Abstract
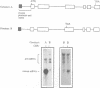
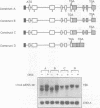
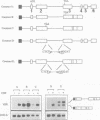
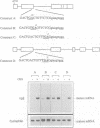
Premature termination codons (PTCs) can cause the decay of mRNAs in the nuclear fraction of mammalian cells. This enigmatic nuclear response is of interest because it suggests that translation signals do not restrict their effect to the cytoplasm, where fully assembled ribosomes reside. Here we examined the molecular mechanism for this putative nuclear response by using the T-cell receptor-beta (TCR-beta) gene, which acquires PTCs as a result of programmed rearrangements that occur during normal thymic ontogeny. We found that PTCs had little or no measurable effect on TCR-beta pre-mRNA levels, but they sharply depressed TCR-beta mature mRNA levels in the nuclear fraction of stably transfected cells. A PTC split by an intron was able to trigger the down-regulatory response, implying that PTC recognition occurs after an mRNA is at least partially spliced. However, intron deletion and addition studies demonstrated that a PTC must be followed by at least one functional (spliceable) intron to depress mRNA levels. One explanation for this downstream intron-dependence is that cytoplasmic ribosomes adjacent to nuclear pores scan mRNAs still undergoing splicing as they emerge from the nucleus. We found this explanation to be unlikely because PTCs only 8 or 10 nt upstream of a terminal intron down-regulated mRNA levels, even though this distance is too short to permit PTC recognition in the cytoplasm prior to the splicing of the downstream intron in the nucleus. Collectively, the results suggest that nonsense codon recognition may occur in the nucleus.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aoufouchi S., Yélamos J., Milstein C. Nonsense mutations inhibit RNA splicing in a cell-free system: recognition of mutant codon is independent of protein synthesis. Cell. 1996 May 3;85(3):415–422. doi: 10.1016/s0092-8674(00)81119-8. [DOI] [PubMed] [Google Scholar]
- Atkin A. L., Altamura N., Leeds P., Culbertson M. R. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol Biol Cell. 1995 May;6(5):611–625. doi: 10.1091/mbc.6.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga S. J., Benz E. J., Jr Beta-globin nonsense mutation: deficient accumulation of mRNA occurs despite normal cytoplasmic stability. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2935–2939. doi: 10.1073/pnas.89.7.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann B., Potash M. J., Köhler G. Consequences of frameshift mutations at the immunoglobulin heavy chain locus of the mouse. EMBO J. 1985 Feb;4(2):351–359. doi: 10.1002/j.1460-2075.1985.tb03636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P., Cheng J., Maquat L. E. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):482–486. doi: 10.1073/pnas.90.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P., Cheng J., Zhou X., Stephenson L. S., Maquat L. E. Mammalian nonsense codons can be cis effectors of nuclear mRNA half-life. Mol Cell Biol. 1994 Dec;14(12):8219–8228. doi: 10.1128/mcb.14.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P., Maquat L. E. Nonsense but not missense mutations can decrease the abundance of nuclear mRNA for the mouse major urinary protein, while both types of mutations can facilitate exon skipping. Mol Cell Biol. 1994 Sep;14(9):6326–6336. doi: 10.1128/mcb.14.9.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget S. M. Exon recognition in vertebrate splicing. J Biol Chem. 1995 Feb 10;270(6):2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- Beyer A. L., Osheim Y. N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988 Jun;2(6):754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Beyer D., Skripkin E., Wadzack J., Nierhaus K. H. How the ribosome moves along the mRNA during protein synthesis. J Biol Chem. 1994 Dec 2;269(48):30713–30717. [PubMed] [Google Scholar]
- Blencowe B. J., Issner R., Kim J., Mccaw P., Sharp P. A. New proteins related to the Ser-Arg family of splicing factors. RNA. 1995 Oct;1(8):852–865. [PMC free article] [PubMed] [Google Scholar]
