ABSTRACT
In Pseudomonas aeruginosa, the transcription factor Anr controls the cellular response to low oxygen or anoxia. Anr activity is high in oxygen-limited environments, including biofilms and populations associated with chronic infections, and Anr is necessary for persistence in a model of pulmonary infection. In this study, we characterized the Anr regulon in biofilm-grown cells at 1% oxygen in the laboratory strain PAO1 and in a quorum sensing (QS)-deficient clinical isolate, J215. As expected, transcripts related to denitrification, arginine fermentation, high-affinity cytochrome oxidases, and CupA fimbriae were lower in the Δanr derivatives. In addition, we observed that transcripts associated with quorum sensing regulation, iron acquisition and storage, type VI secretion, and the catabolism of aromatic compounds were also differentially expressed in the Δanr strains. Prior reports have shown that quorum sensing-defective mutants have higher levels of denitrification, and we found that multiple Anr-regulated processes, including denitrification, were strongly inversely proportional to quorum sensing in both transcriptional and protein-based assays. We also found that in LasR-defective strains but not their LasR-intact counterparts, Anr regulated the production of the 4-hydroxy-2-alkylquinolines, which play roles in quorum sensing and interspecies interactions. These data show that Anr was required for the expression of important metabolic pathways in low-oxygen biofilms, and they reveal an expanded and compensatory role for Anr in the regulation of virulence-related genes in quorum sensing mutants, such as those commonly isolated from infections.
IMPORTANCE Pseudomonas aeruginosa causes acute ocular, soft tissue, and pulmonary infections, as well as chronic infections in the airways of cystic fibrosis patients. P. aeruginosa uses quorum sensing (QS) to regulate virulence, but mutations in the gene encoding the master regulator of QS, lasR, are frequently observed in clinical isolates. We demonstrated that the regulon attributed to Anr, an oxygen-sensitive transcription factor, was more highly expressed in lasR mutants. Furthermore, we show that Anr regulates the production of several different secreted factors in lasR mutants. These data demonstrate the importance of Anr in naturally occurring quorum sensing mutants in the context of chronic infections.
INTRODUCTION
Pseudomonas aeruginosa, a notorious pulmonary pathogen, is frequently a causative agent of nosocomial pneumonias (1), is commonly isolated from the lungs of chronic obstructive pulmonary disease (COPD) patients experiencing exacerbation (2), and is a problematic colonizer of the lungs of individuals with cystic fibrosis (CF) (3). By age 20, 80% of CF patients harbor P. aeruginosa in their lungs (4), and the presence of P. aeruginosa in the airway is correlated with accelerated lung function decline and poor patient prognosis (5, 6). Evidence suggests that in the context of infections, P. aeruginosa is often found in a biofilm state, which contributes to its extreme recalcitrance to antibiotic treatments or clearance by surveilling immune cells (7–9).
Multiple lines of evidence show that oxygen concentrations within P. aeruginosa biofilms and Pseudomonas-infected mucus in CF patient airways are low due to factors such as reduced ventilation, chronic inflammation, and the consumption of oxygen by microbes (10–14). P. aeruginosa senses and responds to low levels of environmental oxygen through the activity of the transcription factor Anr, due to the fact that Anr requires an intact, oxygen-labile [4Fe-4S]2+ cluster for dimerization and subsequent DNA binding (15, 16). In addition, Anr activity is stimulated by phosphatidylcholine (PC) catabolic products that are abundant in vivo (17). While required for anoxic growth via denitrification, P. aeruginosa Δanr strains are not impaired in growth under hypoxic (low-oxygen) conditions (18, 19). Anr homologs have been identified as regulators of virulence in other Gram-negative microbes (20–22). The high-level expression of transcripts encoding the denitrification and arginine fermentation machinery, as well as certain high-affinity cytochrome oxidases (23), suggests that Anr activity is high in vivo (11, 18, 24).
In both in vitro (25) and clinical (3) P. aeruginosa biofilms, cells use quorum sensing (QS) cascades to coordinately regulate gene expression (26). P. aeruginosa QS is controlled by three hierarchically arranged systems, with the LasRI system being the regulatory circuit in control of downstream pathways involving RhlRI and Pseudomonas quinoline signaling systems. QS-controlled virulence factors include pyocyanin, hydrogen cyanide, protease, and lipase (27). Because quorum sensing positively regulates virulence factors, it may seem paradoxical that loss-of-function mutations in lasR are frequently observed in strains isolated from the CF airway (28) and that the presence of lasR mutants in a CF infection is associated with a higher rate of lung function decline (29). In addition, lasR mutants have been observed in acute infections at other body sites (30–32) and can arise spontaneously in laboratory-grown biofilms (33). Taken together, these data imply that under certain conditions, the loss of lasR confers a selective advantage. Previous studies have demonstrated that lasR mutants grow to higher cell densities on specific amino acids found in CF sputum (33), resist cell lysis in high-density cultures (34), show increased resistance to oxidative stress and antibiotic treatment (33, 35), and have higher rates of denitrification (36). The prevalence of QS mutants in infections and their relationship with disease progression illustrates the importance of understanding how pathogenesis is regulated in these strains.
Anr activity has been profiled in planktonic cultures grown anoxically with nitrate, an important alternative electron acceptor for P. aeruginosa. In this study, we used transcriptome sequencing (RNA-Seq) to examine the Anr regulon in colony biofilms grown in low oxygen without exogenous nitrate, using two strains of P. aeruginosa: PAO1 (a laboratory strain) and a QS-deficient CF clinical isolate. We observed Anr regulation of the denitrification machinery under these conditions, as well as a role for Anr in regulation of high-affinity cytochromes, the arginine fermentation genes, and transcripts associated with CupA fimbriae. We also observed Anr regulation of the 4-hydroxy-2-alkylquinoline (HAQ)-dependent quorum sensing pathway, iron acquisition and storage, type VI secretion, the catabolism of aromatic compounds, and many hypothetical proteins. We established that production of CupA fimbriae, known to be important for acute and chronic infections (37, 38), was strictly dependent on Anr in both laboratory and clinical isolates and that enhanced Anr activity increases CupA production. Using both constructed and naturally occurring lasR mutants, we showed that Anr activity increased in the absence of LasRI signaling. Furthermore, we identified a role for Anr in production of HAQs in LasRI signaling-deficient strains but not their QS-competent counterparts. We propose that in the absence of LasRI signaling, Anr is an important regulator of pathogenic processes and that increased expression of the Anr regulon when LasR signaling is off may help explain the basis of selection for lasR mutants in vivo.
MATERIALS AND METHODS
Growth conditions.
All strains used in this study are listed in Table S1 in the supplemental material. J215 is a tracheal isolate from an individual with CF at the Dartmouth-Hitchcock Medical Center in Lebanon, NH. P. aeruginosa and Escherichia coli were routinely cultured in lysogeny broth (LB) at 37°C, and the medium was supplemented with gentamicin (60 μg/ml for Pseudomonas and 15 μg/ml for E. coli) and carbenicillin (300 μg/ml and 100 μg/ml) as required. For studies under low-oxygen conditions, strains were grown at 30°C inside a hypoxic cabinet with an O2 controller and CO2 controller (COY Laboratory Products, Grass Lake, MI) at 1% O2 and 5% CO2. Colony biofilms were inoculated with cells from overnight cultures that had been washed and diluted to an optical density at 600 nm (OD600) of 1.0. Five microliters of this suspension was spotted onto the surface of a T-broth (10 g of tryptone and 5 g of NaCl per liter) agar plate, allowed to dry, and then incubated for 12 to 72 h as indicated.
Construction of in-frame deletion mutants and plasmids.
Strains and plasmids were built using a Saccharomyces cerevisiae recombination technique described previously (39). Primers used in the construction of plasmids are listed in Table S1 in the supplemental material. Knockout constructs generated in this study were built using the pMQ30 allelic replacement vector. The cgrABC expression plasmid was built using the PBAD expression vector pMQ70, and the anr expression plasmids were built using the Ptaq expression vector pMQ123.
Cycle sequencing of lasR, lasI, rhlR, and rhlI in J215.
Target genes were PCR amplified from J215 genomic DNA and the products were sequenced at the Molecular Biology Core at the Geisel School of Medicine at Dartmouth. The resulting sequences were aligned to the PAO1 genomic sequence using the NCBI BLAST program (40).
RNA sequencing analysis.
Colony biofilms of wild-type (WT) or Δanr PAO1 and J215 were grown for 12 h, then harvested in 1 ml of phosphate-buffered saline (PBS) applied to the plate, followed by recovery with an angled glass rod. Samples were pelleted by centrifugation and stored at −80°C. RNA was isolated from pelleted cells using the RNeasy minikit (Qiagen), followed by treatment with RQ1 DNase from Promega, both in accordance with the manufacturers' instructions. RNA quality was assessed using a Bioanalyzer (Agilent Technologies). Two biological replicates (samples from separate single colonies) were analyzed for each strain. One microgram of total RNA was treated for rRNA and tRNA removal using the MICROBExpress Bacterial mRNA Enrichment kit (Life Technologies) before sequencing. Single-read RNA-Seq was performed on the HiSeq platform at the Helmholtz Center for Infection Research (Braunschweig, Germany). Raw reads were processed and normalized using the CLC Genomics Workbench platform (v7.5.1) using the default parameter setting installed by the manufacturer. All sequences were trimmed and mapped to the PAO1 (GenBank accession number NC_002516) reference genome from NCBI using the RNA-Seq analysis tool, and mapped reads were quantile normalized to control for any differences in library size. Very-low-abundance transcripts (<10 mapped reads in all samples) were discarded from further analysis, since there is little power to detect expression changes of genes expressed at low levels.
