ABSTRACT
Rhodobacter sphaeroides is a free-living alphaproteobacterium that contains two clusters of functional flagellar genes in its genome: one acquired by horizontal gene transfer (fla1) and one that is endogenous (fla2). We have shown that the Fla2 system is normally quiescent and under certain conditions produces polar flagella, while the Fla1 system is always active and produces a single flagellum at a nonpolar position. In this work we purified and characterized the structure and analyzed the composition of the Fla2 flagellum. The number of polar filaments per cell is 4.6 on average. By comparison with the Fla1 flagellum, the prominent features of the ultra structure of the Fla2 HBB are the absence of an H ring, thick and long hooks, and a smoother zone at the hook-filament junction. The Fla2 helical filaments have a pitch of 2.64 μm and a diameter of 1.4 μm, which are smaller than those of the Fla1 filaments. Fla2 filaments undergo polymorphic transitions in vitro and showed two polymorphs: curly (right-handed) and coiled. However, in vivo in free-swimming cells, we observed only a bundle of filaments, which should probably be left-handed. Together, our results indicate that Fla2 cell produces multiple right-handed polar flagella, which are not conventional but exceptional.
IMPORTANCE R. sphaeroides possesses two functional sets of flagellar genes. The fla1 genes are normally expressed in the laboratory and were acquired by horizontal transfer. The fla2 genes are endogenous and are expressed in a Fla1− mutant grown phototrophically and in the absence of organic acids. The Fla1 system produces a single lateral or subpolar flagellum, and the Fla2 system produces multiple polar flagella. The two kinds of flagella are never expressed simultaneously, and both are used for swimming in liquid media. The two sets of genes are certainly ready for responding to specific environmental conditions. The characterization of the Fla2 system will help us to understand its role in the physiology of this microorganism.
INTRODUCTION
Motility provides microorganisms with a fundamental survival advantage. Flagella are one of the most complex and effective organelles of locomotion, capable of propelling bacteria through liquids (swimming) and through viscous environments or over surfaces (swarming), and are widely used among Bacteria and Archaea. In addition, these organelles play an important role in adhesion to substrates and biofilm formation, and they contribute to the virulence process in pathogenic bacteria (1, 2).
The bacterial flagellum is a rotary motor powered by the electrochemical proton or sodium potential. The morphology of the flagellum is similar among different bacterial species. Nevertheless, there are differences in their substructures, which are not yet clearly understood. The improvement in powerful microscopy techniques has revealed variations in the architecture of the bacterial flagellar motors (3, 4). The bacterial flagellum is a supramolecular complex made of about 30 different proteins with copy numbers that range from a few to thousands. The structure has been divided into three parts: filament, hook, and basal body. The basal body spans the bacterial cell envelope and comprises a rod and a series of rings. In the cytoplasm the basal body forms a bell-like structure, named the C ring, that houses the export apparatus. This rotating structure is also the input for signals that control the direction of rotation and consequently cell movement (for a review, see reference 5). Around the basal body sit the stator subunits that harness the energy provided by the electrochemical gradient (6). The hook is considered to act as a universal joint that transmits torque to the filament. The filament performs as a rigid propeller that undergoes polymorphic transitions under stressful conditions. These two tubular structures are linked together by means of several subunits of two hook-associated proteins (HAP1 and HAP3). It has been shown that HAP3 contributes to control the conformation of the flagellar helix and to counterbalance torque applied at the base of the flagellum (7).
Rhodobacter sphaeroides is a versatile nonsulfur photosynthetic bacterium that belongs to the α-subgroup of the Proteobacteria. In contrast to other bacteria with two flagellar systems, which produce lateral flagella and a polar flagellum, in R. sphaeroides one set of flagellar genes produces flagella that are located at the cell pole (8), and the other set produces a single flagellum located medially or at a near-polar position (9). The presence of a second set of flagellar genes (fla2) in R. sphaeroides was discovered when the genomic sequence of this bacterium was released (10). At that time, no other flagellar structure distinct from the Fla1 flagellum had been observed. The fla2 genes are not expressed under common laboratory conditions in the wild-type strain WS8N. However, we found the conditions to isolate a spontaneous mutant that expressed the fla2 genes (Fla2+) (8). This mutant, isolated in a Fla1− background, showed the presence of several polar flagella, and it was able to swim in liquid medium, in a way similar to that of the wild-type strain (WS8N) that possesses the Fla1 single subpolar flagellum. It should be noted that neither Fla1 nor Fla2 flagella enable swarming of R. sphaeroides on surfaces. The unidirectional rotation of Fla1 produces smooth swimming, and reorientation occurs when flagellar rotation briefly stops and Brownian motion, and probably the slow rotation of the filament, reorients the cell body (9, 11, 12). During stop periods the filament is observed as a high-amplitude helix that coils against the cell body; when swimming is resumed, the filament forms a helix called normal or in fast-swimming cells it appears to be straight by a visual effect due to the rapid rotation of the bundle (11). Thus far, it has been suggested that Fla2 is also controlled by stop-and-go events (13).
Phylogenetic analyses showed that the fla2 cluster contains the native flagellar genes of this photosynthetic bacterium, whereas the genes of the fla1 cluster were acquired by horizontal gene transfer, involving an ancestral gammaproteobacterium as the possible donor (8). Recently, we have shown that expression of the Fla2 system is controlled by the CckA pathway (14).
