b). Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci. Total Environ. 421–422, 173–183. 10.1016/j.scitotenv.2012.01.061 [DOI] [PubMed] [Google Scholar]
- Guerra B., Junker E., Schroeter A., Malorny B., Lehmann S., Helmuth R. (2003). Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coli isolates from cattle, swine and poultry. J. Antimicrob. Chemother. 52 489–492. 10.1093/jac/dkg362 [DOI] [PubMed] [Google Scholar]
- Gullberg E., Cao S., Berg O. G., Ilback C., Sandegren L., Hughes D., et al. (2011). Selection of resistant bacteria at very low antibiotic. PLoS Pathog. 7:e1002158 10.1371/journal.ppat.1002158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerum A. M., Sandvang D., Andersen S. R., Seyfarth A. M., Porsbo L. J., Frimodt-Moller N., et al. (2006). Detection of sul1, sul2 and sul3 in sulphonamide resistant Escherichia coli isolates obtained from healthy humans, pork and pigs in Denmark. Int. J. Food Microbiol. 106 235–237. 10.1016/j.ijfoodmicro.2005.06.023 [DOI] [PubMed] [Google Scholar]
- Heuer H., Focks A., Lamshoft M., Smalla K. (2008). Fate of sulfadiazine administered to pigs and its quantitative effect on the dynamics of bacterial resistance genes in manure and manured soil. Soil Biol. Biochem. 40 1892–1900. 10.1016/j.soilbio.2008.03.014 [DOI] [Google Scholar]
- Heuer H., Smalla K. (2007). Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ. Microbiol. 9 657–666. 10.1111/j.1462-2920.2006.01185.x [DOI] [PubMed] [Google Scholar]
- Hoa P. T. P., Managaki S., Nakada N., Takada H., Shimizu A., Anh D. H., et al. (2011). Antibiotic contamination and occurrence of antibiotic-resistant bacteria in aquatic environments of northern Vietnam. Sci. Total Environ. 409 2894–2901. 10.1016/j.scitotenv.2011.04.030 [DOI] [PubMed] [Google Scholar]
- Hu J., Shi J., Chang H., Li D., Yang M., Kamagata Y. (2008). Phenotyping and genotyping of antibiotic-resistant Escherichia coli isolated from a natural river basin. Environ. Sci. Technol. 42 3415–3420. 10.1021/es7026746 [DOI] [PubMed] [Google Scholar]
- Huang L., Crothers K., Atzori C., Benfield T., Miller R., Rabodonirina M., et al. (2004). Dihydropteroate synthase gene mutations in pneumocystis and sulfa resistance. Emmerg. Infect. Dis. 10 1721–1728. 10.3201/eid1010.030994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iweriebor B. C., Gaqavu S., Obi L. C., Nwodo U. U., Okoh A. I. (2015). Antibiotic susceptibilities of Enterococcus species isolated from hospital and domestic wastewater effluents in Alice, Eastern Cape Province of South Africa. Int. J. Environ. Res. Public Health 12 4231–4246. 10.3390/ijerph120404231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.-J., Hirono I., Kurokawa K., Maki T., Hawke J., Kondo H., et al. (2008). Complete DNA sequence and analysis of the transferable multiple-drug resistance plasmids (R plasmids) from Photobacterium damselae subsp. piscicida isolates collected in Japan and the United States. Antimicrob. Agents Chemother. 52 606–611. 10.1128/AAC.01216-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp C. W., McCluskey S. M., Singh B. K., Campbell C. D., Hudson G., Graham D. W. (2011). Antibiotic resistance gene abundances correlate with metal and geochemical conditions in archived Scottish soils. PLoS ONE 6:e27300 10.1371/journal.pone.0027300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Nonaka L., Maruyama F., Suzuki S. (2007). Molecular evidence for the ancient origin of the ribosomal protection protein that mediates tetracycline resistance in bacteria. J. Mol. Evol. 65 228–235. 10.1007/s00239-007-9006-z [DOI] [PubMed] [Google Scholar]
- Langendijk P. S., Schut F., Jansen G. J., Raangs G. C., Kamphuis G. R., Wilkinson M. H. F., et al. (1995). Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61 3069–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C. W., Loftin K. A., Meyer M. T., Davis J. G., Pruden A. (2010). tet and sul antibiotic resistance genes in livestock lagoons of various operation type, configuration, and antibiotic occurrence. Environ. Sci. Technol. 44 6102–6109. 10.1021/es9038165 [DOI] [PubMed] [Google Scholar]
- Muziasari W. A., Managaki A., Pärnänen K., Karkman A., Lyra C., Tamminen M., et al. (2014). Sulphonamide and trimethoprim resistance genes persist in sediments at Baltic Sea aquaculture farms but are not detected in the surrounding environment. PLoS ONE 9:e92702 10.1371/journal.pone.0092702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka L., Maruyama F., Miyamoto M., Miyakoshi M., Kurokawa K., Masuda M. (2012). Novel conjugative transferable multiple drug resistance plasmid pAQU1 from Photobacterium damselae subsp. damselae isolated from marine aquaculture environment. Microbes Environ. 27 263–272. 10.1264/jsme2.ME11338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omulo S., Thumbi S. M., Njenga M. K., Call D. R. (2015). A review of 40 years of enteric antimicrobial resistance research in Eastern Africa: what can be done better? Antimicrob. Resist. Infect. Cont. 4 1–13. 10.1186/s13756-014-0041-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei R., Kim S.-C., Carlson K. H., Pruden A. (2006). Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Wat. Res. 40 2427–2435. 10.1016/j.watres.2006.04.017 [DOI] [PubMed] [Google Scholar]
