Abstract
For the past 25 years, phage display technology has been an invaluable tool for studies of protein–protein interactions. However, the inherent biological, biochemical, and biophysical properties of filamentous bacteriophage, as well as the ease of its genetic manipulation, also make it an attractive platform outside the traditional phage display canon. This review will focus on the unique properties of the filamentous bacteriophage and highlight its diverse applications in current research. Particular emphases are placed on: (i) the advantages of the phage as a vaccine carrier, including its high immunogenicity, relative antigenic simplicity and ability to activate a range of immune responses, (ii) the phage’s potential as a prophylactic and therapeutic agent for infectious and chronic diseases, (iii) the regularity of the virion major coat protein lattice, which enables a variety of bioconjugation and surface chemistry applications, particularly in nanomaterials, and (iv) the phage’s large population sizes and fast generation times, which make it an excellent model system for directed protein evolution. Despite their ubiquity in the biosphere, metagenomics work is just beginning to explore the ecology of filamentous and non-filamentous phage, and their role in the evolution of bacterial populations. Thus, the filamentous phage represents a robust, inexpensive, and versatile microorganism whose bioengineering applications continue to expand in new directions, although its limitations in some spheres impose obstacles to its widespread adoption and use.
Keywords: filamentous phage, vaccine, therapeutic, antimicrobial, bioconjugation
Introduction
The filamentous bacteriophage (genera Inovirus and Plectrovirus) are non-enveloped, rod-shaped viruses of Escherichia coli whose long helical capsids encapsulate a single-stranded circular DNA genome. Subsequent to the independent discovery of bacteriophage by Twort (1915) and d’Hérelle (1917), the first filamentous phage, f1, was isolated in Loeb (1960) and later characterized as a member of a larger group of phage (Ff, including f1, M13, and fd phage) specific for the E. coli conjugative F pilus (Hofschneider and Mueller-Jensen, 1963; Marvin and Hoffmann-Berling, 1963; Zinder et al., 1963; Salivar et al., 1964). Soon thereafter, filamentous phage were discovered that do not use F-pili for entry (If and Ike; Meynell and Lawn, 1968; Khatoon et al., 1972), and over time the list of known filamentous phage has expanded to over 60 members (Fauquet et al., 2005), including temperate and Gram-positive-tropic species. Work by multiple groups over the past 50 years has contributed to a relatively sophisticated understanding of filamentous phage structure, biology and life cycle (reviewed in Marvin, 1998; Rakonjac et al., 2011; Rakonjac, 2012).
In the mid-1980s, the principle of modifying the filamentous phage genome to display polypeptides as fusions to coat proteins on the virion surface was invented by Smith and colleagues (Smith, 1985; Parmley and Smith, 1988). Based on the ideas described in Parmley and Smith (1988), groups in California, Germany, and the UK developed phage-display platforms to create and screen libraries of peptide and folded-protein variants (Bass et al., 1990; Devlin et al., 1990; McCafferty et al., 1990; Scott and Smith, 1990; Breitling et al., 1991; Kang et al., 1991). This technology allowed, for the first time, the ability to seamlessly connect genetic information with protein function for a large number of protein variants simultaneously, and has been widely and productively exploited in studies of protein–protein interactions. Many excellent reviews are available on phage-display libraries and their applications (Kehoe and Kay, 2005; Bratkovic, 2010; Pande et al., 2010). However, the phage also has a number of unique structural and biological properties that make it highly useful in areas of research that have received far less attention.
Thus, the purpose of this review is to highlight recent and current work using filamentous phage in novel and non-traditional applications. Specifically, we refer to projects that rely on the filamentous phage as a key element, but whose primary purpose is not the generation or screening of phage-displayed libraries to obtain binding polypeptide ligands. These tend to fall into four major categories of use: (i) filamentous phage as a vaccine carrier; (ii) engineered filamentous phage as a therapeutic biologic agent in infectious and chronic diseases; (iii) filamentous phage as a scaffold for bioconjugation and surface chemistry; and (iv) filamentous phage as an engine for evolving variants of displayed proteins with novel functions. A final section is dedicated to recent developments in filamentous phage ecology and phage–host interactions. Common themes shared amongst all these applications include the unique biological, immunological, and physicochemical properties of the phage, its ability to display a variety of biomolecules in modular fashion, and its relative simplicity and ease of manipulation.
Filamentous Phage Display Systems: An Overview
Nearly all applications of the filamentous phage depend on its ability to display polypeptides on the virion’s surface as fusions to phage coat proteins (Table 1). The display mode determines the maximum tolerated size of the fused polypeptide, its copy number on the phage, and potentially, the structure of the displayed polypeptide. Display may be achieved by fusing DNA encoding a polypeptide of interest directly to the gene encoding a coat protein within the phage genome (type 8 display on pVIII, type 3 display on pIII, etc.), resulting in fully recombinant phage. Much more commonly, however, only one copy of the coat protein is modified in the presence of a second, wild-type copy (e.g., type 88 display if both recombinant and wild-type pVIII genes are on the phage genome, type 8+8 display if the recombinant gene 8 is on a plasmid with a phage origin of replication) resulting in a hybrid virion bearing two different types of a given coat protein. Multivalent display on some coat proteins can also be enforced using helper phage bearing non-functional copies of the relevant coat protein gene (e.g., type 3∗+3 display). By far the most commonly used coat proteins for display are the major coat protein, pVIII, and the minor coat protein, pIII, with the major advantage of the former being higher copy number display (up to ~15% of recombinant pVIII molecules in a hybrid virion, at least for short peptide fusions), and of the latter being the ability to display some folded proteins at an appreciable copy number (1–5 per phage particle). While pVIII display of folded proteins on hybrid phage is possible, it typically results in a copy number of much less than 1 per virion (Sidhu et al., 2000). For the purposes of this review, we use the term “phage display” to refer to a recombinant filamentous phage displaying a single polypeptide sequence on its surface (or more rarely, bispecific display achieved via fusion of polypeptides to two different capsid proteins), and the term “phage-displayed library” to refer to a diverse pool of recombinant filamentous phage displaying an array of polypeptide variants (e.g., antibody fragments; peptides). Such libraries are typically screened by iterative cycles of panning against an immobilized protein of interest (e.g., antigen for phage-displayed antibody libraries; antibody for phage-displayed peptide libraries) followed by amplification of the bound phage in E. coli cells.
Table 1.
Filamentous phage display modes and their associated characteristics.
| Phage coat protein | Display mode | Helper phage required? | Polypeptide copy number | Polypeptide size limit | Reference(s) |
|---|---|---|---|---|---|
| pIII | Fully recombinant (type 3 and 3+3∗ systems)1 | Type 3+3∗ system | ~5 | >25 kDa | Parmley and Smith (1988), McConnell et al. (1994),Rondot et al. (2001) |
| Hybrid (type 33 and 3+3 systems) | Type 3+3 system | <12 | Smith and Scott (1993),Smith and Petrenko (1997) | ||
| pVI | Hybrid (type 6+6 system) | Yes | <12 | >25 kDa | Hufton et al. (1999) |
| pVII | Fully recombinant (type 7 system) | No | ~5 | >25 kDa | Kwasnikowski et al. (2005) |
| Hybrid (type 7+7 system) | Yes | <12 | Gao et al. (1999) | ||
| pVIII | Fully recombinant (landscape phage; type 8 system) | No | 27003 | ~5–8 residues | Kishchenko et al. (1994), Petrenko et al. (1996) |
| Hybrid (type 88 and 8+8 systems) | Type 8+8 system | ~1–3002 | >50 kDa | Scott and Smith (1990), Greenwood et al. (1991),Smith and Fernandez (2004) | |
| pIX | Fully recombinant (type 9+9∗ system) | Yes | ~5 | >25 kDa | Gao et al. (2002) |
| Hybrid (type 9+9 system) | No | <12 | Gao et al. (1999), Shi et al. (2010), Tornetta et al. (2010) |
1Asterisks indicate non-functional copies of the coat protein are present in the genome of the helper phage used to rescue a phagemid whose coat protein has been fused to a recombinant polypeptide.
2The copy number depends on polypeptide size; typically <1 copy per phage particle but for pVIII peptide display can be up to ~15% of pVIII molecules in hybrid virions.
3The total number of pVIII molecules depends on the phage genome size; one pVIII molecule is added for every 2.3 nucleotides in the viral genome.
Filamentous Phage as an Immunogenic Vaccine Carrier
Early work with anti-phage antisera generated for species classification purposes demonstrated that the filamentous phage virion is highly immunogenic in the absence of adjuvants (Meynell and Lawn, 1968) and that only the major coat protein, pVIII, and the minor coat protein, pIII, are targeted by antibodies (Pratt et al., 1969; Woolford et al., 1977). Thus, the idea of using the phage as carrier to elicit antibodies against poorly immunogenic haptens or polypeptide was a natural extension of the ability to display recombinant exogenous sequences on its surface, which was first demonstrated by de la Cruz et al. (1988). The phage particle’s low cost of production, high stability and potential for high valency display of foreign antigen (via pVIII display) also made it attractive as a vaccine carrier, especially during the early stages of development of recombinant protein technology.
Antibody Epitope-Based Peptide Vaccines
Building upon existing peptide-carrier technology, the first filamentous phage-based vaccine immunogens displayed short amino acid sequences derived directly from proteins of interest as recombinant fusions to pVIII or pIII (de la Cruz et al., 1988). As library technology was developed and refined, phage-based antigens displaying peptide ligands of monoclonal antibodies (selected from random peptide libraries using the antibody, thus simulating with varying degrees of success the antibody’s folded epitope on its cognate antigen; Geysen et al., 1986; Knittelfelder et al., 2009) were also generated for immunization purposes, with the goal of eliciting anti-peptide antibodies that also recognize the native protein. Some of the pioneering work in this area used peptides derived from infectious disease antigens (or peptide ligands of antibodies against these antigens; Table 2), including malaria and human immunodeficiency virus type 1 (HIV-1). When displayed on phage, peptides encoding the repeat regions of the malarial circumsporozoite protein and merozoite surface protein 1 were immunogenic in mice and rabbits (de la Cruz et al., 1988; Greenwood et al., 1991; Willis et al., 1993; Demangel et al., 1996), and antibodies raised against the latter cross-reacted with the full-length protein. Various peptide determinants (or mimics thereof) of HIV-1 gp120, gp41, gag, and reverse transcriptase were immunogenic when displayed on or conjugated to phage coat proteins (Minenkova et al., 1993; di Marzo Veronese et al., 1994; De Berardinis et al., 1999; Scala et al., 1999; Chen et al., 2001; van Houten et al., 2006, 2010), and in some cases elicited antibodies that were able to weakly neutralize lab-adapted viruses (di Marzo Veronese et al., 1994; Scala et al., 1999). The list of animal and human infections for which phage-displayed peptide immunogens have been developed as vaccine leads continues to expand and includes bacterial, fungal, viral, and parasitic pathogens (Table 2). While in some cases the results of these studies have been promising, antibody epitope-based peptide vaccines are no longer an area of active research for several reasons: (i) in many cases, peptides incompletely or inadequately mimic epitopes on folded proteins (Irving et al., 2010; see below); (ii) antibodies against a single epitope may be of limited utility, especially for highly variable pathogens (Van Regenmortel, 2012); and (iii) for pathogens for which protective immune responses are generated efficiently during natural infection, peptide vaccines offer few advantages over recombinant subunit and live vector vaccines, which have become easier to produce over time.
Table 2.
Studies using filamentous phage as an immunogenic carrier for peptide B-cell epitopes.
| Antigen | Species | Epitope type(s) | Display format(s) | Ab response against peptide | Ab response against native protein | Reference(s) |
|---|---|---|---|---|---|---|
| Alzheimer’s disease β-amyloid fibrils | Mouse (BALB/c, APP transgenic), Guinea pig | Linear | pVIII hybrid, pIII fully recombinant |
+ | + | Frenkel et al. (2000, 2003), Lavie et al. (2004), Esposito et al. (2008), Tanaka et al. (2011) |
| Aspergillus fumigatus MIRB | Mouse (BALB/c) | Linear | Chemical conjugate | + | + | Raymond-Bouchard et al. (2012) |
| Bordetella pertussis toxin | Mouse (BALB/c) | Discontinuous | pVIII hybrid | + | - | Felici et al. (1993) |
| Candida albicans HSP90 | Mouse (C57/BL6, BALB/c) | Linear | pVIII hybrid | + | + | Yang et al. (2005a), Wang et al. (2006, 2014d) |
| Chlamydia trachomatis major outer membrane protein | Mouse (C57/BL6, BALB/c, CBA/j) | Linear | pVIII hybrid | + | + | Zhong et al. (1994) |
| Coronavirus S glycoprotein | Mouse (C57/BL6, BALB/c) | Linear | pVIII hybrid | + | + | Yu et al. (2000) |
| ERBB2/HER2 | Mouse (BALB/c) | Linear | pVIII hybrid, pIII fully recombinant |
+ | + | Yip et al. (2001) |
| Entamoeba histolytica proteophosphoglycan | Mouse (BALB/c) | Discontinuous | pVIII hybrid | + | + | Melzer et al. (2002) |
| Follicle-stimulating hormone | Sheep | Linear | pVIII hybrid | + | + | Abdennebi et al. (1999) |
| Foot-and-mouth disease VP1 | Guinea pig | Linear | pVIII hybrid | ND | + | Kim et al. (2004) |
| Gonadotropin-releasing hormone | Mouse (CD-1) | Linear | Chemical conjugate | ND | ND | Samoylov et al. (2012) |
| HBV surface antigen | Mouse (C57/BL6, BALB/c, B10.M) | Linear | pVIII hybrid, pIII fully recombinant |
+ | + | Folgori et al. (1994), Motti et al. (1994), Meola et al. (1995), Delmastro et al. (1997) |
| HCV core antigen and NS4B protein | Mouse (C57/BL6, BALB/c, MF1) | Linear | pVIII hybrid | + | ND | Prezzi et al. (1996), Delmastro et al. (1997) |
| HCV E2 protein | Mouse (BALB/c) | Linear | pVIII hybrid | + | ND | Puntoriero et al. (1998) |
| Helicobacter pyloriUrease B | Mouse (BALB/c) | Linear | pIII fully recombinant | + | + | Li et al. (2010c) |
| HIV-1 gp120 V3 loop | Mouse (BALB/c) | Linear | pVIII hybrid | + | + | di Marzo Veronese et al. (1994) |
| HIV-1 gp120 CD4-binding site | Mouse (BALB/c) | Discontinuous | Chemical conjugate | + | - | van Houten et al. (2006) |
| HIV-1 gp120 and gp41 | Mouse (C57/BL6, BALB/c), Rhesus macaque | Linear, Discontinuous |
pVIII hybrid, pIII hybrid, Chemical conjugate |
+ | ND | Scala et al. (1999), Chen et al. (2001), van Houten et al. (2010) |
| HIV-1 gag p17 | Rabbit | Linear | pVIII fully recombinant | + | + | Minenkova et al. (1993) |
| HIV-1 reverse transcriptase | Mouse (C57BL/6, HLA-A2 transgenic) | Linear | pVIII hybrid | + | ND | De Berardinis et al. (1999) |
| HPV E7 protein | Mouse (BALB/c) | Linear | pVIII hybrid | + | + | Lidqvist et al. (2008) |
| HSV-2 glycoprotein G | Mouse (BALB/c) | Linear | pVIII hybrid | + | + | Grabowska et al. (2000) |
| Immunoglobulin E | Rabbit | Discontinuous | pVIII hybrid | ND | + | Rudolf et al. (1998) |
| Mycoplasma hyopneumoniae unknown proteins | Mouse (BALB/cByJ) | Linear, Discontinuous |
pIII fully recombinant | + | + | Yang et al. (2005b) |
| Neisseria meningitidis PorA protein | Mouse (BALB/c) | Linear | pVIII hybrid | + | + | Menendez et al. (2001) |
| Plasmodium falciparum CSP | Mouse (BALB/c, C57/BL10, Swiss, Str/ort), rabbit | Linear | pVIII hybrid, pIII fully recombinant |
+ | ND | de la Cruz et al. (1988), Greenwood et al. (1991), Willis et al. (1993) |
| Plasmodium vivax MSP1 | Mouse (C57/BL6, BALB/c, Biozzi) | Linear, Discontinuous |
pIII hybrid | ND | + | Demangel et al. (1996) |
| Rhabdovirus RABVG | Mouse (BALB/c) | Linear | pIII fully recombinant | + | + | Houimel and Dellagi (2009) |
| RSV glycoprotein G | Mouse (BALB/c) | Linear | pIII fully recombinant | + | - | Bastien et al. (1997) |
| Schistosoma japonicum unknown protein(s) | Mouse (Kunming) | Discontinuous | pIII fully recombinant | + | ND | |
| Shigella flexneri LPS | Mouse (BALB/c) | Discontinuous | pVIII hybrid, Chemical conjugate |
+ | - | van Houten et al. (2010), Henry et al. (2011) |
| Sperm | Mouse (CD-1), Pig | Discontinuous | pVIII fully recombinant | + | + | Samoylova et al. (2012a,b) |
| Toxic shock syndrome toxin | Mouse (BALB/c) | Linear | pIII fully recombinant | + | ND | Rubinchik and Chow (2000) |
More recently, peptide-displaying phage have been used in attempts to generate therapeutic antibody responses for chronic diseases, cancer, immunotherapy, and immunocontraception. Immunization with phage displaying Alzheimer’s disease β-amyloid fibril peptides elicited anti-aggregating antibodies in mice and guinea pigs (Frenkel et al., 2000, 2003; Esposito et al., 2008; Tanaka et al., 2011), possibly reduced amyloid plaque formation in mice (Frenkel et al., 2003; Solomon, 2005; Esposito et al., 2008), and may have helped maintain cognitive abilities in a transgenic mouse model of Alzheimer’s disease (Lavie et al., 2004); however, it remains unclear how such antibodies are proposed to cross the blood–brain barrier. Yip et al. (2001) found that antibodies raised in mice against an ERBB2/HER2 peptide could inhibit breast-cancer cell proliferation. Phage displaying peptide ligands of an anti-IgE antibody elicited antibodies that bound purified IgE molecules (Rudolf et al., 1998), which may be useful in allergy immunotherapy. Several strategies for phage-based contraceptive vaccines have been proposed for control of animal populations. For example, immunization with phage displaying follicle-stimulating hormone peptides on pVIII elicited antibodies that impaired the fertility of mice and ewes (Abdennebi et al., 1999). Phage displaying or chemically conjugated to sperm antigen peptides or peptide mimics (Samoylova et al., 2012a,b) and gonadotropin-releasing hormone (Samoylov et al., 2012) are also in development.