In the present study, we describe the isolation of the Fla2 flagellum of R. sphaeroides and for the first time characterize its structure. We also discuss the properties of the filament and the hook, as well as other relevant aspects of this flagellum.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
We used here the wild-type R. sphaeroides strain WS8N (15). AM1, a derivative of SP13, which has a Kanr cassette inserted into the master regulator gene fleQ that blocks the expression of the Fla1 system (16). This strain was selected as a spontaneous swimmer that expresses the Fla2 system under certain growth conditions (8, 17). RSflaA, an AM1 derivative carrying an insertion mutation in the flagellin gene flaA (RSflaA, flaA::aadA). All R. sphaeroides cells were grown in Sistrom's minimal medium at 30°C with orbital shaking (18); when indicated, kanamycin (25 μg/ml) was added to the culture medium. For the isolation of Fla2 filament-hook basal bodies (HBBs), AM1 cells were grown in Sistrom medium without succinate and in the presence of Casamino Acids (0.2%) with orbital shaking (250 rpm) at 30°C as described previously (14).
HBB isolation and purification.
For the preparation of basal bodies, we followed the procedure previously reported (19, 20) with minor modifications. A 5-ml overnight culture of AM1 or RSflaA cells was diluted 1:100 and grown to an optical density at 600 nm of 0.45 to 0.5. Cells were harvested by low speed centrifugation and resuspended by gentle stirring in 10 ml of cold sucrose (0.5 M sucrose, 150 mM Trizma base). Lysozyme (0.1 mg/ml, final concentration) and EDTA (2 mM, final concentration) were gradually added to the cell suspension and incubated at room temperature with gentle stirring. After 50 min of incubation, most cells became spheroplasts, as judged by dark-field microscopy. The cells were lysed with Triton X-100 (1% final concentration), and DNA was degraded by the addition of 600 μl of 0.1 M MgSO4 and 1 mg of DNase I powder. The suspension was incubated at room temperature until it was clear. Cell debris, were removed by centrifugation (9,000 × g) for 10 min at 4°C. The supernatant was centrifuged at 70,000 × g for 35 min, and the pellet containing HBBs was resuspended in TET buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.1% Triton X-100). The HBBs were purified by centrifugation in 40% cesium chloride and centrifuged in a SW40 Ti rotor for 21 h at 70,000 × g at 20°C. A turbid band approximately halfway down the tube was collected and diluted in 25 ml of TET buffer and centrifuged at 70,000 × g for 35 min as described above. The pellet was resuspended in 200 μl of TET buffer and stored at 4°C.
Electron microscopy.
The cell and HBB samples were negatively stained with 1.0 and 2.0% phosphotungstic acid (pH 7.0), respectively, and observed with a JEM-1200EXII electron microscope (JEOL, Tokyo, Japan). Micrographs were taken at an accelerating voltage of 80 or 100 kV. The measurement of the different structures was determined with the help of ImageJ software (21). The numbers of flagella were determined and counted directly from the electron micrographs of cells from independent cultures.
Filament polymorphism and hook length.
Purified filament-HBBs were centrifuged and at 70,000 × g for 35 min, and the pellet containing the filaments was resuspended in TET buffer. Flagellar filaments were incubated in McIlvaine buffer (22) at room temperature. The pH values of the solutions ranged from 3.0 to 8.0, and two concentrations of NaCl (2.5 and 0.2 M) were tested. Samples were observed with an Olympus BH2 microscope with a high-intensity dark field set up. The images were recorded and analyzed using ImageJ software (21). Hook length was determined using ImageJ. The length (L) of the curved structures was obtained by adjusting the curved hooks to a circle from which the angle (θ) and the radius (r) were determined, and the length was calculated using the following equation: L = θ r π/180.
Immunoblotting.
Purified filament-HBBs were precipitated with chloroform-methanol and resuspended in 150 μl of sample buffer. Proteins were separated by SDS–15% PAGE (23), transferred to nitrocellulose membranes (Bio-Rad, Richmond, CA), and incubated for 1 h in TTBS (TBS-Tween 20) buffer (20 mM Tris-Cl [pH 7.5], 0.5 M NaCl, 0.1% [vol/vol] Tween 20) with 5% nonfat milk powder. The membrane was washed three times for 10 min with TTBS, incubated with either anti-FlaA or anti-FlgE2 antibodies at a 1:10,000 dilution for 1 h, and washed again as described above. Detection was performed using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific, Rockford, IL).
Mass spectrometry (MS) and data analysis.
The protein bands were excised from the Coomassie brilliant blue (CBB)-stained 15% SDS-PAGE gels, destained, reduced with 10 mM dithiothreitol, alkylated with 100 mM iodoacetamide, and digested with modified porcine trypsin (Sigma-Aldrich, St. Louis, MO). The resulting peptides were concentrated and desalted using Zip Tips C18 (Millipore, Billerica, MA) and spotted with alpha-cyano-4-hydroxycinnamic acid (Sigma-Aldrich) on a stainless steel plate. The spots generated were analyzed in a MALDI-TOF/TOF 4800 Plus mass spectrometer (AB Sciex, Foster City, CA). Each MS spectrum was acquired by cumulative 1,000 shots in a mass range of 850 to 4,000 Da with a laser intensity of 3,100. The MS/MS spectra for each precursor selected were fragmented and acquired by cumulative 3,000 shots with a laser intensity of 3,800. The fragmentation spectra were compared using Protein Pilot software (v2.0.1; AB Sciex) against a R. sphaeroides database downloaded from NCBI using the Paragon algorithm (24). The search parameters allowed were as follows: cysteine modification by iodoacetamide (carbamidomethylcysteine), trypsin as a cutting enzyme, and biological modifications set by the algorithm. The detected protein threshold in the software was set to 1.3 to achieve 95% confidence. The identified proteins were grouped by the ProGroup algorithm in the software to minimize redundancy.
RESULTS
AM1 cells display several Fla2 flagella at the cell pole.