- Pruden A. (2014). Balancing water sustainability and public health goals in the face of growing concerns about antibiotic resistance. Environ. Sci. Technol. 48 5–14. 10.1021/es403883p [DOI] [PubMed] [Google Scholar]
- Pruden A., Joakim Larsson D. G., Amézquita A., Collignon P., Brandt K. K., Graham D. W., et al. (2013). Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environ. Health Perspect. 121 878–885. 10.1289/ehp.1206446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radstrom P., Swedberg G. (1988). RSF1010 and a conjugative plasmid contain sulII, one of two known genes for plasmid-borne sulfonamide resistance dihydropteroate synthase. Antimicrob. Agents Chemother. 32 1684–1692. 10.1128/AAC.32.11.1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo L., Manaia C., Merlin C., Schwartz T., Dagot C., Ploy M. C., et al. (2013). Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci. Total Environ. 447 345–360. 10.1016/j.scitotenv.2013.01.032 [DOI] [PubMed] [Google Scholar]
- Rizzotti L., La Gioia F., Dellaglio F., Torriani S. (2009). Molecular diversity and transferability of the tetracycline resistance gene tet(M), carried on Tn916-1545 family transposons, in enterococci from a total food chain. Antonie Van Leeuwenhoek 96 43–52. 10.1007/s10482-009-9334-7 [DOI] [PubMed] [Google Scholar]
- Roberts M. C., Schwarz S., Aarts H. J. M. (2012). Erratum: acquired antibiotic resistance genes: an overview. Front. Microbiol 3:384 10.3389/fmicb.2012.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato-Takabe Y., Suzuki S., Shishikura R., Hamasaki K., Tada Y., Kataoka T., et al. (2015). Spatial distribution and cell size of aerobic anoxygenic phototrophic bacteria in the Uwa Sea. Japan. J. Oceanogr. 71 151–159. 10.1007/s10872-014-0267-z [DOI] [Google Scholar]
- Segura P. A., Takada H., Correa J. A., Saadi K. E., Koike T., Onwona-Agyeman S., et al. (2015). Global occurrence of anti-infectives in contaminated surface waters: impact of income inequality between countries. Environ. Int. 80 89–97. 10.1016/j.envint.2015.04.001 [DOI] [PubMed] [Google Scholar]
- Shimizu A., Takada H., Koike T., Takeshita T., Saha M., Rinawati, et al. (2013). Ubiquitous occurrence of sulfonamides in tropical Asian waters. Sci. Total Environ. 45 108–115. 10.1016/j.scitotenv.2013.02.027 [DOI] [PubMed] [Google Scholar]
- Smith C. J., Rogers M. B., McKee M. L. (1992). Heterologous gene expression in Bacteroides fragilis. Plasmid 27 141–154. 10.1016/0147-619X(92)90014-2 [DOI] [PubMed] [Google Scholar]
- Snyman H. G., Van Niekerk A. M., Rajasakran N. (2006). “Sustainable wastewater treatment – what has gone wrong and how do we get back on track,” in Proceedings of the water Institute of South Africa (WISA) Conference, 21–25 May 2006 Durban. [Google Scholar]
- Suzuki M. T., Taylor L. T., Delong E. F. (2000). Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5-nuclease assays. Appl. Environ. Microbiol. 66 4605–4614. 10.1128/AEM.66.11.4605-4614.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Hoa P. T. P. (2012). Distribution of quinolone, sulfonamides, tetracyclines in aquatic environment and antibiotic resistance in Indochina. Front. Microbiol. 3:67 10.3389/fmicb.2012.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Ogo M., Miller T. W., Shimizu A., Takada H., Sringan M. A. T. (2013). Who possesses drug resistance genes in the aquatic environment? :sulfamethoxazole (SMX) resistance genes among the bacterial community in water environment of Metro-Manila, Philippines. Front. Microbiol 4:102 10.3389/fmicb.2013.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami H., Hanada S., Sekiguchi Y., Tanaka Y., Kamagata Y. (2009). Effect of gelling agent on colony formation in solid cultivation of microbial community in lake sediment. Environ. Microbiol. 11 1827–1834. 10.1111/j.1462-2920.2009.01907.x [DOI] [PubMed] [Google Scholar]
- Takasu H., Suzuki S., Reungsang A., Viet P. H. (2011). Fluoroquinolone (FQ) Contamination does not correlate with occurrence of FQ-resistant bacteria in aquatic environments of Vietnam and Thailand. Microbes Environ. 26 135–143. 10.1264/jsme2.ME10204 [DOI] [PubMed] [Google Scholar]
- Tamminen M., Karkman A., Lõhmus A., Muziasari W. I., Takasu H., Wada S., et al. (2011). Tetracycline resistance genes persist at aquaculture farms in the absence of selection pressure. Environ. Sci. Technol. 45 386–391. 10.1021/es102725n [DOI] [PubMed] [Google Scholar]
- United States Environmental Protection and Agency. (2004). Primer for Municipal Wastewater Treatment Systems. EPA 832-R-04-001. Washington DC: United States Environmental Protection and Agency; 25 Available at: http://www.epa.gov/npdes/pubs/primer.pdf [Google Scholar]
- Water Research and Commission. (2006). Wastewater Treatment in South Africa: From Crisis to Compliance. WRC Report, Report No. 8001/8295/3/P. Pretoria: Water Research and Commission. [Google Scholar]
- Zachariah R., Harries A. D., Luo C., Bachman G., Graham S. M. (2007). Scaling-up co-trimoxazole prophylaxis in HIV-exposed and HIV-infected children in high HIV-prevalence countries. Lancet Inf. Dis. 7 686–693. 10.1016/S1473-3099(07)70239-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.