Western blot analysis of CupA1 and OprF.
Cells grown as colony biofilms were harvested as described for RNA extraction. Cells were pelleted by centrifugation and boiled in SDS loading buffer for 10 min to generate a whole-cell lysate. Protein concentrations were determined using a bicinchoninic acid (BCA) protein assay reagent (Pierce Biotechnology, Inc.). Proteins were separated on a 15% acrylamide gel via SDS-PAGE for 1 h at 180 V. Proteins were transferred to a polyvinylidene fluoride membrane, washed, and probed with polyclonal serum directed against CupA1 as the primary antibody (41) and a peroxidase-conjugated goat anti-rabbit antibody as the secondary antibody (Sigma-Aldrich). Bound antibodies were visualized by SuperSignal West Pico chemiluminescent substrate (Pierce). Densitometry measurements of CupA1 were conducted using ImageJ (42).
NanoString analysis of P. aeruginosa transcripts.
The NanoString nCounter analysis system (NanoString Technologies) was used to analyze the transcript abundance for 75 transcripts and was used with a custom-designed codeset. Each reaction mixture contained 80 ng of RNA in 5 μl of hybridization buffer containing reporter probes, capture probes, and 6 positive and 8 negative controls. Overnight hybridization of RNA with reporter and capture probes was conducted at 65°C and was followed by sample preparation using the NanoString prep station. Finally, targets were counted on the nCounter using 255 fields of view per sample. Data were analyzed using nSolver Analysis software v1.1. Raw counts for all transcripts were normalized to the arithmetic mean of six positive controls and to the geometric mean of three P. aeruginosa housekeeping genes (fbp, ppiD, and rpoD).
Identification of J215 pqsA::TnMar, pqsB::TnMar, and pqsH::TnMar mutants.
Overnight cultures of E. coli S17-1 λpir carrying the pBT20 plasmid and J215 recipient strain were subcultured and grown to an OD600 of 1.0, at which point 1 ml of each culture was washed and suspended in 1 ml LB, and the J215 culture was incubated at 42°C for 10 min. Five hundred microliters from each culture was combined, pelleted, and suspended in 40 μl of LB. This mixture was spotted on LB agar and incubated at 30°C for 22 h. The entire colony was collected, suspended in 100 μl of LB, spread on LB agar containing gentamicin (60 μg/ml) and nalidixic acid (40 μg/ml), and incubated at 30°C and 1% O2. After 3 days, colonies that failed to produce an iridescent sheen were identified and analyzed by arbitrarily primed PCR as previously described (43). Returned sequences were mapped using the Pseudomonas Genome Database BLAST function (44). These three mutants were among those identified.
S. aureus inhibition assays.
P. aeruginosa strains to be tested were grown overnight in LB at 37°C, then washed, and suspended to an OD600 of 1.0. This suspension (5 μl) was spotted on Whatman paper discs on T-broth agar and incubated for 24 h at the desired oxygen concentration. S. aureus strain 8325-4 was grown with shaking overnight at 37°C in tryptic soy broth, then washed, and suspended to an OD600 of 0.1. S. aureus suspension (100 μl) was spread on tryptic soy agar plates using glass beads. Whatman paper discs with P. aeruginosa biofilms were transferred to the plates, and the zone of inhibition was observed after an additional 16 h of incubation.
Statistical analyses.
Fold change values and significance statistics between RNA-Seq samples were calculated using the “Empirical analysis of DGE” algorithm in the CLC Genomics Workbench, which is a reimplementation of the “Exact Test” from the EdgeR Bioconductor package (45, 46) and which was conducted between all pairs, with a total count filter cutoff of 5.0. For the comparison between LasR-regulated transcripts in PAO1 and J215 (see Fig. S1 in the supplemental material), significance was determined using a Wilcoxon rank sum test, with a P value of ≤0.05 considered significant. Differences in expression of LasR and Anr-regulated transcripts in lasR mutants (see Fig. 3) were evaluated with a paired t test, and a P value of ≤0.05 was considered significant. In NanoString experiments (see Fig. 5), significance was determined with ratio paired t test, and a P value of ≤0.05 was considered significant.
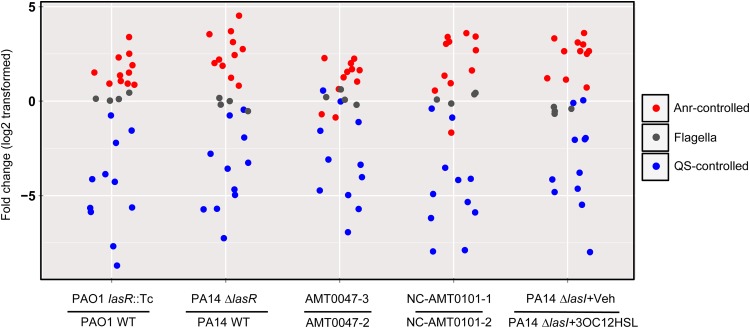
FIG 3.
Abundance of Anr-controlled transcripts is higher in transcript analysis using NanoString in the absence of LasR signaling. Normalized NanoString counts, reflecting mRNA levels, from 12-h colony biofilms grown in 1% O2 are shown. Data are provided in Data set S2 in the supplemental material. Isolate AMT0047-3 (LasR−) evolved from AMT0047-2 (LasR+), and isolate NC-AMT0101-1 (LasR−) evolved from NC-AMT0101-2 (LasR+) (28). The PA14 ΔlasI strain, lacking the 3OC12HSL synthase, was analyzed after growth in the absence or presence of purified 3OC12HSL provided at 25 μM. An equal volume of ethyl acetate was provided as the vehicle control.
FIG 5.
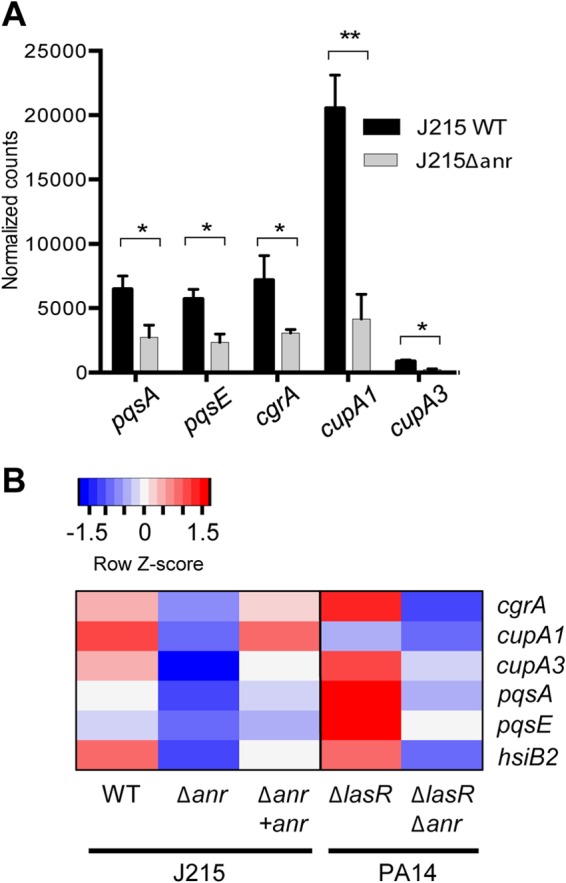
Anr is required for expression of virulence-associated pathways in lasR mutants. NanoString data from colony biofilms grown in 1% O2 for 12 h are shown. (A) Data represent the average number of transcript copies from 3 biological replicates. Bars represent standard deviations. Significance was determined by ratio paired t test.*, P < 0.05; **, P < 0.01. (B) Heat map representation of one of the experiments included in panel A, as well as a separate experiment measuring PA14 ΔlasR and PA14 ΔlasR Δanr. Z-scoring was done by row.
RNA sequencing data accession number.
The raw and processed RNA-Seq data have been deposited into NCBI Gene Expression Omnibus under accession number GSE68534.
RESULTS
Profiling of the Anr regulon in two P. aeruginosa strains grown as colony biofilms in 1% oxygen.
We sought to define the Anr regulon under conditions that relate to those in the mucus plugs that form in CF airways and in clinically relevant biofilms (e.g., low oxygen and high cell density) in two P. aeruginosa strains (PAO1 and a clinical isolate, J215) and their Δanr derivatives. PAO1 is a commonly used laboratory strain with intact quorum sensing. Clinical isolate J215 had colony morphology characteristics of lasR loss-of-function mutants, including the lack of pyocyanin production and the presence of an iridescent colony sheen (33). J215 has a lasRG588T allele that encodes LasR E196D, a variant shown previously to lead to decreased LasR activity or LasR loss of function (32). Other synonymous mutations in lasI, rhlR, and rhlI, as well as nonsynonymous mutations in rhlI, were identified (see Table S2 in the supplemental material).