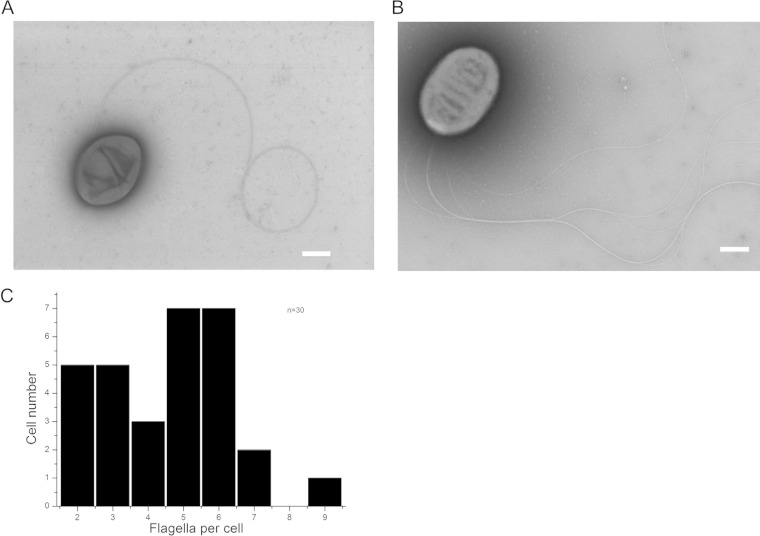
R. sphaeroides has been cultured in the laboratory, and for many years its motility was studied in the wild-type strain WS8N, which produces a single medially located flagellum Fla1. This flagellum differs from typical lateral flagella in several aspects: (i) the singularity of the flagellum is robust, (ii) more than one flagellum per cell has never been observed, and (iii) its location is not fixed at a certain position but is distributed all over the cell body (25–27). Recently, we reported the isolation and characterization of a spontaneous mutant that expresses only the second set of flagellar genes (fla2) (8, 17). Figure 1 shows electron micrographs of WS8N and AM1 flagellated R. sphaeroides cells. Panel A shows the wild-type strain WS8N expressing the single Fla1 flagellum on the cell side, while panel B shows the mutant strain AM1 that produces multiple Fla2 polar flagella. We analyzed 30 AM1 cells to determine the variation in the number of Fla2 flagella at a pole (Fig. 1C). The number of flagella per cell ranges from two to nine and, about a half of the cells that were analyzed had either five or six filaments. The average number was 4.5, with a standard deviation of 1.79. In agreement with previous observations (8), the electron microscopic (EM) images also reveled that the filaments are usually intertwined, suggesting that during swimming the flagellar filaments are arranged in a bundle.
FIG 1.
Electron micrographs of R. sphaeroides strains WS8N and AM1. (A) Wild-type WS8N R. sphaeroides showing a medially located flagellum. (B) AM1 strain of R. sphaeroides displaying a bundle of polar flagella. Scale bars, 500 nm. (C) Graph showing the number of flagella displayed by AM1 cells. A total of 30 cells were scrutinized.
Hook-filament junction of the Fla2 flagellum.
The dimensions of the Fla1 and Fla2 flagellar components were determined from 75 EM images of intact flagella (filament-HBB) and also from previously reported studies (8, 26, 28). Table 1 shows the physical sizes of the flagellar substructures.
TABLE 1.
Physical sizes of flagellar substructures
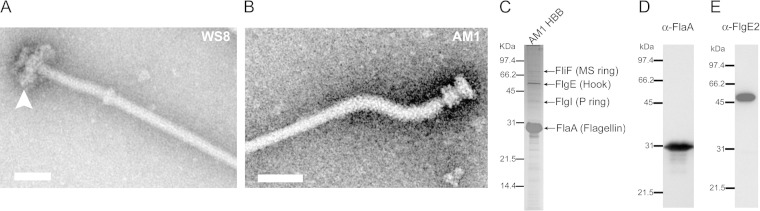
It should be noted that the Fla2 filament is thinner, and the hook is thicker than those of the Fla1 flagellum, and that there is a smooth junction between the filament and the hook. It has been reported previously that one of the prominent characteristics of WS8N Fla1 flagellum is a bulky hook-filament junction (25, 26) (Fig. 2A). We have found that this feature is absent in Fla2 and that the junction looks smooth (Fig. 2B). The difference in thickness between the filament and the hook is larger in the Fla2 system (ca. 5 nm) than in the Fla1 system (ca. 0.6 nm) (Table 1). The filament-hook junction comprises HAP1 (FlgK) and HAP3 (FlgL) (29). The molecular sizes of these two proteins are 1,363 amino acids (aa) and 409 aa for Fla1 and 481 aa and 333 aa for Fla2, respectively, indicating that the bulkiness of the Fla1 junction is due to the larger size of HAP1, which is almost 3-fold larger than the Fla2 HAP1.
FIG 2.
Structure and composition of the Fla2 filament-HBB. (A) Filament-HBB isolated from the wild-type strain WS8N (Fla1). The arrowhead indicates the presence of the H ring. (B) Filament-HBB from AM1 cells (Fla2). Scale bars, 50 nm. (C) SDS-PAGE pattern of Fla2 filament-HBB structures. Arrows indicate the subunits that were identified by SDS-PAGE. (D and E) Western blot analyses of Fla2 filament-HBB using anti-FlaA and anti-Flg2 antibodies, respectively.
Characteristics of the Fla2 hook.