For the most part, peptides displayed on phage elicit antibodies in experimental animals (Table 2), although this depends on characteristics of the peptide and the method of its display: pIII fusions tend toward lower immunogenicity than pVIII fusions (Greenwood et al., 1991) possibly due to copy number differences (pIII: 1–5 copies vs. pVIII: estimated at several hundred copies; Malik et al., 1996). In fact, the phage is at least as immunogenic as traditional carrier proteins such as bovine serum albumin (BSA) and keyhole limpet hemocyanin (KLH; Melzer et al., 2003; Su et al., 2007), and has comparatively few endogenous B-cell epitopes to divert the antibody response from its intended target (Henry et al., 2011). Excepting small epitopes that can be accurately represented by a contiguous short amino acid sequence, however, it has been extremely difficult to elicit antibody responses that cross-react with native protein epitopes using peptides. The overall picture is considerably bleaker than that painted by Table 2, since in several studies either: (i) peptide ligands selected from phage-displayed libraries were classified by the authors as mimics of discontinuous epitopes if they bore no obvious sequence homology to the native protein, which is weak evidence of non-linearity, or (ii) the evidence for cross-reactivity of antibodies elicited by immunization with phage-displayed peptides with native protein was uncompelling. Irving et al. (2010) describe at least one reason for this lack of success: it seems that peptide antigens elicit a set of topologically restricted antibodies that are largely unable to recognize discontinuous or complex epitopes on larger biomolecules. While the peptide may mimic the chemistry of a given epitope on a folded protein (allowing it to cross-react with a targeted antibody), being a smaller molecule, it cannot mimic the topology of that antibody’s full epitope. Despite this, the filamentous phage remains highly useful as a carrier for peptides with relatively simple secondary structures, which may be stablilized via anchoring to the coat proteins (Henry et al., 2011). This may be especially true of peptides with poor inherent immunogenicity, which may be increased by high-valency display and phage-associated adjuvanticity (see Immunological Mechanisms of Vaccination with Filamentous Phage below).
Cytotoxic T-Cell-Based Vaccines
The filamentous phage has been used to a lesser extent as a carrier for T-cell peptide epitopes, primarily as fusion proteins with pVIII (Table 3). Early work, showing that immunization with phage elicited T-cell help (Kölsch et al., 1971; Willis et al., 1993), was confirmed by several subsequent studies (De Berardinis et al., 1999; Ulivieri et al., 2008). From the perspective of vaccination against infectious disease, De Berardinis et al. (2000) showed that a cytotoxic T-cell (CTL) epitope from HIV-1 reverse transcriptase could elicit antigen-specific CTLs in vitro and in vivo without addition of exogenous helper T-cell epitopes, presumably since these are already present in the phage coat proteins (Mascolo et al., 2007). Similarly, efficient priming of CTLs was observed against phage-displayed T-cell epitopes from Hepatitis B virus (Wan et al., 2001) and Candida albicans (Yang et al., 2005a; Wang et al., 2006, 2014d), which, together with other types of immune responses, protected mice against systemic candidiasis. Vaccination with a combination of phage-displayed peptides elicited antigen-specific CTLs that proved effective in reducing porcine cysticercosis in a randomized controlled trial (Manoutcharian et al., 2004; Morales et al., 2008).
Table 3.
Studies using filamentous phage as an immunogenic carrier for peptide T-cell epitopes.
| Antigen | Species | Epitope type(s) | Display format(s) | T-cell response against peptide | Reference(s) |
|---|---|---|---|---|---|
| Taenia crassiceps KETc1, KETc7, and KETc12 | Pig | MHC class I | pVIII hybrid, pIII fully recombinant |
+ | Manoutcharian et al. (2004), Morales et al. (2008) |
| Ovalbumin | Mouse (C57BL/6) | MHC class I | pVIII hybrid | + | Mascolo et al. (2007) |
| HIV-1 reverse transcriptase | Mouse (C57BL/6, HLA-A2 transgenic) | MHC class I, MHC class II |
pVIII hybrid | + | De Berardinis et al. (1999, 2000) |
| PIA tumor antigen | Mouse (DBA/2) | MHC class I | pVIII hybrid | + | Wu et al. (2002) |
| Ovalbumin (B16.OVA model) | Mouse (C57BL/6, tlr4-/-, OT-I) | MHC class I | pVIII hybrid | + | Sartorius et al. (2011) |
| MAGE-A1, MAGE-A10, MAGE-A3 | Mouse (C57BL/6, HLA-A2 transgenic) | MHC class I | pVIII hybrid | + | Fang et al. (2005), Sartorius et al. (2008) |
| Candida albicans HSP90 | Mouse (C57BL6/, BALB/c) | MHC class I, MHC class II |
pVIII hybrid | + | Yang et al. (2005a), Wang et al. (2006, 2014d) |
| HBV | Mouse (BALB/c) | MHC class I | pVIII hybrid | + | Wan et al. (2001) |
While the correlates of vaccine-induced immune protection for infectious diseases, where they are known, are almost exclusively serum or mucosal antibodies (Plotkin, 2010), a primary goal of cancer vaccines is to elicit therapeutic CTL responses and/or suppress CD4+ regulatory T cells against tumor antigens while avoiding triggering adverse autoimmune responses. To this end, Wu et al. (2002) showed that a phage-displayed CTL epitope derived from the P1A tumor antigen was immunogenic and that the CTLs elicited in response to immunization both prevented mastocytoma tumor establishment and prolonged survival in mice with established tumors. Similarly, melanoma tumor CTL epitopes (MAGE-A1, MAGE-A3, and MAGE-A10) elicited CTLs that slowed tumor growth in mice and prolonged their survival (Fang et al., 2005; Sartorius et al., 2008). This work was later extended by Sartorius et al. (2011), who showed using the B16-OVA tumor model that phage double-displaying an ovalbumin CTL epitope on pVIII and an scFv against DEC-205 (a dendritic cell marker) on pIII could elicit much stronger anti-B16 tumor CTL responses that further improved survival rates. Thus, despite their large size, filamentous phage particles can elicit vigorous CTL responses, which can be enhanced in some cases by direct targeting of the phage to antigen-presenting cells. Although no studies have conducted a thorough comparison of the immunogenicity of a single CTL epitope in the context of different carriers, it is possible that the filamentous phage virion may induce stronger and more durable CTL responses compared to soluble exogenous proteins because of its efficient cross-presentation into the MHC class I pathway (see Immunological Mechanisms of Vaccination with Filamentous Phage below).
Recombinant Protein and DNA Vaccines
In certain vaccine applications, the filamentous phage has been used as a carrier for larger molecules that would be immunogenic even in isolation. Initially, the major advantages to phage display of such antigens were speed, ease of purification and low cost of production (Gram et al., 1993). E. coli F17a-G adhesin (Van Gerven et al., 2008), hepatitis B core antigen (Bahadir et al., 2011), and hepatitis B surface antigen (Balcioglu et al., 2014) all elicited antibody responses when displayed on pIII, although none of these studies compared the immunogenicity of the phage-displayed proteins with that of the purified protein alone. Phage displaying Schistosoma mansoni glutathione S-transferase on pIII elicited an antibody response that was both higher in titer and of different isotypes compared to immunization with the protein alone (Rao et al., 2003). Two studies of anti-idiotypic vaccines have used the phage as a carrier for antibody fragments bearing immunogenic idiotypes. Immunization with phage displaying the 1E10 idiotype scFv (mimicking a Vibrio anguillarum surface epitope) elicited antibodies that protected flounder fish from Vibrio anguillarum challenge (Xia et al., 2005). A chemically linked phage-BCL1 tumor-specific idiotype vaccine was weakly immunogenic in mice but extended survival time in a B-cell lymphoma model (Roehnisch et al., 2013), and was well-tolerated and immunogenic in patients with multiple myeloma (Roehnisch et al., 2014). One study of DNA vaccination with an anti-laminarin scFv found that DNA encoding a pIII-scFv fusion protein elicited stronger humoral and cell-mediated immune responses than DNA encoding the scFv alone (Cuesta et al., 2006), suggesting that under some circumstances, endogenous phage T-cell epitopes can enhance the immunogenicity of associated proteins. Taken together, the results of these studies show that as a particulate virus-like particle, the filamentous phage likely triggers different types of immune responses than recombinant protein antigens, and provide additional T-cell help to displayed or conjugated proteins. However, the low copy number of pIII-displayed proteins, as well as potentially unwanted phage-associated adjuvanticity, can make display of recombinant proteins by phage a suboptimal vaccine choice.
Immunological Mechanisms of Vaccination with Filamentous Phage
Although our understanding of the immune response against the filamentous phage pales in comparison to classical model antigens such as ovalbumin, recent work has begun to shed light on the immune mechanisms activated in response to phage vaccination (Figure 1). The phage particle is immunogenic without adjuvant in all species tested to date, including mice (Willis et al., 1993), rats (Dente et al., 1994), rabbits (de la Cruz et al., 1988), guinea pigs (Frenkel et al., 2000; Kim et al., 2004), fish (Coull et al., 1996; Xia et al., 2005), non-human primates (Chen et al., 2001), and humans (Roehnisch et al., 2014). Various routes of immunization have been employed, including oral administration (Delmastro et al., 1997) as well as subcutaneous (Grabowska et al., 2000), intraperitoneal (van Houten et al., 2006), intramuscular (Samoylova et al., 2012a), intravenous (Vaks and Benhar, 2011), and intradermal injection (Roehnisch et al., 2013); no published study has directly compared the effect of administration route on filamentous phage immunogenicity. Antibodies are generated against only three major sites on the virion: (i) the surface-exposed N-terminal ~12 residues of the pVIII monomer lattice (Terry et al., 1997; Kneissel et al., 1999); (ii) the N-terminal N1 and N2 domains of pIII (van Houten et al., 2010); and (iii) bacterial lipopolysaccharide (LPS) embedded in the phage coat (Henry et al., 2011). In mice, serum antibody titers against the phage typically reach 1:105–1:106 after 2–3 immunizations, and are maintained for at least 1 year post-immunization (Frenkel et al., 2000). Primary antibody responses against the phage appear to be composed of a mixture of IgM and IgG2b isotypes in C57BL/6 mice, while secondary antibody responses are composed primarily of IgG1 and IgG2b isotypes, with a lesser contribution of IgG2c and IgG3 isotypes (Hashiguchi et al., 2010). Deletion of the surface-exposed N1 and N2 domains of pIII produces a truncated form of this protein that does not elicit antibodies, but also results in a non-infective phage particle with lower overall immunogenicity (van Houten et al., 2010).
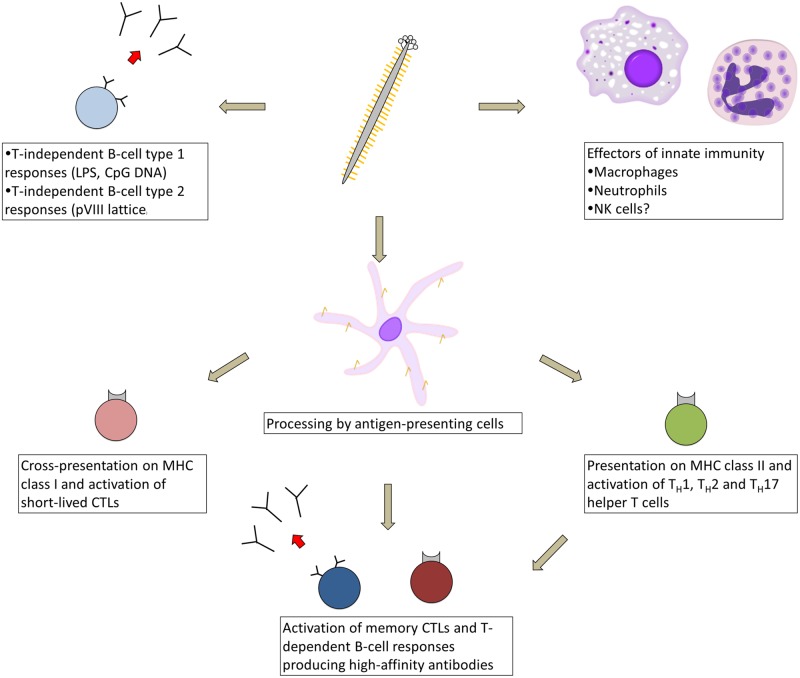
FIGURE 1.