For the RNA-Seq analysis, RNA was harvested from P. aeruginosa strains PAO1 and J215 grown in colony biofilms at 30°C in 1% O2 and 5% CO2 on T-broth agar for 12 h. We have shown previously that Anr activity is high under these conditions (17). It is important to note that the culture medium was not amended with nitrate or other compounds that can support energy generation by denitrification. The wild type (WT) and corresponding Δanr derivative in each background grew similarly, with 1.0E8 ± 0.29E8 CFU and 1.1E8 ± 0.27E8 CFU per colony for WT and Δanr PAO1, respectively, and 5.7E7 ± 0.2E7 and 9.0E7 ± 1.7E7 CFU per colony for WT and Δanr J215, respectively. Consistent with the lasRG588T allele encoding a loss-of-function LasR variant, we saw that 67 of 72 LasR-regulated transcripts were expressed at a significantly lower level in J215 than in PAO1 (P < 0.5) (see Fig. S1 in the supplemental material).
Deletion of anr had considerable effects on transcription in both PAO1 and J215. Two hundred fifty-nine genes were significantly different (P < 0.05), more than 2-fold, between the WT and the anr mutant in both backgrounds (see Data set S1 in the supplemental material). A summary of the major genes and pathways transcriptionally affected by the loss of anr is presented in Fig. 1. Below, we first describe Anr regulation of known Anr-regulated transcripts, many of which are involved in metabolism when oxygen is limiting. In addition, we describe the discovery that loss of Anr influences expression of pathways involved in the production of secreted molecules and factors.
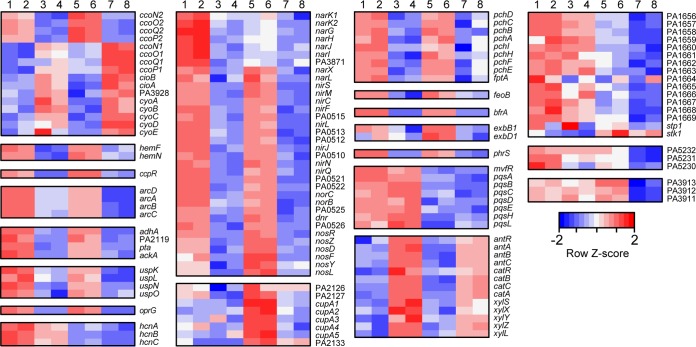
FIG 1.
Heat map showing levels of transcripts regulated by Anr in PAO1 and J215. RNA from wild-type (WT) PAO1 and J215, and their Δanr derivatives, from duplicate colony biofilms grown for 12 h in 1% O2 was sequenced. Total reads for each transcript were quantile normalized, log2 transformed, and Z-scored by row. Data for transcripts significantly affected (>2-fold) are shown. Paired columns represent biological replicates. Columns 1 and 2, WT PAO1; columns 3 and 4, PAO1 Δanr; columns 5 and 6, WT J215; columns 7 and 8, J215 Δanr.
Differential expression of known Anr-regulated pathways in colonies grown in the absence of nitrate at 1% oxygen.
Many of the genes differentially regulated in both strains are known to be under the control of Anr and encode proteins involved in the metabolic response to a low-oxygen environment. For example, Anr impacted the expression of terminal oxidases involved in aerobic respiration. Anr was required for induction of the cbb3-2-oxidase (ccoN2O2Q2P2) and repression of the cyanide-insensitive oxidase (cioAB, PA3928) (47). Levels of ccoN2O2Q2P2 transcript were 12- to 60-fold lower and levels of cioAB were 2- to 8-fold higher in anr mutants from both strains. Transcripts for both the cbb3-1 (ccoN1O1P1) and cytochrome bo3 (cyoABC) oxidases were higher in the anr mutants in both PAO1 and J215 (2- to 3-fold and 2- to 6-fold, respectively). In contrast, the loss of anr did not affect expression of the aa3 oxidase encoded by coxAB-coIII. Together, these data confirm that Anr participates in the control of the adaptation of respiration under low-oxygen conditions. Anr also controlled expression of genes involved in heme biosynthesis. Both hemF and hemN, are known to be controlled by Anr/Dnr (23, 48) and were expressed between 2- and 5-fold lower in the anr mutants. Because high-affinity cytochromes require heme as a cofactor, Anr may mediate their coordinated expression. The diheme cytochrome c551 peroxidase precursor, encoded by ccpR, is appreciated to be regulated by Anr (23), and it was reduced ∼20-fold in both strains.
Transcripts involved in arginine fermentation (arcDABC) were 6- to 23-fold higher in WT strains than in the anr mutants. Our data additionally showed Anr-dependent expression of multiple genes involved in other fermentation pathways, including those for two putative alcohol dehydrogenases that are Anr regulated, adhA (39-fold and 65-fold lower in the Δanr mutants of PAO1 and J215, respectively) and PA2119 (6- and 4-fold lower in the Δanr mutants) (23, 49), as well as a phosphate acetyltransferase pta (5- and 3-fold lower) and an acetate kinase ackA (4- and 3-fold lower) gene. Studies have established a role for P. aeruginosa fermentation pathways and universal stress response proteins in long-term survival within anaerobic biofilms (50, 51). In line with previous work, we found that Anr-dependent expression of the stress response genes uspK, uspL, uspN, and uspO was lower in both Δanr mutants than in their parental strains.
Despite the absence of added nitrate, nitrite or nitric oxide, genes involved in denitrification (dnr, narK1K2GHJ, narXL, nirSMCFLGHJEN, nirQ, norCBD, and nosRZDFLY, as well as co-operonic hypothetical protein genes) were expressed in the WT strains and were much lower in the Δanr derivatives (up to ∼150-fold reduced). Interestingly, the nar genes responsible for the initial reduction of nitrate to nitrite were more highly expressed in PAO1 than in J215 (Fig. 1).
Anr also regulates the production of OprG and of CupA fimbriae; it is not yet known if these factors influence metabolism when oxygen is limiting. The gene that encodes outer membrane protein OprG was reduced 18- and 35-fold in the anr mutants from PAO1 and J215, respectively, consistent with previous reports (52). The chaperone usher pili, including the CupA fimbrial appendages, are expressed on the cell surface and have been implicated in biofilm formation and disease (37, 38, 53, 54). Anr positively regulates the expression of CupA-encoding genes through the activity of a trimeric regulator encoded by the 3-gene operon cgrABC (55, 56). The cupA1-5 transcripts were regulated by Anr in both PAO1 and J215. We also noted that expression of the cupA1-5 genes in J215 was 2- to 29-fold higher than in PAO1 (Fig. 1).
Loss of Anr impacts the expression of genes involved in iron acquisition and quorum sensing.
A notable signature from our data sets reflected a change in expression levels of iron acquisition and storage pathways upon the loss of Anr. The pchDCBA and pchEFHI pyochelin biosynthesis and transport genes showed lower expression in the anr mutant than in the WT for both PAO1 and J215 (between 2- and 14-fold); the pyochelin outer membrane receptor gene fptA had a similar expression pattern. We also saw decreased expression of feoB and bfrA, which encode a ferrous iron transporter and bacterioferritin, respectively. Genes encoding the other siderophore produced by P. aeruginosa, pyoverdine, were not differentially expressed in PAO1 but showed slightly higher expression in the J215 Δanr mutant than in the wild type (pvdA, pvdN, pvdM, and pvdS were induced 2- to 5-fold). However, the transcript encoding ExbB1, involved in pyoverdine uptake, was more than 5-fold lower in the anr mutants from both strains, and exbD1, also involved in pyoverdine uptake, was 16-fold lower in J215 Δanr than in the wild type but not differentially expressed in PAO1.
Our analysis also revealed a role for Anr in regulating expression of multiple pathways related to quorum sensing. Among these were hcnABC, involved in production of hydrogen cyanide. The hcn operon is regulated by both LasR and Anr (57), and while transcripts from this operon were much lower in J215 than in PAO1, we observed a reduction of expression in both Δanr mutants (Fig. 1). Our data set also showed a strong role for Anr in regulation of the small RNA PhrS, which is part of the Pseudomonas quinoline system and has been shown to be controlled by Anr previously (58). PhrS levels were reduced 200-fold in PAO1 and 100-fold in J215. The role for Anr in regulating other LasR-controlled transcripts is discussed further below.
In both strains, a very strong feature of the RNA-Seq data set was the upregulation of genes involved in degradation of aromatic compounds in the anr mutants. Such transcripts included antABC and catBCA, which were expressed between 50- and 150-fold higher in Δanr strains. AntR, the positive regulator of antABC, was expressed at approximately 10-fold-higher levels. Anr also repressed the gene for the hypothetical protein adjacent to catA, PA2506.The ant and cat gene products degrade anthranilate, which is a precursor to the QS molecule HHQ (59). The list of additional genes that were differentially expressed upon the loss of Anr in both strains includes numerous genes that encode hypothetical proteins (see Data set S1 in the supplemental material).
Anr-dependent regulation of CupA is greater in the absence of LasR.