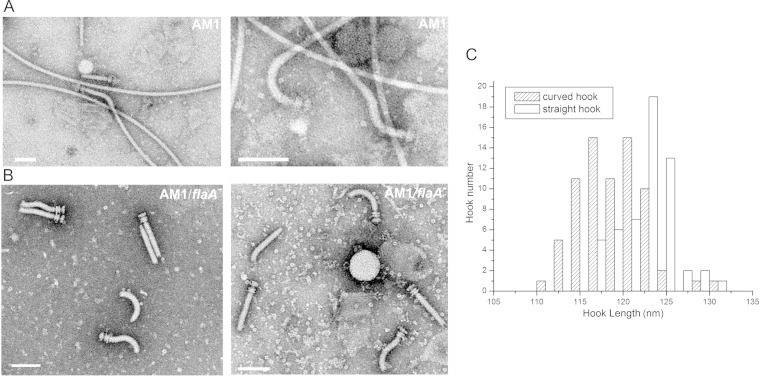
The morphology of the Fla2 hook was clearly different from that of the Fla1 hook. The Fla1 hook is invariably straight (30), whereas the Fla2 hook showed three distinctive shapes: curved, straight, and S-shaped (Fig. 3). These morphologies have also been observed in Selenomonas ruminantium (see Discussion) (31). Images of isolated Fla2 filament-HBB and HBB structures are shown in Fig. 3A and B. We plotted the length of the curved and straight hooks against the hook number (Fig. 3C) and found that the majority of the hooks are between 115 and 125 nm long. The Fla2 hook length was found to be 120.8 ± 6.6 nm, whereas the Fla1 hook was 94 ± 1.8 nm (25) (Table 1). It has been proposed that the hook length is proportional to the molecular length of FliK (32–35). The molecular sizes of Fla1 FliK and Fla2 FliK are 700 aa and 772 aa, respectively, supporting the idea that the length of FliK is proportional to the hook length. However, this correlation seems to have some exceptions since it has been reported that FliK of S. ruminantium (817 aa) has a hook length (105 ± 12 nm) (31) that is shorter than the Fla2 hook. An alignment of these FliK proteins revealed a low similarity between these proteins (data not shown); this result was not unexpected since it has been observed that FliK proteins show a low similarity degree, even in closely related species (36). This low similarity is more pronounced in the N-terminal region, which has been suggested to have an intrinsically disordered structure (37, 38).
FIG 3.
Hook shapes of purified Fla2 filament-HBB and HBB structures. (A) Filament-HBB isolated from AM1 cells; (B) HBB from ΔflaA AM1 mutant cells. Scale bars, 100 nm. The left and right panels show different fields in order to better illustrate the various shapes displayed by the Fla2 hooks. (C) Graph showing the length of curved and straight hooks (n = 60).
Analysis of the components of the Fla2 filament-HBB.
Filament-HBBs from WS8N or AM1 cells were purified and observed by electron microscopy (Fig. 2). Interestingly, the H ring (see arrowhead in Fig. 2A) that is observed in Fla1 filament-HBB structures from WS8N (28) is absent in the Fla2 filament-HBB from AM1 cells (Fig. 2B). The H ring is the outer ring of the LP ring and probably contributes to make a single flagellum more robust (39). The sizes of the other ring structures are similar between the two flagella within measurement errors (Table 1).
We analyzed the protein composition of Fla2 filament-HBBs by SDS-PAGE and found two major and many minor bands in CBB-stained gels (Fig. 2C). The two major bands were assumed to be flagellin FlaA and hook protein FlgE from the molecular masses and their abundance in the flagellum. This assumption was confirmed by Western blotting with specific antibodies against FlaA and FlgE (Fig. 2D and E, respectively). For identification of minor bands, we used mass spectroscopy. In summary, we identified the following 11 proteins: 68-kDa FliF2 (MS ring), 62-kDa FlgK2 (HAP1), 46-kDa FlgE2 (hook protein), 38-kDa FlgI2 (P ring), 34-kDa FlgL2 (HAP3), 31-kDa FlaA (flagellin), 27-kDa FlgG2 (rod protein), 25-kDa FlgF2 (rod protein), 14-kDa FlgC2 (rod protein), 10-kDa FliE2 (rod protein), and 13-kDa FlgB2 (rod protein) (Table 2).
TABLE 2.
Components of Fla2 flagellum
| Substructure | Function | Protein | Molecular mass (kDa) |
|
|---|---|---|---|---|
| Observed | Calculated | |||
| MS ring | Construction base | FliF2 | 60 | 56.2 |
| Filament-hook junction | HAP1 | FlgK2 | NDa | 49.1 |
| Hook | Universal joint | FlgE2 | 54.66 | 48.3 |
| P ring | Bushing in the OM | FlgI2 | 43 | 38.2 |
| Filament-hook junction | HAP3 | FlgL2 | ND | 34.3 |
| Filament | Screw | FlaA | 31.6 | 28.3 |
| Distal rod | Rod | FlgG2 | ND | 27.3 |
| Proximal rod 1 | Rod | FlgF2 | ND | 25.4 |
| Proximal rod 2 | Rod | FlgB2 | ND | 13.6 |
| Proximal rod 3 | Rod | FlgC2 | ND | 14 |
| Proximal rod 4 | Rod | FliE2 | ND | 10.9 |
ND, not determined.
Polymorphism of the Fla2 filaments.
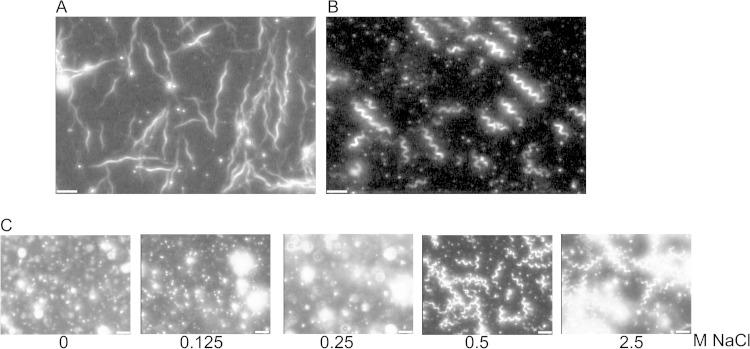
We observed Fla1 and Fla2 filaments by high intensity dark-field microscopy immediately after purification by CsCl density gradient centrifugation (Fig. 4A and B) and determined the handedness by moving the sample stage of the microscope up and down as described previously (40) and also the helical pitch and the diameter of each kind of filament. The pitch values were 2.64 μm for the Fla2 filaments and 4.29 μm for the Fla1 filaments; both filaments were found to be right-handed (Table 3).