Types of immune responses elicited in response to immunization with filamentous bacteriophage. As a virus-like particle, the filamentous phage engages multiple arms of the immune system, beginning with cellular effectors of innate immunity (macrophages, neutrophils, and possibly natural killer cells), which are recruited to tumor sites by phage displaying tumor-targeting moieties. The phage likely activates T-cell independent antibody responses, either via phage-associated TLR ligands or cross-linking by the pVIII lattice. After processing by antigen-presenting cells, phage-derived peptides are presented on MHC class II and cross-presented on MHC class I, resulting in activation of short-lived CTLs and an array of helper T-cell types, which help prime memory CTL and high-affinity B-cell responses.
Although serum anti-phage antibody titers appear to be at least partially T-cell dependent (Kölsch et al., 1971; Willis et al., 1993; De Berardinis et al., 1999; van Houten et al., 2010), many circulating pVIII-specific B cells in the blood are devoid of somatic mutation even after repeated biweekly immunizations, suggesting that under these conditions, the phage activates T-cell-independent B-cell responses in addition to high-affinity T-cell-dependent responses (Murira, 2014). Filamentous phage particles can be processed by antigen-presenting cells and presented on MHC class II molecules (Gaubin et al., 2003; Ulivieri et al., 2008) and can activate TH1, TH2, and TH17 helper T cells (Yang et al., 2005a; Wang et al., 2014d). Anti-phage TH2 responses were enhanced through display of CTLA-4 peptides fused to pIII (Kajihara et al., 2000). Phage proteins can also be cross-presented on MHC class I molecules (Wan et al., 2005) and can prime two waves of CTL responses, consisting first of short-lived CTLs and later of long-lived memory CTLs that require CD4+ T-cell help (Del Pozzo et al., 2010). The latter CTLs mediate a delayed-type hypersensitivity reaction (Fang et al., 2005; Del Pozzo et al., 2010).
The phage particle is self-adjuvanting through multiple mechanisms. Host cell wall-derived LPS enhances the virion’s immunogenicity, and its removal by polymyxin B chromatography reduces antibody titers against phage coat proteins (Grabowska et al., 2000). The phage’s single-stranded DNA genome contains CpG motifs and may also have an adjuvant effect. The antibody response against the phage is entirely dependent on MyD88 signaling and is modulated by stimulation of several Toll-like receptors (Hashiguchi et al., 2010), indicating that innate immunity plays an important but largely uncharacterized role in the activation of anti-phage adaptive immune responses. Biodistribution studies of the phage after intravenous injection show that it is cleared from the blood within hours through the reticuloendothelial system (Molenaar et al., 2002), particularly of the liver and spleen, where it is retained for days (Zou et al., 2004), potentially activating marginal-zone B-cell responses. Thus, the filamentous phage is not only a highly immunogenic carrier, but by virtue of activating a range of innate and adaptive immune responses, serves as an excellent model virus-like particle antigen.
Filamentous Phage as a Therapeutic and Prophylactic Agent in Bacterial Infection, Cancer, and Chronic Disease
Long before the identification of filamentous phage, other types of bacteriophage were already being used for antibacterial therapy in the former Soviet Union and Eastern Europe (reviewed in Sulakvelidze et al., 2001). The filamentous phage, with its non-lytic life cycle, has less obvious clinical uses, despite the fact that the host specificity of Inovirus and Plectrovirus includes many pathogens of medical importance, including Salmonella, E. coli, Shigella, Pseudomonas, Clostridium, and Mycoplasma species. In an effort to enhance their bactericidal activity, genetically modified filamentous phage have been used as a “Trojan horse” to introduce various antibacterial agents into cells. M13 and Pf3 phage engineered to express either BglII restriction endonuclease (Hagens and Blasi, 2003; Hagens et al., 2004), lambda phage S holin (Hagens and Blasi, 2003) or a lethal catabolite gene activator protein (Moradpour et al., 2009) effectively killed E. coli and Pseudomonas aeruginosa cells, respectively, with no concomitant release of LPS (Hagens and Blasi, 2003; Hagens et al., 2004). Unfortunately, the rapid emergence of resistant bacteria with modified F pili represents a major and possibly insurmountable obstacle to this approach. However, there are some indications that filamentous phage can exert useful but more subtle effects upon their bacterial hosts that may not result in the development of resistance to infection. Several studies have reported increased antibiotic sensitivity in bacterial populations simultaneously infected with either wild type filamentous phage (Hagens et al., 2006) or phage engineered to repress the cellular SOS response (Lu and Collins, 2009). Filamentous phage f1 infection inhibited early stage, but not mature, biofilm formation in E. coli (May et al., 2011). Thus, unmodified filamentous phage may be of future interest as elements of combination therapeutics against certain drug-resistant infections.
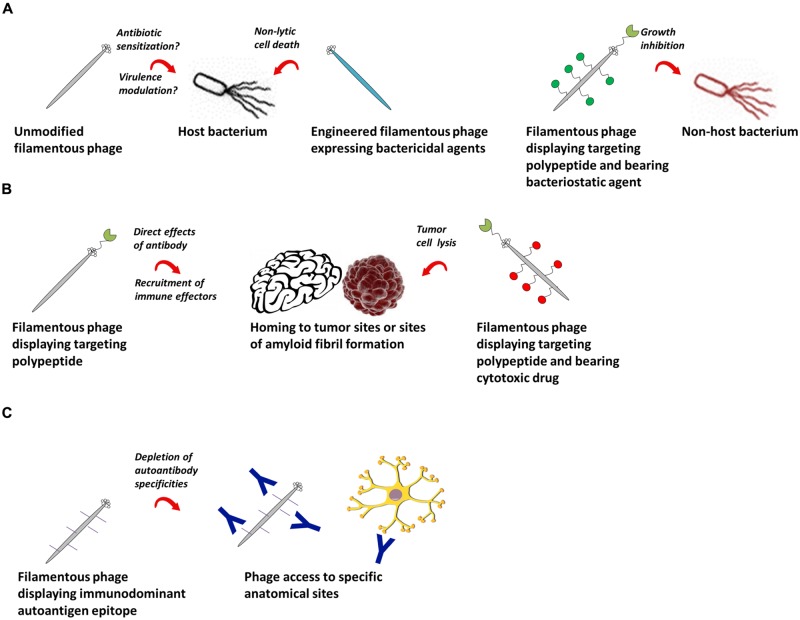
More advanced therapeutic applications of the filamentous phage emerge when it is modified to express a targeting moiety specific for pathogenic cells and/or proteins for the treatment of infectious diseases, cancer and autoimmunity (Figure 2). The first work in this area showed as proof-of-concept that phage encoding a GFP expression cassette and displaying a HER2-specific scFv on all copies of pIII were internalized into breast tumor cells, resulting in GFP expression (Poul and Marks, 1999). M13 or fd phage displaying either a targeting peptide or antibody fragment and tethered to chloramphenicol by a labile crosslinker were more potent inhibitors of Staphylococcus aureus growth than high-concentration free chloramphenicol (Yacoby et al., 2006; Vaks and Benhar, 2011). M13 phage loaded with doxorubicin and displaying a targeting peptide on pIII specifically killed prostate cancer cells in vitro (Ghosh et al., 2012a). Tumor-specific peptide:pVIII fusion proteins selected from “landscape” phage (Romanov et al., 2001; Abbineni et al., 2010; Fagbohun et al., 2012, 2013; Lang et al., 2014; Wang et al., 2014a) were able to target and deliver siRNA-, paclitaxel-, and doxorubicin-containing liposomes to tumor cells (Jayanna et al., 2010a; Wang et al., 2010a,b,c, 2014b,c; Bedi et al., 2011, 2013, 2014); they were non-toxic and increased tumor remission rates in mouse models (Jayanna et al., 2010b; Wang et al., 2014b,c). Using the B16-OVA tumor model, Eriksson et al. (2007) showed that phage displaying peptides and/or Fabs specific for tumor antigens delayed tumor growth and improved survival, owing in large part to activation of tumor-associated macrophages and recruitment of neutrophils to the tumor site (Eriksson et al., 2009). Phage displaying an scFv against β-amyloid fibrils showed promise as a diagnostic (Frenkel and Solomon, 2002) and therapeutic (Solomon, 2008) reagent for Alzheimer’s disease and Parkinson’s disease due to the unanticipated ability of the phage to penetrate into brain tissue (Ksendzovsky et al., 2012). Similarly, phage displaying an immunodominant peptide epitope derived from myelin oligodendrocyte glycoprotein depleted pathogenic demyelinating antibodies in brain tissue in the murine experimental autoimmune encephalomyelitis model of multiple sclerosis (Rakover et al., 2010). The advantages of the filamentous phage in this context over traditional antibody-drug or protein–peptide conjugates are (i) its ability to carry very high amounts of drug or peptide, and (ii) its ability to access anatomical compartments that cannot generally be reached by systemic administration of a protein.
FIGURE 2.
Potential therapeutic applications of filamentous bacteriophage. (A) Filamentous phage as a therapeutic against bacterial infections. Left: wild-type or engineered phage bearing genes encoding antibacterial agents can be used as therapeutics against their natural bacterial hosts. Right: filamentous phage displaying antibodies or polypeptides can target bacterial cells outside their natural species tropism. (B) Filamentous phage as a therapeutic in cancer and chronic diseases. Phage encoding targeting peptides home specifically to tumor or amyloid fibril sites, where they can have direct effects, recruit immune effector cells, or deliver conjugated cytotoxic drugs. (C) Filamentous phage as a therapeutic for autoantibody-mediated autoimmune conditions. Phage displaying immunodominant autoantigen epitopes can selectively deplete autoantibodies of defined specificities.
Unlike most therapeutic biologics, the filamentous phage’s production in bacteria complicates its use in humans in several ways. First and foremost, crude preparations of filamentous phage typically contain very high levels of contaminating LPS, in the range of ~102–104 endotoxin units (EU)/mL (Boratynski et al., 2004; Branston et al., 2015), which have the potential to cause severe adverse reactions. LPS is not completely removed by polyethylene glycol precipitation or cesium chloride density gradient centrifugation (Smith and Gingrich, 2005; Branston et al., 2015), but its levels can be reduced dramatically using additional purification steps such as size exclusion chromatography (Boratynski et al., 2004; Zakharova et al., 2005), polymyxin B chromatography (Grabowska et al., 2000), and treatment with detergents such as Triton X-100 or Triton X-114 (Roehnisch et al., 2014; Branston et al., 2015). These strategies routinely achieve endotoxin levels of <1 EU/mL as measured by the limulus amebocyte lysate (LAL) assay, well below the FDA limit for parenteral administration of 5 EU/kg body weight/dose, although concerns remain regarding the presence of residual virion-associated LPS which may be undetectable. A second and perhaps unavoidable consequence of the filamentous phage’s bacterial production is inherent heterogeneity of particle size and the spectrum of host cell-derived virion-associated and soluble contaminants, which may be cause for safety concerns and restrict its use to high-risk groups.
Many types of bacteriophage and engineered phage variants, including filamentous phage, have been proposed for prophylactic use ex vivo in food safety, either in the production pipeline (reviewed in Dalmasso et al., 2014) or for detection of foodborne pathogens post-production (reviewed in Schmelcher and Loessner, 2014). Filamentous phage displaying a tetracysteine tag on pIII were used to detect E. coli cells through staining with biarsenical dye (Wu et al., 2011). M13 phage functionalized with metallic silver were highly bactericidal against E. coli and Staphylococcus epidermidis (Mao et al., 2010). Biosensors based on surface plasmon resonance (Nanduri et al., 2007), piezoelectric transducers (Olsen et al., 2006), linear dichroism (Pacheco-Gomez et al., 2012), and magnetoelastic sensor technology (Lakshmanan et al., 2007; Huang et al., 2009) were devised using filamentous phage displaying scFv or conjugated to whole IgG against E. coli, Listeria monocytogenes, Salmonella typhimurium, and Bacillus anthracis with limits of detection on the order of 102–106 bacterial cells/mL. Proof of concept has been demonstrated for use of such phage-based biosensors to detect bacterial contamination of live produce (Li et al., 2010b) and eggs (Chai et al., 2012).
Filamentous Phage as a Scaffold for Bioconjugation and Surface Chemistry
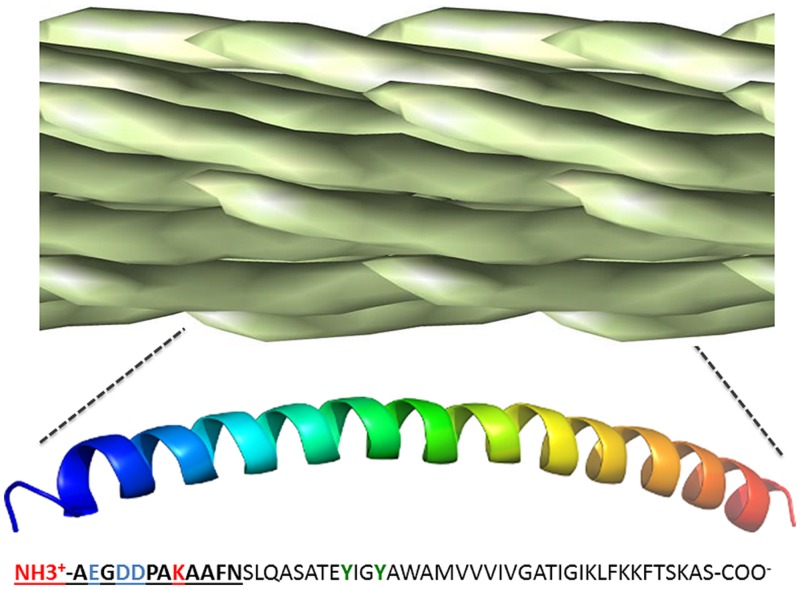
The filamentous phage particle is enclosed by a rod-like protein capsid, ~1000 nm long and 5 nm wide, made up almost entirely of overlapping pVIII monomers, each of which lies ~27 angstroms from its nearest neighbor and exposes two amine groups as well as at least three carboxyl groups (Henry et al., 2011). The regularity of the phage pVIII lattice and its diversity of chemically addressable groups make it an ideal scaffold for bioconjugation (Figure 3). The most commonly used approach is functionalization of amine groups with NHS esters (van Houten et al., 2006, 2010; Yacoby et al., 2006), although this can result in unwanted acylation of pIII and any displayed biomolecules. Carboxyl groups and tyrosine residues can also be functionalized using carbodiimide coupling and diazonium coupling, respectively (Li et al., 2010a). Carrico et al. (2012) developed methods to specifically label pVIII N-termini without modification of exposed lysine residues through a two-step transamination-oxime formation reaction. Specific modification of phage coat proteins is even more easily accomplished using genetically modified phage displaying peptides (Ng et al., 2012) or enzymes (Chen et al., 2007; Hess et al., 2012), but this can be cumbersome and is less general in application.
FIGURE 3.
Chemically addressable groups of the filamentous bacteriophage major coat protein lattice. The filamentous phage virion is made up of ~2,500–4,000 overlapping copies of the 50-residue major coat protein, pVIII, arranged in a shingle-type lattice. Each monomer has an array of chemically addressable groups available for bioorthogonal conjugation, including two primary amine groups (shown in red), three carboxyl groups (show in blue) and two hydroxyl groups (show in green). The 12 N-terminal residues generally exposed to the immune system for antibody binding are in bold underline. Figure adapted from structural data of Marvin, 1990, freely available in PDB and SCOPe databases.