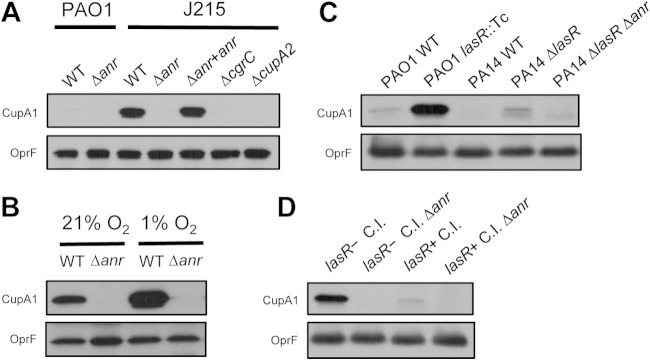
Visual inspection of transcripts strongly regulated by Anr (e.g., ccoN2O2Q2P2, arcDABC, and cupA1-A5) suggested a larger difference in expression between the WT and the Δanr mutant for J215 compared to PAO1. To test this, we performed a Western blot analysis of Anr-regulated CupA fimbriae using an anti-CupA1 antibody (41). We found that CupA protein was strikingly more abundant in colony biofilms formed by J215 than in those formed by PAO1 (Fig. 2A). Levels of outer membrane porin OprF, used as a reference protein, were similar in both strains. CupA1 protein production was completely absent in anr mutants for both strains and was restored in J215 Δanr upon complementation with anr at the native locus (Fig. 2A). Consistent with published data showing that Anr regulates CupA production through its control of cgrABC expression (55), we found that an isogenic ΔcgrC mutant produced no detectable CupA1 (Fig. 2A). A ΔcupA2 mutant is also shown for comparison. While only minute amounts of CupA1 were found in PAO1 colonies grown at 1% oxygen, CupA1 levels were strongly increased by cgrABC overexpression from a plasmid, indicating that the lack of CupA1 in PAO1 was likely due to a regulatory difference rather than another type of defect in CupA production itself (see Fig. S2A in the supplemental material).
FIG 2.
CupA fimbria production is higher in J215 than in PAO1 and is regulated by oxygen, Anr, and LasR. Levels of CupA1 and OprF, a reference protein, were determined by Western blotting of whole-cell lysates from 48-h colony biofilms grown on T-broth agar. (A) The wild type (WT) and mutant derivatives of strains PAO1 and J215 grown in 1% O2. (B) Comparison of CupA1 production in WT and Δanr J215 after growth at 21% and 1% O2. (C) PAO1 and PA14 wild types compared to their corresponding lasR mutant derivatives, after growth in 1% O2. A PA14 lasR anr double mutant is also shown. (D) A lasR-negative clinical isolate (lasR− C.I.; NC-AMT0101-1), a lasR loss-of-function mutant isolated from a CF airway infection, and its lasR+ C.I. parental strain (NC-AMT0101-2) (28) are shown along with their isogenic Δanr derivatives after growth at 1% O2.
We sought to further characterize the connection between Anr activity and CupA levels in biofilms grown in low oxygen in order to evaluate CupA as an indicator of Anr activity. We found that CupA1 levels were higher in colonies grown at 1% oxygen than in those grown at atmospheric oxygen levels (21%), indicating that increased Anr activity stimulated CupA production (Fig. 2B). The CupA1 produced in 21% oxygen is likely due to depletion of oxygen by cell respiration, a process that is well characterized in P. aeruginosa biofilms and colonies (12, 13). A direct relationship between Anr activity and CupA production was confirmed upon expression of an oxygen-resistant allele of Anr, D149A, in 21% oxygen. Activity of this Anr variant strongly induced CupA (see Fig. S2B in the supplemental material). We also explored the possibility that lower CupA levels in the Anr background were due to altered levels of the bacterial second messenger cyclic di-GMP, which positively regulates CupA fimbria production in small-colony variants of a Pseudomonas clinical isolate (41). Deletion of PA2133, a putative phosphodiesterase within the Anr-regulated cupA operon (Fig. 1), did not affect the amount of CupA1 produced (see Fig. S2C). In addition, we found that overexpression of two constitutively active variants of the Pseudomonas fluorescens diguanylate cyclase gcbC in J215 did not affect CupA1 production (see Fig. S2D). Together, these data suggest that decreased CupA production upon loss of Anr is consistent with Anr regulation of cgrABC influencing CupA production in this setting, and not likely due solely to changes in other regulatory signals, such as cyclic di-GMP.
The loss of LasR signaling causes an Anr-dependent increase in CupA fimbriae.
The observation that CupA1 levels were higher in J215 than in PAO1 suggested a connection between the loss of LasR/N-3-oxo-dodecanoyl-homoserine lactone (3OC12HSL) quorum sensing and increased Anr activity. In order to test this model further, we measured production of CupA fimbriae in strain PAO1, strain PA14 (another laboratory strain), and their respective ΔlasR derivatives. In both instances the lasR mutant had higher levels of CupA fimbriae than the wild-type parental strain (10-fold higher in PAO1 ΔlasR than in the PAO1 wild type and 4-fold higher in PA14 ΔlasR than in the PA14 wild type) (Fig. 2C). We also measured CupA production in a pair of genetically related CF clinical strains isolated from the same subject (28). Previous genomic analyses revealed that one isolate, NC-AMT0101-2, has a functional allele of lasR, while NC-AMT0101-1 had acquired a lasR mutation. The lasR-defective clinical isolate produced 7.5-fold more CupA fimbriae than the parental strain (Fig. 2D), and in both NC-AMT0101-1 and NC-AMT0101-2, deletion of anr abolished CupA1 production (Fig. 2D). Taken together, these results suggest that lasR mutants have higher levels of Anr activity and that this leads to higher production of CupA fimbriae.
Anr is repressed by LasR/3OC12HSL.
To further test the hypothesis that QS signaling is inversely correlated with Anr activity, we used a multiplex method for the simultaneous analysis of multiple transcripts with the nCounter NanoString technology. NanoString mRNA quantitation uses fluorescent probes to capture and count specific mRNA targets (60), and we developed a NanoString codeset that included a number of QS and Anr-regulated transcripts. The genes directly controlled by Anr in our codeset were arcA, arcD, adhA, ccoN2, ccoP2, dnr, narG, nirS, norC, cgrA, and PA1673. The QS-controlled genes were lasI, lasB, rhlI, rhlA, pqsA, pqsE, pqsH, phzA2, phzC, phzH, and phzM. We analyzed mRNA levels of these transcripts in PAO1 and PA14 and their lasR mutant derivatives, the paired clinical isolates NC-AMT0101-2 and NC-AMT0101-1 (described above), and another set of genetically related clinical isolates with and without functional LasR (AMT0047-2 and AMT0047-3, respectively) (28). We observed that nearly all Anr-regulated genes were more highly expressed in the lasR mutants than in their cognate lasR-intact strains (Fig. 3). For example, expression of Anr-regulated ccoN2 was 4- to 12-fold higher in the lasR mutants and PA1673 expression was 5- to 25-fold higher in the lasR mutants. As expected, QS-regulated transcripts were uniformly lower in the absence of functional LasR. In addition to comparing wild-type and LasR-defective strains, we examined Anr activity in a ΔlasI mutant, which lacks the 3OC12HSL synthase, in the absence and presence of 3OC12HSL (61). We found that complementation of the ΔlasI strain with exogenous 3OC12HSL was sufficient to rescue the expression of QS-controlled genes and led to decreased expression of Anr-regulated genes (Fig. 3). Across all five pairs of samples, 7 of 11 Anr-regulated transcripts and 8 of 11 QS-regulated transcripts were significantly different (P < 0.05, paired t test). We did observe that in both AMT0047-3 and NC-AMT0101-1 (carrying natural lasR defective alleles) but not the other lasR or lasI mutants, narG expression was reduced. There is a potential LasR binding motif (CTCACTGTTTTAAAAG) 150 bp upstream of narK1 translational start (the first gene in the operon containing narG), and our data may indicate a positive role for LasR in regulating expression of this operon in these strain backgrounds. This is consistent with reduced expression of the narK1-PA3871 operon in J215 compared with that in PAO1 (Fig. 1). Expression of norC was also decreased in AMT0047-3 compared to its parental strain, AMT0047-2. In contrast, four flagellar transcripts (flgD, flgG, flgK, and fliC) did not vary between the pairs (Fig. 3). The repression of Anr activity upon addition of 3OC12HSL to a ΔlasI strain indicated that Anr was directly responsive to LasR signaling and that increased Anr activity was not due to a secondary effect common in the absence of LasR. The complete NanoString data set from these experiments is provided in Data set S2 in the supplemental material. LasR regulation of Anr is likely indirect, as there is no evidence that LasR binds the anr promoter (62), and anr transcript levels are not altered in transcriptional profiling analyses of the LasR regulon (63, 64) (see Data set S2). LasRI positively regulates 3,4-dihydroxy-2-heptylquinolone (PQS) production (65), and PQS has been shown to inhibit denitrification (66). To determine if increased Anr activity is due to a decrease in PQS, we tested whether CupA1 production was greater in three mutants in the PQS pathway in J215. There were no detectable differences (see Fig. S2E in the supplemental material), indicating that HAQ production does not likely effect Anr activity in J215.
Anr partially compensates for the loss of LasR signaling in the regulation of HAQs.