FIG 4.
Filament polymorphism. (A) Fla1 filaments from the wild-type strain WS8N. (B) Filaments from the Fla2 strain AM1. Filaments were observed with a high-intensity dark-field microscope. (C) Fla2 filaments in the presence of increasing concentrations of NaCl. Scale bars, 5 μm.
TABLE 3.
Helical parameters of filaments
| Helical parameter | Value or characteristic for WS8N (Fla1) |
Value or characteristic for AM1 (Fla2) |
|||||
|---|---|---|---|---|---|---|---|
| Filaments on cellsc | Filaments isolated previouslya | Filaments isolated in this studyd | Filaments on cellse | Filaments isolatedf | Filaments in 0.5 M NaClg | Filaments in 2.5 M NaClg | |
| Pitch (μm) | ND | 3.04 | 4.29 | 2.64 | 2.64 | 2.64 | 2.60 |
| Diam (πD, μm) | 4.28 | 1.38 | 1.79 | 2.70 | 3.18 | 3.13 | 3.37 |
| Handedness | NDb | Right | ND | Right | Right | Right | Right |
We also analyzed changes in pitch length of the Fla2 filaments in response to increasing concentrations of NaCl at pH 8.0. Figure 4C shows the transition from coiled filaments at low salt concentration to curly filaments at 0.5 and 2.5 M NaCl. We have not observed normal Fla2 filaments that are left-handed; we have only observed polymorphic transitions induced by motor rotation in vivo.
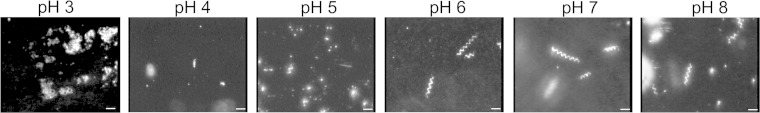
It is known that filament coiling is dependent on the ionic strength and also on the pH of the medium. Therefore, we tested the effect of different pH values at a NaCl concentration of 2.5 M. Figure 5 shows that at this NaCl concentration the filaments form coils at an acidic pH of 4 to 5 and curly filaments at pH 6 to 8. These transitions do not occur at a lower concentration of NaCl (data not shown).
FIG 5.
Effect of pH on the polymorphic transitions of Fla2 filaments. Fla2 filaments isolated from AM1 cells were incubated in the presence of 2.5 M NaCl at the indicated pH values and analyzed by high-intensity dark-field microscopy (see Materials and Methods). Scale bars, 5 μm.
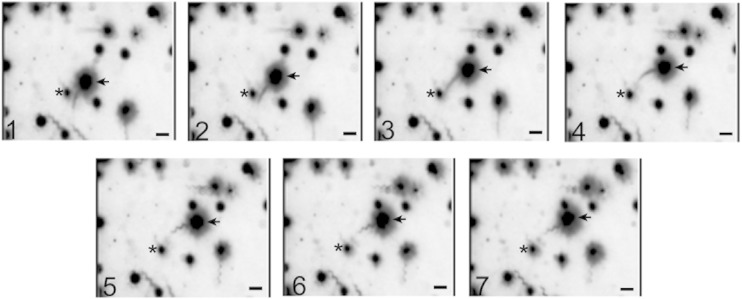
Figure 6 depicts a time series of seven consecutive frames from a movie showing a free-swimming cell where flagella can be easily observed. In cells swimming at full speed the helical waveform of the filament could not be detected, possibly because of its small amplitude and rapid rotation (frames 1 to 4). When the swimming cell stops on frames 5 to 7, a bundle of curly filaments was clearly observed. We could observe left-handed normal filaments, which work by counterclockwise rotation of the motor.
FIG 6.
Polymorphic transitions of Fla2 flagella in swimming AM1 cells. Continuous frames from a short movie (30 frames/s). Arrows show a moving cell (frames 1 to 4) and when the same cell stops (frames 5 to 7). The asterisk indicates a fixed object that was used as a static reference. Scale bars, 5 μm.
DISCUSSION
In situ structures of flagellar motors from several bacteria have been recently characterized by electron cryotomography (3, 4, 41–43). These findings have revealed common features and also important differences, hinting at the great diversity of motor structures in bacteria. The rotary function is conserved with variations in the components of the system. Some flagella are more complex than others perhaps due to adaptation to the particular conditions in which the different microorganisms thrive.
R. sphaeroides WS8N contains two different sets of flagellar genes that are expressed independently, and its presence on the cell appears to be mutually exclusive (14). Both Fla1 and Fla2 flagella are capable of propelling the cell quite efficiently (8, 9, 17). In the wild-type WS8N strain the expression of the Fla1 system always predominates when this strain is grown in Sistrom's minimal medium under either phototrophic or photoheterotrophic conditions. Recently, we have shown that the mutant strain AM1 that expresses the Fla2 system in a Fla1− background carries a single point mutation in the histidine kinase CckA that allows the expression of fla2 genes (14).