For more than a decade, interest in the filamentous phage as a building block for nanomaterials has been growing because of its unique physicochemical properties, with emerging applications in magnetics, optics, and electronics. It has long been known that above a certain concentration threshold, phage can form ordered crystalline suspensions (Welsh et al., 1996). Lee et al. (2002) engineered M13 phage to display a ZnS-binding peptide on pIII and showed that, in the presence of ZnS nanoparticles, they self-assemble into highly ordered film biomaterials that can be aligned using magnetic fields. Taking advantage of the ability to display substrate-specific peptides at known locations on the phage filament (Huang et al., 2005; Hess et al., 2012), this pioneering work became the basis for construction of two- and three-dimensional nanomaterials with more advanced architectures, including semiconducting nanowires (Mao et al., 2003, 2004), nanoparticles (Yoo et al., 2006), and nanocomposites (Oh et al., 2012; Chen et al., 2014). Using hybrid M13 phage displaying Co3O4- and gold-binding peptides on pVIII as a scaffold to assemble nanowires on polyelectrolyte multilayers, Nam et al. (2006) produced a thin, flexible lithium ion battery, which could be stamped onto platinum microband current collectors (Nam et al., 2008). The electrochemical properties of such batteries were further improved through pIII-display of single-walled carbon nanotube-binding peptides (Lee et al., 2009), offering an approach for sustainable production of nanostructured electrodes from poorly conductive starting materials. Phage-based nanomaterials have found applications in cancer imaging (Ghosh et al., 2012b; Yi et al., 2012), photocatalytic water splitting (Nam et al., 2010a; Neltner et al., 2010), light harvesting (Nam et al., 2010b; Chen et al., 2013), photoresponsive technologies (Murugesan et al., 2013), neural electrodes (Kim et al., 2014), and piezoelectric energy generation (Murugesan et al., 2013).
Thus, the unique physicochemical properties of the phage, in combination with modular display of peptides and proteins with known binding specificity, have spawned wholly novel materials with diverse applications. It is worth noting that the unusual biophysical properties of the filamentous phage can also be exploited in the study of structures of other macromolecules. Magnetic alignment of high-concentration filamentous phage in solution can partially order DNA, RNA, proteins, and other biomolecules for measurement of dipolar coupling interactions (Hansen et al., 1998, 2000; Dahlke Ojennus et al., 1999) in NMR spectroscopy.
Filamentous Phage as an Engine for Experimental Protein Evolution
Because of their large population sizes, short generation times, small genome sizes and ease of manipulation, various filamentous and non-filamentous bacteriophages have been used as models of experimental evolution (reviewed in Husimi, 1989; Wichman and Brown, 2010; Kawecki et al., 2012; Hall et al., 2013). The filamentous phage has additional practical uses in protein engineering and directed protein evolution, due to its unique tolerance of genetic modifications that allow biomolecules to be displayed on the virion surface. First and foremost among these applications is in vitro affinity maturation of antibody fragments displayed on pIII. Libraries of variant Fabs and single chain antibodies can be generated via random or site-directed mutagenesis and selected on the basis of improved or altered binding, roughly mimicking the somatic evolution strategy of the immune system (Marks et al., 1992; Bradbury et al., 2011). However, other in vitro display systems, such as yeast display, have important advantages over the filamentous phage for affinity maturation (although each display technology has complementary strengths; Koide and Koide, 2012), and regardless of the display method, selection of “improved” variants can be slow and cumbersome. Iterative methods have been developed to combine computationally designed mutations (Lippow et al., 2007) and circumvent the screening of combinatorial libraries, but these have had limited success to date.
Recently, Esvelt et al. (2011) developed a novel strategy for directed evolution of filamentous phage-displayed proteins, called phage-assisted continuous evolution (PACE), which allows multiple rounds of evolution per day with little experimental intervention. The authors engineered M13 phage to encode an exogenous protein (the subject for directed evolution), whose functional activity triggers gene III expression from an accessory plasmid; variants of the exogenous protein arise by random mutagenesis during phage replication, the rate of which can be increased by inducible expression of error-prone DNA polymerases. By supplying limiting amounts of receptive E. coli cells to the engineered phage variants, Esvelt et al. (2011) elegantly linked phage infectivity and production of offspring with the presence of a desired protein phenotype. Carlson et al. (2014) later showed that PACE selection stringency could be modulated by providing small amounts of pIII independently of protein phenotype, and undesirable protein functions negatively selected by linking them to expression of a truncated pIII variant that impairs infectivity in a dominant negative fashion. PACE is currently limited to protein functions that can be linked in some way to the expression of a gene III reporter, such as protein–protein interaction, recombination, DNA or RNA binding, and enzymatic catalysis (Meyer and Ellington, 2011). This approach represents a promising avenue for both basic research in molecular evolution (Dickinson et al., 2013) and synthetic biology, including antibody engineering.
Filamentous Phage Ecology
Filamentous bacteriophage have been recovered from diverse environmental sources, including soil (Murugaiyan et al., 2011), coastal fresh water (Xue et al., 2012), alpine lakes (Hofer and Sommaruga, 2001) and deep sea bacteria (Jian et al., 2012), but not, perhaps surprisingly, the human gut (Kim et al., 2011). The environmental “phageome” in soil and water represent the largest source of replicating DNA on the planet, and is estimated to contain upward of 1030 viral particles (Ashelford et al., 2003; Chibani-Chennoufi et al., 2004; Suttle, 2005). The few studies attempting to investigate filamentous phage environmental ecology using classical environmental microbiology techniques (typically direct observation by electron microscopy) found that filamentous phage made up anywhere from 0 to 100% of all viral particles (Demuth et al., 1993; Pina et al., 1998; Hofer and Sommaruga, 2001). There was some evidence of seasonal fluctuation of filamentous phage populations in tandem with the relative abundance of free-living heterotrophic bacteria (Hofer and Sommaruga, 2001). Environmental metagenomics efforts are just beginning to unravel the composition of viral ecosystems. The existing data suggest that filamentous phage comprise minor constituents of viral communities in freshwater (Roux et al., 2012) and reclaimed and potable water (Rosario et al., 2009) but have much higher frequencies in wastewater and sewage (Cantalupo et al., 2011; Alhamlan et al., 2013), with the caveat that biases inherent to the methodologies for ascertaining these data (purification of viral particles, sequencing biases) have not been not well validated. There are no data describing the population dynamics of filamentous phage and their host species in the natural environment.
At the individual virus-bacterium level, it is clear that filamentous phage can modulate host phenotype, including the virulence of important human and crop pathogens. This can occur either through direct effects of phage replication on cell growth and physiology, or, more typically, by horizontal transfer of genetic material contained within episomes and/or chromosomally integrated prophage. Temperate filamentous phage may also play a role in genome evolution (reviewed in Canchaya et al., 2003). Perhaps the best-studied example of virulence modulation by filamentous phage is that of Vibrio cholerae, whose full virulence requires lysogenic conversion by the cholera toxin-encoding CTXϕ phage (Waldor and Mekalanos, 1996). Integration of CTXϕ phage occurs at specific sites in the genome; these sequences are introduced through the combined action of another filamentous phage, fs2ϕ, and a satellite filamentous phage, TLC-Knϕ1 (Hassan et al., 2010). Thus, filamentous phage species interact and coevolve with each other in addition to their hosts. Infection by filamentous phage has been implicated in the virulence of Yersinia pestis (Derbise et al., 2007), Neisseria meningitidis (Bille et al., 2005, 2008), Vibrio parahaemolyticus (Iida et al., 2001), E. coli 018:K1:H7 (Gonzalez et al., 2002), Xanthomonas campestris (Kamiunten and Wakimoto, 1982), and P. aeruginosa (Webb et al., 2004), although in most of these cases, the specific mechanisms modulating virulence are unclear. Phage infection can both enhance or repress virulence depending on the characteristics of the phage, the host bacterium, and the environmental milieu, as is the case for the bacterial wilt pathogen Ralstonia solanacearum (Yamada, 2013). Since infection results in downregulation of the pili used for viral entry, filamentous phage treatment has been proposed as a hypothetical means of inhibiting bacterial conjugation and horizontal gene transfer, so as to prevent the spread of antibiotic resistance genes (Lin et al., 2011).
Finally, the filamentous phage may also play a future role in the preservation of biodiversity of other organisms in at-risk ecosystems. Engineered phage have been proposed for use in bioremediation, either displaying antibody fragments of desired specificity for filtration of toxins and environmental contaminants (Petrenko and Makowski, 1993), or as biodegradable polymers displaying peptides selected for their ability to aggregate pollutants, such as oil sands tailings (Curtis et al., 2011, 2013). Engineered phage displaying peptides that specifically bind inorganic materials have also been proposed for use in more advanced and less intrusive mineral separation technologies (Curtis et al., 2009).
Conclusion and Future Perspectives
The filamentous phage represents a highly versatile organism whose uses extend far beyond traditional phage display and affinity selection of antibodies and polypeptides of desired specificity. Its high immunogenicity and ability to display a variety of surface antigens make the phage an excellent particulate vaccine carrier, although its bacterial production and preparation heterogeneity likely limits its applications in human vaccines at present, despite being apparently safe and well-tolerated in animals and people. Unanticipated characteristics of the phage particle, such as crossing of the blood–brain barrier and formation of highly ordered liquid crystalline phases, have opened up entirely new avenues of research in therapeutics for chronic disease and the design of nanomaterials. Our comparatively detailed understanding of the interactions of model filamentous phage with their bacterial hosts has allowed researchers to harness the phage life cycle to direct protein evolution in the lab. Hopefully, deeper knowledge of phage–host interactions at an ecological level may produce novel strategies to control bacterial pathogenesis. While novel applications of the filamentous phage continue to be developed, the phage is likely to retain its position as a workhorse for therapeutic antibody discovery for many years to come, even with the advent of competing technologies.
Author Contributions
KH and JS conceived and wrote the manuscript. MA-G read the manuscript and commented on the text.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by funding from the National Research Council of Canada (KH, MA-G) and the Canada Research Chair Program (JS). We thank Jyothi Kumaran and Roger MacKenzie for critical appraisal of the manuscript, and Jasna Rakonjac for inviting us to contribute it. This is National Research Council Canada publication number 53282.
References
- Abbineni G., Modali S., Safiejko-Mroczka B., Petrenko V. A., Mao C. (2010). Evolutionary selection of new breast cancer cell-targeting peptides and phages with the cell-targeting peptides fully displayed on the major coat and their effects on actin dynamics during cell internalization. Mol. Pharm. 7 1629–1642. 10.1021/mp100052y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdennebi L., Couture L., Grebert D., Pajot E., Salesse R., Remy J. J. (1999). Generating FSH antagonists and agonists through immunization against FSH receptor N-terminal decapeptides. J. Mol. Endocrinol. 22 151–159. 10.1677/jme.0.0220151 [DOI] [PubMed] [Google Scholar]
- Alhamlan F. S., Ederer M. M., Brown C. J., Coats E. R., Crawford R. L. (2013). Metagenomics-based analysis of viral communities in dairy lagoon wastewater. J. Microbiol. Methods 92 183–188. 10.1016/j.mimet.2012.11.016 [DOI] [PubMed] [Google Scholar]
- Ashelford K. E., Day M. J., Fry J. C. (2003). Elevated abundance of bacteriophage infecting bacteria in soil. Appl. Environ. Microbiol. 69 285–289. 10.1128/AEM.69.1.285-289.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahadir A. O., Balcioglu B. K., Uzyol K. S., Hatipoglu I., Sogut I., Basalp A., et al. (2011). Phage displayed HBV core antigen with immunogenic activity. Appl. Biochem. Biotechnol. 165 1437–1447. 10.1007/s12010-011-9365-1 [DOI] [PubMed] [Google Scholar]
- Balcioglu B. K., Ozdemir-Barahir A., Hinc D., Tamerler C., Erdag B. (2014). Cost effective filamentous phage based immunization nanoparticles displaying a full-length hepatitis B virus surface antigen. Adv. Biosci. Biotechnol. 5 7 10.4236/abb.2014.51008 [DOI] [Google Scholar]
- Bass S., Greene R., Wells J. A. (1990). Hormone phage: an enrichment method for variant proteins with altered binding properties. Proteins 8 309–314. 10.1002/prot.340080405 [DOI] [PubMed] [Google Scholar]
- Bastien N., Trudel M., Simard C. (1997). Protective immune responses induced by the immunization of mice with a recombinant bacteriophage displaying an epitope of the human respiratory syncytial virus. Virology 234 118–122. 10.1006/viro.1997.8632 [DOI] [PubMed] [Google Scholar]