We showed above that mutants or strains with lower levels of LasR activity have increased Anr activity. Our studies also showed that in LasR-defective backgrounds, Anr plays roles that are not evident in las+ laboratory strains. When active, LasR regulates production of 4-hydroxy-2-alkylquinolines (HAQs), including PQS and its direct precursor 4-hydroxy-2-heptylquinoline (HHQ). The iridescent sheen that is characteristic of lasR mutants is caused by an accumulation of HHQ, which is due to an inability to properly induce the pqsH product, the enzyme that converts HHQ to PQS, in the absence of LasR (33). In contrast to the J215 WT, J215 Δanr did not make HHQ sheen when grown in 1% oxygen (Fig. 4A). These visual phenotypes were supported by the RNA-Seq data, which showed that three of the five transcripts in the HHQ biosynthetic operon (pqsA, pqsC, and pqsE) were 2- to 8-fold lower in J215 Δanr than in the J215 WT. In contrast, the loss of anr had no effect on colony morphology in PAO1 (see Fig. S3A in the supplemental material), and pqs transcripts were not different between the PAO1 WT and PAO1 Δanr. Interestingly, anr was not required for HHQ production when cells were grown at 21% oxygen, a condition correlated with lower Anr activity (see Fig. S3B). Consistent with the RNA-Seq analysis of HAQ-related transcripts and phenotypic data, a targeted analysis of pqsA and pqsE in J215 was performed. In the absence of Anr, pqsA and pqsE were both reduced in the J215 background (Fig. 5A; see also Data set S3 in the supplemental material). The cgrA, cupA1, and cupA3 transcripts, as expected based on the data shown above, also followed this pattern. Expression of these genes was restored upon complementation with anr at the native locus. Anr-dependent regulation of the pqs and cupA genes was also observed in the PA14 ΔlasR and PA14 ΔlasR Δanr pair (Fig. 5B; see also Data set S3). Together, these data suggest that Anr activity impacts HAQ production in strains with low or absent LasR activity.
FIG 4.
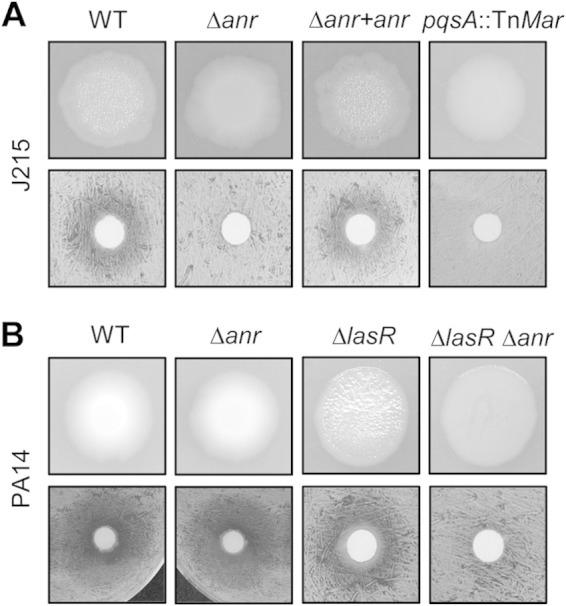
Anr is required for HHQ-dependent colony sheen and HQNO-dependent S. aureus inhibition at 1% oxygen in lasR mutants. (A) J215; (B) PA14. Top row, colony biofilms at 5 days on T-broth agar. Bottom row, Whatman paper discs with 24-h biofilms grown on T-broth agar were transferred to tryptic soy agar plates spread with a lawn of S. aureus and grown for an additional 16 h.
In addition to impacting pqs transcription and HHQ production, deletion of Anr also affected production of the HHQ-derived exoproduct 4-hydroxy-2 heptylquinoline N-oxide (HQNO), which can slow the growth of Staphylococcus aureus and other Gram-positive organisms by inhibiting electron transport (67, 68). We tested the role for anr in this interaction by exposing a lawn of S. aureus to J215 biofilms and observing the zone of growth inhibition. We saw that biofilms formed by J215 WT inhibited S. aureus in 1% oxygen and that this ability required anr and pqsA (Fig. 4A). To further study Anr regulation of HAQs in lasR mutant backgrounds, we examined the role for Anr in PA14 ΔlasR. (Strain PAO1 ΔlasR was not used because it does not overproduce HHQ, a fact that has been previously published [33].) Deletion of anr in a PA14 lasR mutant led to a marked decrease in HHQ production and an inability to inhibit S. aureus at 1% oxygen, while deletion of anr in a lasR intact background had no effect on either colony morphology or S. aureus inhibition (Fig. 4B). As with J215, anr was not required for HHQ production in PA14 ΔlasR at 21% oxygen (see Fig. S3B in the supplemental material). It is important to note that Anr activity was not sufficient to maintain the same level of HAQ production as was seen in lasR-intact strains; transcriptional data showed that pqs transcripts were less abundant in J215 than in PAO1 (Fig. 1), and the zone of S. aureus inhibition is larger in PA14 than in PA14 ΔlasR (Fig. 4B). However, these data indicate that Anr supports biologically active levels of HAQ production in low-oxygen biofilms in the absence of LasR.
We tested a number of potential mechanisms by which Anr could impact HAQ production in lasR mutants. RhlR has been shown to activate HAQ production in lasR mutants under certain conditions (69, 70), but as anr is required for the production of HAQs in both the PA14 ΔlasR mutant and the PA14 ΔlasR ΔrhlR double mutant, we conclude that Anr-dependent regulation of HAQ production is not through RhlR (see Fig. S4A in the supplemental material). Sonnleitner et al. described a connection between Anr and HAQs through PhrS, a small ncRNA that activates translation of pqsR and is induced by Anr (58). In our RNA-Seq experiment, both PAO1 and J215 anr mutants showed a strong reduction in phrS expression (200-fold and 100-fold, respectively). However, a J215 ΔphrS mutant did not show a decrease in the HHQ-dependent colony phenotype that is lost upon mutation of anr (see Fig. S4B). The antABC, catBCA, and xylXYZL gene products are involved in degradation of aromatic compounds, including the HHQ/HQNO precursor anthranilate. These genes were induced up to 140-fold in both Δanr mutants, and we reasoned that overactivity of these pathways could deplete intracellular anthranilate and lead to an inability to synthesize HAQs. However, deletion of antA in J215 Δanr did not restore HHQ production, as colony biofilms from this strain remained smooth (see Fig. S4B). Because Anr is necessary for expression of arcD, which encodes an arginine-ornithine antiporter, and because arginine has been linked with HHQ-mediated modulation of P. aeruginosa swarming motility (71), we hypothesized that the anr defect may be linked to an inability to acquire or synthesize arginine. However, growth on media supplemented with 0.4% arginine did not restore colony sheen in J215 Δanr (see Fig. S4B). Finally, Dnr, a major downstream regulator under the control of Anr, does not participate in Anr regulation of HAQs, as a Δdnr strain retains HHQ production, while the Anr mutant does not (see Fig. S4B). Taken together, these data indicate that the Anr effect on HAQ production is likely due to a confluence of factors or through an unrecognized pathway.
Analysis of links between LasR and Anr in the P. aeruginosa genome.
In the RNA-Seq data, Anr-dependent expression of genes in the H2-type VI secretion system was more pronounced in J215 than in PAO1 (Fig. 1). The H2-type VI secretion system delivers a phospholipase with activity against bacterial membranes, PldA, directly into target cells (72) and has been shown to contribute to P. aeruginosa virulence in eukaryotic models of infection (73, 74). Expression of the H2-type VI secretion locus is controlled by LasR/3OC12HSL (73, 74), and a putative Anr-binding site has been identified at bp −174 relative to the start of transcription of the operon (23). Further analysis confirmed that expression of hsiB2, a gene within the H2-type VI secretion system operon, was dependent on Anr in the absence of functional LasR (Fig. 5B).
In order to identify additional genes potentially regulated by both QS and Anr, we cross-referenced data sets from a LasR-ChIP experiment (62) and a microarray experiment comparing a ΔlasI ΔrhlI strain of PAO1 grown in the presence and absence of 3OC12HSL and N-butyryl-homoserine lactone (C4HSL) (63) against a list of all P. aeruginosa genes with a putative Anr binding sequence in their upstream region (23). This analysis returned hcnA, nosR, narK1, and ccpR as well as hsiA2, the first gene in the H2-type VI secretion locus. It also returned the hypothetical genes PA3662, PA3913 (a putative collagenase), and PA5232 (part of a putative ABC transporter). Comparing the expression profiles of these genes and co-operonic genes in our RNA-Seq experiment supports the hypothesis that they are regulated by quorum sensing and Anr, in that in J215, a strain without functional LasR, Anr is necessary for strong expression (Fig. 1).