- Bond V. C., Wold B. Nucleolar localization of myc transcripts. Mol Cell Biol. 1993 Jun;13(6):3221–3230. doi: 10.1128/mcb.13.6.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M. S., Doskow J., Morris P., Li S., Nhim R. P., Sandstedt S., Wilkinson M. F. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995 Dec 1;270(48):28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- Cheng J., Belgrader P., Zhou X., Maquat L. E. Introns are cis effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance. Mol Cell Biol. 1994 Sep;14(9):6317–6325. doi: 10.1128/mcb.14.9.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Fogel-Petrovic M., Maquat L. E. Translation to near the distal end of the penultimate exon is required for normal levels of spliced triosephosphate isomerase mRNA. Mol Cell Biol. 1990 Oct;10(10):5215–5225. doi: 10.1128/mcb.10.10.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Maquat L. E. Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life of cytoplasmic mRNA. Mol Cell Biol. 1993 Mar;13(3):1892–1902. doi: 10.1128/mcb.13.3.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H. S., Anderson S. J., Louie M. C., Godambe S. A., Pozzi M. R., Behlke M. A., Huppi K., Loh D. Y. Tandem linkage and unusual RNA splicing of the T-cell receptor beta-chain variable-region genes. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1992–1996. doi: 10.1073/pnas.84.7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor A., Wiersma E., Shulman M. J. On the linkage between RNA processing and RNA translatability. J Biol Chem. 1994 Oct 7;269(40):25178–25184. [PubMed] [Google Scholar]
- Dietz H. C., Kendzior R. J., Jr Maintenance of an open reading frame as an additional level of scrutiny during splice site selection. Nat Genet. 1994 Oct;8(2):183–188. doi: 10.1038/ng1094-183. [DOI] [PubMed] [Google Scholar]
- Enssle J., Kugler W., Hentze M. W., Kulozik A. E. Determination of mRNA fate by different RNA polymerase II promoters. Proc Natl Acad Sci U S A. 1993 Nov 1;90(21):10091–10095. doi: 10.1073/pnas.90.21.10091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W., Mueller D. L., Pennell C. A., Rivard J. J., Li Y. S., Hardy R. R., Schlissel M. S., Behrens T. W. Frequent aberrant immunoglobulin gene rearrangements in pro-B cells revealed by a bcl-xL transgene. Immunity. 1996 Mar;4(3):291–299. doi: 10.1016/s1074-7613(00)80437-9. [DOI] [PubMed] [Google Scholar]
- Gascoigne N. R., Chien Y., Becker D. M., Kavaler J., Davis M. M. Genomic organization and sequence of T-cell receptor beta-chain constant- and joining-region genes. Nature. 1984 Aug 2;310(5976):387–391. doi: 10.1038/310387a0. [DOI] [PubMed] [Google Scholar]
- Harpold M. M., Evans R. M., Salditt-Georgieff M., Darnell J. E. Production of mRNA in Chinese hamster cells: relationship of the rate of synthesis to the cytoplasmic concentration of nine specific mRNA sequences. Cell. 1979 Aug;17(4):1025–1035. doi: 10.1016/0092-8674(79)90341-6. [DOI] [PubMed] [Google Scholar]
- Hawkins J. D. A survey on intron and exon lengths. Nucleic Acids Res. 1988 Nov 11;16(21):9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995 Feb 15;9(4):437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- He F., Peltz S. W., Donahue J. L., Rosbash M., Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick S. M., Nielsen E. A., Kavaler J., Cohen D. I., Davis M. M. Sequence relationships between putative T-cell receptor polypeptides and immunoglobulins. Nature. 1984 Mar 8;308(5955):153–158. doi: 10.1038/308153a0. [DOI] [PubMed] [Google Scholar]
- Jäck H. M., Berg J., Wabl M. Translation affects immunoglobulin mRNA stability. Eur J Immunol. 1989 May;19(5):843–847. doi: 10.1002/eji.1830190510. [DOI] [PubMed] [Google Scholar]
- Kadowaki T., Schneiter R., Hitomi M., Tartakoff A. M. Mutations in nucleolar proteins lead to nucleolar accumulation of polyA+ RNA in Saccharomyces cerevisiae. Mol Biol Cell. 1995 Sep;6(9):1103–1110. doi: 10.1091/mbc.6.9.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler O., Jiang Y., Chasin L. A. Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Mol Cell Biol. 1993 Oct;13(10):6211–6222. doi: 10.1128/mcb.13.10.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg M., Siu G., Hood L. E., Shastri N. The molecular genetics of the T-cell antigen receptor and T-cell antigen recognition. Annu Rev Immunol. 1986;4:529–591. doi: 10.1146/annurev.iy.04.040186.002525. [DOI] [PubMed] [Google Scholar]