- Bedi D., Gillespie J. W., Petrenko V. A. (2014). Selection of pancreatic cancer cell-binding landscape phages and their use in development of anticancer nanomedicines. Protein Eng. Des. Sel. 27 235–243. 10.1093/protein/gzu020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedi D., Gillespie J. W., Petrenko V. A., Jr., Ebner A., Leitner M., Hinterdorfer P., et al. (2013). Targeted delivery of siRNA into breast cancer cells via phage fusion proteins. Mol. Pharm. 10 551–559. 10.1021/mp3006006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedi D., Musacchio T., Fagbohun O. A., Gillespie J. W., Deinnocentes P., Bird R. C., et al. (2011). Delivery of siRNA into breast cancer cells via phage fusion protein-targeted liposomes. Nanomedicine 7 315–323. 10.1016/j.nano.2010.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bille E., Ure R., Gray S. J., Kaczmarski E. B., Mccarthy N. D., Nassif X., et al. (2008). Association of a bacteriophage with meningococcal disease in young adults. PLoS ONE 3:e3885 10.1371/journal.pone.0003885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bille E., Zahar J. R., Perrin A., Morelle S., Kriz P., Jolley K. A., et al. (2005). A chromosomally integrated bacteriophage in invasive meningococci. J. Exp. Med. 201 1905–1913. 10.1084/jem.20050112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratynski J., Syper D., Weber-Dabrowska B., Lusiak-Szelachowska M., Pozniak G., Gorski A. (2004). Preparation of endotoxin-free bacteriophages. Cell Mol. Biol. Lett. 9 253–259. [PubMed] [Google Scholar]
- Bradbury A. R., Sidhu S., Dubel S., Mccafferty J. (2011). Beyond natural antibodies: the power of in vitro display technologies. Nat. Biotechnol. 29 245–254. 10.1038/nbt.1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branston S. D., Wright J., Keshavarz-Moore E. (2015). A non-chromatographic method for the removal of endotoxins from bacteriophages. Biotechnol. Bioeng. 112 1714–1719. 10.1002/bit.25571 [DOI] [PubMed] [Google Scholar]
- Bratkovic T. (2010). Progress in phage display: evolution of the technique and its application. Cell. Mol. Life Sci. 67 749–767. 10.1007/s00018-009-0192-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling F., Dubel S., Seehaus T., Klewinghaus I., Little M. (1991). A surface expression vector for antibody screening. Gene 104 147–153. 10.1016/0378-1119(91)90244-6 [DOI] [PubMed] [Google Scholar]
- Canchaya C., Proux C., Fournous G., Bruttin A., Brussow H. (2003). Prophage genomics. Microbiol. Mol. Biol. Rev. 67 238–276. 10.1128/MMBR.67.2.238-276.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo P. G., Calgua B., Zhao G., Hundesa A., Wier A. D., Katz J. P., et al. (2011). Raw sewage harbors diverse viral populations. MBio 2:e00180–11. 10.1128/mBio.00180-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson J. C., Badran A. H., Guggiana-Nilo D. A., Liu D. R. (2014). Negative selection and stringency modulation in phage-assisted continuous evolution. Nat. Chem. Biol. 10 216–222. 10.1038/nchembio.1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrico Z. M., Farkas M. E., Zhou Y., Hsiao S. C., Marks J. D., Chokhawala H., et al. (2012). N-Terminal labeling of filamentous phage to create cancer marker imaging agents. ACS Nano 6 6675–6680. 10.1021/nn301134z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y., Li S., Horikawa S., Park M. K., Vodyanoy V., Chin B. A. (2012). Rapid and sensitive detection of Salmonella Typhimurium on eggshells by using wireless biosensors. J. Food. Prot. 75 631–636. 10.4315/0362-028X.JFP-11-339 [DOI] [PubMed] [Google Scholar]
- Chen I., Choi Y. A., Ting A. Y. (2007). Phage display evolution of a peptide substrate for yeast biotin ligase and application to two-color quantum dot labeling of cell surface proteins. J. Am. Chem. Soc. 129 6619–6625. 10.1021/ja071013g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. Y., Dang X., Klug M. T., Qi J., Courchesne N.-M. D., Burpo F. J., et al. (2013). Versatile three-dimensional virus-based template for dye-sensitized solar cells with improved electron transport and light harvesting. ACS Nano 7 6563–6574. 10.1021/nn4014164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. Y., Hyder M. N., Mackanic D., Courchesne N. M., Qi J., Klug M. T., et al. (2014). Assembly of viral hydrogels for three-dimensional conducting nanocomposites. Adv. Mater. 26 5101–5107. 10.1002/adma.201400828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Scala G., Quinto I., Liu W., Chun T. W., Justement J. S., et al. (2001). Protection of rhesus macaques against disease progression from pathogenic SHIV-89.6PD by vaccination with phage-displayed HIV-1 epitopes. Nat. Med. 7 1225–1231. 10.1038/nm1101-1225 [DOI] [PubMed] [Google Scholar]
- Chibani-Chennoufi S., Bruttin A., Dillmann M. L., Brussow H. (2004). Phage-host interaction: an ecological perspective. J. Bacteriol. 186 3677–3686. 10.1128/JB.186.12.3677-3686.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coull J. J., Melvin W. T., Labus M. B., Raynard R. S., King J., Munro A. L. (1996). Evaluation of filamentous bacteriophage as immunogens in Atlantic salmon. Biochem. Soc. Trans. 24 255S. [DOI] [PubMed] [Google Scholar]
- Cuesta A. M., Suarez E., Larsen M., Jensen K. B., Sanz L., Compte M., et al. (2006). Enhancement of DNA vaccine potency through linkage of antigen to filamentous bacteriophage coat protein III domain I. Immunology 117 502–506. 10.1111/j.1365-2567.2006.02325.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis S. B., Dunbar W. S., Macgillivray R. T. (2013). Bacteriophage-induced aggregation of oil sands tailings. Biotechnol. Bioeng. 110 803–811. 10.1002/bit.24745 [DOI] [PubMed] [Google Scholar]
- Curtis S. B., Hewitt J., Macgillivray R. T., Dunbar W. S. (2009). Biomining with bacteriophage: selectivity of displayed peptides for naturally occurring sphalerite and chalcopyrite. Biotechnol. Bioeng. 102 644–650. 10.1002/bit.22073 [DOI] [PubMed] [Google Scholar]
- Curtis S. B., Macgillivray R. T., Dunbar W. S. (2011). Effects of bacteriophage on the surface properties of chalcopyrite (CuFeS(2)), and phage-induced flocculation of chalcopyrite, glacial till, and oil sands tailings. Biotechnol. Bioeng. 108 1579–1590. 10.1002/bit.23097 [DOI] [PubMed] [Google Scholar]
- Dahlke Ojennus D., Mitton-Fry R. M., Wuttke D. S. (1999). Induced alignment and measurement of dipolar couplings of an SH2 domain through direct binding with filamentous phage. J. Biomol. NMR 14 175–179. 10.1023/A:1008304332574 [DOI] [PubMed] [Google Scholar]
- Dalmasso M., Hill C., Ross R. P. (2014). Exploiting gut bacteriophages for human health. Trends Microbiol. 22 399–405. 10.1016/j.tim.2014.02.010 [DOI] [PubMed] [Google Scholar]
- De Berardinis P., D’apice L., Prisco A., Ombra M. N., Barba P., Del Pozzo G., et al. (1999). Recognition of HIV-derived B and T cell epitopes displayed on filamentous phages. Vaccine 17 1434–1441. 10.1016/S0264-410X(98)00377-6 [DOI] [PubMed] [Google Scholar]
- De Berardinis P., Sartorius R., Fanutti C., Perham R. N., Del Pozzo G., Guardiola J. (2000). Phage display of peptide epitopes from HIV-1 elicits strong cytolytic responses. Nat. Biotechnol. 18 873–876. 10.1038/78490 [DOI] [PubMed] [Google Scholar]
- de la Cruz V. F., Lal A. A., Mccutchan T. F. (1988). Immunogenicity and epitope mapping of foreign sequences via genetically engineered filamentous phage. J. Biol. Chem. 263 4318–4322. [PubMed] [Google Scholar]
- Del Pozzo G., Mascolo D., Sartorius R., Citro A., Barba P., D’Apice L., et al. (2010). Triggering DTH and CTL activity by fd filamentous bacteriophages: role of CD4+ T cells in memory responses. J. Biomed. Biotechnol. 2010:894971 10.1155/2010/894971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmastro P., Meola A., Monaci P., Cortese R., Galfre G. (1997). Immunogenicity of filamentous phage displaying peptide mimotopes after oral administration. Vaccine 15 1276–1285. 10.1016/S0264-410X(97)00072-8 [DOI] [PubMed] [Google Scholar]
- Demangel C., Lafaye P., Mazie J. C. (1996). Reproducing the immune response against the Plasmodium vivax merozoite surface protein 1 with mimotopes selected from a phage-displayed peptide library. Mol. Immunol. 33 909–916. 10.1016/S0161-5890(96)00058-2 [DOI] [PubMed] [Google Scholar]
- Demuth J., Neve H., Witzel K. P. (1993). Direct electron microscopy study on the morphological diversity of bacteriophage populations in lake Pluβsee. Appl. Environ. Microbiol. 59 3378–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Micheli G., Felici F., Folgori A., Luzzago A., et al. (1994). Monoclonal antibodies that recognise filamentous phage: tools for phage display technology. Gene 148 7–13. 10.1016/0378-1119(94)90227-5 [DOI] [PubMed] [Google Scholar]
- Derbise A., Chenal-Francisque V., Pouillot F., Fayolle C., Prevost M. C., Medigue C., et al. (2007). A horizontally acquired filamentous phage contributes to the pathogenicity of the plague bacillus. Mol. Microbiol. 63 1145–1157. 10.1111/j.1365-2958.2006.05570.x [DOI] [PubMed] [Google Scholar]
- Devlin J. J., Panganiban L. C., Devlin P. E. (1990). Random peptide libraries: a source of specific protein binding molecules. Science 249 404–406. 10.1126/science.2143033 [DOI] [PubMed] [Google Scholar]
- Dickinson B. C., Leconte A. M., Allen B., Esvelt K. M., Liu D. R. (2013). Experimental interrogation of the path dependence and stochasticity of protein evolution using phage-assisted continuous evolution. Proc. Natl. Acad. Sci. U.S.A. 110 9007–9012. 10.1073/pnas.1220670110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Marzo Veronese F., Willis A. E., Boyer-Thompson C., Appella E., Perham R. N. (1994). Structural mimicry and enhanced immunogenicity of peptide epitopes displayed on filamentous bacteriophage. The V3 loop of HIV-1 gp120. J. Mol. Biol. 243 167–172. 10.1006/jmbi.1994.1643 [DOI] [PubMed] [Google Scholar]
- d’Hérelle F. (1917). Sur un microbe invisible antagoniste des bacilles dysentérique. Acad. Sci. Paris 165 3. [Google Scholar]
- Eriksson F., Culp W. D., Massey R., Egevad L., Garland D., Persson M. A., et al. (2007). Tumor specific phage particles promote tumor regression in a mouse melanoma model. Cancer Immunol. Immunother. 56 677–687. 10.1007/s00262-006-0227-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson F., Tsagozis P., Lundberg K., Parsa R., Mangsbo S. M., Persson M. A., et al. (2009). Tumor-specific bacteriophages induce tumor destruction through activation of tumor-associated macrophages. J. Immunol. 182 3105–3111. 10.4049/jimmunol.0800224 [DOI] [PubMed] [Google Scholar]
- Esposito M., Luccarini I., Cicatiello V., De Falco D., Fiorentini A., Barba P., et al. (2008). Immunogenicity and therapeutic efficacy of phage-displayed beta-amyloid epitopes. Mol. Immunol. 45 1056–1062. 10.1016/j.molimm.2007.07.023 [DOI] [PubMed] [Google Scholar]
- Esvelt K. M., Carlson J. C., Liu D. R. (2011). A system for the continuous directed evolution of biomolecules. Nature 472 499–503. 10.1038/nature09929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagbohun O. A., Bedi D., Grabchenko N. I., Deinnocentes P. A., Bird R. C., Petrenko V. A. (2012). Landscape phages and their fusion proteins targeted to breast cancer cells. Protein Eng. Des. Sel. 25 271–283. 10.1093/protein/gzs013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagbohun O. A., Kazmierczak R. A., Petrenko V. A., Eisenstark A. (2013). Metastatic prostate cancer cell-specific phage-like particles as a targeted gene-delivery system. J. Nanobiotechnol. 11 31 10.1186/1477-3155-11-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J., Wang G., Yang Q., Song J., Wang Y., Wang L. (2005). The potential of phage display virions expressing malignant tumor specific antigen MAGE-A1 epitope in murine model. Vaccine 23 4860–4866. 10.1016/j.vaccine.2005.05.024 [DOI] [PubMed] [Google Scholar]
- Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.) (2005). Virus Taxonomy. 8th report of the International Committee on the Taxonomy of Viruses. London: Elsevier/Academic Press. [Google Scholar]
- Felici F., Luzzago A., Folgori A., Cortese R. (1993). Mimicking of discontinuous epitopes by phage-displayed peptides, II. Selection of clones recognized by a protective monoclonal antibody against the Bordetella pertussis toxin from phage peptide libraries. Gene 128 21–27. 10.1016/0378-1119(93)90148-V [DOI] [PubMed] [Google Scholar]
- Folgori A., Tafi R., Meola A., Felici F., Galfre G., Cortese R., et al. (1994). A general strategy to identify mimotopes of pathological antigens using only random peptide libraries and human sera. EMBO J. 13 2236–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel D., Dewachter I., Van Leuven F., Solomon B. (2003). Reduction of β-amyloid plaques in brain of transgenic mouse model of Alzheimer’s disease by EFRH-phage immunization. Vaccine 21 1060–1065. 10.1016/S0264-410X(02)00609-6 [DOI] [PubMed] [Google Scholar]
- Frenkel D., Katz O., Solomon B. (2000). Immunization against Alzheimer’s β-amyloid plaques via EFRH phage administration. Proc. Natl. Acad. Sci. U.S.A. 97 11455–11459. 10.1073/pnas.97.21.11455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel D., Solomon B. (2002). Filamentous phage as vector-mediated antibody delivery to the brain. Proc. Natl. Acad. Sci. U.S.A. 99 5675–5679. 10.1073/pnas.072027199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Mao S., Kaufmann G., Wirsching P., Lerner R. A., Janda K. D. (2002). A method for the generation of combinatorial antibody libraries using pIX phage display. Proc. Natl. Acad. Sci. U.S.A. 99 12612–12616. 10.1073/pnas.192467999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Mao S., Lo C. H., Wirsching P., Lerner R. A., Janda K. D. (1999). Making artificial antibodies: a format for phage display of combinatorial heterodimeric arrays. Proc. Natl. Acad. Sci. U.S.A. 96 6025–6030. 10.1073/pnas.96.11.6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubin M., Fanutti C., Mishal Z., Durrbach A., De Berardinis P., Sartorius R., et al. (2003). Processing of filamentous bacteriophage virions in antigen-presenting cells targets both HLA class I and class II peptide loading compartments. DNA Cell Biol. 22 11–18. 10.1089/104454903321112451 [DOI] [PubMed] [Google Scholar]
- Geysen H. M., Rodda S. J., Mason T. J. (1986). A priori delineation of a peptide which mimics a discontinuous antigenic determinant. Mol. Immunol. 23 709–715. 10.1016/0161-5890(86)90081-7 [DOI] [PubMed] [Google Scholar]
- Ghosh D., Kohli A. G., Moser F., Endy D., Belcher A. M. (2012a). Refactored M13 bacteriophage as a platform for tumor cell imaging and drug delivery. ACS Synth. Biol. 1 576–582. 10.1021/sb300052u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D., Lee Y., Thomas S., Kohli A. G., Yun D. S., Belcher A. M., et al. (2012b). M13-templated magnetic nanoparticles for targeted in vivo imaging of prostate cancer. Nat. Nanotechnol. 7 677–682. 10.1038/nnano.2012.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M. D., Lichtensteiger C. A., Caughlan R., Vimr E. R. (2002). Conserved filamentous prophage in Escherichia coli O18:K1:H7 and Yersinia pestis biovar orientalis. J. Bacteriol. 184 6050–6055. 10.1128/JB.184.21.6050-6055.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowska A. M., Jennings R., Laing P., Darsley M., Jameson C. L., Swift L., et al. (2000). Immunisation with phage displaying peptides representing single epitopes of the glycoprotein G can give rise to partial protective immunity to HSV-2. Virology 269 47–53. 10.1006/viro.2000.0185 [DOI] [PubMed] [Google Scholar]