DISCUSSION
In the present study, we analyzed the transcriptomes of WT and Δanr strains in colony biofilms grown in a low-oxygen environment without nitrate. We analyzed two distinct strain backgrounds, including one lacking activity of the QS regulator LasR, and found that the absence of LasR correlates with higher Anr activity. This is consistent with reports that have noted that the metabolism of lasR mutants differs from that of WT strains, particularly in that there is increased nitrate utilization and higher expression of the denitrification machinery (35, 36, 63). Our data show that Anr is necessary for induction of pathways that promote the generation of energy under low-oxygen conditions, including genes encoding high-affinity cytochrome oxidases, the machinery necessary for denitrification, and arginine fermentation enzymes (18, 23). Additionally, our conditions revealed a previously unobserved role for Anr in the control of expression of pathways related to iron acquisition and storage, HAQ production, the catabolism of aromatic compounds, and H2-type VI secretion (Fig. 6).
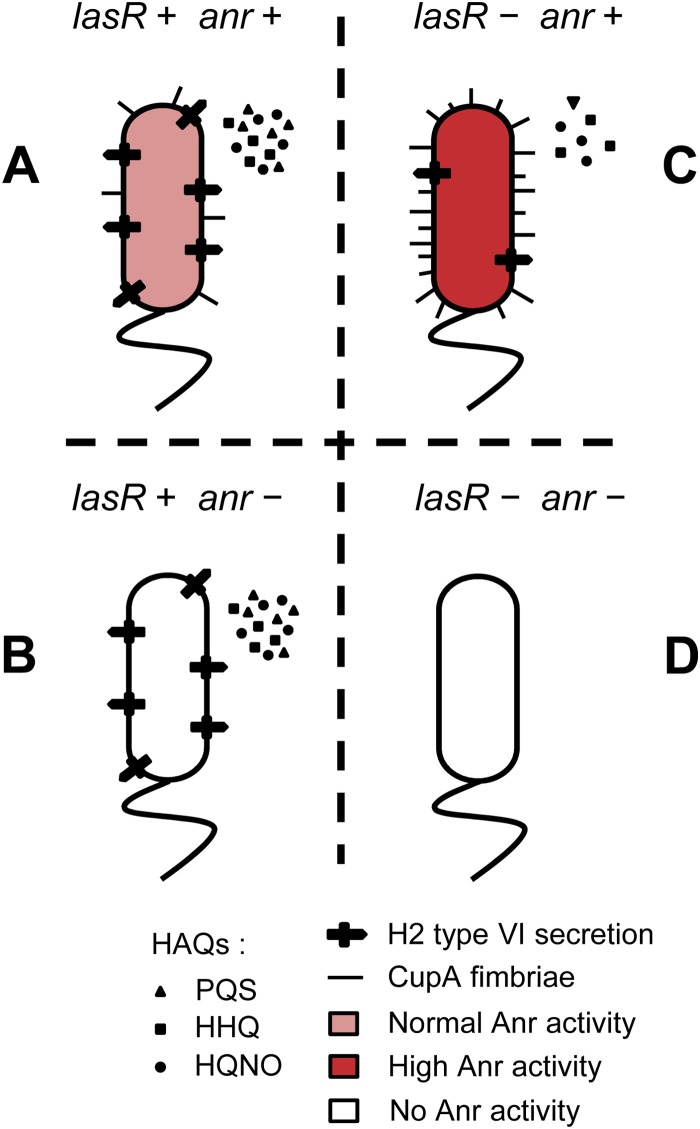
FIG 6.
Model for the role of Anr in lasR mutant biofilms. In the presence of LasR-mediated QS (A and B), Anr is active in biofilms and is required for production of CupA fimbriae, but absence of Anr does not affect HAQ production, H2-type VI secretion, or expression of the operons PA5232-PA5230 (containing a putative collagenase gene) and PA3913-PA3911 (encoding a putative ABC transporter). In the absence of lasR signaling (C and D), increased Anr activity leads to increased production of CupA fimbriae, and Anr is required for HHQ/HQNO production and H2-type VI secretion, as well as expression of the operons described above.
Because isolates with loss-of-function mutations in lasR are common in CF, it is interesting to speculate that increased Anr activity contributes to the fitness of these strains. We have shown a role for anr in a model of pulmonary infection, and there is evidence to suggest that Anr-regulated pathways are an important part of the long-term adaptation of P. aeruginosa to the CF airway (17, 18, 50, 51, 75). Additionally, our data showing that Anr activity can be reduced in a ΔlasI strain by addition of exogenous 3OC12HSL raise the possibility that Anr has an important role in the regulation of virulence factors in low-cell-density environments, when the concentration of 3OC12HSL is low.
The activities of Anr and also the E. coli Anr homolog Fnr are redox sensitive, due to the requirement of an assembled [4Fe-4S]2+ cluster for dimerization and DNA binding (76, 77). Anr is also likely affected by iron availability, as has been shown to be the case for Fnr (78). It is possible that either of these factors is altered by LasR activity. Future studies will determine specifically how LasR affects Anr activity in P. aeruginosa.
The role for Anr in regulation of HAQ production may be both direct and indirect. The antABC and catBCA aromatic compound degradation pathways are strongly activated by intracellular anthranilate, suggesting that the increased expression of these pathways in anr mutants could reflect accumulated anthranilate due to inactivity of the pqsABCD operon. Additionally, AntR, when bound to anthranilate, has been shown to inhibit the activity of PqsR, and PqsR represses antA (59). Although deletion of antA was not sufficient to restore HHQ production in J215 Δanr, it is possible that the Δanr phenotype is due to repressive effects of AntR. Another intriguing possibility is that HAQ production is reduced in the J215 anr mutant as a result of either an increase in or an inability to appropriately respond to oxidative stress. Multiple genes involved in the oxidative stress response (katB, which encodes a catalase, as well as ahpB and ahpCF, which encode hydroperoxide reductases) were expressed at a level >2-fold higher in both the PAO1 and J215 Δanr mutants. Quinolines have been shown to sensitize P. aeruginosa to oxidative stress (79), and pyochelin can promote oxidative stress (80) and is regulated in response to oxidative stress (81).
The requirement for Anr in HHQ production in LasR-defective strains is interesting in light of data which showed that HHQ, rather than its derivative PQS, is required for infection in a murine model (82). P. aeruginosa cannot produce PQS anaerobically, due to the fact that PqsH (the enzyme that oxidizes HHQ to PQS) requires oxygen as a cofactor (83), and HHQ is readily detected in CF airway secretions (84). We believe that the relationship between Anr and HHQ in quorum sensing mutants may functionally compensate for effects of losing LasR, and this could help explain the ability for P. aeruginosa lasR mutants to thrive in infections. A recent study demonstrated equal infectivity between WT PAO1 and a lasRI rhlRI quadruple mutant in a mouse lung model, suggesting that homoserine lactone signaling in general may be dispensable for infection in this context (85). Future studies will be aimed at measuring the role for Anr regulation of HHQ production in infections.
Thus, LasR loss-of-function mutants show increased expression of metabolic pathways that are valuable in low oxygen, increased production of CupA fimbriae, and functionally active levels of HHQ and HQNO (Fig. 6), all of which are dependent on Anr. We propose that Anr-regulated pathways may contribute significantly to virulence and fitness in lasR mutant isolates, and future studies will be aimed at measuring the specific role for Anr-regulated pathways in lasR mutants in infections.
Supplementary Material
ACKNOWLEDGMENTS
Research reported in this publication was supported by National Institutes of Health (NIH) grant RO1A109170 to D.A.H. and the Renal Function and Disease training grant NIGMS of the NIH under award number 5P20-RR018787 (Stanton) to J.H.H. This work was also supported by grants from the Cystic Fibrosis Foundation Research Development Program (STANTO07R0) and the National Institute of General Medical Sciences (NIGMS) of the NIH as P20GM103413, which supported the Translational Research Core.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
We thank Susanne Häussler (Helmholtz Centre for Infections Research) for providing CupA1 antibody and Michael Jarek for RNA sequencing work. We acknowledge Thomas Hampton and Sven Willger (Geisel School of Medicine at Dartmouth) for helpful discussions regarding data analysis and presentation and Nicholas Jacobs for comments on the manuscript. We thank Simon Dove (Harvard Medical School) and George O'Toole (Geisel School of Medicine at Dartmouth) for providing plasmids.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00182-15.