Based on the helical values of the Fla2 filaments, we determined that this flagellum does not belong to any of the families described previously (27). This observation agrees with the idea that neither Fla1 or Fla2 can be considered lateral flagella (laf) such as those present in several bacterial species: Vibrio parahaemolyticus, Vibrio shilonii, Azospirillum brasilense, and Bradyrhizobium japonicum (27, 44–47).
We also observed that Fla2 filament-HBBs show significant structural differences compared to Fla1 filament-HBBs. These differences were expected given the independent evolutionary origin of these two flagella (8). For instance, we detected that the 120.8-nm long Fla2 hook is longer than the Fla1 hook that has a reported length of 94 nm (26). The Fla1 hook is straight (30), and the Fla2 hooks display a variety of shapes: curved, S shaped, and straight. This morphology of the Fla2 hook does not seem to depend on the presence of the filament given that HBB structures isolated from a FlaA mutant show the same variations in shape. Similar long S-shaped hooks were described in the lateral flagella of S. ruminantium (31). The fact that the curved hooks are present in Fla2 flagella could be related to the fact that these hooks are more flexible than the straight Fla1 hook, and this characteristic may allow several filaments to rotate together in a bundle (48).
The Fla2 filament-HBB lacks an extra ring structure named the H ring. The presence of the H ring, which is formed by the protein FlgT, has been reported in different species of Vibrio and recently in the Fla1 flagellum of R. sphaeroides (28). The evolutionary advantage conferred by this structure is believed to be associated with the fast rotation of the flagellar motor (39). In this regard, it should be noted that proteobacteria commonly do not have an flgT homolog, even though many of them have polar flagella, which is similar to the situation observed for the Fla2 flagella. Another important difference between Fla1 and Fla2 is found at the hook-filament junction. The Fla2 filament HBB shows a smooth transition in contrast to the bulky HAP region of Fla1 that was previously described (25, 26, 49). It has been suggested that the large size of the HAP1 (FlgK) protein present in the Fla1 flagellum (1363 residues) accounts for the bulky interphase between the hook and the filament. Whereas HAP1 of the Fla2 flagellum shows a size that conforms well with most of the FlgK proteins thus far reported (481 residues). The relevance of a large HAP1 in the Fla1 flagellum has been correlated with efficient swimming (49). However, the fundamental reason for such a large HAP1 protein in Fla1 remains an open question.
Filament-HBB structures were analyzed by MS, and 11 characteristic flagellar proteins were identified. This result confirms that Fla2 is assembled with proteins encoded by the fla2 gene cluster (8) (see Fig. S1 in the supplemental material).
The Fla1 filament belongs to the exceptions among the flagellum families, as does the Fla2 filament. This type of filament does not obey the conventional rule of the polymorphism. Accordingly, Fla2 filaments are not the “typical” polar flagella but “exceptional” polar flagella that are right-handed at rest. Nevertheless, the possibility exists that it changes into a left-handed helix when rotating, as seen in the case of the Fla1 filament (50). Furthermore, this has been also described for mutant filaments of Escherichia coli (51). We have not found any conditions under which the filament takes on left-handed helix. If the motor rotates clockwise, it could solve the current problem, but this raises a question regarding the direction of motor rotation. We leave the question open and present the results as observed.
Similar to Fla1 (9, 27), the Fla2 filaments are right-handed helices at rest, but the pitch of the two filaments differs greatly. It is not clear whether these different pitch values could affect the performance of these flagella. Although it is known that a shorter pitch/radius produces more torque (52), the lack of information regarding the efficiency of the Fla1 and Fla2 motors make impossible to reach a conclusion at this point. Previously, it was shown that WS8N cells have a swimming speed of ∼50 μm/s (53) and that AM1 cells have a swimming speed of 60 μm/s (17). Therefore, these two flagellar systems that work on a stop-go mode (9, 17) achieve similar swimming speeds regardless of the particular properties of the filaments.
Supplementary Material
ACKNOWLEDGMENTS
We thank Teresa Ballado, Aurora Osorio, and Saori Higaki for helpful technical assistance and Fernando García from the Imaging Unit of the Instituto de Fisiología Celular and Emmanuel Rios-Castro, who carried out mass spectrometry analyses at the Genomics, Proteomics, and Metabolomics Unit of the National Experimental Services (LaNSE-CINVESTAV-IPN Mexico).
This study was partially supported by grant IN204614 from PAPIIT and PASPA-UNAM.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00170-15.