- Gram H., Strittmatter U., Lorenz M., Gluck D., Zenke G. (1993). Phage display as a rapid gene expression system: production of bioactive cytokine-phage and generation of neutralizing monoclonal antibodies. J. Immunol. Methods 161 169–176. 10.1016/0022-1759(93)90292-F [DOI] [PubMed] [Google Scholar]
- Greenwood J., Willis A. E., Perham R. N. (1991). Multiple display of foreign peptides on a filamentous bacteriophage. Peptides from Plasmodium falciparum circumsporozoite protein as antigens. J. Mol. Biol. 220 821–827. 10.1016/0022-2836(91)90354-9 [DOI] [PubMed] [Google Scholar]
- Hagens S., Blasi U. (2003). Genetically modified filamentous phage as bactericidal agents: a pilot study. Lett. Appl. Microbiol. 37 318–323. 10.1046/j.1472-765X.2003.01400.x [DOI] [PubMed] [Google Scholar]
- Hagens S., Habel A., Blasi U. (2006). Augmentation of the antimicrobial efficacy of antibiotics by filamentous phage. Microb. Drug Resist. 12 164–168. 10.1089/mdr.2006.12.164 [DOI] [PubMed] [Google Scholar]
- Hagens S., Habel A., Von Ahsen U., Von Gabain A., Blasi U. (2004). Therapy of experimental Pseudomonas infections with a nonreplicating genetically modified phage. Antimicrob. Agents Chemother. 48 3817–3822. 10.1128/AAC.48.10.3817-3822.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. P., Harrison E., Brockhurst M. A. (2013). Viral host-adaptation: insights from evolution experiments with phages. Curr. Opin. Virol. 3 572–577. 10.1016/j.coviro.2013.07.001 [DOI] [PubMed] [Google Scholar]
- Hansen M. R., Hanson P., Pardi A. (2000). Pf1 filamentous phage as an alignment tool for generating local and global structural information in nucleic acids. J. Biomol. Struct. Dyn. 17(Suppl. 1) 365–369. 10.1080/07391102.2000.10506642 [DOI] [PubMed] [Google Scholar]
- Hansen M. R., Mueller L., Pardi A. (1998). Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 5 1065–1074. 10.1038/4176 [DOI] [PubMed] [Google Scholar]
- Hashiguchi S., Yamaguchi Y., Takeuchi O., Akira S., Sugimura K. (2010). Immunological basis of M13 phage vaccine: regulation under MyD88 and TLR9 signaling. Biochem. Biophys. Res. Commun. 402 19–22. 10.1016/j.bbrc.2010.09.094 [DOI] [PubMed] [Google Scholar]
- Hassan F., Kamruzzaman M., Mekalanos J. J., Faruque S. M. (2010). Satellite phage TLCϕ enables toxigenic conversion by CTX phage through dif site alteration. Nature 467 982–985. 10.1038/nature09469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry K. A., Murira A., Van Houten N. E., Scott J. K. (2011). Developing strategies to enhance and focus humoral immune responses using filamentous phage as a model antigen. Bioengineered 2 275–283. 10.4161/bbug.2.5.16559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess G. T., Cragnolini J. J., Popp M. W., Allen M. A., Dougan S. K., Spooner E., et al. (2012). M13 bacteriophage display framework that allows sortase-mediated modification of surface-accessible phage proteins. Bioconjug. Chem. 23 1478–1487. 10.1021/bc300130z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer J. S., Sommaruga R. (2001). Seasonal dynamics of viruses in an alpine lake: importance of filamentous forms. Aquat. Microb. Ecol. 26 11 10.3354/ame026001 [DOI] [Google Scholar]
- Hofschneider P. H., Mueller-Jensen K. (1963). On infectious substructures from E. coli bacteriophages. 3. Demonstration and properties of “Ht2” particles. Z. Naturforsch B 18 922–927. [PubMed] [Google Scholar]
- Houimel M., Dellagi K. (2009). Peptide mimotopes of rabies virus glycoprotein with immunogenic activity. Vaccine 27 4648–4655. 10.1016/j.vaccine.2009.05.055 [DOI] [PubMed] [Google Scholar]
- Huang S., Yang H., Lakshmanan R. S., Johnson M. L., Wan J., Chen I. H., et al. (2009). Sequential detection of Salmonella typhimurium and Bacillus anthracis spores using magnetoelastic biosensors. Biosens. Bioelectron. 24 1730–1736. 10.1016/j.bios.2008.09.006 [DOI] [PubMed] [Google Scholar]
- Huang Y., Chiang C. Y., Lee S. K., Gao Y., Hu E. L., De Yoreo J., et al. (2005). Programmable assembly of nanoarchitectures using genetically engineered viruses. Nano Lett. 5 1429–1434. 10.1021/nl050795d [DOI] [PubMed] [Google Scholar]
- Hufton S. E., Moerkerk P. T., Meulemans E. V., De Bruine A., Arends J. W., Hoogenboom H. R. (1999). Phage display of cDNA repertoires: the pVI display system and its applications for the selection of immunogenic ligands. J. Immunol. Methods 231 39–51. 10.1016/S0022-1759(99)00139-8 [DOI] [PubMed] [Google Scholar]
- Husimi Y. (1989). Selection and evolution of bacteriophages in cellstat. Adv. Biophys. 25 1–43. 10.1016/0065-227X(89)90003-8 [DOI] [PubMed] [Google Scholar]
- Iida T., Hattori A., Tagomori K., Nasu H., Naim R., Honda T. (2001). Filamentous phage associated with recent pandemic strains of Vibrio parahaemolyticus. Emerg. Infect. Dis. 7 477–478. 10.3201/eid0703.010325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving M. B., Craig L., Menendez A., Gangadhar B. P., Montero M., van Houten N. E., et al. (2010). Exploring peptide mimics for the production of antibodies against discontinuous protein epitopes. Mol. Immunol. 47 1137–1148. 10.1016/j.molimm.2009.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayanna P. K., Bedi D., Deinnocentes P., Bird R. C., Petrenko V. A. (2010a). Landscape phage ligands for PC3 prostate carcinoma cells. Protein Eng. Des. Sel. 23 423–430. 10.1093/protein/gzq011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayanna P. K., Bedi D., Gillespie J. W., Deinnocentes P., Wang T., Torchilin V. P., et al. (2010b). Landscape phage fusion protein-mediated targeting of nanomedicines enhances their prostate tumor cell association and cytotoxic efficiency. Nanomedicine 6 538–546. 10.1016/j.nano.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian H., Xu J., Xiao X., Wang F. (2012). Dynamic modulation of DNA replication and gene transcription in deep-sea filamentous phage SW1 in response to changes of host growth and temperature. PLoS ONE 7:e41578 10.1371/journal.pone.0041578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajihara Y., Hashiguchi S., Ito Y., Sugimura K. (2000). Th2-type immune response induced by a phage clone displaying a CTLA4-binding domain mimic-motif. Gene Ther. Mol. Biol. 5 6. [Google Scholar]
- Kamiunten H., Wakimoto S. (1982). Effect of infection with filamentous phage xf on the growth, ultrastructure and virulence of Xanthomonas campestris pv. oryzae N 5850. Jpn. J. Phytopatholo. 48 642–647. 10.3186/jjphytopath.48.642 [DOI] [Google Scholar]
- Kang A. S., Barbas C. F., Janda K. D., Benkovic S. J., Lerner R. A. (1991). Linkage of recognition and replication functions by assembling combinatorial antibody Fab libraries along phage surfaces. Proc. Natl. Acad. Sci. U.S.A. 88 4363–4366. 10.1073/pnas.88.10.4363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawecki T. J., Lenski R. E., Ebert D., Hollis B., Olivieri I., Whitlock M. C. (2012). Experimental evolution. Trends Ecol. Evol. 27 547–560. 10.1016/j.tree.2012.06.001 [DOI] [PubMed] [Google Scholar]
- Kehoe J. W., Kay B. K. (2005). Filamentous phage display in the new millennium. Chem. Rev. 105 4056–4072. 10.1021/cr000261r [DOI] [PubMed] [Google Scholar]
- Khatoon H., Iyer R. V., Iyer V. N. (1972). A new filamentous bacteriophage with sex-factor specificity. Virology 48 145–155. 10.1016/0042-6822(72)90122-5 [DOI] [PubMed] [Google Scholar]
- Kim M. S., Park E. J., Roh S. W., Bae J. W. (2011). Diversity and abundance of single-stranded DNA viruses in human feces. Appl. Environ. Microbiol. 77 8062–8070. 10.1128/AEM.06331-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Jin Y. H., Salieb-Beugelaar G. B., Nam C. H., Stieglitz T. (2014). Genetically engineered bacteriophage delivers a tumor necrosis factor alpha antagonist coating on neural electrodes. Biomed. Mater. 9:015009 10.1088/1748-6041/9/1/015009 [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Lebreton F., Kaiser C., Cruciere C., Remond M. (2004). Expression of a foot-and-mouth disease virus immunodominant epitope by a filamentous bacteriophage vector. Arch. Virol. 149 365–377. 10.1007/s00705-003-0192-198 [DOI] [PubMed] [Google Scholar]
- Kishchenko G., Batliwala H., Makowski L. (1994). Structure of a foreign peptide displayed on the surface of bacteriophage M13. J. Mol. Biol. 241 208–213. 10.1006/jmbi.1994.1489 [DOI] [PubMed] [Google Scholar]
- Kneissel S., Queitsch I., Petersen G., Behrsing O., Micheel B., Dubel S. (1999). Epitope structures recognised by antibodies against the major coat protein (g8p) of filamentous bacteriophage fd (Inoviridae). J. Mol. Biol. 288 21–28. 10.1006/jmbi.1999.2676 [DOI] [PubMed] [Google Scholar]
- Knittelfelder R., Riemer A. B., Jensen-Jarolim E. (2009). Mimotope vaccination–from allergy to cancer. Expert. Opin. Biol. Ther. 9 493–506. 10.1517/14712590902870386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide A., Koide S. (2012). Affinity maturation of single-domain antibodies by yeast surface display. Methods Mol. Biol. 911 431–443. 10.1007/978-1-61779-968-6-26 [DOI] [PubMed] [Google Scholar]
- Kölsch E., Diller E., Weber G., Davies A. J. (1971). Genetics of the immune response. I. The immune response to the phage fd in high and low responding inbred strains of mice. Eur. J. Immunol. 1 201–210. 10.1002/eji.1830010310 [DOI] [PubMed] [Google Scholar]
- Ksendzovsky A., Walbridge S., Saunders R. C., Asthagiri A. R., Heiss J. D., Lonser R. R. (2012). Convection-enhanced delivery of M13 bacteriophage to the brain. J. Neurosurg. 117 197–203. 10.3171/2012.4.JNS111528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwasnikowski P., Kristensen P., Markiewicz W. T. (2005). Multivalent display system on filamentous bacteriophage pVII minor coat protein. J. Immunol. Methods 307 135–143. 10.1016/j.jim.2005.10.002 [DOI] [PubMed] [Google Scholar]
- Lakshmanan R. S., Guntupalli R., Hu J., Kim D. J., Petrenko V. A., Barbaree J. M., et al. (2007). Phage immobilized magnetoelastic sensor for the detection of Salmonella typhimurium. J. Microbiol. Methods 71 55–60. 10.1016/j.mimet.2007.07.012 [DOI] [PubMed] [Google Scholar]
- Lang Q., Wang F., Yin L., Liu M., Petrenko V. A., Liu A. (2014). Specific probe selection from landscape phage display library and its application in enzyme-linked immunosorbent assay of free prostate-specific antigen. Anal. Chem. 86 2767–2774. 10.1021/ac404189k [DOI] [PubMed] [Google Scholar]
- Lavie V., Becker M., Cohen-Kupiec R., Yacoby I., Koppel R., Wedenig M., et al. (2004). EFRH-phage immunization of Alzheimer’s disease animal model improves behavioral performance in Morris water maze trials. J. Mol. Neurosci. 24 105–113. 10.1385/JMN:24:1105 [DOI] [PubMed] [Google Scholar]
- Lee S. W., Mao C., Flynn C. E., Belcher A. M. (2002). Ordering of quantum dots using genetically engineered viruses. Science 296 892–895. 10.1126/science.1068054 [DOI] [PubMed] [Google Scholar]
- Lee Y. J., Yi H., Kim W. J., Kang K., Yun D. S., Strano M. S., et al. (2009). Fabricating genetically engineered high-power lithium-ion batteries using multiple virus genes. Science 324 1051–1055. 10.1126/science.1171541 [DOI] [PubMed] [Google Scholar]
- Li K., Chen Y., Li S., Nguyen H. G., Niu Z., You S., et al. (2010a). Chemical modification of M13 bacteriophage and its application in cancer cell imaging. Bioconjug. Chem. 21 1369–1377. 10.1021/bc900405q [DOI] [PubMed] [Google Scholar]
- Li S., Li Y., Chen H., Horikawa S., Shen W., Simonian A., et al. (2010b). Direct detection of Salmonella typhimurium on fresh produce using phage-based magnetoelastic biosensors. Biosens. Bioelectron. 26 1313–1319. 10.1016/j.bios.2010.07.029 [DOI] [PubMed] [Google Scholar]
- Li Y., Ning Y., Wang Y., Peng D., Jiang Y., Zhang L., et al. (2010c). Mimotopes selected with a neutralizing antibody against urease B from Helicobacter pylori induce enzyme inhibitory antibodies in mice upon vaccination. BMC Biotechnol. 10:84 10.1186/1472-6750-10-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidqvist M., Nilsson O., Holmgren J., Hall C., Fermer C. (2008). Phage display for site-specific immunization and characterization of high-risk human papillomavirus specific E7 monoclonal antibodies. J. Immunol. Methods 337 88–96. 10.1016/j.jim.2008.06.002 [DOI] [PubMed] [Google Scholar]
- Lin A., Jimenez J., Derr J., Vera P., Manapat M. L., Esvelt K. M., et al. (2011). Inhibition of bacterial conjugation by phage M13 and its protein g3p: quantitative analysis and model. PLoS ONE 6:e19991 10.1371/journal.pone.0019991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippow S. M., Wittrup K. D., Tidor B. (2007). Computational design of antibody-affinity improvement beyond in vivo maturation. Nat. Biotechnol. 25 1171–1176. 10.1038/nbt1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb T. (1960). Isolation of a bacteriophage specific for the F plus and Hfr mating types of Escherichia coli K-12. Science 131 932–933. 10.1126/science.131.3404.932 [DOI] [PubMed] [Google Scholar]
- Lu T. K., Collins J. J. (2009). Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc. Natl. Acad. Sci. U.S.A. 106 4629–4634. 10.1073/pnas.0800442106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik P., Terry T. D., Gowda L. R., Langara A., Petukhov S. A., Symmons M. F., et al. (1996). Role of capsid structure and membrane protein processing in determining the size and copy number of peptides displayed on the major coat protein of filamentous bacteriophage. J. Mol. Biol. 260 9–21. 10.1006/jmbi.1996.0378 [DOI] [PubMed] [Google Scholar]
- Manoutcharian K., Diaz-Orea A., Gevorkian G., Fragoso G., Acero G., Gonzalez E., et al. (2004). Recombinant bacteriophage-based multiepitope vaccine against Taenia solium pig cysticercosis. Vet. Immunol. Immunopathol. 99 11–24. 10.1016/j.vetimm.2003.12.009 [DOI] [PubMed] [Google Scholar]
- Mao C., Flynn C. E., Hayhurst A., Sweeney R., Qi J., Georgiou G., et al. (2003). Viral assembly of oriented quantum dot nanowires. Proc. Natl. Acad. Sci. U.S.A. 100 6946–6951. 10.1073/pnas.0832310100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C., Solis D. J., Reiss B. D., Kottmann S. T., Sweeney R. Y., Hayhurst A., et al. (2004). Virus-based toolkit for the directed synthesis of magnetic and semiconducting nanowires. Science 303 213–217. 10.1126/science.1092740 [DOI] [PubMed] [Google Scholar]