REFERENCES
- 1.Weber DJ, Rutala WA, Sickbert-Bennett EE, Samsa GP, Brown V, Niederman MS. 2007. Microbiology of ventilator-associated pneumonia compared with that of hospital-acquired pneumonia. Infect Control Hosp Epidemiol 28:825–831. doi: 10.1086/518460. [DOI] [PubMed] [Google Scholar]
- 2.Murphy TF. 2009. Pseudomonas aeruginosa in adults with chronic obstructive pulmonary disease. Curr Opin Pulm Med 15:138–142. doi: 10.1097/MCP.0b013e328321861a. [DOI] [PubMed] [Google Scholar]
- 3.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 4.Rajan S, Saiman L. 2002. Pulmonary infections in patients with cystic fibrosis. Semin Respir Infect 1:47–56. [DOI] [PubMed] [Google Scholar]
- 5.Kosorok MR, Zeng L, West SEH, Rock MJ, Splaingard ML, Laxova A, Green CG, Collins J, Farrell PM. 2001. Acceleration of lung disease in children with cystic fibrosis after Pseudomonas aeruginosa acquisition. Pediatr Pulmonol 32:277–287. doi: 10.1002/ppul.2009.abs. [DOI] [PubMed] [Google Scholar]
- 6.Gaspar MC, Couet W, Olivier JC, Pais AA, Sousa JJ. 2013. Pseudomonas aeruginosa infection in cystic fibrosis lung disease and new perspectives of treatment: a review. Eur J Clin Microbiol Infect Dis 32:1231–1252. doi: 10.1007/s10096-013-1876-y. [DOI] [PubMed] [Google Scholar]
- 7.Høiby N, Johansen HK, Moser C, Song Z, Ciofu O, Kharazmi A. 2001. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microbes Infect 3:23–35. doi: 10.1016/S1286-4579(00)01349-6. [DOI] [PubMed] [Google Scholar]
- 8.Tran CS, Rangel SM, Almblad H, Kierbel A, Givskov M, Tolker-Nielsen T, Hauser AR, Engel JN. 2014. The Pseudomonas aeruginosa type III translocon is required for biofilm formation at the epithelial barrier. PLoS Pathog 10:e1004479. doi: 10.1371/journal.ppat.1004479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaber JA, Triffo WJ, Suh SJ, Oliver JW, Hastert MC, Griswold JA, Auer M, Hamood AN, Rumbaugh KP. 2007. Pseudomonas aeruginosa forms biofilms in acute infection independent of cell-to-cell signaling. Infect Immun 75:3715–3721. doi: 10.1128/IAI.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Weiss T, Botzenhart K, Yankaskas JR, Randell S, Boucher RC, Doring G. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest 109:317–325. doi: 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassett DJ, Sutton MD, Schurr MJ, Herr AB, Caldwell CC, Matu JO. 2009. Pseudomonas aeruginosa hypoxic or anaerobic biofilm infections within cystic fibrosis airways. Trends Microbiol 17:130–138. doi: 10.1016/j.tim.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Wessel AK, Arshad TA, Fitzpatrick M, Connell JL, Bonnecaze RT, Shear JB, Whiteley M. 2014. Oxygen limitation within a bacterial aggregate. mBio 5(2):e00992-14. doi: 10.1128/mBio.00992-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Werner E, Roe F, Bugnicourt A, Franklin MJ, Heydorn A, Molin S, Pitts B, Stewart PS. 2004. Stratified growth in Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 70:6188–6196. doi: 10.1128/AEM.70.10.6188-6196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon SS, Hennigan RF, Hilliard GM, Ochsner UA, Parvatiyar K, DeKievit TR, Gardner PR, Schwab U, Rowe JJ, Iglewski BH, McDermott TR, Mason RP, Wozniak DJ, Hancock RE, Parsek MR, Noah TL, Boucher RC, Hassett DJ. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev Cell 3:593–603. doi: 10.1016/S1534-5807(02)00295-2. [DOI] [PubMed] [Google Scholar]
- 15.Lazazzera BA, Beinert H, Khoroshilova N, Kennedy MC, Kiley PJ. 1996. DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J Biol Chem 271:2762–2768. doi: 10.1074/jbc.271.5.2762. [DOI] [PubMed] [Google Scholar]
- 16.Ye RW, Haas D, Ka JO, Krishnapillai V, Zimmermann A, Baird C, Tiedje JM. 1995. Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosa requires Anr, an analog of FNR. J Bacteriol 177:3606–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson AA, Gross MJ, Daniels EF, Hampton TH, Hammond JH, Vallet-Gely I, Dove SL, Stanton BA, Hogan DA. 2013. Anr, and its activation by PlcH activity, in Pseudomonas aeruginosa host colonization and virulence. J Bacteriol doi: 10.1128/JB.02169-12. [DOI] [PMC free article] [PubMed]
- 18.Alvarez-Ortega C, Harwood CS. 2007. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol Microbiol 65:153–165. doi: 10.1111/j.1365-2958.2007.05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson AA, Daniels EF, Hammond JH, Willger SD, Hogan DA. 29 July 2014. The global regulator Anr represses PlcH phospholipase activity in Pseudomonas aeruginosa when oxygen is limiting. Microbiology doi: 10.1099/mic.0.081158-0. [DOI] [PMC free article] [PubMed]
- 20.Marteyn B, West NP, Browning DF, Cole JA, Shaw JG, Palm F, Mounier J, Prevost M-C, Sansonetti P, Tang CM. 2010. Modulation of Shigella virulence in response to available oxygen in vivo. Nat Lett 465:355–358. doi: 10.1038/nature08970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baltes N, N′Diaye M, Jacobsen ID, Maas A, Buettner FF, Gerlach GF. 2005. Deletion of the anaerobic regulator HlyX causes reduced colonization and persistence of Actinobacillus pleuropneumoniae in the porcine respiratory tract. Infect Immun 73:4614–4619. doi: 10.1128/IAI.73.8.4614-4619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fink RC, Evans MR, Porwollik S, Vazquez-Torres A, Jones-Carson J, Troxell B, Libby SJ, McClelland M, Hassan HM. 2007. FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028s). J Bacteriol 189:2262–2273. doi: 10.1128/JB.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trunk K, Benkert B, Quack N, Munch R, Scheer M, Garbe J, Jansch L, Trost M, Wehland J, Buer J, Jahn M, Schobert M, Jahn D. 2010. Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ Microbiol 12:1719–1733. [DOI] [PubMed] [Google Scholar]
- 24.Son MS, Matthews WJ Jr, Kang Y, Nguyen DT, Hoang TT. 2007. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect Immun 75:5313–5324. doi: 10.1128/IAI.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Kievit TR, Gillis R, Marx S, Brown C, Iglewski BH. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl Environ Microbiol 67:1865–1873. doi: 10.1128/AEM.67.4.1865-1873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubern JF, Diggle SP. 2008. Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Mol Biosyst 4:882–888. doi: 10.1039/b803796p. [DOI] [PubMed] [Google Scholar]
- 27.Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. 2012. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev 76:46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman LR, Kulasekara HD, Emerson J, Houston LS, Burns JL, Ramsey BW, Miller SI. 2009. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cyst Fibros 8:66–70. doi: 10.1016/j.jcf.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dénervaud V, TuQuoc P, Blanc D, Favre-Bonté S, Krishnapillai V, Reimmann C, Haas D, van Delden C. 2004. Characterization of cell-to-cell signaling-deficient Pseudomonas aeruginosa strains colonizing intubated patients. J Clin Microbiol 42:554–562. doi: 10.1128/JCM.42.2.554-562.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaber JA. 2004. Analysis of quorum sensing-deficient clinical isolates of Pseudomonas aeruginosa. J Med Microbiol 53:841–853. doi: 10.1099/jmm.0.45617-0. [DOI] [PubMed] [Google Scholar]
- 32.Köhler T, Buckling A, van Delden C. 2009. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc Natl Acad Sci U S A 106:6339–6344. doi: 10.1073/pnas.0811741106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D'Argenio DA, Wu M, Hoffman LR, Kulasekara HD, Deziel E, Smith EE, Nguyen H, Ernst RK, Larson Freeman TJ, Spencer DH, Brittnacher M, Hayden HS, Selgrade S, Klausen M, Goodlett DR, Burns JL, Ramsey BW, Miller SI. 2007. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol 64:512–533. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heurlier K, Denervaud V, Haenni M, Guy L, Krishnapillai V, Haas D. 2005. Quorum-sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. J Bacteriol 187:4875–4883. doi: 10.1128/JB.187.14.4875-4883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffman LR, Richardson AR, Houston LS, Kulasekara HD, Martens-Habbena W, Klausen M, Burns JL, Stahl DA, Hassett DJ, Fang FC, Miller SI. 2010. Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS Pathog 6:e1000712. doi: 10.1371/journal.ppat.1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toyofuku M, Nomura N, Fujii T, Takaya N, Maseda H, Sawada I, Nakajima T, Uchiyama H. 2007. Quorum sensing regulates denitrification in Pseudomonas aeruginosa PAO1. J Bacteriol 189:4969–4972. doi: 10.1128/JB.00289-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skurnik D, Roux D, Aschard H, Cattoir V, Yoder-Himes D, Lory S, Pier GB. 2013. A comprehensive analysis of in vitro and in vivo genetic fitness of Pseudomonas aeruginosa using high-throughput sequencing of transposon libraries. PLoS Pathog 9:e1003582. doi: 10.1371/journal.ppat.1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winstanley C, Langille MG, Fothergill JL, Kukavica-Ibrulj I, Paradis-Bleau C, Sanschagrin F, Thomson NR, Winsor GL, Quail MA, Lennard N, Bignell A, Clarke L, Seeger K, Saunders D, Harris D, Parkhill J, Hancock RE, Brinkman FS, Levesque RC. 2009. Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool epidemic strain of Pseudomonas aeruginosa. Genome Res 19:12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from Gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 41.Meissner A, Wild V, Simm R, Rohde M, Erck C, Bredenbruch F, Morr M, Romling U, Haussler S. 2007. Pseudomonas aeruginosa cupA-encoded fimbriae expression is regulated by a GGDEF and EAL domain-dependent modulation of the intracellular level of cyclic diguanylate. Environ Microbiol 9:2475–2485. doi: 10.1111/j.1462-2920.2007.01366.x. [DOI] [PubMed] [Google Scholar]