REFERENCES
- 1.Kirov SM. 2003. Bacteria that express lateral flagella enable dissection of the multifunctional roles of flagella in pathogenesis. FEMS Microbiol Lett 224:151–159. doi: 10.1016/S0378-1097(03)00445-2. [DOI] [PubMed] [Google Scholar]
- 2.Belas R. 2014. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol 22:517–527. doi: 10.1016/j.tim.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Muller A, Dobro MJ, Jensen GJ. 2011. Structural diversity of bacterial flagellar motors. EMBO J 30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao X, Norris SJ, Liu J. 2014. Molecular architecture of the bacterial flagellar motor in cells. Biochemistry 53:4323–4333. doi: 10.1021/bi500059y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minamino T, Imada K, Namba K. 2008. Molecular motors of the bacterial flagella. Curr Opin Struct Biol 18:693–701. doi: 10.1016/j.sbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Sowa Y, Berry RM. 2008. Bacterial flagellar motor. Q Rev Biophys 41:103–132. doi: 10.1017/S0033583508004691. [DOI] [PubMed] [Google Scholar]
- 7.Fahrner KA, Block SM, Krishnaswamy S, Parkinson JS, Berg HC. 1994. A mutant hook-associated protein (HAP3) facilitates torsionally induced transformations of the flagellar filament of Escherichia coli. J Mol Biol 238:173–186. doi: 10.1006/jmbi.1994.1279. [DOI] [PubMed] [Google Scholar]
- 8.Poggio S, Abreu-Goodger C, Fabela S, Osorio A, Dreyfus G, Vinuesa P, Camarena L. 2007. A complete set of flagellar genes acquired by horizontal transfer coexists with the endogenous flagellar system in Rhodobacter sphaeroides. J Bacteriol 189:3208–3216. doi: 10.1128/JB.01681-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armitage JP, Macnab RM. 1987. Unidirectional, intermittent rotation of the flagellum of Rhodobacter sphaeroides. J Bacteriol 169:514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mackenzie CCM, Larimer FW, Predki PF, Stilwagen S, Armitage JP, Barber RD, Donohue TJ, Hosler JP, Newman JE, Shapleigh JP, Sockett RE, Zeilstra-Ryalls J, Kaplan S. 2001. The home stretch, a first analysis of the nearly completed genome of Rhodobacter sphaeroides 2.4.1. Photosynth Res 70:19–41. doi: 10.1023/A:1013831823701. [DOI] [PubMed] [Google Scholar]
- 11.Armitage JP, Pitta TP, Vigeant MA, Packer HL, Ford RM. 1999. Transformations in flagellar structure of Rhodobacter sphaeroides and possible relationship to changes in swimming speed. J Bacteriol 181:4825–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosser G, Baker RE, Armitage JP, Fletcher AG. 2014. Modelling and analysis of bacterial tracks suggest an active reorientation mechanism in Rhodobacter sphaeroides. J R Soc Interface 11:20140320. doi: 10.1098/rsif.2014.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-del Campo A, Ballado T, Camarena L, Dreyfus G. 2011. In Rhodobacter sphaeroides, chemotactic operon 1 regulates rotation of the flagellar system 2. J Bacteriol 193:6781–6786. doi: 10.1128/JB.05933-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vega-Baray B, Domenzain C, Rivera A, Alfaro-Lopez R, Gomez-Cesar E, Poggio S, Dreyfus G, Camarena L. 2014. The flagellar set Fla2 in Rhodobacter sphaeroides is controlled by the CckA pathway and is repressed by organic acids and the expression of Fla1. J Bacteriol 197:833–847. doi: 10.1128/JB.02429-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sockett RE, Foster JCA, Armitage JP. 1990. Molecular biology of the Rhodobacter sphaeroides flagellum. FEMS Symp 53:473–479. [Google Scholar]
- 16.Poggio S, Osorio A, Dreyfus G, Camarena L. 2005. The flagellar hierarchy of Rhodobacter sphaeroides is controlled by the concerted action of two enhancer-binding proteins. Mol Microbiol 58:969–983. doi: 10.1111/j.1365-2958.2005.04900.x. [DOI] [PubMed] [Google Scholar]
- 17.del Campo AM, Ballado T, de la Mora J, Poggio S, Camarena L, Dreyfus G. 2007. Chemotactic control of the two flagellar systems of Rhodobacter sphaeroides is mediated by different sets of CheY and FliM proteins. J Bacteriol 189:8397–8401. doi: 10.1128/JB.00730-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sistrom WR. 1962. The kinetics of the synthesis of photopigments in Rhodopseudomonas sphaeroides. J Gen Microbiol 28:607–616. doi: 10.1099/00221287-28-4-607. [DOI] [PubMed] [Google Scholar]
- 19.Aizawa SI. 2014. Protocols for purification of flagella, p 106 In The flagellar world. Academic Press, Inc, New York, NY. [Google Scholar]
- 20.Aizawa SI, Dean GE, Jones CJ, Macnab RM, Yamaguchi S. 1985. Purification and characterization of the flagellar hook-basal body complex of Salmonella typhimurium. J Bacteriol 161:836–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McIlvaine T. 1921. A buffer solution for colorimetric comparison. J Biol Chem 49:183–186. [Google Scholar]
- 23.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. 2007. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics 6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi K, Saitoh T, Shah DS, Ohnishi K, Goodfellow IG, Sockett RE, Aizawa SI. 2003. Purification and characterization of the flagellar basal body of Rhodobacter sphaeroides. J Bacteriol 185:5295–5300. doi: 10.1128/JB.185.17.5295-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.West MA, Dreyfus G. 1997. Isolation and ultrastructural study of the flagellar basal body complex from Rhodobacter sphaeroides WS8 (wild type) and a polyhook mutant PG. Biochem Biophys Res Commun 238:733–737. doi: 10.1006/bbrc.1997.7359. [DOI] [PubMed] [Google Scholar]
- 27.Fujii M, Shibata S, Aizawa S. 2008. Polar, peritrichous, and lateral flagella belong to three distinguishable flagellar families. J Mol Biol 379:273–283. doi: 10.1016/j.jmb.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Fabela S, Domenzain C, De la Mora J, Osorio A, Ramirez-Cabrera V, Poggio S, Dreyfus G, Camarena L. 2013. A distant homologue of the FlgT protein interacts with MotB and FliL and is essential for flagellar rotation in Rhodobacter sphaeroides. J Bacteriol 195:5285–5296. doi: 10.1128/JB.00760-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Homma M, DeRosier DJ, Macnab RM. 1990. Flagellar hook and hook-associated proteins of Salmonella typhimurium and their relationship to other axial components of the flagellum. J Mol Biol 213:819–832. doi: 10.1016/S0022-2836(05)80266-9. [DOI] [PubMed] [Google Scholar]