- Mao J. Y., Belcher A. M., Van Vliet K. J. (2010). Genetically engineered phage fibers and coatings for antibacterial applications. Adv. Funct. Mater. 20 5 10.1002/adfm.200900782 [DOI] [Google Scholar]
- Marks J. D., Hoogenboom H. R., Griffiths A. D., Winter G. (1992). Molecular evolution of proteins on filamentous phage. Mimicking the strategy of the immune system. J. Biol. Chem. 267 16007–16010. [PubMed] [Google Scholar]
- Marvin D. A. (1990). Model-building studies of Inovirus: genetic variations on a geometric theme. Int. J. Biol. Macromol. 12 125–138. 10.1016/0141-8130(90)90064-H [DOI] [PubMed] [Google Scholar]
- Marvin D. A. (1998). Filamentous phage structure, infection and assembly. Curr. Opin. Struct. Biol. 8 150–158. 10.1016/S0959-440X(98)80032-8 [DOI] [PubMed] [Google Scholar]
- Marvin D. A., Hoffmann-Berling H. (1963). A fibrous DNA phage (Fd) and a spherical RNA phage (Fr) specific for male strains of E. coli. II. Physical characteristics. Z. Naturforsch. B 18 884–893. 10.1515/znb-1963-1106 [DOI] [PubMed] [Google Scholar]
- Mascolo D., Barba P., De Berardinis P., Di Rosa F., Del Pozzo G. (2007). Phage display of a CTL epitope elicits a long-term in vivo cytotoxic response. FEMS Immunol. Med. Microbiol. 50 59–66. 10.1111/j.1574-695X.2007.00229.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- May T., Tsuruta K., Okabe S. (2011). Exposure of conjugative plasmid carrying Escherichia coli biofilms to male-specific bacteriophages. ISME J. 5 771–775. 10.1038/ismej.2010.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCafferty J., Griffiths A. D., Winter G., Chiswell D. J. (1990). Phage antibodies: filamentous phage displaying antibody variable domains. Nature 348 552–554. 10.1038/348552a0 [DOI] [PubMed] [Google Scholar]
- McConnell S. J., Kendall M. L., Reilly T. M., Hoess R. H. (1994). Constrained peptide libraries as a tool for finding mimotopes. Gene 151 115–118. 10.1016/0378-1119(94)90640-8 [DOI] [PubMed] [Google Scholar]
- Melzer H., Baier K., Felici F., Von Specht B. U., Wiedermann G., Kollaritsch H., et al. (2003). Humoral immune response against proteophosphoglycan surface antigens of Entamoeba histolytica elicited by immunization with synthetic mimotope peptides. FEMS Immunol. Med. Microbiol. 37 179–183. 10.1016/S092882440300074-9 [DOI] [PubMed] [Google Scholar]
- Melzer H., Fortugno P., Mansouri E., Felici F., Marinets A., Wiedermann G., et al. (2002). Antigenicity and immunogenicity of phage library-selected peptide mimics of the major surface proteophosphoglycan antigens of Entamoeba histolytica. Parasite Immunol. 24 321–328. 10.1046/j.1365-3024.2002.00463.x [DOI] [PubMed] [Google Scholar]
- Menendez T., De Haz I., Delgado M., Garay H., Martin A., Vispo N. S. (2001). Immunisation with phage-displayed variable region 2 from meningococcal PorA outer membrane protein induces bactericidal antibodies against Neisseria meningitidis. Immunol. Lett. 78 143–148. 10.1016/S0165-2478(01)00245-0 [DOI] [PubMed] [Google Scholar]
- Meola A., Delmastro P., Monaci P., Luzzago A., Nicosia A., Felici F., et al. (1995). Derivation of vaccines from mimotopes. Immunologic properties of human hepatitis B virus surface antigen mimotopes displayed on filamentous phage. J. Immunol. 154 3162–3172. [PubMed] [Google Scholar]
- Meyer A. J., Ellington A. D. (2011). Molecular evolution picks up the PACE. Nat. Biotechnol. 29 502–503. 10.1038/nbt.1884 [DOI] [PubMed] [Google Scholar]
- Meynell G. G., Lawn A. M. (1968). Filamentous phages specific for the I sex factor. Nature 217 1184–1186. 10.1038/2171184a0 [DOI] [PubMed] [Google Scholar]
- Minenkova O. O., Ilyichev A. A., Kishchenko G. P., Petrenko V. A. (1993). Design of specific immunogens using filamentous phage as the carrier. Gene 128 85–88. 10.1016/0378-1119(93)90157-X [DOI] [PubMed] [Google Scholar]
- Molenaar T. J., Michon I., De Haas S. A., Van Berkel T. J., Kuiper J., Biessen E. A. (2002). Uptake and processing of modified bacteriophage M13 in mice: implications for phage display. Virology 293 182–191. 10.1006/viro.2001.1254 [DOI] [PubMed] [Google Scholar]
- Moradpour Z., Sepehrizadeh Z., Rahbarizadeh F., Ghasemian A., Yazdi M. T., Shahverdi A. R. (2009). Genetically engineered phage harbouring the lethal catabolite gene activator protein gene with an inducer-independent promoter for biocontrol of Escherichia coli. FEMS Microbiol. Lett. 296 67–71. 10.1111/j.1574-6968.2009.01620.x [DOI] [PubMed] [Google Scholar]
- Morales J., Martinez J. J., Manoutcharian K., Hernandez M., Fleury A., Gevorkian G., et al. (2008). Inexpensive anti-cysticercosis vaccine: S3Pvac expressed in heat inactivated M13 filamentous phage proves effective against naturally acquired Taenia solium porcine cysticercosis. Vaccine 26 2899–2905. 10.1016/j.vaccine.2008.03.042 [DOI] [PubMed] [Google Scholar]
- Motti C., Nuzzo M., Meola A., Galfre G., Felici F., Cortese R., et al. (1994). Recognition by human sera and immunogenicity of HBsAg mimotopes selected from an M13 phage display library. Gene 146 191–198. 10.1016/0378-1119(94)90292-5 [DOI] [PubMed] [Google Scholar]
- Murira A. (2014). Characterization of Molecular Correlates of the Chronic Humoral Immune Response: Clues towards Eliciting Broadly Neutralizing Antibodies against HIV. Ph.D. thesis, Simon Fraser University; Burnaby, BC. [Google Scholar]
- Murugaiyan S., Bae J. Y., Wu J., Lee S. D., Um H. Y., Choi H. K., et al. (2011). Characterization of filamentous bacteriophage PE226 infecting Ralstonia solanacearum strains. J. Appl. Microbiol. 110 296–303. 10.1111/j.1365-2672.2010.04882.x [DOI] [PubMed] [Google Scholar]
- Murugesan M., Abbineni G., Nimmo S. L., Cao B., Mao C. (2013). Virus-based photo-responsive nanowires formed by linking site-directed mutagenesis and chemical reaction. Sci. Rep. 3:1820 10.1038/srep01820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K. T., Kim D. W., Yoo P. J., Chiang C. Y., Meethong N., Hammond P. T., et al. (2006). Virus-enabled synthesis and assembly of nanowires for lithium ion battery electrodes. Science 312 885–888. 10.1126/science.1122716 [DOI] [PubMed] [Google Scholar]
- Nam K. T., Wartena R., Yoo P. J., Liau F. W., Lee Y. J., Chiang Y. M., et al. (2008). Stamped microbattery electrodes based on self-assembled M13 viruses. Proc. Natl. Acad. Sci. U.S.A. 105 17227–17231. 10.1073/pnas.0711620105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y. S., Magyar A. P., Lee D., Kim J. W., Yun D. S., Park H., et al. (2010a). Biologically templated photocatalytic nanostructures for sustained light-driven water oxidation. Nat. Nanotechnol. 5 340–344. 10.1038/nnano.2010.57 [DOI] [PubMed] [Google Scholar]
- Nam Y. S., Shin T., Park H., Magyar A. P., Choi K., Fantner G., et al. (2010b). Virus-templated assembly of porphyrins into light-harvesting nanoantennae. J. Am. Chem. Soc. 132 1462–1463. 10.1021/ja908812b [DOI] [PubMed] [Google Scholar]
- Nanduri V., Bhunia A. K., Tu S. I., Paoli G. C., Brewster J. D. (2007). SPR biosensor for the detection of L. monocytogenes using phage-displayed antibody. Biosens. Bioelectron. 23 248–252. 10.1016/j.bios.2007.04.007 [DOI] [PubMed] [Google Scholar]
- Neltner B., Peddie B., Xu A., Doenlen W., Durand K., Yun D. S., et al. (2010). Production of hydrogen using nanocrystalline protein-templated catalysts on m13 phage. ACS Nano 4 3227–3235. 10.1021/nn100346h [DOI] [PubMed] [Google Scholar]
- Ng S., Jafari M. R., Matochko W. L., Derda R. (2012). Quantitative synthesis of genetically encoded glycopeptide libraries displayed on M13 phage. ACS Chem. Biol. 7 1482–1487. 10.1021/cb300187t [DOI] [PubMed] [Google Scholar]
- Oh D., Dang X., Yi H., Allen M. A., Xu K., Lee Y. J., et al. (2012). Graphene sheets stabilized on genetically engineered M13 viral templates as conducting frameworks for hybrid energy-storage materials. Small 8 1006–1011. 10.1002/smll.201102036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen E. V., Sorokulova I. B., Petrenko V. A., Chen I. H., Barbaree J. M., Vodyanoy V. J. (2006). Affinity-selected filamentous bacteriophage as a probe for acoustic wave biodetectors of Salmonella typhimurium. Biosens. Bioelectron. 21 1434–1442. 10.1016/j.bios.2005.06.004 [DOI] [PubMed] [Google Scholar]
- Pacheco-Gomez R., Kraemer J., Stokoe S., England H. J., Penn C. W., Stanley E., et al. (2012). Detection of pathogenic bacteria using a homogeneous immunoassay based on shear alignment of virus particles and linear dichroism. Anal. Chem. 84 91–97. 10.1021/ac201544h [DOI] [PubMed] [Google Scholar]
- Pande J., Szewczyk M. M., Grover A. K. (2010). Phage display: concept, innovations, applications and future. Biotechnol. Adv. 28 849–858. 10.1016/j.biotechadv.2010.07.004 [DOI] [PubMed] [Google Scholar]
- Parmley S. F., Smith G. P. (1988). Antibody-selectable filamentous fd phage vectors: affinity purification of target genes. Gene 73 305–318. 10.1016/0378-1119(88)90495-7 [DOI] [PubMed] [Google Scholar]
- Petrenko V. A., Makowski L. (1993). “Potential applications of phage display to bioremediation,” in Bioremediation of Chlorinated and Polycyclic Aromatic Hydrocarbon Compounds eds Hinchee R. E., Semprini L., Ong S. K. (Columbus, OH: Taylor and Francis Group; ) 266–280. [Google Scholar]
- Petrenko V. A., Smith G. P., Gong X., Quinn T. (1996). A library of organic landscapes on filamentous phage. Protein Eng. 9 797–801. 10.1093/protein/9.9.797 [DOI] [PubMed] [Google Scholar]
- Pina S., Creus A., Gonzaalez N., Gironees R., Felip M., Sommaruga R. (1998). Abundance, morphology and distribution of planktonic virus-like particles in two high-mountain lakes. J. Plankton Res. 20:8 10.1093/plankt/20.12.2413 [DOI] [Google Scholar]
- Plotkin S. A. (2010). Correlates of protection induced by vaccination. Clin. Vacc. Immunol. 17 1055–1065. 10.1128/CVI.00131-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poul M. A., Marks J. D. (1999). Targeted gene delivery to mammalian cells by filamentous bacteriophage. J. Mol. Biol. 288 203–211. 10.1006/jmbi.1999.2678 [DOI] [PubMed] [Google Scholar]
- Pratt D., Tzagoloff H., Beaudoin J. (1969). Conditional lethal mutants of the small filamentous coliphage M13. II. Two genes for coat proteins. Virology 39 42–53. 10.1016/0042-6822(69)90346-8 [DOI] [PubMed] [Google Scholar]
- Prezzi C., Nuzzo M., Meola A., Delmastro P., Galfre G., Cortese R., et al. (1996). Selection of antigenic and immunogenic mimics of hepatitis C virus using sera from patients. J. Immunol. 156 4504–4513. [PubMed] [Google Scholar]
- Puntoriero G., Meola A., Lahm A., Zucchelli S., Ercole B. B., Tafi R., et al. (1998). Towards a solution for hepatitis C virus hypervariability: mimotopes of the hypervariable region 1 can induce antibodies cross-reacting with a large number of viral variants. EMBO J. 17 3521–3533. 10.1093/emboj/17.13.3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakonjac J. (2012). “Filamentous bacteriophages: biology and applications,” in eLS (the Encyclopaedia of Life Sciences) Chichester: John Wiley & Sons, Ltd; 10.1002/9780470015902.a0000777 [DOI] [Google Scholar]
- Rakonjac J., Bennett N. J., Spagnuolo J., Gagic D., Russel M. (2011). Filamentous bacteriophage: biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol. 13 51–76. [PubMed] [Google Scholar]
- Rakover I. S., Zabavnik N., Kopel R., Paz-Rozner M., Solomon B. (2010). Antigen-specific therapy of EAE via intranasal delivery of filamentous phage displaying a myelin immunodominant epitope. J. Neuroimmunol. 225 68–76. 10.1016/j.jneuroim.2010.04.014 [DOI] [PubMed] [Google Scholar]
- Rao K. V., He Y. X., Kalyanasundaram R. (2003). Expression of a 28-kilodalton glutathione S-transferase antigen of Schistosoma mansoni on the surface of filamentous phages and evaluation of its vaccine potential. Clin. Diagn. Lab. Immunol. 10 536–541. 10.1128/cdli.10.4.536-541.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond-Bouchard I., Carroll C. S., Nesbitt J. R., Henry K. A., Pinto L. J., Moinzadeh M., et al. (2012). Structural requirements for the activity of the MirB ferrisiderophore transporter of Aspergillus fumigatus. Eukaryot. Cell 11 1333–1344. 10.1128/EC.00159-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehnisch T., Then C., Nagel W., Blumenthal C., Braciak T., Donzeau M., et al. (2013). Chemically linked phage idiotype vaccination in the murine B cell lymphoma 1 model. J. Transl. Med. 11:267 10.1186/1479-5876-11-267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehnisch T., Then C., Nagel W., Blumenthal C., Braciak T., Donzeau M., et al. (2014). Phage idiotype vaccination: first phase I/II clinical trial in patients with multiple myeloma. J. Transl. Med. 12 119 10.1186/1479-5876-12-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov V. I., Durand D. B., Petrenko V. A. (2001). Phage display selection of peptides that affect prostate carcinoma cells attachment and invasion. Prostate 47 239–251. 10.1002/pros.1068 [DOI] [PubMed] [Google Scholar]
- Rondot S., Koch J., Breitling F., Dubel S. (2001). A helper phage to improve single-chain antibody presentation in phage display. Nat. Biotechnol. 19 75–78. 10.1038/83567 [DOI] [PubMed] [Google Scholar]
- Rosario K., Nilsson C., Lim Y. W., Ruan Y., Breitbart M. (2009). Metagenomic analysis of viruses in reclaimed water. Environ. Microbiol. 11 2806–2820. 10.1111/j.1462-2920.2009.01964.x [DOI] [PubMed] [Google Scholar]
- Roux S., Enault F., Robin A., Ravet V., Personnic S., Theil S., et al. (2012). Assessing the diversity and specificity of two freshwater viral communities through metagenomics. PLoS ONE 7:e33641 10.1371/journal.pone.0033641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinchik E., Chow A. W. (2000). Recombinant expression and neutralizing activity of an MHC class II binding epitope of toxic shock syndrome toxin-1. Vaccine 18 2312–2320. 10.1016/S0264-410X(99)00554-X [DOI] [PubMed] [Google Scholar]
- Rudolf M. P., Vogel M., Kricek F., Ruf C., Zurcher A. W., Reuschel R., et al. (1998). Epitope-specific antibody response to IgE by mimotope immunization. J. Immunol. 160 3315–3321. [PubMed] [Google Scholar]
- Salivar W. O., Tzagoloff H., Pratt D. (1964). Some physical-chemical and biological properties of the rod-shaped coliphage M13. Virology 24 359–371. 10.1016/0042-6822(64)90173-4 [DOI] [PubMed] [Google Scholar]
- Samoylov A., Cox N., Cochran A., Wolfe K., Donovan C., Kutzler M., et al. (2012). Generation and characterization of phage-GnRH chemical conjugates for potential use in cat and dog immunocontraception. Reprod. Domest. Anim. 47(Suppl. 6) 406–411. 10.1111/rda.12061 [DOI] [PubMed] [Google Scholar]
- Samoylova T. I., Cochran A. M., Samoylov A. M., Schemera B., Breiteneicher A. H., Ditchkoff S. S., et al. (2012a). Phage display allows identification of zona pellucida-binding peptides with species-specific properties: novel approach for development of contraceptive vaccines for wildlife. J. Biotechnol. 162 311–318. 10.1016/j.jbiotec.2012.10.006 [DOI] [PubMed] [Google Scholar]