- 42.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 44.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 39:D596–D600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson MD, Smyth GK. 2008. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9:321–332. [DOI] [PubMed] [Google Scholar]
- 46.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawakami T, Kuroki M, Ishii M, Igarashi Y, Arai H. 2010. Differential expression of multiple terminal oxidases for aerobic respiration in Pseudomonas aeruginosa. Environ Microbiol 12:1399–1412. [DOI] [PubMed] [Google Scholar]
- 48.Rompf A, Hungerer C, Hoffman T, Lindenmeyer M, Romling U, Gross U, Doss MO, Arai H, Igarashi Y, Jahn D. 1998. Regulation of Pseudomonas aeruginosa hemF and hemN by the dual action of the redox response regulators Anr and Dnr. Mol Microbiol 29:985–997. doi: 10.1046/j.1365-2958.1998.00980.x. [DOI] [PubMed] [Google Scholar]
- 49.Platt MD, Schurr MJ, Sauer K, Vazquez G, Kukavica-Ibrulj I, Potvin E, Levesque RC, Fedynak A, Brinkman FS, Schurr J, Hwang SH, Lau GW, Limbach PA, Rowe JJ, Lieberman MA, Barraud N, Webb J, Kjelleberg S, Hunt DF, Hassett DJ. 2008. Proteomic, microarray, and signature-tagged mutagenesis analyses of anaerobic Pseudomonas aeruginosa at pH 6.5, likely representing chronic, late-stage cystic fibrosis airway conditions. J Bacteriol 190:2739–2758. doi: 10.1128/JB.01683-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eschbach M, Schreiber K, Trunk K, Buer J, Jahn D, Schobert M. 2004. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J Bacteriol 186:4596–4604. doi: 10.1128/JB.186.14.4596-4604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schreiber K, Boes N, Eschbach M, Jaensch L, Wehland J, Bjarnsholt T, Givskov M, Hentzer M, Schobert M. 2006. Anaerobic survival of Pseudomonas aeruginosa by pyruvate fermentation requires an Usp-type stress protein. J Bacteriol 188:659–668. doi: 10.1128/JB.188.2.659-668.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McPhee JB, Tamber S, Bains M, Maier E, Gellatly S, Lo A, Benz R, Hancock RE. 2009. The major outer membrane protein OprG of Pseudomonas aeruginosa contributes to cytotoxicity and forms an anaerobically regulated, cation-selective channel. FEMS Microbiol Lett 296:241–247. doi: 10.1111/j.1574-6968.2009.01651.x. [DOI] [PubMed] [Google Scholar]
- 53.Vallet I, Olson JW, Lory S, Lazdunski A, Filloux A. 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc Natl Acad Sci U S A 98:6911–6916. doi: 10.1073/pnas.111551898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Friedman L, Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol 51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 55.Vallet-Gely I, Sharp JS, Dove SL. 2007. Local and global regulators linking anaerobiosis to cupA fimbrial gene expression in Pseudomonas aeruginosa. J Bacteriol 189:8667–8676. doi: 10.1128/JB.01344-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McManus HR, Dove SL. 2011. The CgrA and CgrC proteins form a complex that positively regulates cupA fimbrial gene expression in Pseudomonas aeruginosa. J Bacteriol 193:6152–6161. doi: 10.1128/JB.05904-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pessi G, Haas D. 2000. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J Bacteriol 182:6940–6949. doi: 10.1128/JB.182.24.6940-6949.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sonnleitner E, Gonzalez N, Sorger-Domenigg T, Heeb S, Richter AS, Backofen R, Williams P, Huttenhofer A, Haas D, Blasi U. 2011. The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Mol Microbiol 80:868–885. doi: 10.1111/j.1365-2958.2011.07620.x. [DOI] [PubMed] [Google Scholar]
- 59.Oglesby AG, Farrow JM III, Lee JH, Tomaras AP, Greenberg EP, Pesci EC, Vasil ML. 2008. The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J Biol Chem 283:15558–15567. doi: 10.1074/jbc.M707840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K. 2008. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 61.Schuster M, Greenberg EP. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol 296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 62.Gilbert KB, Kim TH, Gupta R, Greenberg EP, Schuster M. 2009. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol Microbiol 73:1072–1085. doi: 10.1111/j.1365-2958.2009.06832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol 185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schuster M, Lostroh CP, Ogi T, Greenberg EP. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol 185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pesci EC, Milbank JB, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Toyofuku M, Nomura N, Kuno E, Tashiro Y, Nakajima T, Uchiyama H. 2008. Influence of the Pseudomonas quinolone signal on denitrification in Pseudomonas aeruginosa. J Bacteriol 190:7947–7956. doi: 10.1128/JB.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mashburn LM, Jett AM, Akins DR, Whiteley M. 2005. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol 187:554–566. doi: 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoffman LR, Deziel E, D'Argenio DA, Lepine F, Emerson J, McNamara S, Gibson RL, Ramsey BW, Miller SI. 2006. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 103:19890–19895. doi: 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cugini C, Morales DK, Hogan DA. 2010. Candida albicans-produced farnesol stimulates Pseudomonas quinolone signal production in LasR-defective Pseudomonas aeruginosa strains. Microbiology 156:3096–3107. doi: 10.1099/mic.0.037911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dekimpe V, Deziel E. 2009. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 155:712–723. doi: 10.1099/mic.0.022764-0. [DOI] [PubMed] [Google Scholar]
- 71.Ha DG, Merritt JH, Hampton TH, Hodgkinson JT, Janecek M, Spring DR, Welch M, O'Toole GA. 2011. 2-Heptyl-4-quinolone, a precursor of the Pseudomonas quinolone signal molecule, modulates swarming motility in Pseudomonas aeruginosa. J Bacteriol 193:6770–6780. doi: 10.1128/JB.05929-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD. 2013. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496:508–512. doi: 10.1038/nature12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lesic B, Starkey M, He J, Hazan R, Rahme LG. 2009. Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology 155:2845–2855. doi: 10.1099/mic.0.029082-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sana TG, Hachani A, Bucior I, Soscia C, Garvis S, Termine E, Engel J, Filloux A, Bleves S. 2012. The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and Fur and modulates internalization in epithelial cells. J Biol Chem 287:27095–27105. doi: 10.1074/jbc.M112.376368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schobert M, Jahn D. 2010. Anaerobic physiology of Pseudomonas aeruginosa in the cystic fibrosis lung. Int J Med Microbiol 300:549–556. doi: 10.1016/j.ijmm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 76.Yoon SS, Karabulut AC, Lipscomb JD, Hennigan RF, Lymar SV, Groce SL, Herr AB, Howell ML, Kiley PJ, Schurr MJ, Gaston B, Choi KH, Schweizer HP, Hassett DJ. 2007. Two-pronged survival strategy for the major cystic fibrosis pathogen, Pseudomonas aeruginosa, lacking the capacity to degrade nitric oxide during anaerobic respiration. EMBO J 26:3662–3672. doi: 10.1038/sj.emboj.7601787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Unden G, Trageser M, Duchene A. 1990. Effect of positive redox potentials (>+400mV) on the expression of anaerobic respiratory enzymes in Escherichia coli. Mol Microbiol 4:315–319. doi: 10.1111/j.1365-2958.1990.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 78.Engel P, Trageser M, Unden G. 1991. Reversible interconversion of the functional state of the gene regulator FNR from Escherichia coli in vivo by O2 and iron availability. Arch Microbiol 156:463–470. [DOI] [PubMed] [Google Scholar]
- 79.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Adler C, Corbalan NS, Seyedsayamdost MR, Pomares MF, de Cristobal RE, Clardy J, Kolter R, Vincent PA. 2012. Catecholate siderophores protect bacteria from pyochelin toxicity. PLoS One 7:e46754. doi: 10.1371/journal.pone.0046754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vinckx T, Wei Q, Matthijs S, Noben JP, Daniels R, Cornelis P. 2011. A proteome analysis of the response of a Pseudomonas aeruginosa oxyR mutant to iron limitation. Biometals 24:523–532. doi: 10.1007/s10534-010-9403-4. [DOI] [PubMed] [Google Scholar]
- 82.Xiao G, Deziel E, He J, Lepine F, Lesic B, Castonguay MH, Milot S, Tampakaki AP, Stachel SE, Rahme LG. 2006. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol Microbiol 62:1689–1699. doi: 10.1111/j.1365-2958.2006.05462.x. [DOI] [PubMed] [Google Scholar]
- 83.Schertzer JW, Brown SA, Whiteley M. 2010. Oxygen levels rapidly modulate Pseudomonas aeruginosa social behaviours via substrate limitation of PqsH. Mol Microbiol 77:1527–1538. doi: 10.1111/j.1365-2958.2010.07303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Machan ZA, Taylor G, Pitt TL, Cole PJ, Wilson R. 1992. 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J Antimicrob Chemother 30:615–623. [DOI] [PubMed] [Google Scholar]
- 85.Lazenby JJ, Griffin PE, Kyd J, Whitchurch CB, Cooley MA. 2013. A quadruple knockout of lasIR and rhlIR of Pseudomonas aeruginosa PAO1 that retains wild-type twitching motility has equivalent infectivity and persistence to PAO1 in a mouse model of lung infection. PLoS One 8:e60973. doi: 10.1371/journal.pone.0060973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.