- Lejbkowicz F., Goyer C., Darveau A., Neron S., Lemieux R., Sonenberg N. A fraction of the mRNA 5' cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9612–9616. doi: 10.1073/pnas.89.20.9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S. K., Sigmund C. D., Gross K. W., Maquat L. E. Nonsense codons in human beta-globin mRNA result in the production of mRNA degradation products. Mol Cell Biol. 1992 Mar;12(3):1149–1161. doi: 10.1128/mcb.12.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano F., Maertzdorf B., Pannell R., Milstein C. Low cytoplasmic mRNA levels of immunoglobulin kappa light chain genes containing nonsense codons correlate with inefficient splicing. EMBO J. 1994 Oct 3;13(19):4617–4622. doi: 10.1002/j.1460-2075.1994.tb06783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat L. E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995 Jul;1(5):453–465. [PMC free article] [PubMed] [Google Scholar]
- Miriami E., Sperling J., Sperling R. Heat shock affects 5' splice site selection, cleavage and ligation of CAD pre-mRNA in hamster cells, but not its packaging in InRNP particles. Nucleic Acids Res. 1994 Aug 11;22(15):3084–3091. doi: 10.1093/nar/22.15.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeger L. K., Schoborg R. V., Zhao Q., Tullis G. E., Pintel D. J. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 1992 Jun;6(6):1107–1119. doi: 10.1101/gad.6.6.1107. [DOI] [PubMed] [Google Scholar]
- Peltz S. W., Brown A. H., Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 1993 Sep;7(9):1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- Peltz S. W., He F., Welch E., Jacobson A. Nonsense-mediated mRNA decay in yeast. Prog Nucleic Acid Res Mol Biol. 1994;47:271–298. doi: 10.1016/s0079-6603(08)60254-8. [DOI] [PubMed] [Google Scholar]
- Piñol-Roma S., Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992 Feb 20;355(6362):730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Pulak R., Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993 Oct;7(10):1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- Qian L., Theodor L., Carter M., Vu M. N., Sasaki A. W., Wilkinson M. F. T cell receptor-beta mRNA splicing: regulation of unusual splicing intermediates. Mol Cell Biol. 1993 Mar;13(3):1686–1696. doi: 10.1128/mcb.13.3.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L., Vu M. N., Carter M. S., Doskow J., Wilkinson M. F. T cell receptor-beta mRNA splicing during thymic maturation in vivo and in an inducible T cell clone in vitro. J Immunol. 1993 Dec 15;151(12):6801–6814. [PubMed] [Google Scholar]
- Shyu A. B., Greenberg M. E., Belasco J. G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989 Jan;3(1):60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- Simpson S. B., Stoltzfus C. M. Frameshift mutations in the v-src gene of avian sarcoma virus act in cis to specifically reduce v-src mRNA levels. Mol Cell Biol. 1994 Mar;14(3):1835–1844. doi: 10.1128/mcb.14.3.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani T., Derby R. J., Hiraoka Y., Spector D. L. Nucleolar accumulation of poly (A)+ RNA in heat-shocked yeast cells: implication of nucleolar involvement in mRNA transport. Mol Biol Cell. 1995 Nov;6(11):1515–1534. doi: 10.1091/mbc.6.11.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G., Mitchell P. J., Ciudad C. J., Chasin L. A. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989 Jul;9(7):2868–2880. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verner K., Schatz G. Protein translocation across membranes. Science. 1988 Sep 9;241(4871):1307–1313. doi: 10.1126/science.2842866. [DOI] [PubMed] [Google Scholar]
- Visa N., Alzhanova-Ericsson A. T., Sun X., Kiseleva E., Björkroth B., Wurtz T., Daneholt B. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996 Jan 26;84(2):253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- Visa N., Puvion-Dutilleul F., Harper F., Bachellerie J. P., Puvion E. Intranuclear distribution of poly(A) RNA determined by electron microscope in situ hybridization. Exp Cell Res. 1993 Sep;208(1):19–34. doi: 10.1006/excr.1993.1218. [DOI] [PubMed] [Google Scholar]
- Wilkinson M. F., MacLeod C. L. Induction of T-cell receptor-alpha and -beta mRNA in SL12 cells can occur by transcriptional and post-transcriptional mechanisms. EMBO J. 1988 Jan;7(1):101–109. doi: 10.1002/j.1460-2075.1988.tb02788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M., Doskow J., Lindsey S. RNA blots: staining procedures and optimization of conditions. Nucleic Acids Res. 1991 Feb 11;19(3):679–679. doi: 10.1093/nar/19.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Ruiz-Echevarria M. J., Quan Y., Peltz S. W. Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol Cell Biol. 1995 Apr;15(4):2231–2244. doi: 10.1128/mcb.15.4.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]