- Samoylova T. I., Norris M. D., Samoylov A. M., Cochran A. M., Wolfe K. G., Petrenko V. A., et al. (2012b). Infective and inactivated filamentous phage as carriers for immunogenic peptides. J. Virol. Methods 183 63–68. 10.1016/j.jviromet.2012.03.032 [DOI] [PubMed] [Google Scholar]
- Sartorius R., Bettua C., D’apice L., Caivano A., Trovato M., Russo D., et al. (2011). Vaccination with filamentous bacteriophages targeting DEC-205 induces DC maturation and potent anti-tumor T-cell responses in the absence of adjuvants. Eur. J. Immunol. 41 2573–2584. 10.1002/eji.201141526 [DOI] [PubMed] [Google Scholar]
- Sartorius R., Pisu P., D’apice L., Pizzella L., Romano C., Cortese G., et al. (2008). The use of filamentous bacteriophage fd to deliver MAGE-A10 or MAGE-A3 HLA-A2-restricted peptides and to induce strong antitumor CTL responses. J. Immunol. 180 3719–3728. 10.4049/jimmunol.180.6.3719 [DOI] [PubMed] [Google Scholar]
- Scala G., Chen X., Liu W., Telles J. N., Cohen O. J., Vaccarezza M., et al. (1999). Selection of HIV-specific immunogenic epitopes by screening random peptide libraries with HIV-1-positive sera. J. Immunol. 162 6155–6161. [PubMed] [Google Scholar]
- Schmelcher M., Loessner M. J. (2014). Application of bacteriophages for detection of foodborne pathogens. Bacteriophage 4:e28137 10.4161/bact.28137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. K., Smith G. P. (1990). Searching for peptide ligands with an epitope library. Science 249 386–390. 10.1126/science.1696028 [DOI] [PubMed] [Google Scholar]
- Shi L., Wheeler J. C., Sweet R. W., Lu J., Luo J., Tornetta M., et al. (2010). De novo selection of high-affinity antibodies from synthetic Fab libraries displayed on phage as pIX fusion proteins. J. Mol. Biol. 397 385–396. 10.1016/j.jmb.2010.01.034 [DOI] [PubMed] [Google Scholar]
- Sidhu S. S., Weiss G. A., Wells J. A. (2000). High copy display of large proteins on phage for functional selections. J. Mol. Biol. 296 487–495. 10.1006/jmbi.1999.3465 [DOI] [PubMed] [Google Scholar]
- Smith G. P. (1985). Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228 1315–1317. 10.1126/science.4001944 [DOI] [PubMed] [Google Scholar]
- Smith G. P., Fernandez A. M. (2004). Effect of DNA copy number on genetic stability of phage-displayed peptides. Biotechniques 36 610–614. [DOI] [PubMed] [Google Scholar]
- Smith G. P., Gingrich T. R. (2005). Hydroxyapatite chromatography of phage-display virions. BioTechniques 39 879–884. 10.2144/000112032 [DOI] [PubMed] [Google Scholar]
- Smith G. P., Petrenko V. A. (1997). Phage display. Chem. Rev. 97 391–410. 10.1021/cr960065d [DOI] [PubMed] [Google Scholar]
- Smith G. P., Scott J. K. (1993). Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 217 228–257. 10.1016/0076-6879(93)17065-D [DOI] [PubMed] [Google Scholar]
- Solomon B. (2005). Generation of anti-β-amyloid antibodies via phage display technology towards Alzheimer’s disease vaccination. Vaccine 23 2327–2330. 10.1016/j.vaccine.2005.01.034 [DOI] [PubMed] [Google Scholar]
- Solomon B. (2008). Filamentous bacteriophage as a novel therapeutic tool for Alzheimer’s disease treatment. J. Alzheimers Dis. 15 193–198. [DOI] [PubMed] [Google Scholar]
- Su Q. P., Wen D. Z., Yang Q., Zhang Y. H., Liu C., Wang L. (2007). Comparison of phage pVIII and KLH as vector in inducing the production of cytokines in C57BL/6J mice. Vaccine 25 970–975. 10.1016/j.vaccine.2006.08.045 [DOI] [PubMed] [Google Scholar]
- Sulakvelidze A., Alavidze Z., Morris J. G., Jr. (2001). Bacteriophage therapy. Antimicrob. Agents Chemother. 45 649–659. 10.1128/AAC.45.3.649-659.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle C. A. (2005). Viruses in the sea. Nature 437 356–361. 10.1038/nature04160 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Nishimura M., Yamaguchi Y., Hashiguchi S., Takiguchi S., Yamaguchi M., et al. (2011). A mimotope peptide of Aβ42 fibril-specific antibodies with Aβ42 fibrillation inhibitory activity induces anti-Aβ42 conformer antibody response by a displayed form on an M13 phage in mice. J. Neuroimmunol. 236 27–38. 10.1016/j.jneuroim.2011.04.010 [DOI] [PubMed] [Google Scholar]
- Terry T. D., Malik P., Perham R. N. (1997). Accessibility of peptides displayed on filamentous bacteriophage virions: susceptibility to proteinases. Biol. Chem. 378 523–530. 10.1515/bchm.1997.378.6.523 [DOI] [PubMed] [Google Scholar]
- Tornetta M., Baker S., Whitaker B., Lu J., Chen Q., Pisors E., et al. (2010). Antibody Fab display and selection through fusion to the pIX coat protein of filamentous phage. J. Immunol. Methods 360 39–46. 10.1016/j.jim.2010.06.001 [DOI] [PubMed] [Google Scholar]
- Twort F. W. (1915). An investigation on the nature of ultra-microscopic viruses. Lancet 186:3 10.1016/S0140-6736(01)20383-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulivieri C., Citro A., Ivaldi F., Mascolo D., Ghittoni R., Fanigliulo D., et al. (2008). Antigenic properties of HCMV peptides displayed by filamentous bacteriophages vs. synthetic peptides. Immunol. Lett. 119 62–70. 10.1016/j.imlet.2008.04.004 [DOI] [PubMed] [Google Scholar]
- Vaks L., Benhar I. (2011). In vivo characteristics of targeted drug-carrying filamentous bacteriophage nanomedicines. J. Nanobiotechnol. 9 58 10.1186/1477-3155-9-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gerven N., De Greve H., Hernalsteens J. P. (2008). Presentation of the functional receptor-binding domain of the bacterial adhesin F17a-G on bacteriophage M13. Antonie Van Leeuwenhoek 93 219–226. 10.1007/s10482-007-9195-x [DOI] [PubMed] [Google Scholar]
- van Houten N. E., Henry K. A., Smith G. P., Scott J. K. (2010). Engineering filamentous phage carriers to improve focusing of antibody responses against peptides. Vaccine 28 2174–2185. 10.1016/j.vaccine.2009.12.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houten N. E., Zwick M. B., Menendez A., Scott J. K. (2006). Filamentous phage as an immunogenic carrier to elicit focused antibody responses against a synthetic peptide. Vaccine 24 4188–4200. 10.1016/j.vaccine.2006.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Regenmortel M. H. (2012). Basic research in HIV vaccinology is hampered by reductionist thinking. Front. Immunol. 3:194 10.3389/fimmu.2012.00194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldor M. K., Mekalanos J. J. (1996). Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272 1910–1914. 10.1126/science.272.5270.1910 [DOI] [PubMed] [Google Scholar]
- Wan Y., Wu Y., Bian J., Wang X. Z., Zhou W., Jia Z. C., et al. (2001). Induction of hepatitis B virus-specific cytotoxic T lymphocytes response in vivo by filamentous phage display vaccine. Vaccine 19 2918–2923. 10.1016/S0264-410X(00)00561-2 [DOI] [PubMed] [Google Scholar]
- Wan Y., Wu Y., Zhou J., Zou L., Liang Y., Zhao J., et al. (2005). Cross-presentation of phage particle antigen in MHC class II and endoplasmic reticulum marker-positive compartments. Eur. J. Immunol. 35 2041–2050. 10.1002/eji.200425322 [DOI] [PubMed] [Google Scholar]
- Wang F., Liu P., Sun L., Li C., Petrenko V. A., Liu A. (2014a). Bio-mimetic nanostructure self-assembled from Au@Ag heterogeneous nanorods and phage fusion proteins for targeted tumor optical detection and photothermal therapy. Sci. Rep. 4:6808 10.1038/srep06808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Hartner W. C., Gillespie J. W., Praveen K. P., Yang S., Mei L. A., et al. (2014b). Enhanced tumor delivery and antitumor activity in vivo of liposomal doxorubicin modified with MCF-7-specific phage fusion protein. Nanomedicine 10 421–430. 10.1016/j.nano.2013.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Yang S., Mei L. A., Parmar C. K., Gillespie J. W., Praveen K. P., et al. (2014c). Paclitaxel-loaded PEG-PE-based micellar nanopreparations targeted with tumor specific landscape phage fusion protein enhance apoptosis and efficiently reduce tumors. Mol. Cancer Ther. 10.1158/1535-7163.MCT-14-0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Su Q., Dong S., Shi H., Gao X., Wang L. (2014d). Hybrid phage displaying SLAQVKYTSASSI induces protection against Candida albicans challenge in BALB/c mice. Hum. Vaccin. Immunother. 10 1057–1063. 10.4161/hv.27714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Sun M., Fang J., Yang Q., Tong H., Wang L. (2006). Protective immune responses against systemic candidiasis mediated by phage-displayed specific epitope of Candida albicans heat shock protein 90 in C57BL/6J mice. Vaccine 24 6065–6073. 10.1016/j.vaccine.2006.05.022 [DOI] [PubMed] [Google Scholar]
- Wang T., D’souza G. G., Bedi D., Fagbohun O. A., Potturi L. P., Papahadjopoulos-Sternberg B., et al. (2010a). Enhanced binding and killing of target tumor cells by drug-loaded liposomes modified with tumor-specific phage fusion coat protein. Nanomedicine 5 563–574. 10.2217/nnm.10.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Petrenko V. A., Torchilin V. P. (2010b). Paclitaxel-loaded polymeric micelles modified with MCF-7 cell-specific phage protein: enhanced binding to target cancer cells and increased cytotoxicity. Mol. Pharm. 7 1007–1014. 10.1021/mp1001125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Yang S., Petrenko V. A., Torchilin V. P. (2010c). Cytoplasmic delivery of liposomes into MCF-7 breast cancer cells mediated by cell-specific phage fusion coat protein. Mol. Pharm. 7 1149–1158. 10.1021/mp1000229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb J. S., Lau M., Kjelleberg S. (2004). Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186 8066–8073. 10.1128/JB.186.23.8066-8073.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh L. C., Symmons M. F., Nave C., Perham R. N., Marseglia E. A., Marvin D. A. (1996). Evidence for tilted smectic liquid crystalline packing of fd inovirus from X-ray fiber diffraction. Macromolecules 29 8 10.1021/ma9605614 [DOI] [Google Scholar]
- Wichman H. A., Brown C. J. (2010). Experimental evolution of viruses: Microviridae as a model system. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365 2495–2501. 10.1098/rstb.2010.0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis A. E., Perham R. N., Wraith D. (1993). Immunological properties of foreign peptides in multiple display on a filamentous bacteriophage. Gene 128 79–83. 10.1016/0378-1119(93)90156-W [DOI] [PubMed] [Google Scholar]
- Woolford J. L., Jr., Steinman H. M., Webster R. E. (1977). Adsorption protein of bacteriophage fl: solubilization in deoxycholate and localization in the fl virion. Biochemistry 16 2694–2700. 10.1021/bi00631a017 [DOI] [PubMed] [Google Scholar]
- Wu L., Huang T., Yang L., Pan J., Zhu S., Yan X. (2011). Sensitive and selective bacterial detection using tetracysteine-tagged phages in conjunction with biarsenical dye. Angew. Chem. Int. Ed. Engl. 50 5873–5877. 10.1002/anie.201100334 [DOI] [PubMed] [Google Scholar]
- Wu Y., Wan Y., Bian J., Zhao J., Jia Z., Zhou L., et al. (2002). Phage display particles expressing tumor-specific antigens induce preventive and therapeutic anti-tumor immunity in murine p815 model. Int. J. Cancer 98 748–753. 10.1002/ijc.10260 [DOI] [PubMed] [Google Scholar]
- Xia Y. J., Wen W. H., Huang W. Q., Huang B. C. (2005). Development of a phage displayed disulfide-stabilized Fv fragment vaccine against Vibrio anguillarum. Vaccine 23 3174–3180. 10.1016/j.vaccine.2004.12.018 [DOI] [PubMed] [Google Scholar]
- Xue H., Xu Y., Boucher Y., Polz M. F. (2012). High frequency of a novel filamentous phage, VCYϕ, within an environmental Vibrio cholerae population. Appl. Environ. Microbiol. 78 28–33. 10.1128/AEM.06297-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacoby I., Shamis M., Bar H., Shabat D., Benhar I. (2006). Targeting antibacterial agents by using drug-carrying filamentous bacteriophages. Antimicrob. Agents Chemother. 50 2087–2097. 10.1128/AAC.00169-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T. (2013). Filamentous phages of Ralstonia solanacearum: double-edged swords for pathogenic bacteria. Front. Microbiol. 4:325 10.3389/fmicb.2013.00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Wang L., Lu D. N., Gao R. J., Song J. N., Hua P. Y., et al. (2005a). Prophylactic vaccination with phage-displayed epitope of C. albicans elicits protective immune responses against systemic candidiasis in C57BL/6 mice. Vaccine 23 4088–4096. 10.1016/j.vaccine.2004.07.005 [DOI] [PubMed] [Google Scholar]
- Yang W. J., Lai J. F., Peng K. C., Chiang H. J., Weng C. N., Shiuan D. (2005b). Epitope mapping of Mycoplasma hyopneumoniae using phage displayed peptide libraries and the immune responses of the selected phagotopes. J. Immunol. Methods 304 15–29. 10.1016/j.jim.2005.05.009 [DOI] [PubMed] [Google Scholar]
- Yi H., Ghosh D., Ham M. H., Qi J., Barone P. W., Strano M. S., et al. (2012). M13 phage-functionalized single-walled carbon nanotubes as nanoprobes for second near-infrared window fluorescence imaging of targeted tumors. Nano Lett. 12 1176–1183. 10.1021/nl2031663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip Y. L., Smith G., Ward R. L. (2001). Comparison of phage pIII, pVIII and GST as carrier proteins for peptide immunisation in Balb/c mice. Immunol. Lett. 79 197–202. 10.1016/S0165-2478(01)00281-4 [DOI] [PubMed] [Google Scholar]
- Yoo P. J., Nam K. T., Qi J., Lee S. K., Park J., Belcher A. M., et al. (2006). Spontaneous assembly of viruses on multilayered polymer surfaces. Nat. Mater. 5 234–240. 10.1038/nmat1596 [DOI] [PubMed] [Google Scholar]
- Yu M. W., Scott J. K., Fournier A., Talbot P. J. (2000). Characterization of murine coronavirus neutralization epitopes with phage-displayed peptides. Virology 271 182–196. 10.1006/viro.2000.0310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharova M. Y., Kozyr A. V., Ignatova A. N., Vinnikov I. A., Shemyakin I. G., Kolesnikov A. V. (2005). Purification of filamentous bacteriophage for phage display using size-exclusion chromatography. Biotechniques 38:194 10.2144/05382BM04 [DOI] [PubMed] [Google Scholar]
- Zhong G., Smith G. P., Berry J., Brunham R. C. (1994). Conformational mimicry of a chlamydial neutralization epitope on filamentous phage. J. Biol. Chem. 269 24183–24188. [PubMed] [Google Scholar]
- Zinder N. D., Valentine R. C., Roger M., Stoeckenius W. (1963). f1, a rod-shaped male-specific bacteriophage that contains DNA. Virology 20 638–640. 10.1016/0042-6822(63)90290-3 [DOI] [PubMed] [Google Scholar]
- Zou J., Dickerson M. T., Owen N. K., Landon L. A., Deutscher S. L. (2004). Biodistribution of filamentous phage peptide libraries in mice. Mol. Biol. Rep. 31 121–129. 10.1023/B:MOLE.0000031459.14448.af [DOI] [PubMed] [Google Scholar]