- 30.Ballado T, Camarena L, Gonzalez-Pedrajo B, Silva-Herzog E, Dreyfus G. 2001. The hook gene (flgE) is expressed from the flgBCDEF operon in Rhodobacter sphaeroides: study of an flgE mutant. J Bacteriol 183:1680–1687. doi: 10.1128/JB.183.5.1680-1687.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haya S, Tokumaru Y, Abe N, Kaneko J, Aizawa S. 2011. Characterization of lateral flagella of Selenomonas ruminantium. Appl Environ Microbiol 77:2799–2802. doi: 10.1128/AEM.00286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shibata S, Takahashi N, Chevance FF, Karlinsey JE, Hughes KT, Aizawa S. 2007. FliK regulates flagellar hook length as an internal ruler. Mol Microbiol 64:1404–1415. doi: 10.1111/j.1365-2958.2007.05750.x. [DOI] [PubMed] [Google Scholar]
- 33.Aizawa S. 2012. Mystery of FliK in length control of the flagellar hook. J Bacteriol 194:4798–4800. doi: 10.1128/JB.06239-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes KT. 2012. Rebuttal: mystery of FliK in length control of the flagellar hook. J Bacteriol 194:4801. doi: 10.1128/JB.06454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uchida K, Aizawa S. 2014. The flagellar soluble protein FliK determines the minimal length of the hook in Salmonella enterica serovar Typhimurium. J Bacteriol 196:1753–1758. doi: 10.1128/JB.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawagishi I, Homma M, Williams AW, Macnab RM. 1996. Characterization of the flagellar hook length control protein FliK of Salmonella typhimurium and Escherichia coli. J Bacteriol 178:2954–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizuno S, Amida H, Kobayashi N, Aizawa S, Tate S. 2011. The NMR structure of FliK, the trigger for the switch of substrate specificity in the flagellar type III secretion apparatus. J Mol Biol 409:558–573. doi: 10.1016/j.jmb.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Kodera N, Uchida K, Ando T, Aizawa S. 2015. Two-ball structure of the flagellar hook-length control protein FliK as revealed by high-speed atomic force microscopy. J Mol Biol 427:406–414. doi: 10.1016/j.jmb.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Minamino T, Imada K. 2015. The bacterial flagellar motor and its structural diversity. Trends Microbiol 23:267–274. doi: 10.1016/j.tim.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 40.Shimada K, Kamiya R, Asakura S. 1975. Left-handed to right-handed helix conversion in Salmonella flagella. Nature 254:332–334. doi: 10.1038/254332a0. [DOI] [PubMed] [Google Scholar]
- 41.Murphy GE, Leadbetter JR, Jensen GJ. 2006. In situ structure of the complete Treponema primitia flagellar motor. Nature 442:1062–1064. doi: 10.1038/nature05015. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, Lin T, Botkin DJ, McCrum E, Winkler H, Norris SJ. 2009. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol 191:5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kudryashev M, Cyrklaff M, Wallich R, Baumeister W, Frischknecht F. 2010. Distinct in situ structures of the Borrelia flagellar motor. J Struct Biol 169:54–61. doi: 10.1016/j.jsb.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalez Y, Camarena L, Dreyfus G. 2015. Induction of the lateral flagellar system of Vibrio shilonii is an early event after inhibition of the sodium ion flux in the polar flagellum. Can J Microbiol 61:183–191. doi: 10.1139/cjm-2014-0579. [DOI] [PubMed] [Google Scholar]
- 45.Shinoda S, Okamoto K. 1977. Formation and function of Vibrio parahaemolyticus lateral flagella. J Bacteriol 129:1266–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tarrand JJ, Krieg NR, Dobereiner J. 1978. A taxonomic study of the Spirillum lipoferum group, with descriptions of a new genus, Azospirillum gen. nov. and two species, Azospirillum lipoferum (Beijerinck) comb. nov. and Azospirillum brasilense sp. nov. Can J Microbiol 24:967–980. [DOI] [PubMed] [Google Scholar]
- 47.Kanbe M, Yagasaki J, Zehner S, Gottfert M, Aizawa S. 2007. Characterization of two sets of subpolar flagella in Bradyrhizobium japonicum. J Bacteriol 189:1083–1089. doi: 10.1128/JB.01405-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown MT, Steel BC, Silvestrin C, Wilkinson DA, Delalez NJ, Lumb CN, Obara B, Armitage JP, Berry RM. 2012. Flagellar hook flexibility is essential for bundle formation in swimming Escherichia coli cells. J Bacteriol 194:3495–3501. doi: 10.1128/JB.00209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castillo DJ, Ballado T, Camarena L, Dreyfus G. 2009. Functional analysis of a large non-conserved region of FlgK (HAP1) from Rhodobacter sphaeroides. Antonie Van Leeuwenhoek 95:77–90. doi: 10.1007/s10482-008-9290-7. [DOI] [PubMed] [Google Scholar]
- 50.Pilizota T, Brown MT, Leake MC, Branch RW, Berry RM, Armitage JP. 2009. A molecular brake, not a clutch, stops the Rhodobacter sphaeroides flagellar motor. Proc Natl Acad Sci U S A 106:11582–11587. doi: 10.1073/pnas.0813164106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang W, Jiang Z, Westermann M, Ping L. 2012. Three mutations in Escherichia coli that generate transformable functional flagella. J Bacteriol 194:5856–5863. doi: 10.1128/JB.01102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang HP, Liu B, Rodenborn B, Swinney HL. 2014. Propulsive matrix of a helical flagellum. Chin Phys B 23:114703. doi: 10.1088/1674-1056/23/11/114703. [DOI] [Google Scholar]
- 53.Youle M, Rohwer F, Stacy A, Whiteley M, Steel BC, Delalez NJ, Nord AL, Berry RM, Armitage JP, Kamoun S, Hogenhout S, Diggle SP, Gurney J, Pollitt EJ, Boetius A, Cary SC. 2012. The microbial olympics. Nat Rev Microbiol 10:583–588. doi: 10.1038/nrmicro2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.